Abstract
Salivary and pancreatic amylases (encoded by AMY1 and AMY2 genes, respectively) are responsible for digesting starchy foods. AMY1 and AMY2 show copy number variations that affect differences in amylase amount and activity, and AMY1 copies have been associated with adiposity. We investigated whether genetic variants determining amylase gene copies are associated with 2-year changes in adiposity among 692 overweight and obese individuals who were randomly assigned to diets varying in macronutrient content. We found that changes in body weight (BW) and waist circumference (WC) were significantly different according to the AMY1-AMY2 rs11185098 genotype. Individuals carrying the A allele (indicating higher amylase amount and activity) showed a greater reduction in BW and WC at 6, 12, 18, and 24 months than those without the A allele (P < 0.05 for all). The association was stronger for long-term changes compared with short-term changes of these outcomes. The genetic effects on these outcomes did not significantly differ across diet groups. In conclusion, the genetic variant determining starch metabolism influences the response to weight-loss dietary intervention. Overweight and obese individuals carrying the AMY1-AMY2 rs11185098 genotype associated with higher amylase activity may have greater loss of adiposity during low-calorie diet interventions.
Introduction
Dietary carbohydrates account for the greatest source of energy intake in most human diets (1) and play an important role in determining energy balance, which regulates body weight (BW) and adiposity (2,3). The digestion of polysaccharide carbohydrates begins in the mouth by action of salivary α-amylase, which catalyzes the hydrolysis of the α-1,4-glycosidic bonds of starch, followed by the action of pancreatic amylase. Salivary α-amylase is encoded by the salivary amylase gene (AMY1), which shows extensive copy number variations (4–7) that directly affect individual variability in salivary amylase amount and activity (8–10) as well as serum concentrations of total and pancreatic amylase (11). AMY1 is located in a gene cluster (1p21.1) that also includes pancreatic amylase genes (AMY2A and AMY2B), and AMY1 and AMY2A copy numbers are significantly correlated (6).
Increasing evidence suggests that low AMY1 gene copy numbers are associated with obesity (5,12–14), although the findings are not entirely consistent (7,15). In epidemiological studies, low serum concentrations of amylase have been associated with elevated BMI (16–19), waist circumference (WC) (16,20), or visceral adipose tissue area (21). Furthermore, studies have suggested that AMY1 copy number also plays a role in glucose uptake and glucose metabolism (11,22). A novel study compared AMY1 copy number of individuals of European ancestry from the 1000 Genomes Project with their single nucleotide polymorphism (SNP) genotypes and identified genetic variants associated with AMY1 copies (7). Such copy number variation of AMY1 likely is due to adaptation to diets rich in starchy food (4). We hypothesized that starch digestion–related amylase genetic variants affect the regulation of body adiposity, particularly when the intake of polysaccharide, the substrate for amylase, is changed. We examined associations of copy number related to AMY1-AMY2 genotype and 2-year changes of adiposity in participants of the Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trial. POUNDS Lost is thus far one of the largest randomized dietary intervention trials on long-term weight loss among overweight and obese individuals (23). We investigated whether the AMY1-AMY2 genotype rs11185098, which determines starch metabolism, influences long-term weight-loss response to dietary intervention among overweight and obese individuals. We also investigated associations of the AMY1-AMY2 genotype with changes in glucose metabolism in relation to weight loss.
Research Design and Methods
Study Participants
The sample comprised 811 overweight and obese individuals randomly assigned to one of four energy-reduced diets varying in macronutrient composition of fat, protein, and carbohydrates to compare the diets’ effects on BW change over 2 years in the POUNDS Lost trial (23). The study was conducted from October 2004 through December 2007 at two sites: the Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital in Boston, Massachusetts, and the Pennington Biomedical Research Center of Louisiana State University System, in Baton Rouge, Louisiana. All participants gave written informed consent. The study was approved by the human subjects committee at each institution and by a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute.
The targeted percentages of energy derived from fat, protein, and carbohydrates in the four diets were 1) 20%, 15%, and 65%; 2) 20%, 25%, and 55%; 3) 40%, 15%, and 45%; and 4) 40%, 25%, and 35%, respectively. Two diets were low fat (20%) and the other two high fat (40%), and two diets were average protein (15%) and the other two high protein (25%), which constituted a 2 × 2 factorial design. Major exclusion criteria were the presence of diabetes or unstable cardiovascular disease, the use of medications that affect BW, and insufficient motivation (23). More details of the study have been described in detail elsewhere (23).
The current study included 692 individuals with data on the AMY1 variant SNP rs11185098 located in amylase genes (AMY1, AMY2A, and AMY2B) at the baseline examination. Most of the individuals were considered normoglycemic on the basis of fasting glucose levels (93.8% had fasting glucose <6.1 mmol/L, and 82.8% had <5.5 mmol/L), and none of the study participants were regularly taking insulin or oral hypoglycemic agents at the baseline examination. Of the 692 participants, 29.5% were taking antihypertensive medication, and 18.8% were taking lipid-lowering medication at the baseline examination. Individuals who remained in the trial and with data on BW or WC at 6, 12, 18, or 24 months or data on fasting glucose at 6 or 24 months were included in the current analysis. The genotype frequency was not significantly different between individuals with and without follow-up data.
Measurements and Selection of Amylase Gene SNP
Height was measured at the baseline examination, and BW and WC were measured in the morning before breakfast at baseline and at 6, 12, 18, and 24 months during the intervention. BW was measured by calibrated hospital scales, and WC was measured by using a nonstretchable tape measure 4 cm above the iliac crest (24). BMI was calculated as weight in kilograms divided by the square of height in meters. Fasting blood samples were obtained at baseline and 6 and 24 months, and fasting serum glucose and insulin levels (Diagnostic Products Corporation) were measured at the clinical laboratory at the Pennington Biomedical Research Center. HOMA for insulin resistance (HOMA-IR) was calculated as described previously (25). Blood pressure was measured with the use of an automated device (HEM-907XL; Omron). A random sample of ∼50% of the total study participants were selected to undergo DEXA scans for the assessment of body composition (26). The DEXA scan was performed with a Hologic QDR 4500A after an overnight fast; total fat mass, total lean mass, whole-body total percent fat mass, and trunk fat percent were measured at baseline and 6 and 24 months during the intervention.
DNA was extracted from the buffy coat fraction of centrifuged blood by using the QIAamp DNA Blood Mini Kit (QIAGEN, Chatsworth, CA). A total of 799 participants were genotyped for 715,481 SNPs by using an Illumina Infinium OmniExpress-24 Kit. The genotype success rate was 99%. After quality control (sample call rate ≥95%, SNP call rate ≥95%, P > 1.0E-6 for Hardy-Weinberg equilibrium test, minor allele frequency ≥0.01), the genotyped data set had 727 samples and 660,673 SNPs. Because of the potential population stratification and limited sample size of non-European Hispanics (n = 24) and Asians (n = 11), 692 participants (583 whites and 109 blacks) had available data for the current study. We used the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html) to impute SNPs on chromosomes 1–22, with 1000G Phase 3 v5 as the reference panel. In the current study, we used an imputed SNP, rs11185098, with high imputation quality (R2 ≥ 0.9). Selection of this SNP was based on the variant that showed the strongest association (among all the SNPs presented) with obesity in the previous study (7) and was the only SNP located near all three amylase genes (AMY1, AMY2A, and AMY2B). In addition, the minor allele frequency of this SNP was common, which provided better power in the analysis (7). The minor (A) allele of SNP rs11185098 was reported to be positively associated with AMY1 gene copies (7). A higher number of AMY1 gene copies was associated with high salivary amylase activity in other studies (8–10), and three copy numbers of AMY1 and AMY2A have been shown to be numerically correlated (6). Therefore, the A allele of SNP rs11185098 was used in the current study as a marker of having high amylase activity.
Statistical Analysis
General linear models for continuous variables and the χ2 test for categorical variables were used for comparison of characteristics at baseline examination across the genotype. Primary outcomes in the current study were changes in BW and WC during the 2-year intervention. We used Quanto 1.2.4 software (http://biostats.usc.edu/index.html; University of Southern California, Los Angeles) to estimate the detectable effect sizes of the genetic effect. The current study was estimated to have >80% power to detect associations of the genotype and the outcomes. General linear models were used to assess genotype effect on the outcomes. Covariates in the multivariate-adjusted model included age, sex, ethnicity, and diet group (model 1), with additional adjustment of value for the respective outcome traits at the baseline examination (either BW or WC) (model 2). We also performed a multivariate-adjusted model including age, sex, ethnicity, diet group, BMI, WC, and fasting glucose at baseline (model 3), with additional adjustment for use of antihypertensive and lipid-lowering medication (model 4). We performed a sensitivity analysis for concurrent changes in fasting glucose in model 4. Additive genetic models were used in the analysis.
To test potential gene-diet interactions, a genotype-by-diet product term was included in the models. We used linear mixed models with the variance components structure, including time × genotype interaction terms to test the genotype effect on the trajectory of changes in BW and WC. We analyzed associations of the genotype and fasting glucose changes at 6 months as a secondary outcome by using multivariate-adjusted models that included age, sex, ethnicity, and diet group, with additional adjustment for fasting glucose at baseline. We also performed a fully adjusted model further including BMI, WC, medication use, and concurrent weight changes as covariates in the model. Statistical analyses were performed with SAS 9.3 software (SAS Institute). Statistical significance was considered for P < 0.05.
Results
No significant differences in age, blood pressure measures, triglyceride levels, HbA1c concentrations, fasting insulin concentrations, HOMA-IR, body composition, and dietary intake were found according to the AMY1-AMY2 rs11185098 genotype at the baseline examination (Table 1). The genotype distribution did not differ by sex, diet intervention group, or use of medications, whereas a significant difference was observed for ethnicity (P < 0.001). At the baseline examination, an increasing number of the A allele was significantly correlated with higher BMI (P = 0.04) and WC (P = 0.03). We observed that carrying the A allele was associated with lower glucose concentrations at the baseline examination (P = 0.03). Table 2 shows nutrient intake and biomarkers of adherence at 6 months and 2 years according to the AMY1-AMY2 rs11185098 genotype. No significant differences were found in mean values of nutrient intake and biomarkers of adherence at 6 months and 2 years across the genotype (P > 0.05).
Table 1.
Characteristics of study participants according to the AMY1-AMY2 rs11185098 genotype
| GG (n = 380) | GA (n = 257) | AA (n = 55) | P value | |
|---|---|---|---|---|
| Age (years) | 51.9 (9.0) | 51.1 (8.8) | 50.0 (10.2) | 0.11 |
| Male sex, n (%) | 145 (38.2) | 106 (41.2) | 18 (32.7) | 0.46 |
| White race, n (%) | 338 (88.9) | 200 (77.8) | 45 (81.8) | <0.001 |
| Low-carbohydrate/high-fat diet (yes), n (%) | 190 (50.0) | 131 (51.0) | 27 (49.1) | 0.95 |
| Low-protein diet (yes), n (%) | 194 (51.1) | 130 (50.6) | 26 (47.3) | 0.87 |
| BMI (kg/m2) | 32.5 (3.8) | 32.8 (3.9) | 33.7 (4.1) | 0.04 |
| BW (kg) | 92.8 (15.1) | 93.7 (15.6) | 97.4 (17.8) | 0.07 |
| WC (cm) | 103.1 (12.8) | 104.4 (13.1) | 106.9 (14.1) | 0.03 |
| Systolic blood pressure (mmHg) | 120 (13) | 120 (14) | 118 (13) | 0.64 |
| Diastolic blood pressure (mmHg) | 76 (9) | 76 (10) | 75 (9) | 0.71 |
| Antihypertensive medication use (yes) | 105 (27.6) | 81 (31.5) | 18 (32.7) | 0.49 |
| Fasting glucose (mmol/L) | 5.2 (0.7) | 5.1 (0.6) | 5.0 (0.5) | 0.03 |
| Fasting insulin (pmol/L) | 82 (47) | 86 (62) | 96 (53) | 0.16 |
| HbA1c (%) | 5.4 (0.4) | 5.4 (0.4) | 5.4 (0.4) | 0.79 |
| HbA1c (mmol/mol) | 35 (4) | 35 (4) | 35 (4) | — |
| HOMA-IR | 2.8 (1.8) | 2.8 (2.2) | 3.2 (1.9) | 0.4 |
| Triglycerides (mmol/L) | 1.36 (1.02, 2.10) | 1.39 (0.92, 1.89) | 1.47 (1.02, 1.99) | 0.41 |
| Lipid-lowering medication (yes) | 65 (17.1) | 54 (21.0) | 11 (20.0) | 0.45 |
| Body composition† | ||||
| Whole-body fat mass (%) | 37.1 (6.9) | 36.7 (7.0) | 37.1 (6.4) | 0.72 |
| Trunk fat (%) | 38.0 (6.2) | 37.9 (6.1) | 38.4 (6.0) | 0.83 |
| Whole-body total fat mass (kg) | 35.0 (8.1) | 35.3 (7.9) | 36.0 (8.1) | 0.53 |
| Whole-body lean mass (kg) | 59.7 (12.7) | 61.4 (13.1) | 62.0 (15.4) | 0.19 |
| Dietary intake per day* | ||||
| Total energy intake (kcal) | 1,938 (529) | 2,006 (580) | 1,784 (525) | 0.67 |
| Carbohydrate intake (% of total energy intake) | 44.6 (7.7) | 44 (7.7) | 47.4 (6.8) | 0.32 |
| Fat intake (% of total energy intake) | 36.7 (6.0) | 37.7 (6.1) | 35.5 (5.1) | 0.94 |
| Protein intake (% of total energy intake) | 18.3 (3.3) | 18.1 (3.4) | 18.1 (4.1) | 0.75 |
| Biomarkers of adherence | ||||
| Respiratory quotient | 0.84 (0.04) | 0.84 (0.04) | 0.84 (0.04) | 0.14 |
| Urinary nitrogen (mg/day) | 12.2 (4.3) | 12.1 (4.6) | 12.1 (4.6) | 0.75 |
Data are mean (SD) or median (25th, 75th), unless otherwise stated. Data on triglycerides, insulin, and HOMA-IR were log-transformed before analysis.
†Data were available for 211, 135, and 24 individuals across the GG, GA, and AA genotypes, respectively.
*Data were available for 202, 131, and 31 individuals across the GG, GA, and AA genotypes, respectively.
Table 2.
Nutrient intake and biomarkers of adherence according to the AMY1-AMY2 rs11185098 genotype during the intervention
| 6 months |
24 months |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | GG | n | GA | n | AA | n | GG | n | GA | n | AA | |
| Dietary intake per day | ||||||||||||
| Energy (kcal) | 180 | 1,600 (493) | 116 | 1,653 (548) | 21 | 1,524 (532) | 88 | 1,494 (467) | 64 | 1,576 (464) | 11 | 1,459 (718) |
| Carbohydrate (%) | 180 | 50.2 (10.4) | 116 | 50.8 (10.8) | 21 | 50.1 (7.1) | 88 | 49.2 (10.3) | 64 | 48.9 (11.3) | 11 | 51.7 (5.3) |
| Fat (%) | 180 | 30.3 (8.5) | 116 | 30.1 (8.0) | 21 | 31.2 (7.2) | 88 | 30.2 (7.7) | 64 | 32.3 (9.7) | 11 | 26.4 (6.8) |
| Protein (%) | 180 | 20.2 (4.4) | 116 | 19.9 (4.9) | 21 | 20.3 (4.1) | 88 | 20.6 (4.7) | 64 | 19.3 (4.1) | 11 | 22.1 (6.2) |
| Biomarkers of adherence | ||||||||||||
| Respiratory quotient | 318 | 0.84 (0.04) | 198 | 0.84 (0.05) | 47 | 0.84 (0.04) | 237 | 0.83 (0.04) | 165 | 0.83 (0.04) | 38 | 0.83 (0.05) |
| Urinary nitrogen (mg/day) | 288 | 11.3 (4.2) | 181 | 11.9 (5.0) | 43 | 11.4 (5.1) | 191 | 11.8 (4.8) | 133 | 12.1 (4.0) | 32 | 11.3 (4.6) |
Data are n or mean (SD).
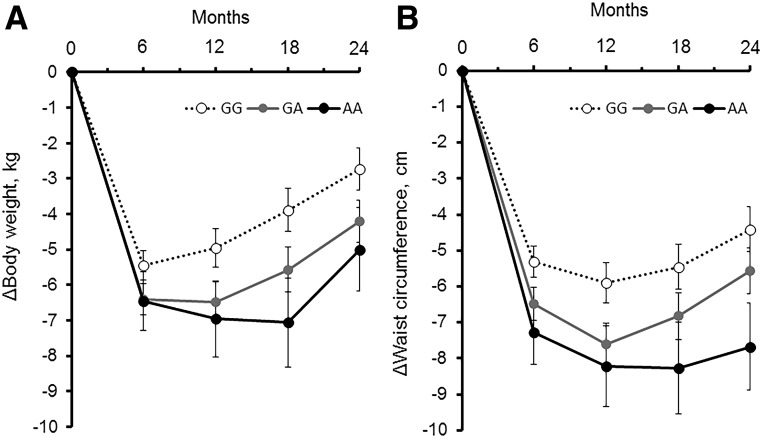
We found that the AMY1-AMY2 genotype was significantly associated with longitudinal changes in BW and WC, regardless of diet intervention group (Table 3). Increasing number of the A allele was significantly associated with greater loss of BW at 6, 12, 18, and 24 months during the dietary intervention (in model 1, P < 0.05 for all). The association was independent of age, sex, ethnicity, diet group, and BW at baseline (model 2). The association on 6-month changes of BW was slightly attenuated in the adjusted model that included BMI, WC, and fasting glucose at baseline (model 3), although changes at 12, 18, and 24 months remained significant. These results were not altered after accounting for medication use in model 4. Similarly, we found significant associations for changes in WC: Carrying the A allele of AMY1-AMY2 rs11185098 was associated with greater improvement of abdominal obesity at 6, 12, 18, and 24 months of the intervention after adjustment for covariates in model 4. After adjustment for covariates in model 4 and concurrent (6-month or 2-year) changes in fasting glucose, the AMY1-AMY2 rs11185098 genotype was significantly associated with a greater loss of BW at 6 months (β [SE] per A allele –0.89 [0.34]; P = 0.01) and 24 months (–1.21 [0.47]; P = 0.01) as well as a greater reduction in WC at 6 months (–1.25 [0.36]; P < 0.001) and 24 months (–1.28 [0.49]; P = 0.01). Results were fundamentally the same in an analysis of only whites (data not shown). We did not find a significant (P < 0.05) interaction effect between diet group (fat/carbohydrate or protein diet group) and genotype for these outcomes.
Table 3.
Changes in BW and WC according to the AMY1-AMY2 rs11185098 genotype
| Model 1 |
Model 2 |
Model 3 |
Model 4 |
|||||
|---|---|---|---|---|---|---|---|---|
| Outcome | β (SE) | P value | β (SE) | P value | β (SE) | P value | β (SE) | P value |
| 6 months | ||||||||
| Δ BW | −0.78 (0.35) | 0.027 | −0.70 (0.35) | 0.044 | −0.64 (0.35) | 0.067 | −0.67 (0.35) | 0.056 |
| Δ WC | −1.21 (0.38) | 0.002 | −1.06 (0.37) | 0.005 | −1.01 (0.37) | 0.007 | −1.02 (0.37) | 0.006 |
| 12 months | ||||||||
| Δ BW | −1.30 (0.47) | 0.005 | −1.21 (0.46) | 0.009 | −1.09 (0.46) | 0.018 | −1.15 (0.46) | 0.014 |
| Δ WC | −1.55 (0.49) | 0.002 | −1.38 (0.48) | 0.004 | −1.31 (0.48) | 0.007 | −1.35 (0.48) | 0.005 |
| 18 months | ||||||||
| Δ BW | −1.70 (0.52) | 0.001 | −1.63 (0.52) | 0.002 | −1.55 (0.52) | 0.003 | −1.59 (0.52) | 0.003 |
| Δ WC | −1.51 (0.54) | 0.005 | −1.39 (0.53) | 0.009 | −1.31 (0.53) | 0.014 | −1.39 (0.53) | 0.009 |
| 24 months | ||||||||
| Δ BW | −1.34 (0.49) | 0.006 | −1.27 (0.49) | 0.01 | −1.19 (0.49) | 0.015 | −1.23 (0.49) | 0.012 |
| Δ WC | −1.57 (0.52) | 0.003 | −1.43 (0.51) | 0.005 | −1.39 (0.51) | 0.006 | −1.41 (0.51) | 0.006 |
β represents changes in outcomes by increasing number of A alleles. Model 1: age, sex, ethnicity, and diet group. Model 2: age, sex, ethnicity, diet group, and value for the respective outcome traits at the baseline examination (either BW or WC). Model 3: age, sex, ethnicity, diet group, BMI at baseline, WC at baseline, and glucose at baseline. Model 4: model 3 + antihypertensive medication use at baseline and lipid-lowering medication use at baseline.
Two-year trajectories of BW (Fig. 1A) and WC (Fig. 1B) according to the AMY1-AMY2 rs11185098 genotype showed a significant time × genotype interaction for BW (Ptime × genotype interaction = 0.01) and WC (Ptime × genotype interaction = 0.01) after adjustment for age, sex, ethnicity, BMI, and WC at baseline. Results remained significant even after additional adjustment for fasting glucose concentrations and medication use at baseline (Ptime × genotype interaction = 0.01 for BW, Ptime × genotype interaction = 0.01 for WC). The effect of the genotype was stronger for long-term changes than for short-term changes (6 months) in these outcomes. Individuals carrying the G allele regained weight after 6–24 months, whereas those without the G allele continued to lose weight after 6 months; in addition, these individuals did not show a marked regain in WC after 6–24 months. We observed that changes in BW were significantly correlated with changes in body composition, including total fat mass, total lean mass, whole-body total percent fat mass, and trunk fat percent. On examination of possible genetic effects on changes in body fat composition, we also observed that increasing number of the A allele tended to be related to greater loss of whole-body total percent fat mass and trunk fat percent, although the results were not statistically significant (data not shown).
Figure 1.
Trajectories of BW (A) and WC (B) over 2 years in participants according to the AMY1-AMY2 rs11185098 genotype. Data are after adjustment for age, sex, ethnicity, diet group, and value for the respective outcome traits at the baseline examination (either BW or WC).
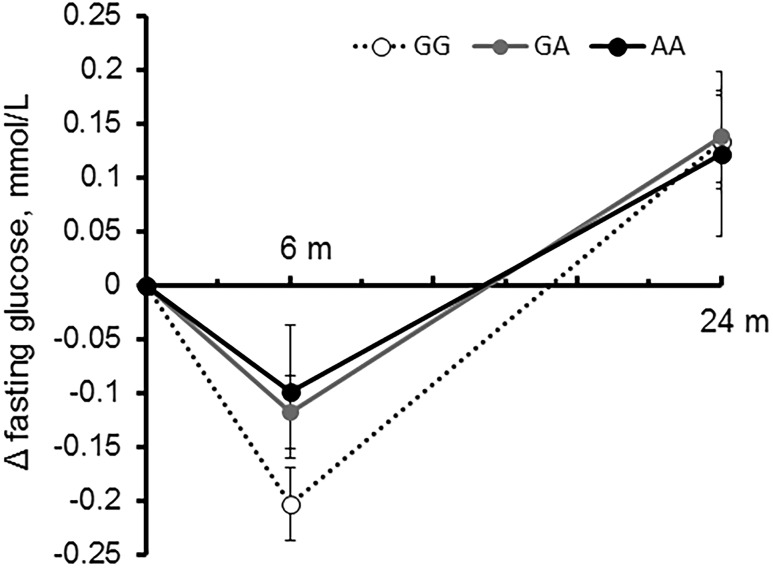
In addition, we observed a significant association of the AMY1-AMY2 rs11185098 genotype with improving fasting glucose concentrations, especially at 6 months because blood glucose levels rebounded between 6 months and 2 years (Fig. 2). No significant interaction was observed between diet group and genotype for changes in fasting glucose. Of note, opposite to changes in obesity measurement, an increasing number of the G allele was associated with greater decreases in fasting glucose concentrations at 6 months after adjustment for age, sex, ethnicity, and diet group (P = 0.005) as well as after additional adjustment for fasting glucose at baseline (P = 0.03). The association remained significant even after further adjustment for BMI, WC, medication use, and concurrent (6-month) weight changes in the fully adjusted model (β [SE] –0.07 [0.03] per G allele; P = 0.01) (Fig. 2); individuals with GG genotype had the largest reduction (mean ± SE, –0.20 ± 0.03 mmol/L; P < 0.001) at 6 months compared with those with other genotypes (GA genotype: –0.12 ± 0.03 mmol/L [P < 0.001]; AA genotype: –0.1 ± 0.06 mmol/L [P = 0.1]). No significant association was found between 24-month glucose changes and genotype (data not shown). We observed similar results only among whites; increasing number of the G allele was associated with greater decreases of fasting glucose at 6 months (β [SE] for 6-month fasting glucose changes –0.07 [0.03] per G allele; P = 0.009).
Figure 2.
Effects of AMY1-AMY2 rs11185098 genotype on trajectories of changes in fasting glucose concentrations over 2 years in response to weight-loss diets. Data are after adjustment for age; sex; ethnicity; diet group; baseline values of BMI, WC, and fasting glucose; antihypertensive medication use; lipid-lowering medication use; and concurrent weight changes. m, months.
Discussion
In this 2-year randomized diet intervention trial among overweight and obese individuals, we show that the effect of dietary intervention on weight loss and improvement of abdominal obesity were significantly different according to the genetic variant regulating starch metabolism. We also showed that the variant in the amylase gene region significantly influenced changes in fasting glucose concentrations. To our knowledge, this study is the first to demonstrate that the genetic variant associated with more-complete starch digestion is also associated with more long-term weight loss during the intervention among overweight and obese individuals. The overweight and obese individuals carrying the AMY1-AMY2 rs11185098 genotype, which is associated with more gene copies and higher amylase activity, benefited from greater weight loss and decrease in central adiposity in response to low-calorie diet interventions.
The results show that the high amylase activity–related A allele of AMY1-AMY2 rs11185098 was a marker for greater weight loss over 2 years during the intervention, regardless of BMI and WC values at baseline. The A allele of SNP rs11185098 was nominally associated with a lower risk of obesity in the Estonian population; however, no relation was reported between the variant and BMI in a well-powered analysis (7). Previous studies reported that low AMY1 gene copies were associated with obesity (5,12–14), although the findings were not consistent (7,15). Studies have shown that the AMY1 gene copy number is positively correlated with salivary amylase activity and serum amylase levels (8–10), and observational studies have consistently reported that low serum amylase concentrations are associated with obesity, insulin resistance, and metabolic abnormalities (16–21,27). Studies linking AMY genotypes, amylase activity, and adiposity are still sparse, and the precise mechanisms underlying the current findings need further investigations. However, a previous animal study reported that increases in body fat in response to a high-fat and high-sucrose diet was associated with a genetic variant near three amylase genes (Amy1, Amy2b, and Amy2a5) (28). AMY1 also is actively expressed in adipose tissue (5), and the different genotype may affect adipose tissue metabolism. The amylase gene locus at chromosome 1p21.1 is structurally complex, and this region contains salivary (AMY1) and pancreatic (AMY2A and AMY2B) amylase genes. Previous studies show that copy numbers of AMY1 and AMY2A (6) as well as serum levels of the two enzymes (5) are significantly correlated and that salivary and pancreatic amylase proportions are approximately equal in serum (5). In addition, production of pancreatic amylase is regulated by insulin (29,30), and obese individuals with insulin resistance may have reduced pancreatic amylase levels (5). Because the SNP rs11185098 is located in the AMY1, AMY2A, and AMY2B gene region, we could not distinguish between salivary and pancreatic gene effects or whether both genes account for the observations. Although a causal relation between starch digestion and the development of obesity has not yet been verified, the current study suggests that genetic signature associated with amylase activity affects long-term weight changes induced by dietary modifications.
In addition, the current analysis, which used linear mixed models, indicated a significant time × genotype interaction, supporting that the genetic effect became stronger in the longer-term intervention. In the POUNDS Lost trial, most participants regained BW from 6 months to 2 years (23), and participants carrying distinct AMY1-AMY2 rs11185098 genotypes showed different patterns of weight regain during this period. Therefore, the results may also help to predict maintenance of weight loss according to individuals’ genomic makeup.
In line with previous studies, we observed that participants with the G allele, which indicates lower amylase activity, had higher fasting glucose concentrations at baseline; however, we observed that these individuals showed a greater reduction of fasting glucose at 6 months independent of concurrent weight changes and baseline fasting glucose concentrations. In an experimental study, individuals with fewer AMY1 copy numbers and lower amylase activity showed higher glucose concentrations after starch ingestion (22) because the low amylase activity group did not exhibit preabsorptive insulin release in response to starch and consequently had a higher glycemic response (22). On the other hand, several studies did not show a significant association of AMY gene copies or salivary amylase activity on glycemic response after carbohydrate/starchy food intake (8,31). A recent metabolomics study on AMY1 copy number suggested that fewer AMY1 gene copies may be associated with delayed glucose uptake after starch digestion (11). The design of these studies was cross-sectional or experimental, and in the current study, we assessed the short-term changes of fasting glucose among overweight and obese individuals during the intervention. The current results suggest that the improvement of glucose metabolism after the dietary intervention differ according to the starch digestion–related genotype. Although detailed mechanisms need to be further investigated, we speculate that the G allele, which is associated with lower AMY1 copy numbers, is associated with baseline impaired fasting glucose metabolism and may show better improvement of glucose metabolism when long-term dietary intake is changed, despite less improvement in adiposity. Nonetheless, postprandial concentrations of glucose and insulin were not available in this study, and individuals’ glucose tolerance status (after an oral glucose tolerance test) was not determined. Additional studies with detailed measurements of glucose metabolism would be warranted to investigate relations between amylase genetic variant and impaired glucose tolerance.
Compelling evidence has shown that diets varying in macronutrient composition are effective in promoting weight loss, but extensive interindividual variation exists in the effectiveness of dietary intervention to achieve weight loss and weight stability, and individual genetic makeup has long been suggested to affect individual responses to weight-loss intervention (32–34). In the current study, we demonstrated that the genotype associated with carbohydrate metabolism partly accounts for such variance. The data indicate that the relation between the AMY1-AMY2 rs11185098 genotype and weight loss is not modified by different types of diets. In our previous studies among the participants in the POUNDS Lost trial, we found that variants in several genes, such as those related to obesity (FTO) (35,36), type 2 diabetes (TCF7L2 and IRS1) (37,38), and amino acid metabolism (PPM1K) (39), modify the effect of weight-loss diets with varying macronutrient content on changes in adiposity and metabolic risk factors. In addition, we previously reported that the genetic variant determining preference to carbohydrate intake is associated with improving obesity in the POUNDS Lost trial (40). However, more studies are necessary to confirm these findings. The results from our series of studies emphasize the importance of considering genetic background in dietary interventions, and the current study suggests a novel approach to considering the genetic variants associated with individual carbohydrate digestion in obesity management.
This study has several strengths, including the robust genetic associations with improvement of obesity at multiple time points during the follow-up in a large and long-term randomized trial. Growing evidence suggests the important implications of amylase gene and copy number variation in obesity and metabolic diseases (30,41), and we show that amylase genetic regulation might have a substantial impact on improving obesity. Limitations of this study were unavailability of data on glucose tolerance tests and that we did not measure AMY1-AMY2 copy numbers, serum, or salivary amylase concentrations, which limited our ability to explore potential underlying mechanisms. We were not able to examine relations between the genotype and impaired glucose tolerance. In addition, the study participants were mainly white, and the genotype distribution differed between blacks and whites. Although the subgroup analysis in whites showed similar results, whether the findings are generalizable to other ethnicities needs to be investigated further. The study included only overweight and obese individuals who participated in the clinical trial, and carrying the A allele was associated with higher BMI at the baseline examination but with greater improvement of adiposity. Whether this variant could be a marker for obesity needs to be confirmed in other studies. The genetic variant may not be causal, and use of a single SNP may not capture overall variation of AMY1-AMY2 copies. In addition, body composition was measured in some (∼50%) of the study participants, and the relatively small sample size limited the statistical power to detect a potential genetic effect on changes in body composition.
In conclusion, this study showed that a genetic variant responsible for starch metabolism significantly influences response to weight-loss dietary interventions. Overweight and obese individuals carrying the AMY1-AMY2 rs11185098 genotype, which is associated with higher amylase activity, may benefit from greater loss of BW and central adiposity during a low-calorie diet intervention.
Article Information
Funding. The study is supported by National Institutes of Health grants from the National Heart, Lung, and Blood Institute (HL-071981, HL-034594, HL-126024), the National Institute of Diabetes and Digestive and Kidney Diseases (DK-091718, DK-100383, DK-078616), and the Boston Obesity Nutrition Research Center (DK-46200) and by the United States-Israel Binational Science Foundation (grant 2011036). Y.H. is a recipient of a Grant-in-Aid for Scientific Research and Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science. L.Q. was a recipient of the American Heart Association Scientist Development Award (0730094N). The sponsors played no role in the design or conduct of the study.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.H. contributed to the study concept and design; analysis and interpretation of data; drafting, revision, and final approval of the manuscript; statistical analysis; and study supervision. D.S., T.W., and T.H. contributed to the analysis and interpretation of data and drafting, revision, and final approval of the manuscript. G.A.B. and F.M.S. contributed to the acquisition of data; interpretation of data; and drafting, revision, and final approval of the manuscript. L.Q. contributed to the study concept and design; acquisition of data; analysis and interpretation of data; drafting, revision, and final approval of the manuscript; statistical analysis; and funding and study supervision. L.Q. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Clinical trial reg. no. NCT00072995, clinicaltrials.gov.
References
- 1.Tucci SA, Boyland EJ, Halford JC. The role of lipid and carbohydrate digestive enzyme inhibitors in the management of obesity: a review of current and emerging therapeutic agents. Diabetes Metab Syndr Obes 2010;3:125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto Y, Fukuda T, Oyabu C, et al. Impact of low-carbohydrate diet on body composition: meta-analysis of randomized controlled studies. Obes Rev 2016;17:499–509 [DOI] [PubMed] [Google Scholar]
- 3.Tobias DK, Chen M, Manson JE, Ludwig DS, Willett W, Hu FB. Effect of low-fat diet interventions versus other diet interventions on long-term weight change in adults: a systematic review and meta-analysis. Lancet Diabetes Endocrinol 2015;3:968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007;39:1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falchi M, El-Sayed Moustafa JS, Takousis P, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat Genet 2014;46:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter D, Dhar S, Mitchell LM, et al. Obesity, starch digestion and amylase: association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum Mol Genet 2015;24:3472–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usher CL, Handsaker RE, Esko T, et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet 2015;47:921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan VM, Ooi DS, Kapur J, et al. The role of digestive factors in determining glycemic response in a multiethnic Asian population. Eur J Nutr 2016;55:1573–1581 [DOI] [PubMed] [Google Scholar]
- 9.Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA. Individual differences in AMY1 gene copy number, salivary α-amylase levels, and the perception of oral starch. PLoS One 2010;5:e13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang ZM, Lin J, Chen LH, Zhang M, Chen WW, Yang XR. The roles of AMY1 copies and protein expression in human salivary α-amylase activity. Physiol Behav 2015;138:173–178 [DOI] [PubMed] [Google Scholar]
- 11.Arredouani A, Stocchero M, Culeddu N, et al.; D.E.S.I.R. Study Group . Metabolomic profile of low-copy number carriers at the salivary α-amylase gene suggests a metabolic shift toward lipid-based energy production. Diabetes 2016;65:3362–3368 [DOI] [PubMed] [Google Scholar]
- 12.Viljakainen H, Andersson-Assarsson JC, Armenio M, et al. Low copy number of the AMY1 locus is associated with early-onset female obesity in Finland. PLoS One 2015;10:e0131883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mejía-Benítez MA, Bonnefond A, Yengo L, et al. Beneficial effect of a high number of copies of salivary amylase AMY1 gene on obesity risk in Mexican children. Diabetologia 2015;58:290–294 [DOI] [PubMed] [Google Scholar]
- 14.Marcovecchio ML, Florio R, Verginelli F, et al. Low AMY1 gene copy number is associated with increased body mass index in prepubertal boys. PLoS One 2016;11:e0154961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yong RY, Mustaffa SB, Wasan PS, et al. Complex copy number variation of AMY1 does not associate with obesity in two East Asian cohorts. Hum Mutat 2016;37:669–678 [DOI] [PubMed] [Google Scholar]
- 16.Lee JG, Park SW, Cho BM, et al. Serum amylase and risk of the metabolic syndrome in Korean adults. Clin Chim Acta 2011;412:1848–1853 [DOI] [PubMed] [Google Scholar]
- 17.Nakajima K, Muneyuki T, Munakata H, Kakei M. Revisiting the cardiometabolic relevance of serum amylase. BMC Res Notes 2011;4:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muneyuki T, Nakajima K, Aoki A, et al. Latent associations of low serum amylase with decreased plasma insulin levels and insulin resistance in asymptomatic middle-aged adults. Cardiovasc Diabetol 2012;11:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang L, Su JB, Zhang XL, et al. Serum amylase levels in relation to islet β cell function in patients with early type 2 diabetes. PLoS One 2016;11:e0162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakajima K, Nemoto T, Muneyuki T, Kakei M, Fuchigami H, Munakata H. Low serum amylase in association with metabolic syndrome and diabetes: a community-based study. Cardiovasc Diabetol 2011;10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dias JP, Schrack JA, Shardell MD, Egan JM, Studenski S. Association of abdominal fat with serum amylase in an older cohort: the Baltimore Longitudinal Study of Aging. Diabetes Res Clin Pract 2016;116:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandel AL, Breslin PA. High endogenous salivary amylase activity is associated with improved glycemic homeostasis following starch ingestion in adults. J Nutr 2012;142:853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sacks FM, Bray GA, Carey VJ, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma W, Huang T, Zheng Y, et al. Weight-loss diets, adiponectin, and changes in cardiometabolic risk in the 2-year POUNDS Lost trial. J Clin Endocrinol Metab 2016;101:2415–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Huang T, Zheng Y, et al. Genetic variation of fasting glucose and changes in glycemia in response to 2-year weight-loss diet intervention: the POUNDS Lost trial. Int J Obes 2016;40:1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza RJ, Bray GA, Carey VJ, et al. Effects of 4 weight-loss diets differing in fat, protein, and carbohydrate on fat mass, lean mass, visceral adipose tissue, and hepatic fat: results from the POUNDS Lost trial. Am J Clin Nutr 2012;95:614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YJ, Nam YS, Yun JM, et al. Association between salivary amylase (AMY1) gene copy numbers and insulin resistance in asymptomatic Korean men. Diabet Med 2015;32:1588–1595 [DOI] [PubMed] [Google Scholar]
- 28.Parks BW, Nam E, Org E, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 2013;17:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mössner J, Logsdon CD, Goldfine ID, Williams JA. Regulation of pancreatic acinar cell insulin receptors by insulin. Am J Physiol 1984;247:G155–G160 [DOI] [PubMed] [Google Scholar]
- 30.Peyrot des Gachons C, Breslin PA. Salivary amylase: digestion and metabolic syndrome. Curr Diab Rep 2016;16:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alberti G, Parada J, Cataldo LR, et al. Glycemic response after starch consumption in relation to salivary amylase activity and copy-number variation of AMY1 gene. J Food Nutr Res 2015;3:558–563 [Google Scholar]
- 32.Moreno-Aliaga MJ, Santos JL, Marti A, Martínez JA. Does weight loss prognosis depend on genetic make-up? Obes Rev 2005;6:155–168 [DOI] [PubMed] [Google Scholar]
- 33.Qi L, Cho YA. Gene-environment interaction and obesity. Nutr Rev 2008;66:684–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L. Gene-diet interaction and weight loss. Curr Opin Lipidol 2014;25:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Qi Q, Zhang C, et al. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS Lost trial (published correction appears in Diabetes 2013;62:662). Diabetes 2012;61:3005–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Huang T, Zhang X, et al. Dietary fat modifies the effects of FTO genotype on changes in insulin sensitivity. J Nutr 2015;145:977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trial. Circulation 2011;124:563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattei J, Qi Q, Hu FB, Sacks FM, Qi L. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention. Am J Clin Nutr 2012;96:1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M, Qi Q, Liang J, et al. Genetic determinant for amino acid metabolites and changes in body weight and insulin resistance in response to weight-loss diets: the Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trial. Circulation 2013;127:1283–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heianza Y, Ma W, Huang T, et al. Macronutrient intake-associated FGF21 genotype modifies effects of weight-loss diets on 2-year changes of central adiposity and body composition: the POUNDS Lost trial. Diabetes Care 2016;39:1909–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakajima K. Low serum amylase and obesity, diabetes and metabolic syndrome: a novel interpretation. World J Diabetes 2016;7:112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]