Abstract
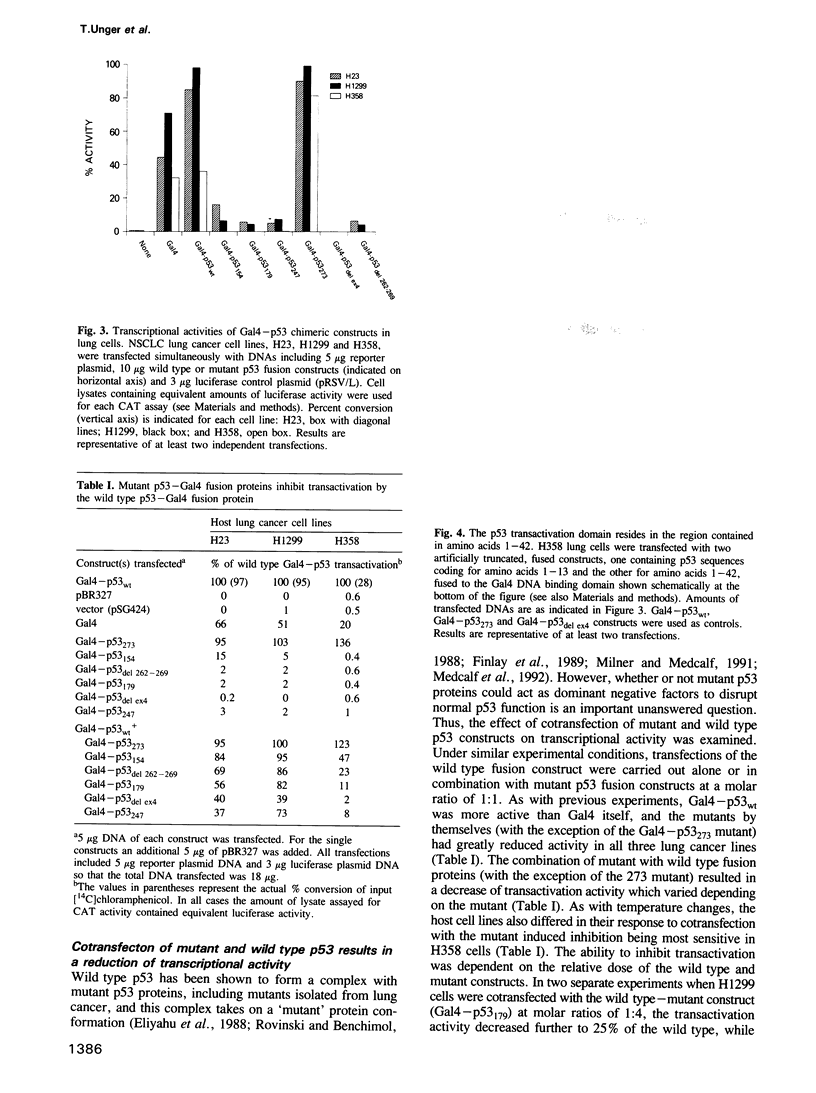
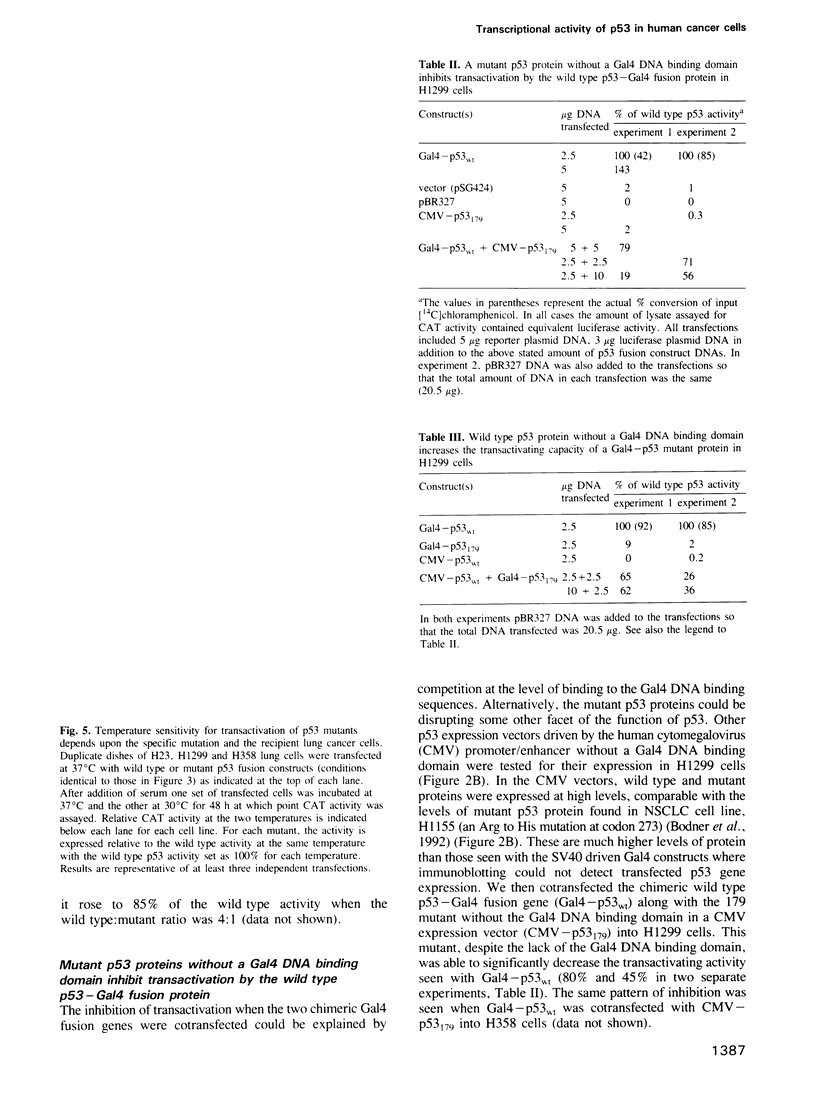
Gal4-p53 fusion constructs demonstrate that wild type p53 is a potent transactivator in human lung cancer cells with the transactivation domain for p53 residing in amino acids 1-42. Strikingly, a variety of lung cancer derived p53 mutations occurring outside this domain disrupt this activity. Temperature sensitive conformational shifts of p53 mutant proteins to the wild type form exist and, with a temperature downshift, several mutants become transcriptionally active. Wild type p53 protein is known to form oligomers with mutant p53 and cotransfection of wild type and mutant genes shows that p53 acts in a transdominant manner that is independent of the DNA binding specificity. Transcription is either increased or decreased depending on whether the wild type is more or less abundant than the mutant form. Finally, lung cancers differ in their ability to support the transactivation related functions, providing evidence of other abnormalities of the p53 system in human cancer.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banks L., Matlashewski G., Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986 Sep 15;159(3):529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- Bargonetti J., Friedman P. N., Kern S. E., Vogelstein B., Prives C. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991 Jun 14;65(6):1083–1091. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- Berger U., Bettelheim R., Mansi J. L., Easton D., Coombes R. C., Neville A. M. The relationship between micrometastases in the bone marrow, histopathologic features of the primary tumor in breast cancer and prognosis. Am J Clin Pathol. 1988 Jul;90(1):1–6. doi: 10.1093/ajcp/90.1.1. [DOI] [PubMed] [Google Scholar]
- Brower M., Carney D. N., Oie H. K., Gazdar A. F., Minna J. D. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986 Feb;46(2):798–806. [PubMed] [Google Scholar]
- Chiba I., Takahashi T., Nau M. M., D'Amico D., Curiel D. T., Mitsudomi T., Buchhagen D. L., Carbone D., Piantadosi S., Koga H. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Lung Cancer Study Group. Oncogene. 1990 Oct;5(10):1603–1610. [PubMed] [Google Scholar]
- Cuttitta F., Fedorko J., Gu J. A., Lebacq-Verheyden A. M., Linnoila R. I., Battey J. F. Gastrin-releasing peptide gene-associated peptides are expressed in normal human fetal lung and small cell lung cancer: a novel peptide family found in man. J Clin Endocrinol Metab. 1988 Sep;67(3):576–583. doi: 10.1210/jcem-67-3-576. [DOI] [PubMed] [Google Scholar]
- Eliyahu D., Goldfinger N., Pinhasi-Kimhi O., Shaulsky G., Skurnik Y., Arai N., Rotter V., Oren M. Meth A fibrosarcoma cells express two transforming mutant p53 species. Oncogene. 1988 Sep;3(3):313–321. [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Jang S. K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990 Aug 31;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Finlay C. A., Hinds P. W., Levine A. J. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989 Jun 30;57(7):1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Gannon J. V., Greaves R., Iggo R., Lane D. P. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 1990 May;9(5):1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Kaelin W. G., Jr, Pallas D. C., DeCaprio J. A., Kaye F. J., Livingston D. M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991 Feb 8;64(3):521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S. E., Kinzler K. W., Bruskin A., Jarosz D., Friedman P., Prives C., Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991 Jun 21;252(5013):1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Levine A. J., Momand J., Finlay C. A. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Michalovitz D., Halevy O., Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990 Aug 24;62(4):671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- Milner J., Medcalf E. A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991 May 31;65(5):765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- Milner J., Medcalf E. A. Temperature-dependent switching between "wild-type" and "mutant" forms of p53-Val135. J Mol Biol. 1990 Dec 5;216(3):481–484. doi: 10.1016/0022-2836(90)90371-R. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- O'Rourke R. W., Miller C. W., Kato G. J., Simon K. J., Chen D. L., Dang C. V., Koeffler H. P. A potential transcriptional activation element in the p53 protein. Oncogene. 1990 Dec;5(12):1829–1832. [PubMed] [Google Scholar]
- Raycroft L., Wu H. Y., Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990 Aug 31;249(4972):1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinski B., Benchimol S. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene. 1988 May;2(5):445–452. [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Soussi T., Caron de Fromentel C., May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene. 1990 Jul;5(7):945–952. [PubMed] [Google Scholar]
- Takahashi T., D'Amico D., Chiba I., Buchhagen D. L., Minna J. D. Identification of intronic point mutations as an alternative mechanism for p53 inactivation in lung cancer. J Clin Invest. 1990 Jul;86(1):363–369. doi: 10.1172/JCI114710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Nau M. M., Chiba I., Birrer M. J., Rosenberg R. K., Vinocour M., Levitt M., Pass H., Gazdar A. F., Minna J. D. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989 Oct 27;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J., LaMarco K. L., McKnight S. L. Evidence of DNA: protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988 Jun;2(6):730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Hauschka S., Tapscott S. J. The MCK enhancer contains a p53 responsive element. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4570–4571. doi: 10.1073/pnas.88.11.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakut-Houri R., Bienz-Tadmor B., Givol D., Oren M. Human p53 cellular tumor antigen: cDNA sequence and expression in COS cells. EMBO J. 1985 May;4(5):1251–1255. doi: 10.1002/j.1460-2075.1985.tb03768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]