Abstract
A genome-wide screen of 4168 homozygous diploid yeast deletion strains has been performed to identify nonessential genes that participate in the bipolar budding pattern. By examining bud scar patterns representing the sites of previous cell divisions, 127 mutants representing three different phenotypes were found: unipolar, axial-like, and random. From this screen, 11 functional classes of known genes were identified, including those involved in actin-cytoskeleton organization, general bud site selection, cell polarity, vesicular transport, cell wall synthesis, protein modification, transcription, nuclear function, translation, and other functions. Four characterized genes that were not known previously to participate in bud site selection were also found to be important for the haploid axial budding pattern. In addition to known genes, we found 22 novel genes (20 are designated BUD13-BUD32) important for bud site selection. Deletion of one resulted in unipolar budding exclusively from the proximal pole, suggesting that this gene plays an important role in diploid distal budding. Mutations in 20 other novel BUD genes produced a random budding phenotype and one produced an axial-like budding defect. Several of the novel Bud proteins were fused to green fluorescence protein; two proteins were found to localize to sites of polarized cell growth (i.e., the bud tip in small budded cells and the neck in cells undergoing cytokinesis), similar to that postulated for the bipolar signals and proteins that target cell division site tags to their proper location in the cell. Four others localized to the nucleus, suggesting that they play a role in gene expression. The bipolar distal marker Bud8 was localized in a number of mutants; many showed an altered Bud8-green fluorescence protein localization pattern. Through the genome-wide identification and analysis of different mutants involved in bipolar bud site selection, an integrated pathway for this process is presented in which proximal and distal bud site selection tags are synthesized and localized at their appropriate poles, thereby directing growth at those sites. Genome-wide screens of defined collections of mutants hold significant promise for dissecting many biological processes in yeast.
INTRODUCTION
Polarized cell division is a fundamental process in which cells divide along specific cleavage planes; this process is essential for the development of both eukaryotes and prokaryotes. Polarized cell divisions can mediate appropriate cell-cell contacts and partition cytoplasmic components asymmetrically between daughter cells. Polarized cell divisions occur during the life cycle of many organisms, including early embryogenesis in Caenorhabditis elegans (Hyman and White, 1987), neurogenesis in Drosophila and mammals (Kraut et al., 1996), spore development in Bacillus subtilus (Shapiro, 1993), and development of the snail body plan (Freeman and Lundelius, 1982). The mechanisms for selecting sites for polarized growth and division as well as for directing growth toward these sites are only beginning to be understood.
The budding yeast Saccharomyces cerevisiae is an excellent model for studying polarized cell division. S. cerevisiae exists in different forms, each with a specific cell morphology and division pattern (Roemer et al., 1996b; Costigan and Snyder, 1998; Madden and Snyder, 1998). During growth in rich media, both haploid and diploid yeast cells are ellipsoid and select bud sites according to their mating locus (MAT) and pedigree (Freifelder, 1960; Snyder, 1989; Chant and Pringle, 1995). Haploid MATa and MATα cells use an axial budding pattern in which cells bud adjacent to the preceding site of cytokinesis (i.e., the proximal poles). Diploid MATa/MATα cells exhibit a more complex bipolar budding pattern: daughter cells bud at distal poles (180° from the birth site), whereas mother cells bud either distal or proximal to the birth site, with new mothers exhibiting a bias for budding at the distal site (i.e., near the previous site of cytokinesis). The different budding patterns are thought to fulfill different purposes. The diploid bipolar pattern is thought to maximize exposure of a growing microcolony to nutrients (Gimeno and Fink, 1992; Gimeno et al., 1992; Madden et al., 1992); in contrast, the haploid pattern is thought to position cells in a microcolony derived from a homothallic cell that switches its mating type close to one another to facilitate mating and diploid formation (Nasmyth, 1982). It is thought that cortical tags mark future bud sites and serve as spatial cues for the initiation of new buds (Chant et al., 1991; Snyder et al., 1991; Flescher et al., 1993; Chant and Pringle, 1995).
Three distinct classes of proteins important for bud site selection have been identified (Snyder, 1989; Chant and Herskowitz, 1991). The first class is specifically required for the axial pattern. Mutations in genes encoding any of these proteins result in bipolar budding in haploid cells, but they do not disrupt the diploid budding pattern. It is thought that these genes are important for tagging the axial pole and/or recognizing the axial tag (Chant and Herskowitz, 1991; Flescher et al., 1993; Fugita et al., 1994; Chant et al., 1995; Halme et al., 1996; Roemer et al., 1996a; Sanders and Herskowitz, 1996). The second class is specifically required for the bipolar pattern. It is likely that these proteins are involved in marking the poles of diploid cells at sites for budding or helping direct those tags to their appropriate location (Snyder, 1989; Bauer et al., 1993; Zahner et al., 1996; Chen et al., 2000; Sheu et al., 2000). The third class is required for both the axial and bipolar patterns. These gene products are believed to recognize the axial or bipolar tags provided by the first two classes of gene products and convey this information to the machinery involved in establishing cell growth (Bender and Pringle, 1989; Chant and Herskowitz, 1991; Chant et al., 1991; Park et al., 1993). Although much is known about the molecular mechanisms that help mediate the axial pattern of budding, much less is known concerning the mechanisms of the bipolar pattern.
A number of components important for bipolar bud site selection have been identified and characterized (Herskowitz et al., 1995; Pringle et al., 1995; Drubin and Nelson, 1996; Costigan and Snyder, 1998;). Three genes, BUD8, BUD9, and STE20, when mutated, cause diploid cells to form buds at one pole (Zahner et al., 1996; Sheu et al., 2000). bud8Δ/bud8Δ and ste20Δ/ste20Δ cells bud at the proximal pole of the daughter cell, whereas bud9Δ/bud9Δ cells bud at the distal pole. Bud8 and Bud9 have been proposed to act as bipolar landmarks or tags that recruit components involved in bud formation (Zahner et al., 1996). Consistent with this hypothesis, Bud8 localizes at the distal pole; two different localizations of Bud9 have been reported, one at the distal pole (where it would most likely be a repressor of bud formation) and the other at the proximal poles (where it might serve as a tag) (Taheri et al., 2000; Harkins et al., 2001). Ste20 is a PAK protein kinase homologue that has been shown to lie in the same genetic pathway as Bud8 (Sheu et al., 2000). How tags are produced, processed, and localized to the poles has not been investigated.
The actin cytoskeleton is essential for both polarized growth and bipolar bud site selection and is thought to mediate directional transport of secretory vesicles (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Novick and Botstein, 1985; Mulholland et al., 1994; Drubin and Nelson, 1996; Pruyne et al., 1998). Both cortical actin patches and components of the secretory apparatus localize to sites of growth (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Finger and Novick, 1998). Mutations in genes encoding actin, several actin-associated proteins, or proteins involved in actin organization cause defects in bipolar budding similar to those of act1 cells (Drubin et al., 1990; Adams et al., 1991; Crouzet et al., 1991; Vojtek et al., 1991; Bauer et al., 1993; Holtzman et al., 1993; Sivadon et al., 1995; Zahner et al., 1996).
A variety of other nonessential polarity proteins are also required for bipolar bud site selection. Deletion of SPA2, PEA2, BUD6, or BNI1 cause a random budding defect. Spa2, Pea2, Bud6, and Bni1 form a complex, the 12S polarisome, that localizes to sites of polarized growth (Fujiwara et al., 1998; Sheu et al., 1998); these proteins are present as a patch at the incipient bud site, at the tip of the growing bud, and at the mother-bud neck region before cytokinesis (Snyder, 1989; Gehrung and Snyder, 1990; Snyder et al., 1991; Kohno et al., 1996; Valtz and Herskowitz, 1996; Zahner et al., 1996; Amberg et al., 1997; Evangelista et al., 1997). Spa2p, Pea2p, Bud6p, and Bni1p are each required for apical growth (Sheu et al., 2000), which is the initial phase of bud growth in which cells grow at the bud tip (Lew and Reed, 1993). It has been suggested that these proteins are required for the polarized deposition of the distal tag during this period (Sheu et al., 2000).
Understanding bipolar bud site selection and identifying the molecules that participate in this process will provide insight into the molecular mechanisms of how this process is carried out and coordinated with other cellular events. Recently, a set of diploid strains containing homozygous deletions of nonessential yeast open reading frames (ORFs) was generated; this set allows a genome-wide investigation into the bipolar bud site selection (Winzeler et al., 1999). Here we report a genome-wide systematic screening of 4168 homozygous diploid mutants (affecting 84% of the total nonessential annotated ORFs) for those defective in the bipolar budding pattern. These studies led to the identification of 127 genes involved in this process, of which only 16 have been reported to affect bipolar budding previously. Based on the classes of mutants identified and the subsequent analysis of these mutants, we have identified a variety of cellular processes involved in the bipolar pattern and present a global model of how this process occurs.
MATERIALS AND METHODS
Yeast Strains and Media
All strains used in this study are congenic derivatives of FY2, which is itself a direct descendant of S288C (Brachmann et al., 1998). Most of the strains contain precise deletions in which the entire ORF after the ATG is replaced with a kanMX4 marker. Two independent transformants were prepared and MATa and MATα segregants were recovered from each transformant and mated to form the homozygous diploid strains (Winzeler et al., 1999). The different strains are available from Research Genetics (Huntsville, AL). Growth media and genetic manipulation have been described previously (Sherman et al., 1986; Guthrie and Fink, 1991).
Budding Pattern Analysis
Mutant strains were grown individually in 0.8 ml of liquid YPAD in 96-well format boxes (Dot Scientific, Burton, MI) and incubated at 30°C with shaking. Each well of the box contained one 3.5-mm glass bead to facilitate mixing. Cells were first grown to stationary phase and then diluted into another 96-well box with glass beads; cells were grown for at least six generations until mid-log-phase. Mid-log–phase cells were then fixed by the addition of formaldehyde to a final concentration of 3.7%. Cells were subsequently incubated for 1 h at 30°C, washed with phosphate-buffered saline, and resuspended in phosphate-buffered saline. Calcofluor White (Sigma, St. Louis, MO) was added to a final concentration of 2 μg/ml, bud scars and birth scars were visualized by fluorescence microscopy. Birth scars are chitin-depleted structures that mark the region where mother cells separated from their daughter cells. A bud site is defined as proximal, distal, or medial depending on whether the bud or bud scar resides in the one-third portion of the cell closest to the birth scar, the one-third of the cell opposite the birth scar, or the middle-third of the cell, respectively (Flescher et al., 1993). Bud site selection mutants were identified and retested. All of the unipolar and axial-like mutants were tested for mating type and appropriate auxotropic markers and examined for cell size to ascertain whether they are diploid or haploid. Twenty-eight strains were found to be haploids. Only mutants that reproducibly exhibited a budding pattern defect and were not haploids were chosen for further analysis.
To quantify the fidelity of bipolar bud site selection, only cells that had experienced at least three budding events were examined. Cells with bud scars exclusively at one pole were classified as unipolar; cells with medial bud scars were scored as random budding. The percentage of random budding cells was then determined for each sample. For each sample, 100 cells were counted from at least four independent fields. For some mutant strains, bud site selection was scored for the first, second, and third bud divisions with the use of the criteria described (Flescher et al., 1993; Zahner et al., 1996). For these analyses a total of 200–600 cells from each sample were scored for each division type.
To determine whether the genes required for the bipolar pattern are also important for the haploid axial pattern, 104 MATa deletion strains were examined by Calcofluor staining. For the genes whose loss in diploids caused a unipolar pattern, both MATa and MATα strains were examined.
Examination of Cell Morphology
The cells used for morphological analysis were grown as described above. Cells were examined by phase-contrast and differential interference contrast microscopy, and mutants were classified as described in RESULTS.
Construction of GFP-tagged Yeast Strain
GFP-tagged fusion proteins were constructed in MATa and MATα haploid wild-type yeast strains with the use of the strategy described by Schneider et al. (1995). Primers were designed with the use of the N-terminal coding sequence of the gene of interest and used to amplify a region of the plasmid pMPY-GFP (a generous gift from Jun-Yi Leu). The resulting ∼2.0-kb polymerase chain reaction (PCR) product contains the complete URA3 gene flanked by direct repeats encoding the GFP gene; the entire cassette is flanked by 45 bp of the gene sequence. These DNA fragments were transformed into haploid yeast, and transformants that had integrated the fragment correctly at the chromosomal locus were identified by PCR analysis. The integrants were allowed to grow overnight in YPAD and plated onto medium containing 5-fluorolorotic acid; resistant colonies that had lost the URA3 gene through homologous recombination between the two GFP-coding sequences were identified by PCR. This recombination event leaves a single in-frame GFP-coding sequence after the ATG codon. MATa and MATα strains containing the in-frame GFP cassette were then mated, and homozygous tagged diploid strains were analyzed to determined whether the GFP allele was functional. Homozygous ORF-GFP/ORF-GFP diploid strains were examined for growth rate and bipolar budding pattern defects.
GFP-Fluorescence Microscopy
Yeast strains harboring the GFP-Bud8 plasmid or strains with GFP-tagged Bud proteins were grown in synthetic complete medium lacking particular nutrients or synthetic complete medium to mid-log–phase. Cells from 3-ml cultures were harvested and immediately viewed with the use of a fluorescence microscope (Leica, Deerfield, IL) equipped with a GFP filter set (41017, Endow GFP, Chroma Technology, Brattleboro, VT). For similar experiments, all images were processed identically with the use of Photoshop 5.0 (Adobe Systems, Mountain View, CA).
RESULTS
Identification of Genes Important for Bipolar Budding in Yeast
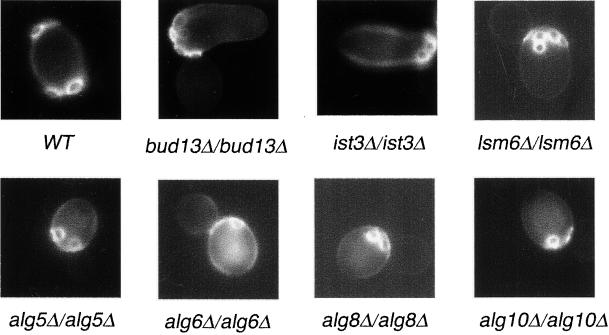
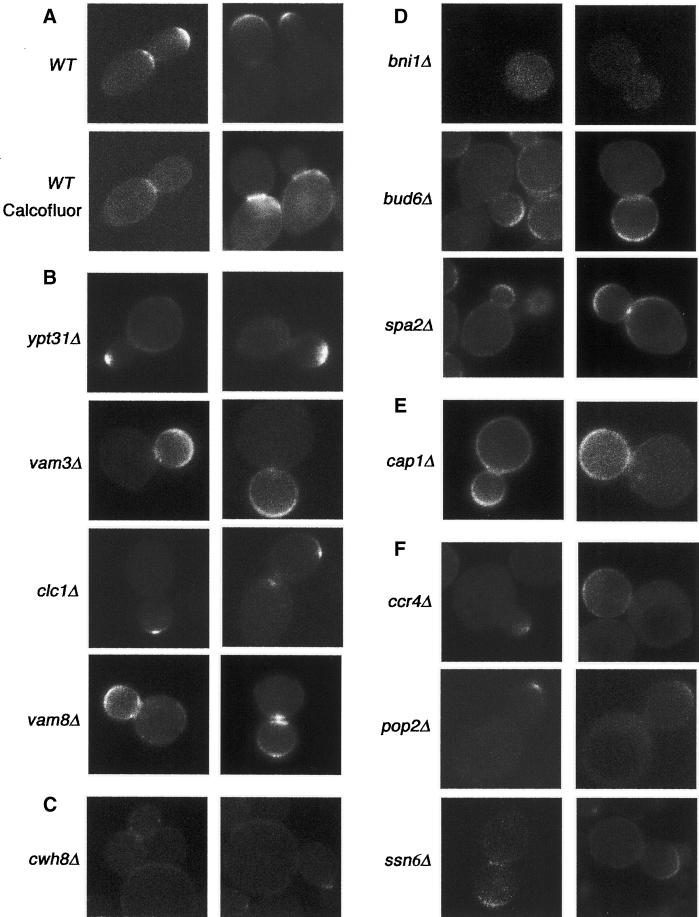
To identify mutants that specifically affect the bipolar budding pattern, we examined 4168 homozygous diploid deletion strains disrupted in different nonessential ORFs by staining with Calcofluor White. Calcofluor binds the chitin-rich bud scars that remain on the cell surface after cytokinesis and thus serve as markers of sites of previous cell divisions (Hayashibe and Katohda, 1973). Yeast cells were grown to mid-log phase, fixed, and stained with Calcofluor, and the budding pattern and cell morphology of each strain were examined. Wild-type diploid yeast cells undergo bipolar budding and bud scars are usually found at both poles of the cell (Figure 1). We identified 127 mutants that reproducibly displayed altered budding patterns; these mutants fell into three categories: unipolar, axial-like, and random (Table 1). In the unipolar mutants, bud scars lay at either the proximal or the distal pole of the cell. In the axial-like mutants, cells displayed chains of adjacent bud scars reminiscent of those observed for haploid cells; the bud scars in these classes usually initiated at the proximal pole but sometimes occurred in the equatorial region or at the distal pole. In the random mutants, bud scars appeared distributed over the entire surface of the cells. Of the 127 mutants identified, 105 are disrupted for genes that have been previously characterized; 22 affected uncharacterized ORFs. Twenty of the new genes were named BUD13-BUD32 (Tables 2–4).
Figure 1.
The budding pattern of diploid wild-type (WT) and unipolar mutant strains. Cells of the indicated strains were stained with Calcofluor to visualize bud scars.
Table 1.
Budding pattern mutants detected during this screen
| Phenotype | No. | % |
|---|---|---|
| Unipolar | 10 | 0.24 |
| Axial-like | 5 | 0.14 |
| Random | ||
| Strong | 74 | 1.78 |
| Mild | 38 | 0.91 |
| Total mutants | 127 | |
| Total strains | 4168 | 3.07 |
Table 2.
Homozygous diploid deletion strains with unipolar budding pattern
| Gene (ORF) | Budding pattern | Morphological phenotypea | Haploid budding pattern | Bud8p localization |
|---|---|---|---|---|
| BUD8 | Proximal | Round 2 | WT | WT |
| BUD9 | Distal | WT | WT | WT |
| STE20 | Proximal | WT | WT | WT |
| IST3 | Proximal | Elongated 3 | WT | Slightly reduced |
| BUD13/YGL174w | Proximal | Elongated 4 | WT | Slightly reduced |
| ALG5 | 74% distal | Round 3 | WT | More neck |
| ALG6 | 85% distal | Round 3 | WT | More neck |
| ALG8 | 56% distal | Round 3 | WT | More neck |
| ALG10 | 53% distal | Round 2 | WT | More neck |
| LSM6 | 69.5% distal | WT | WT | Not tested |
Cells were scored from 1–4: 1, wild type (WT); 2, weak; 3, moderate; 4, exhibited a severe defect.
Table 4.
Homozygous diploid deletion strains with random budding pattern
| Category | Gene (ORF) | Budding patterna | Morphological phenotype | Mat a budding pattern | Bud8p localization |
|---|---|---|---|---|---|
| Ribosome proteins | |||||
| Strong phenotype | RPS27B | >50% R | Round 2 | WT | |
| RPL14A | >50% R | WT | WT | ||
| RPS0B | >50% R | Round 2 | WT | ||
| RPL22A | >50% R | Round 3, clumpy 2 | WT | ||
| RPS17A | >50% R | Small 2 | WT | ||
| RPS7A | >50% R | Round 2+ | WT | ||
| RPL7A | >50% R | WT | WT | ||
| RPL39 | >50% R | Round 2, small 2 | NT | ||
| Weak phenotype | RPL12B | <30% R | Small 2 | WT | |
| RPL27A | <40% R | Round 2 | WT | ||
| RPS28B | <50% R | Round 2 | WT | ||
| RPS30A | <30% R | WT | WT | ||
| RPS29A | <30% R | WT | WT | ||
| RPS1B | <50% R | WT | WT | ||
| RPS18B | <30% R | Small 3 | WT | ||
| Vesicular transport proteins | |||||
| Strong phenotype | BST1 | >50% R | Round 4 | WT | WT |
| CLC1 | >50% R | Round 3, clumpy 3 | WT | Weak or ND | |
| YPT31 | >50% | Round 3 | WT | More patch | |
| VMA5 | >50% R | Round 2 | WT | WT | |
| VAM8 | >50% R | Large 2 | WT | Diffuse at bud and more at neck | |
| VPS34 | >50% R | Round 3, large 3 | WT | WT | |
| VAM3 | >50% R | Round 3 | WT | Most or all of the bud periphery | |
| VAC7 | >50% R | Large 2 | NT | WT | |
| END3 | >50% R | Small 3, round 2 | NT | More concentrated at the tip | |
| COS16 | >50% R | Rounds 2+ | NT | NT | |
| Weak phenotype | LUV1 | <50% R | Round 2 | WT | |
| CUP5 | <50% R | Round 3 | WT | ||
| VPS45 | <20% R, <20% U | Elongate 3, branch | NT | ||
| SEC22 | 15% R | Large 2 | WT | ||
| SNC2 | 50% R | Round 3 | WT | ||
| Actin-Cytoskeleton | |||||
| Strong phenotype | SLA1 | >50% R | Round 3 | WT | |
| RVS167 | >50% R | Round 2, clumpy 3 | WT | ||
| CAP1 | >50% R | Large 3, round 3 | WT | Diffuse | |
| YKE2 | >50% R | Round 2 | WT | ||
| MDM20 | >50% R | Round 3 | WT | ||
| RVS161 | >50% R | Round 3 | WT | ||
| Bud site selection and cell polarity proteins | |||||
| Strong phenotype | RSR1 | >50% R | Small 2, round 2 | Random | |
| BUD2 | >50% R | WT | Random | ||
| SPA2 | >50% R | Round 3 | WT | Diffuse | |
| BUD6 | >50% R | Round 3 | WT | Diffuse | |
| ROM2 | >50% R | Large 3, round 3 | WT | ||
| BEM4 | >50% R | Round 3 | R; U; B | WT | |
| BNI1 | >50% R | Round 3 | WT | ND | |
| Cell wall proteins | |||||
| Strong phenotype | CWH8 | >50% R | Round 3, clumpy 2 | Random | ND |
| FKS1 | >50% R | Small/round 3− | WT | WT | |
| GAS1 | >50% R | Round 3, clumpy 3 | WT | 17% of the cells | |
| Weak phenotype | SLG1 | <20% R | Elongate 2 | WT | |
| KRE6 | <50% R | Round 2, clumpy 2 | WT | ||
| CCW12 | 40% R | Round 2 | NT | ||
| Lipid metabolism | |||||
| Strong phenotype | GUP1 | >50% R | Round 2+, small 2+ | WT | |
| SUR4 | >50% R | Round 2 | B; U; R | ||
| Weak phenotype | FEN1 | 23% U, 7% R | WT | NT | |
| ERG4 | 30% R, 20% U | Round 3 | WT | ||
| ERG3 | 40% U | Round 2 | WT | ||
| Protein modification | |||||
| Strong phenotype | PMT2 | >50% R | Small 2, round 2 | WT | |
| MAP1 | >50% R | Round 3 | WT | ||
| OST3 | >50% R | Small 3, round 3 | WT | ||
| LAS21 | >50% R | Round 4 | WT | ||
| NAT3 | >50% R | Round 3 | WT | ||
| RAD6 | >50% R | Large 3 or large/round 3 | WT | ||
| Weak phenotype | MNN2 | 30% R | Small 3, round 3 | NT | |
| Transcriptional proteins | |||||
| Strong phenotype | CCR4 | >50% R | Round 3 | WT | Weak or ND |
| NOT5 | >50% R | Round 2, clumpy 2 | WT | Weak or ND | |
| POP2 | >50% R | Round 3 | NT | Weak or ND | |
| RLR1 | >50% R | Elongate 2 | NT | ||
| CTK1 | >50% R | Round 3 | NT | ||
| SSN6 | >50% R | Round 3, clumpy 3 | NT | Weak or ND | |
| TUP1 | >50% R | Round 3, clumpy 3 | WT | Weak or ND | |
| HCR1 | >50% R | Small 3 | NT | ||
| Weak phenotype | SIN4 | 50% U, 50% B | Clumpy 3+ | WT | |
| CTK3 | 20% R, few U | WT | WT | ||
| GCR3 | 43% U | WT | WT | ||
| RPB4 | 30% U, few R | Elongate 2 | NT | ||
| Nuclear proteins | |||||
| Strong phenotype | SPO7 | >50% R | Round 3, large 2 | WT | |
| NPL3 | >50% R | WT | WT | ||
| NSR1 | >50% R | Football 2 | WT | ||
| SFP1 | >50% R | Small 2 | WT | ||
| NEM1 | >50% R | Round 2 | WT | ||
| HMO1 | >50% R | NT | NT | ||
| RAI1 | >50% R | Round 2 | NT | ||
| LSM1 | >50% R | Small 3, round 3 | WT | ||
| LOC1 | >50% R | Round 3 | WT | ||
| Weak phenotype | TOP3 | 40% R | Large 2, elongate 2 | WT | |
| TRF4 | 20% U or R | Large 3 | WT | ||
| ZUO1 | <50% U | Round 2 | NT | ||
| Other proteins | |||||
| Strong phenotype | HEM14 | >50% R | Round 3 | WT | |
| ATP14 | >50% R | Small 2 | WT | ||
| PRS3 | >50% R | Round 3 | WT | ||
| RIB4 | >50% R | Round 3 | WT | ||
| Weak phenotype | YPS7 | 30% R | Small 3/round 3 | WT | |
| PRO1 | 50% R | Large 3, football 3 | WT | ||
| ILM1 | 50% R | Round 3 | NT | ||
| Unknown protein | |||||
| Strong phenotype | BUD14/YAR014c | >50% R | Elongate 3 | WT | |
| BUD15/YBL047c | >50% R | WT | WT | ||
| BUD16/YEL029c | >50% R | Large 2, round 3 | WT | ||
| BUD17/YNR027w | >50% R | Round 3 | NT | ||
| BUD18/YER044c | >50% R | Small/round 3, clumpy 2 | WT | ||
| YGR151ca | >50% R | Round 3 | R | ||
| BUD19/YJL188c | >50% R | Round 3 | WT | ||
| BUD20/YLR074c | >50% R | Small/round 3 | WT | ||
| BUD21/YOR078w | >50% R | WT | WT | ||
| BUD22/YMR014w | >50% R | Small 2 | NT | ||
| BUD23/YCR047c | >50% R | NT | NT | ||
| Weak phenotype | BUD24/YDR320c | 50% R | Small/round 2+ | WT | |
| BUD25/YER014c-a | <50% R | WT | WT | ||
| BUD26/YDR241w | 40% R | Small 2 | WT | ||
| BUD27/YFL023w | 40% R | Football 3 | WT | ||
| BUD28/YLR062c | <50% R, 15% U | Round 3 | WT | ||
| BUD29/YOL072w | <40% R | Round 2 | WT | ||
| BUD30/YDL151c | 45% R, 10% U | Round 2 | NT | ||
| BUD31/YCR063w | 50% R, few U | Large 3, round 2 | NT | ||
| BUD32/YGR262c | 50% R | Large 2 | NT |
WT, wild-type; ND, not detected; NT, not tested; R, random; U, unipolar; B, bipolar.
Overlaps with BUD1.
Unipolar Mutants
Ten mutants were found that exhibited a unipolar budding pattern (Table 2). Seven of the mutants have a strong phenotype in which >70% of the cells exhibited a unipolar budding pattern when cells with more than three bud scars were scored. Three exhibit a partial phenotype in which 50–70% of the cells exhibit the defect (Table 2).
BUD8, BUD9, and STE20.
Of the 10 mutants three, bud8Δ/bud8Δ, bud9Δ/bud9Δ, and ste20Δ/ste20Δ, were known previously to exhibit a unipolar budding pattern (Zahner et al., 1996; Sheu et al., 2000; see INTRODUCTION). bud8Δ/bud8Δ and ste20Δ/ste20Δ cells bud at the proximal pole, whereas bud9Δ/bud9Δ mutants bud at the distal pole.
IST3 and BUD13/YGL174w.
IST3 is a gene that has been described to have a role in mediating sodium tolerance; however, its molecular role is not known (Entian et al., 1999). YGL174w is an unknown gene that we named BUD13. These two proteins were recently reported to interact in vivo by the yeast two-hybrid assay (Uetz et al., 2000). Mutants lacking these genes display a unipolar budding pattern, with bud scars clustered adjacent to the birth scar at the proximal pole (Figure 1). In these mutants, individual bud sites can be observed adjacent to, overlapping, or within the birth scar and in the vicinity of other bud scars; however, these bud scars are not always immediately adjacent to the bud site of the previous cell cycle and are more loosely distributed in the polar region. This distribution is typical of the bipolar pattern at the proximal pole and is distinct from the axial budding pattern used by wild-type haploid cells. In addition, ist3Δ/ist3Δ and ygl174wΔ/ygl174wΔ exhibit elongated cell shapes; this phenotype is distinct from bud8Δ/bud8Δ and ste20Δ/ste20Δ mutants, which are round and exhibit an apical growth defect (Sheu et al., 2000).
ALG5, ALG6, ALG8, and ALG10.
ALG5, ALG6, ALG8, and ALG10 are a group of genes whose products function early in the yeast dolichol pathway that synthesizes the dolichol-linked oligosaccharide precursor for N-linked protein glycosylation (Heesen et al., 1994; Stagljar et al., 1994; Reiss et al., 1996; Burda and Aebi, 1998). ALG5 encodes a dolichol-P-glucose synthetase, and ALG6, ALG8, and ALG10 are genes encoding glucosyltransferases that, respectively, transfer the first, second, and third α-1,3-linked glucose to Dol-PP-GlcNac2Man9 in the final modification of outer chain elongation of N-linked oligosaccharides. Mutations of these genes block the addition of glucose residues to the lipid-linked oligosaccharide, but the nonglucosylated oligosaccharides are still transferred to proteins (Huffaker and Robbins, 1983). Homozygous deletion strains lacking ALG5, ALG6, ALG8, or ALG10 have a round cell shape and a partial unipolar distal budding pattern (74, 85, 56, and 53%, respectively) (Table 2; Figure 1). These results indicate that N-linked glycosylation is required for proper bud site selection.
LSM6.
Lsm6 is a member of the Sm-like group of proteins and has been implicated in mRNA decay and the function of the U6 snRNP (small nuclear ribonucleoprotein particles), which is involved in mRNA splicing (Tharun et al., 2000). Deletion of LSM6 causes most cells to undergo unipolar distal budding (69.5% distal budding; 23.4% bipolar budding; 7.1% random budding; Figure 1). Interestingly, deletion of LSM1, which only affects mRNA decay and not U6 function (Tharun et al., 2000), causes a random budding phenotype (see below). Presumably, Lsm6 regulates the stability or splicing of an RNA transcript important for mediating the bipolar budding pattern, such as Bud8.
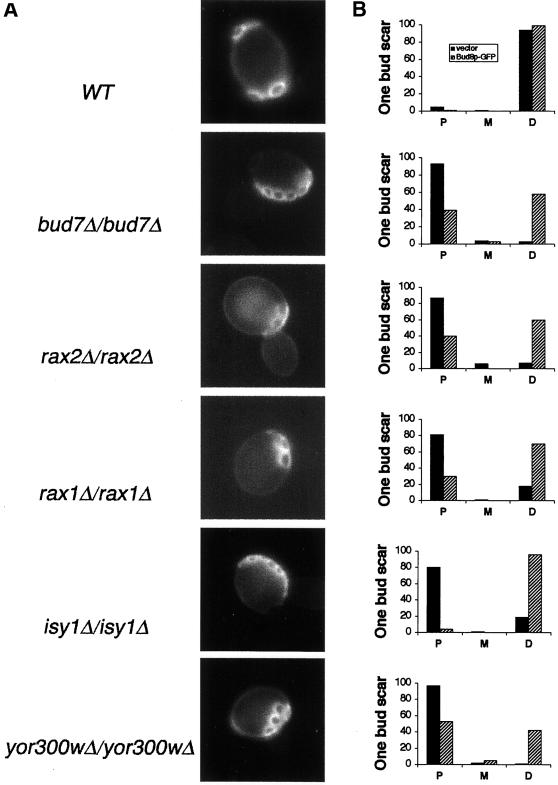
Axial-like Mutants
Five mutants were identified that exhibit an axial-like budding pattern (Table 3). This pattern differs from the unipolar pattern in that the cells had long chains of bud scars, very similar to those seen on axial budding cells (Chant and Pringle, 1995), indicating that new buds formed next to the previous site, rather than simply clustering at one end of the cell. However, in contrast to normal axially budding cells, these mutants produced chains of bud sites starting at either the distal pole or the proximal pole, as well as in the equatorial region, although there was a strong bias in most cases for one end of the chain to originate from the proximal pole. The bud scar chains can be very long and sometimes extend from one pole to another.
Table 3.
Homozygous diploid deletion strains with axial-like budding pattern
| Gene (ORF) | First bud proximal (%) | Morphological phenotype | MATa budding pattern |
|---|---|---|---|
| BUD7 | 93 | WT | WT |
| RAX2 | 87 | WT | WT |
| RAX1 | 80 | WT | WT |
| ISY1 | 81 | WT | WT |
| YOR300wa | 97 | WT | WT |
Overlaps with BUD7.
BUD7.
BUD7 was previously identified in a visual screen for mutants with a defect in the bipolar budding pattern; it was described as having a heterogeneous budding pattern phenotype (Zahner et al., 1996). We found that bud7Δ/bud7Δ cells exhibit an axial-like phenotype and that 75% of the deletion mutants use the proximal pole for the first two divisions (Figure 2A).
Figure 2.
Budding pattern of axial-like diploid mutant strains. (A) Bud scar (Calcofluor) staining of wild-type (WT), bud7Δ/bud7Δ, rax2Δ/rax2Δ, rax1Δ/rax1Δ, isy1Δ/isy1Δ, and yor300wΔ/yor300wΔ cells. (B) Quantitative evaluation of first bud position in wild-type (WT) strains, axial-like mutants, and strains overexpressing Bud8. P,XXX; M, XXX; D, XXX. In all panels, >200 cells were counted.
RAX2.
Chen et al. (2000) recently reported that RAX2 is required for the maintenance of the bipolar budding pattern, and their published data suggest a random budding pattern. We independently found that rax2Δ/rax2Δ mutants are defective in bipolar budding, but the divisions are usually in an axial-like budding pattern (Figure 2A). Furthermore, we find that 78% of the first three budding events occurred at the proximal pole (Ni and Snyder, unpublished result). Further analysis of rax2Δ mutants is described below.
RAX1.
Previous researchers have found that mutation of RAX1 converts the bipolar budding pattern of axl1Δ haploids into an axial budding pattern (Fujita et al., 1994), but the phenotype of rax1Δ/rax1Δ cells has not been described. We found that homozygous rax1Δ/rax1Δ cells bud in an axial-like pattern (Figure 2A); in their first two divisions they exhibit a strong bias to use the proximal pole.
ISY1 and YOR300w.
Two other proteins were identified that exhibit an axial budding pattern (Figure 2A). Isy1p is a pre-mRNA–splicing factor, required for optimal splicing in vivo (Dix et al., 1999). YOR300w is an unknown gene. Quantitative analysis of the first three budding events in these mutants reveals that each possesses a bias to use the proximal pole during the first several cell cycles. yor300wΔ/yor300wΔ cells exhibit the most severe defect of all of the axial-like mutants; the cells often contain bud scars that span from one pole to the other (Figure 2). The analysis of YOR300w is complicated by the fact that the ORF overlaps BUD7 by 100 bp and lies immediately upstream of RAX1 (within 100 bp). Thus, the deletion removes carboxy-terminal coding sequence from BUD7 and might affect the expression of RAX1, thereby affecting two genes involved in the bipolar pattern. This combined defect might account for a phenotype that is more severe than either the bud7Δ/bud7Δ or rax1Δ/rax1Δ single mutants.
Overexpression of Bud8p in each of the axial budding mutants, rax1Δ/rax1Δ, rax2Δ/rax2Δ, bud7Δ/bud7Δ, isy1Δ/isy1Δ, and yor300wΔ/yor300wΔ, induces many of the first buds to form distally, similarly to wild-type cells (Figure 2B). However, the degree of suppression varies with the individual mutants, and Bud8p overexpression usually causes subsequent budding events to occur more randomly, indicating that Bud8p cannot completely compensate for the bipolar budding defect of these mutant strains. Bud8p-GFP localization appears normal in these mutants.
Random Budding Mutants
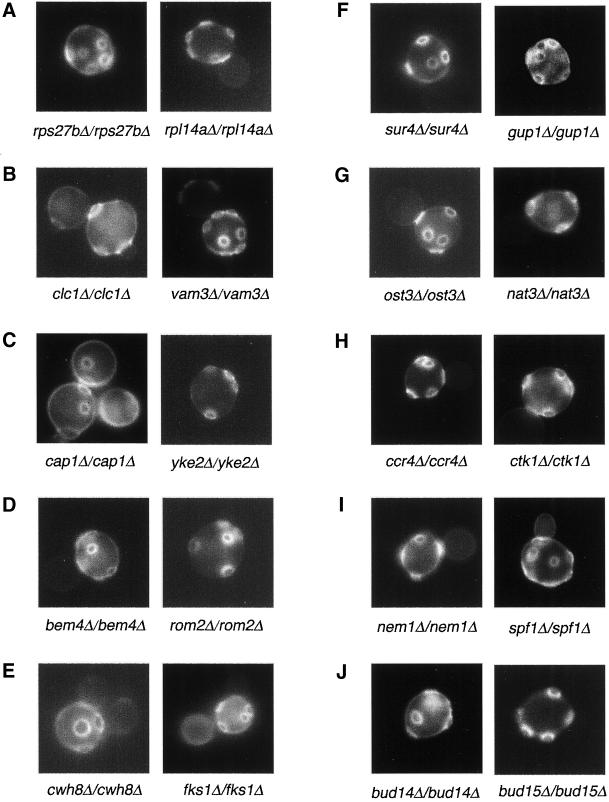
Seventy-four mutants were found to display a strong phenotype in which >50% of the cells budded randomly. An additional 38 mutants were found to exhibit a mild defect in which <50% of the cells budded randomly or sometimes random plus unipolar. The genes mutated in these strains were grouped into 11 functional categories (Table 4).
Ribosomal Proteins.
The first category is ribosomal proteins. The yeast genome is predicted to encode 32 different small subunit proteins and 46 large subunit proteins. Genes for 59 of the ribosomal proteins are duplicated, resulting in 137 total ribosomal protein genes (Planta and Mager, 1998). We found that a number of the duplicated ribosomal protein genes are important for bud site selection (Table 4). rps0bΔ/rps0bΔ, rps7aΔ/rps7aΔ, rps27bΔ/rps27bΔ, rpl7aΔ/rpl7aΔ, and rpl22aΔ/rpl22aΔ null mutants displayed a random budding pattern and reduced growth rate (Figure 3A). rpl12bΔ/rpl12bΔ, rps28bΔ/rps28bΔ, rps29aΔ/rps29aΔ, rps30aΔ/rps30aΔ, rps1bΔ/rps1bΔ, and rps18bΔ/rps18bΔ strains exhibit a random budding pattern but grow similarly to wild-type cells; the percentage of cells exhibiting a random budding defect is reduced relative to the other strains. rpl27aΔ/rpl27aΔ has a reduced growth rate and also exhibits a weaker random budding defect similar to the latter group of mutants. Interestingly, strains containing deletion mutations in the homologues of the affected genes (rps0aΔ/rps0aΔ, rps7bΔ/rps7bΔ, rps27aΔ/rps27aΔ, rpl17bΔ/rpl17bΔ, rpl22bΔ/rpl22bΔ, rpl12aΔ/rpl12aΔ, rps30bΔ/rps30bΔ, rps1aΔ/rps1aΔ, rps28aΔ/rps28aΔ) have no obvious phenotype (i.e., budding pattern or growth rate) even though the affected proteins are often >90% identical. Thus, the duplicated copies of the ribosomal proteins are not equivalent; the copy affected in the first group is more important for the bud site selection than the copy mutated the latter group. These results suggest that inefficient translation of particular genes involved in bipolar budding affects bud site selection. Alternatively, because many of the strains have a reduced growth rate, perhaps impaired growth of these particular strains results in a loss of bipolar budding.
Figure 3.
Micrographs of random budding mutants stained with Calcofluor. A–J are examples of 10 different functional classes. (A) Ribosomal protein mutants. (B) Vesicular transport mutants. (C) Actin-cytoskeleton mutants. (D) Bud site selection and cell polarity mutants. (E) Cell wall mutants. (F) Lipid metabolism mutants. (G) Protein modification mutants. (H) Transcription factor mutants. (I) Nuclear proteins mutants. (J) Uncharacterized proteins mutants.
Vesicular Transport Proteins.
Mutants for 15 proteins involved in vesicular transport exhibited a random budding pattern (Table 4). Four proteins have been implicated in exocytosis. Ypt31 is a small GTPase that belongs to the Ypt/rab family that is involved in trans-Golgi vesicle formation and localizes to sites of polarized cell growth (Jedd et al., 1997; Gerst, 1999). Bst1 was identified as a suppressor of the COPII gene SEC13, which is involved in transport from the endoplasmic reticulum to the Golgi (Elrod-Erickson and Kaiser, 1996). Sec22 and Snc2 are v-SNARE homologues involved in different steps of the secretory pathway; Sec22 is important for transport to the Golgi from the endoplasmic reticulum and Snc2 is important for late secretory steps (Woodman et al., 1996; Spang and Schekman, 1998; Gerst, 1999). Loss of SEC22 and SNC2 in homozygous diploid deletion strains result in modest bud site selection defects. Yeast haploid strains deleted for any of these four genes are not affected in the axial pattern. These results indicate that Ypt31p and Bst1p are important for the transport of membrane-associated bipolar budding components. Possible candidates are Bud8, Bud9, and/or Rax2, which are predicted transmembrane proteins.
The End3 protein, which is required for endocytosis and actin organization, has been described previously to be involved in bipolar budding (Benedetti et al., 1994). We also found that end3Δ/end3Δ mutants exhibit a random budding pattern. In addition, we found that Clc1 (Chu et al., 1996), which is the clathrin light chain, is also important for bipolar budding: clc1Δ/clc1Δ mutants exhibit a random pattern (Figure 3B). These results indicate that endocytosis may play an important role in bud site selection. As described below, we speculate that endocytosis is important for maintaining the localization of bud site selection tags at the bud tip.
Finally, we found that mutations in a variety of other proteins involved in vacuolar targeting or function are also defective in bud site selection. These include the proteins Vps34, Vac7, Vam3, Vam8, Vma5, Luv1, Cos16, Vps45, and Cup5. Vam3 and Vam8 are t-SNAREs (Preston et al., 1991; Rieder and Emr, 1997; Ungermann et al., 1999); Vma5 and Cup5 are vacuolar ATPases (Nelson et al., 1989; Bauerle et al., 1993). Cos16 is a vacuolar associated protein that affects ion homeostasis (Paidhungat and Garrett, 1998). Vps45 is a protein of the Sec1p family essential for vacuolar protein sorting (Bryant et al., 1998), and Vps34 is a phosphatidylinositol 3-kinase required for vacuolar protein sorting (De Camilli et al., 1996). These proteins might affect late trans-Golgi network sorting and thereby affect bud site tag targeting. Consistent with this possibility, many of these mutants have been described previously to be defective in endocytosis and/or exocytosis (Gammie et al., 1995; Rieder and Emr, 1997; Bryant et al., 1998; Conboy and Cyert, 2000).
Actin Cytoskeleton.
The actin cytoskeleton is essential for both polarized growth and bud site selection (Drubin, 1990; Adams and Pringle, 1991; Adams et al., 1991; Crouzet et al., 1991; Vojtek et al., 1991; Bauer et al., 1993; Holtzman et al., 1993; Zahner et al., 1996). We identified three mutants with defects in the actin cytoskleton, rvs167Δ/rvs167Δ, sla1Δ/sla1Δ, and rvs161Δ/rvs161Δ, that have defects in bipolar budding similar to several act1 mutants; these had been known previously to be important for bud site selection. In addition to the cytoskeletal components that were previously shown to be important for bud site selection, we found that yeast strains mutant for two additional proteins involved in actin organization, Cap1 and Yke2, are defective in bud site selection in diploid cells (Figure 3C). Cap1 is the α subunit of actin-capping protein. It colocalizes with cortical actin patches at the site of bud emergence and at the tips of growing buds and shmoos; it does not colocalize with actin cables or with actin rings at the site of cytokinesis (Amatruda and Cooper, 1992). Careful analysis of budding patterns revealed that diploid cap1Δ/cap1Δ mutants bud randomly both in the first division and in subsequent divisions (first bud: distal 29.7%; media 32.4%; proximal 29.7%; n = 200), similar to yeast bni1 mutants. Thus, in addition to its role in bipolar budding of mother cells, Cap1 might play an important role in the establishment of the distal bud tag in the daughter cell. Yke2 encodes prefoldin subunit 6, a component of the Gim protein complex that promotes formation of functional α-tubulin, γ-tubulin, and actin (Geissler et al., 1998; Siegers et al., 1999); mutation of YKE2 causes random budding in diploids.
We also found that mdm20Δ/dec1Δ mutants exhibit a random budding defect. mdm20Δ mutants lack actin cables (Hermann et al., 1997). They also are defective in mitochondrial movement and exhibit a reduced transfer of mitochondria into buds, which are actin-dependent processes (Boldogh et al., 1998). Thus, the random budding defect of mdm20Δ/mdm20Δ cells is likely due to a defect in actin organization.
The random budding defect of the different mutants that affect bud site selection and actin organization indicates that the actin cytoskeleton plays an important role in this process. The actin cytoskeleton at the incipient bud site and at the neck might help to localize cortical bud site selection tags to proximal and distal sites and/or maintain them properly at those sites; alternatively, actin filaments might be important for transport of bud site selection or polarity establishment components to these tags.
Bud Site Selection and Cell Polarity Proteins.
Seven deletion mutants affect bud site selection and cell polarity components (Table 4; Figure 3D). Five strains lack genes known to regulate bipolar budding pattern: RSR1, BUD2, SPA2, BUD6, BNI1; mutation of these genes cause random budding either in all cell divisions or after the first division (Snyder, 1989; Ruggieri et al., 1992; Park et al., 1993; Kohno et al., 1996; Zahner et al., 1996). ROM2 and BEM4 had not been reported previously to be involved in bipolar budding. Rom2 is a GDP-GTP exchange factor for Rho1p that is activated by cell wall defects; null mutations of rom2 impair the redistribution of actin patches in response to cell wall stress (Ozaki et al., 1996; Manning et al., 1997). Similarly, Bem4 interacts with Rho-type GTPases that regulate actin cytoskeletal reorganization (Mack et al., 1996; Hirano et al., 1997). Thus, these results indicate that Rho protein regulators and interacting proteins are important for bud site selection in yeast.
Cell Wall Proteins.
Six cell wall proteins were found to affect bipolar budding (Table 4). Three, fks1Δ/fks1Δ, gas1Δ/gas1Δ, and cwh8Δ/cwh8Δ, have a strong defect in which >50% of the cells bud randomly (Figure 3E). Fks1 is a component of β-1,3-glucan synthase; β-1,3-glucan is a major constituent of the yeast cell wall (Douglas et al., 1994). Gas1 is a putative glycosidase important for cell wall maintenance; it is thought to direct β-1,6-glucans to cross-link to β-1,3-glucan rather than to chitin (Vai et al., 1991). Cwh8 is a protein involved in the generation of the mannoprotein layer of the cell wall. Secreted proteins of cwh8 mutants have abnormally low levels of N-glycosylation (van Berkel et al., 1999). Interestingly, we found that fks1Δ and gas1Δ haploid cells do not have a defect in axial budding, whereas the cwh8Δ haploid strain exhibits a random budding defect.
We also found that mutations in three other components affecting cell wall integrity, KRE6 (a glucan synthase subunit; Roemer and Bussey, 1991), SLG1, and CCW12 (which are important for cell wall integrity) (Verna et al., 1997; Mrsa et al., 1999) exhibit a mild budding pattern defect in which <50% of the cells bud randomly. These results suggest that a proper cell wall is important for bud site selection in diploid cells. The cell wall might help anchor the tags and/or, in the case of Cwh8, might help produce a properly glycosylated tag.
Lipid Metabolism.
We also identified a number of genes involved in lipid biosynthesis or metabolism, including SUR4, FEN1, ERG3, ERG4, and GUP1 (Figure 3F). Sur4 and Fen1 are implicated in fatty acid synthesis and post-Golgi transport; mutations in these genes allow for the bypass of V-SNARE requirement in exocytosis (Oh et al., 1997; David et al., 1998). Erg3 and Erg4 are involved in ergosterol biosynthesis (Daum et al., 1998). Gup1 is a diacylglycerol O-acyltransferase-related protein; gup1Δ mutants have reduced lipid synthesis and increased glycerol synthesis (Oelkers et al., 2000). Mutants lacking these genes may have altered lipid compositions that might affect the secretion or function of bipolar markers and/or cell wall components important for the bipolar pattern.
Protein Modification.
We identified seven bud site selection genes whose proteins participate in protein modification (Table 4; Figure 3G). Three are involved in glycosylation. Mnn2 and Pmt2p are mannosyltransferases involved in N-linked and O-linked glycosylation, respectively (Lussier et al., 1995; Rayner and Munro, 1998). Ost3p is an oligosaccharyltransferase γ subunit, which is part of a complex that transfers core oligosaccharide from dolichol carrier to Asn-X-Ser/Thr motif, and helps position the OTase (oligosaccharyltransferase) complex for efficient N-glycosylation of secretory proteins (Knauer and Lehle, 1999). These results indicate that N-linked and O-linked glycosylation play a role in bipolar budding. It is likely that these proteins affect the modification of tags or cell wall components that anchor the tags.
Three other genes important for the bipolar pattern are involved in protein modification. Las21 is a protein required for addition of a side chain to the glycosylphospatidylinositol core structure (Hong et al., 1999). Map1p is a methionine aminopeptidase that cleaves the N-terminal methionine if the second residue has a small side chain (Gly, Pro, Ser, Thr, Val; Walker and Bradshaw, 1999). Nat3p is a protein N-acetyltransferase (Polevoda et al., 1999). Rad6 is a ubiquitin-conjugating enzyme involved in protein degradation (Jentsch et al., 1987). These proteins presumably modify proteins (or for Rad6 affect the turnover of proteins) involved in the bipolar pattern.
Transcription Factors and Chromosomal Proteins.
In this screen we identified 12 mutants lacking transcriptional factors that displayed a random budding pattern phenotype (Table 4; Figure 3H). Eight, ccr4Δ/ccr4Δ, not5Δ/not5Δ, pop2Δ/pop2Δ, rlr1Δ/rlr1Δ, ctk1Δ/ctk1Δ, ssn6Δ/ssn6Δ, tup1Δ/tup1Δ, and hcr1Δ/hcr1Δ, exhibit a strong defect, and four others, sin4Δ/sin4Δ, ctk3Δ/ctk3Δ, gcr3Δ/gcr3Δ, and rpb4Δ/rpb4Δ, displayed a weak defect. Ccr4, Not5, and Pop2 are components of the CCR4 transcriptional complex, which has positive and negative effects on transcription (Draper et al., 1995; Liu et al., 1998; Oberholzer and Collart, 1998). The CCR4 complex regulates transcription during late mitosis. It may function downstream of the Pkc1p-Mpk1p cascade to regulate the expression of a subset of yeast genes (Chang et al., 1999). Ssn6 and Tup1 form a complex and are general repressors of RNA polymerase II transcription (Williams et al., 1991). The Ssn6-Tup1 repressor has several distinguishing features, including its exceedingly efficient repression (>1000-fold for some target genes), the large number of genes repressed (as many as 3% of the S. cerevisiae genes), and its versatility with respect to the nature and number of activators it can prevail against (reviewed by Smith and Johnson, 2000). Because Tup1p-Ssn6 is required for the repression of many genes (including haploid-specific genes; Fujita et al., 1992; Keleher et al., 1992), deletion of Tup1-Ssn6 may induce expression of genes that affect the budding pattern. These results suggest that these transcription factors and chromosomal proteins regulate the expression of genes involved in the bipolar pattern. Ctk1 and Ctk3 are subunits of a protein kinase that phosphorylates the C-terminal domain of the large subunit of RNA polymerase II (Lee and Greenleaf, 1997). Mutation of CTK1 causes defects in both transcriptional repression and activation (Kuchin and Carlson, 1998). This kinase, which affects lexA-Tup1, might exert its activity by regulating gene expression (perhaps through Tup1) or by directly acting on components important for bud site selection. Altogether, our results indicate that gene regulation plays a very important role in bipolar bud site selection.
Nuclear Proteins.
A variety of other genes involved in nuclear function or whose encoded proteins localize to the nucleus have been identified (Table 4; Figure 3I). These include SPO7, NPL3, NSR1, SFP1, NEM1, HMO1, RAI1, LSM1, TOP3, ZUO1, and TRF4. These gene products are involved in nuclear transport (Npl3, Sfp1; Blumberg and Silver, 1991; Bossie et al., 1992), have RNA-binding motifs, are involved in RNA processing/modification (Rai1, Npl3, Lsm1, Nsr1, Zuo1, and Loc1; Kondo and Inouye, 1992; Kadowaki et al., 1994; Yan et al., 1998; Costanzo et al., 2000; Tharun et al., 2000; Xue et al., 2000), are required for nuclear morphology and/or meiosis (Spo7, Nem1; Siniossoglou et al., 1998), or are DNA-binding proteins and topoisomerase (or related) homologues (Top3, Trf4, Hom1; Kim and Wang, 1992; Castano et al., 1996; Lu et al., 1996). These gene products may be involved in the expression, processing, or transport of RNA transcripts for genes involved in bipolar budding. In addition, because some of these affect processing of rRNA (Npl3, Nsr1, Rai1; Kondo and Inouye, 1992; Russell and Tollervey, 1992; Xue et al., 2000), they might function in bud site selection through ribosome biogenesis or translation.
Other Genes and Uncharacterized Genes.
Other genes important for bipolar budding were also identified (Table 4; Figure 3J). These include seven known genes and 20 genes that have not been characterized previously. Deletion of 11 of the uncharacterized genes cause a strong random budding pattern, whereas the rest cause a weaker budding defect. We have named these genes BUD13 through BUD32. Further characterization of some of these genes is described below.
Many Bud Site Selection Mutants Exhibit a Defect in Cell Morphology
In addition to screening for bud site selection defects, we also examined the diploid collection for morphological defects. Wild-type diploid cells are normally ellipsoid. Previous work has demonstrated that mutants defective in apical growth, which is critical for an elongated cell shape, exhibit random budding, presumably through defective localization of tags at the poles of the cell (Sheu et al., 2000). By visually screening 3394 homozygous diploid deletion strains with the use of differential contrast interference microscopy of fixed cells, we identified 459 strains exhibiting slight to strong morphological alterations. Details of the results will be published elsewhere. Some of these shapes are described in Tables 2–4. Of particular interest, >40% of unipolar ist3Δ/ist3Δ and ygl174wΔ/ygl174wΔ mutants exhibited enhanced growth at the end of the cell where budding occurred. These results suggest that both growth sites and division sites reside at one end of the cell in these strains.
We also noticed that 70% of our random budding pattern mutants display a round phenotype suggestive of an apical growth defect. This observation supports the hypothesis that apical growth is important for bipolar bud site selection, as suggested previously (Sheu et al., 2000).
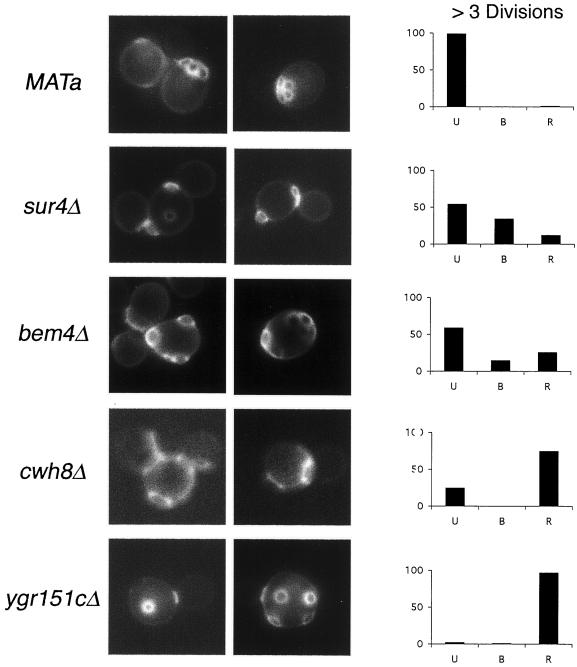
Several Genes Affect the Budding Pattern of Haploids
We expected that mutations affecting the diploid budding pattern would be of two types. The first type would be defective in both haploid and diploid budding patterns, such as rsr1 or bud2; these mutants are thought to be defective in the general bud site selection machinery (Chant et al., 1991; Ruggieri et al., 1992; Park et al., 1993). The second would be defective in functions specifically required for the bipolar budding pattern. To determine whether any of the mutations that affected the bipolar pattern also affected the axial pattern, we analyzed the budding patterns of 103 haploid MATa deletion strains with the use of Calcofluor staining.
Of the 103 mutants, 98 appeared to display normal or near normal axial budding, suggesting that these defects were indeed specific to the bipolar budding pattern. These include genes involved actin cytoskeleton organization, vesicular trafficking, ribosomal synthesis, transcriptional factors, and most cell polarity components (Tables 2–4). However, six haploid deletion mutants bud randomly; these are rsr1Δ, bud2Δ, bem4Δ, cwh8Δ, sur4Δ, and ygr151cΔ (Figure 4). Rsr1 and Bud2 were previously known to be important for the budding patterns of both haploid and diploid strains (Ruggieri et al., 1992; Park et al., 1993, 1999). ORF YGR151c partially overlaps Rsr1, and thus deletion of this ORF might affect Rsr1 function, thereby accounting for the random budding phenotype. cwh8Δ, sur4Δ, and bem4Δ have not been described previously to have a bud site selection defect in haploid cells. The Sur4, Cwh8, and Bem4 proteins might therefore function with either Rsr1, Bud2, or Bud5 to help select bud sites or with the polarity establishment proteins that are thought to be targeted to bud sites by the Rsr1 machinery.
Figure 4.
Budding patterns of haploid deletion mutants. Left, bud scar staining is presented for each sample. Right, cells with more than three bud scars were scored and classified as unipolar (U) if all bud scars were concentrated at one pole, bipolar (B) if one or more scars were at each pole, and random (R) if one of more scars were in the midsection of the cell (Flescher et al., 1993). In all panels, >200 cells were counted.
Bud8-GFP Subcellular Localization in Diploid Wild-Type and Budding Mutant Strains
To further understand how the different bipolar bud site selection mutants function, we analyzed the localization of Bud8 in wild-type and many of the mutant strains. Bud8 has been proposed to act as a distal tag for bipolar bud site selection; it localizes to the bud tip (Taheri et al., 2000; Harkins, et al., 2001). As noted above, bud8Δ/bud8Δ mutants are unable to bud at the distal pole and instead produce a unipolar proximal budding pattern in diploids (Zahner et al., 1996).
A high copy plasmid construct of the GFP:Bud8 gene (Harkins, et al., 2001) was transformed in a bud8Δ/bud8Δ deletion strain and found to complement the bud8Δ/ bud8Δ mutations (Ni and Snyder, unpublished results). When transformed into a wild-type diploid strain, the cells budded in a bipolar pattern; however, there is an increase in use of distal bud sites. This bias is especially apparent in the later budding events. For example, we found that in vegetatively growing wild-type strains containing Bud8-GFP, 95% of the second buds, and 91% of third buds, formed at distal poles, compared with only 68% of the second buds and 40% of the third buds for wild-type cells with vector alone. These results indicate that Bud8 activity is increased when overexpressed.
We next examined the localization of Bud8-GFP in the diploid wild-type strain. The Bud8-GFP was observed in ∼30% of the cells. As shown in Figure 5A, Bud8-GFP localized to the presumptive bud sites in unbudded cells, the distal tips of growing buds, and the proximal pole of some daughter cells, in agreement with previous results (Harkins, et al., 2001). In addition Bud8-GFP is also localized to the mother-daughter neck in some cells and was biased toward the mother side of the neck. Double staining with Calcofluor White revealed that neck staining was observed in only the daughter cells undergoing their first division (Figure 5A). As described in DISCUSSION, we propose that Bud8 at the distal tip helps serve as a tag for the distal site, and Bud8 at the neck in new mothers might also help select these sites.
Figure 5.
Localization of Bud8-GFP in diploid wild-type (WT) and mutant strains. Cells of the indicated strains carrying the plasmid Bud8-GFP were visualized by fluorescence microscopy. (A) Wild-type strains were viewed for both GFP and Calcofluor fluorescence (same cells). (B) Bud8-GFP in vesicular transport mutants. (C) Bud8-GFP in the cwh8Δ/cwh8Δ cell wall mutant. (D) Bud8-GFP in cell polarity mutants. (E) Bud8-GFP in the cap1Δ/cap1Δ actin-cytoskeleton mutant. (F) Bud8-GFP in transcription factor mutants.
Bud8-GFP Localization in alg Mutants.
We next examined the Bud8-GFP localization in the alg mutants, which have reduced outer chain carbohydrate modification. In alg5Δ/alg5Δ, alg6Δ/alg6Δ, alg8Δ/alg8Δ, and alg10Δ/alg10Δ mutants, Bud8p-GFP localized to the bud tip and the mother-daughter neck in many of the cells. However, detailed quantitative analysis of the localization of the mother-daughter neck showed an important feature: more cells contained Bud8-GFP localization at the neck in alg mutants than in wild-type cells. In cells that showed any detectable GFP signal, 48.6% of wild type have Bud8-GFP staining at the neck (n = 200); in contrast, for alg5Δ/alg5Δ, alg6Δ/alg6Δ, alg8Δ/alg8Δ, and alg10Δ/alg10Δ the percentage of cells with staining at the neck increases to 68.9, 65.8, 59.5, and 53.2% (n = 200), respectively. Thus, Bud8 outer chain modification might affect Bud8 targeting or degradation. For the latter possibility, in the absence of the outer chain modification Bud8 might be more stable and persist at the distal site longer. Alternatively, modification of other cell wall components might affect distal tag stability. Regardless, the increased presence of Bud8-GFP at the distal site (neck and bud tip) helps account for the partial unipolar budding phenotype of these cells; the cells might use the persistent Bud8 tag in multiple cell divisions.
Bud8-GFP Localization in Vesicular Transport Mutants.
Nine random budding mutants in the vesicular transport category were examined for Bud8-GFP localization. bst1Δ/bst1Δ, vma5Δ/vma5Δ, vps34Δ/vps34Δ, vac7Δ/ vac7Δ, and end3Δ/end3Δ each localize Bud8-GFP at polarized sites (bud tip and neck) similar to wild-type cells. Four mutants were found to have abnormal Bud8-GFP localization patterns. Clc1, the clathrin light chain, plays a role in formation of clathrin coats and clathrin-coated vesicles (Chu et al., 1996). Unlike a wild-type strain, in which >30% of the cells exhibit a polarized localization of Bud8-GFP, in clc1Δ/clc1Δ mutants >90% of the mutant cells lacked detectable Bud8-GFP localization; most of the remainder have a very weak Bud8-GFP signal, often visible as a tiny spot at the bud tip (Figure 5B). This result indicates that Bud8 undergoes intracellular trafficking through a clathrin-coated vesicle pathway or that disruption of that pathway indirectly alters Bud8 localization. In another mutant, ypt31Δ/ypt31Δ, which lacks a small GTPase involved in trans-Golgi vesicle formation, Bud8-GFP localized in a broader patch at the tip or/and neck (Figure 5B). Two other mutants that exhibit a defect in Bud8-GFP localization are in vam3Δ/vam3Δ and vam8Δ/vam8Δ; Vam3 and Vam8 encode syntaxin homologues (t-SNARE) thought to be important for vesicle docking and fusion reactions with the vacuole (Preston et al., 1991; Rieder and Emr, 1997; Ungermann et al., 1999). Bud8-GFP is not concentrated at the bud tip in vam3Δ/vam3Δ and vam8Δ/vam8Δ mutants. Instead, in vam3Δ/vam3Δ mutants it localizes throughout most or all of the bud periphery (Figure 5B). In vam8Δ/vam8Δ mutants, Bud8p is more diffuse at the tip and tends to form double rings at the neck (Figure 5B). These results indicate that Ypt 31, Vam3, and Vam8 may be involved in the targeting of Bud8p to the bud tip.
Bud8-GFP Localization in Cell Wall Mutants.
Bud8-GFP localization was also examined in three mutants that affect cell wall function. In fks1Δ/fks1Δ, Bud8-GFP localization appears normal. However, in cwh8Δ/cwh8Δ, Bud8-GFP cannot be detected, and in gas1Δ/gas1Δ, Bud8 localizes to polarized sites in only 17% of the cells and was not detectable in the remainder (n = 200; Figure 5C). These results indicate that Fks1 is not required for Bud8-GFP localization, whereas Cwh8 and Gas1 are required. Thus, specific components of the cell wall are important for Bud8 localization and/or Cwh8 is important for Bud8 modification.
Bud8-GFP Localization in Budding, Cell Polarity Mutants.
We also investigated Bud8-GFP localization in spa2Δ/spa2Δ, bud6Δ/bud6Δ, and bni1Δ/bni1Δ mutants. Spa2, Bni1, and Pea2 are important for apical growth and have been suggested to help concentrate polarized growth and secretion at bud tips and mother-bud necks (Sheu et al., 2000). In spa2Δ/spa2Δ and bud6Δ/bud6Δ mutants, Bud8-GFP localization is more diffuse at the bud tip compared with wild-type cells (Figure 5D); the Bud8-GFP signal is weaker in bud6Δ/bud6Δ cells than in spa2Δ/spa2Δ. In bni1Δ/bni1Δ, Bud8-GFP localization is not detected, indicating that Bni1 is required for Bud8 localization (Figure 5D). The absence of Bud8-GFP in any cells is consistent with the observation that bni1Δ/bni1Δ mutants bud randomly both in the first division and in subsequent divisions. The diffuse localization of Bud8-GFP in spa2Δ/spa2Δ cells is consistent with the observation that the first bud does not always occur at the apex of the distal tip of the cell (Sheu et al., 2000).
Bud8-GFP Localization in cap1Δ/cap1Δ Mutants.
Bud8-GFP localization was also investigated in cap1Δ/cap1Δ mutants, which lacks Cap1, an actin-capping protein α subunit. Bud8-GFP signal appeared more diffuse in this mutant, and the Bud8-GFP signal is diffuse not only in the bud but also in the mother, in contrast to the signal observed in the spa2Δ/spa2Δ and bud6Δ/bud6Δ mutants (Figure 5E). This result is consistent with the bud tip localization of Cap1 (Amatruda and Cooper, 1992) and implies that the actin cytoskeleton at the incipient bud site might help to localize cortical bud site selection tags to distal sites and/or maintain them properly at those sites.
Bud8-GFP Localization in Transcription Factor Mutants.
Bud8-GFP localization was examined in five transcription factors mutants. Ccr4, Not5, and Pop2 are components of the CCR4 transcriptional complex, which has positive and negative effects on transcription (Draper et al., 1995; Liu et al., 1998). The localization signal of Bud8-GFP in these mutants is very faint; most cells lack a detectable signal and the remainder have only a weak signal at the bud tip (Figure 5F). We presume that these mutants may directly or indirectly affect Bud8p expression. We also analyzed Bud8-GFP localization in ssn6Δ/ssn6Δ and tup1Δ/tup1Δ mutants. Tup1 and Ssn6 form a complex that functions as a transcriptional repressor (Williams et al., 1991). In both mutants, Bud8-GFP localizes to polarized sites in <10% of the cells, but the signal is very weak compared with wild-type cells (Figure 5 F). Thus, the Ssn6-Tup1 complex may also be involved in Bud8 regulation.
The altered localization and/or signal of Bud8 in many of these mutants demonstrate that the activity of Bud8 is influenced by many gene products and that Bud8 plays a critical role in bipolar budding. The mislocalization of Bud8 in many random budding mutants offers a clue as to why these mutants select nonpolar sites for bud formation; presumably other tags or localization components are also mislocalized, leading to further randomization of the budding pattern.
GFP Tagging of Uncharacterized Genes Required for Bipolar Budding
To further understand how the genes that were uncharacterized or had limited characterization function in bipolar bud site selection, subcellular localization studies were performed. We analyzed two proteins whose loss caused a unipolar budding phenotype (Ist3 and Bud13/Ygl174w) and seven uncharacterized proteins whose loss caused a random budding pattern (Bud14/Yar014c, Bud15/Ybl047c, Bud18/Yjl188c, Bud20/Ylr074c, Bud21/Yor078w, Bud22/Ymr014c, Loc1/Yfr001w, and Ygr151c; Table 5).
Table 5.
Localization of GFP fusion protein in tagged strains
| Class | Gene name | ORF | GFP-protein localization | Budding pattern of the tagged strains |
|---|---|---|---|---|
| Unipolar | IST3 | YIR005w | Nuclear | Bipolar |
| BUD13 | YGL174w(C) | Nuclear | Bipolar | |
| Random | BUD14 | YAR014c | Small bud tip, large bud neck | Bipolar |
| BUD15 | YBL047c | Small bud tip, large bud neck | Bipolar | |
| LOC1 | YFR001w | Nuclear | Most bipolar, few random | |
| BUD18 | YJL188c | ND | Random | |
| BUD20 | YLR074c | Nuclear | Bipolar | |
| BUD21 | YOR078w | Nuclear | Bipolar | |
| BUD22 | YMR014w | Nuclear | Bipolar | |
| YGR151ca | ND | Random | ||
| BUD18 | YJL188c | ND, slow growth | Random |
ND, not detected.
Overlaps with RSR1.
A GFP cassette was precisely inserted in the chromosome at the second codon after the initiator ATG for each of the genes, except for BUD13/YGL174w, in which GFP was placed immediately upstream of the translational stop codon. Thus, the different constructs express the GFP-ORF fusion protein from the endogenous promoter (see MATERIALS AND METHODS). Three of the tagged constructs exhibited defects in bipolar budding, and hence, insertion of the GFP cassettes inactivated the target gene; GFP localization was not detected for any of these strains (Table 5). One tagged strain, Loc1p/Yfr001w-GFP, which is a protein predicted to bind double-stranded RNA and is involved in localization of mRNA (Costanzo et al., 2000), displayed a mild defect in bipolar budding and a slow growth phenotype indicative of a partial defect in gene function; the Loc1p/Yfr001w-GFP localized to the nucleus (Figure 6C). Seven tagged strains appeared normal in growth and bipolar budding pattern, indicating that these fusion proteins were functional; these strains were studied further for protein localization.
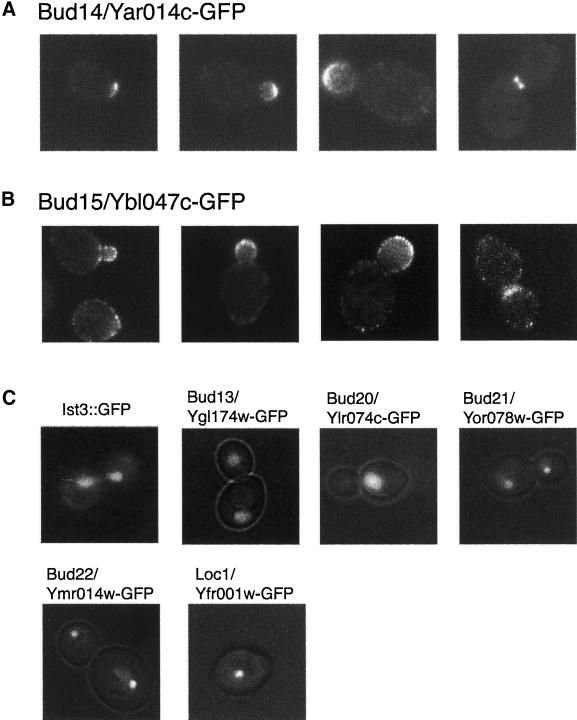
Figure 6.
Localization of Bud-GFP proteins. (A) Bud14/Yar014c-GFP, which is localized as a patch at the presumptive bud sites in unbudded cells, the distal tips of growing buds and the mother-daughter bud neck in large-budded cells. (B) Bud15/Ybl047c-GFP, which at the bud tip in small and large budded cells and at the mother-daughter neck in cells undergoing cytokinesis. (C) Six other GFP fusion proteins localized to nucleus.
BUD14/YAR014c is an unknown ORF, which encodes a predicted SH3 domain protein. Bud14p/Yar014c-GFP was found to be presented at the presumptive bud sites in unbudded cells, the distal tips of growing buds, and the mother-daughter bud neck in large-budded cells (Figure 6A). This polarized localization of Bud14p/Yar014c and the random budding phenotype of bud14Δ/bud14Δ (yar014cΔ/yar014cΔ) are consistent with a role for Bud14p/Yar014 in polarized growth.
Bud15p/Ybl047c is an unknown protein with similarity to the cytoskeletal proteins Uso1p and Pan1p and contains three Eps homology domains in the N-terminal region, which are often associated with regulation of protein transport, protein sorting, and membrane trafficking (Paoluzi et al., 1998). Bud15p/Ybl047c-GFP localized as a patch concentrated in the developing bud tip in cells with small buds and at the mother-daughter neck in cells undergoing cytokinesis (Figure 6B). This cellular localization suggests that Bud15p/Ybl047c may affect bipolar budding through polarized secretion. During the preparation of this report, Bud15p/Ybl047c was reported to play a role in endocytosis and genetically interact with Rsp5, a ubiquitin ligase. These authors reported a localization pattern similar to the one that we describe (Gagny et al., 2000).
GFP fusions for five other proteins, Ist3p, Bud13p/Ygl174w, Bud20/Ylr074c, Bud21/Yor078w, and Bud22/Ymr014w, localize to the nucleus. Three of these proteins, Ist3p, Bud13p/Ygl174w, and Bud20p/Ylr074c, localized throughout the nucleus (for Ist3p the signal was weak; Figure 6C). In contrast, two other proteins, Bud21p/Yor078w and Bud22p/Ymr014w, are highly concentrated in a single region of the nucleus (Figure 6C). In budded cells, the fluorescent signal was in the region of the nucleus closest to the bud. Fluorescence was observed in cells at all different phases of the cell cycle. These results suggest that these six proteins may function in the regulation of expression (transcription, processing, or transport, etc.) of bipolar budding components.
DISCUSSION
In this paper we report the screening of a large collection of homozygous diploid deletion mutants for defects in bipolar bud site selection. From a screen of 4168 mutant strains we found 127 bipolar budding mutants affecting both previously characterized genes as well as uncharacterized genes. The previously characterized genes include those involved in the actin cytoskeleton, bud site selection, cell polarity, exocytosis, and endocytosis, cell wall integrity, protein modification, lipid metabolism, translation, gene transcription, and nuclear functions. We also found a large number of new genes, including several whose encoded proteins are highly polarized in the cell. Analysis of these different mutants generated a comprehensive list of nonessential genes important for bipolar bud site selection and therefore provides a framework for this process in yeast.
An Integrated View of Bud Site Selection in Yeast
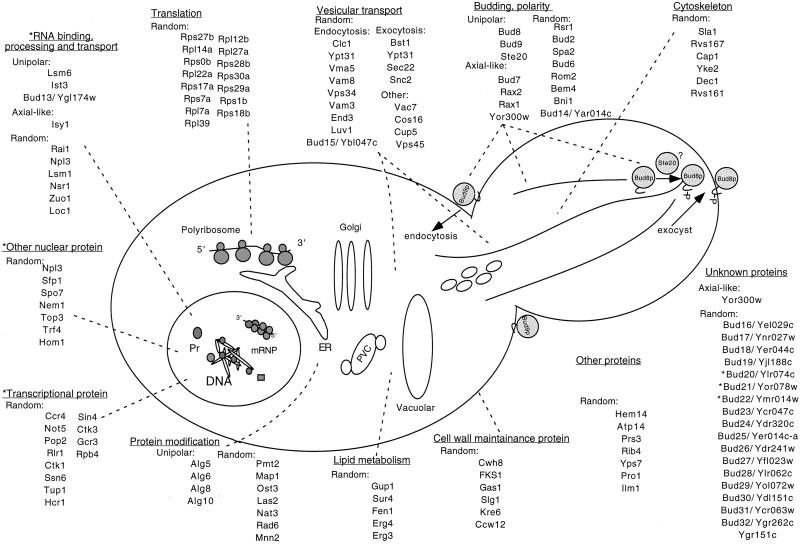
A large number of different types of genes involved in the bipolar pattern were identified. These include those involved in transcription, translation, polarity, bud site selection, lipid metabolism, protein modification, vesicular trafficking, cytoskeleton, and others. Based on our identification of their bud site selection phenotypes, their effect on Bud8-GFP localization, and the localization of their gene products, a model for our genome-wide characterization of this process is presented in Figure 7.
Figure 7.
A model for bipolar bud site selection that integrates the results of this study. As described in the text the different bud site selection proteins are likely to act at the sites indicated in the diagram. The bud genes indicated with a asterisk might either participate in transcription or other nuclear processes. ER, endoplasmic reticulum. mRNP, mRNA-protein; Pr, proteins; PVC, prevacuolar compartment.
We propose that the actin cytoskeleton is important for apical growth and perhaps the transport of the tags to cortical sites and/or their maintenance at those sites. Polarity proteins such as Spa2, Bud6, and Bni1 are required for the proper localization or maintenance of bipolar tags at their proper site (Sheu et al., 2000). Consistent with this interpretation, Bud8-GFP is not found at polarized sites in bni1Δ/bni1Δ strains and is more diffuse in spa2Δ/spa2Δ and bud6Δ/bud6Δ strains. These results account for the observations that bni1Δ/bni1Δ mutants bud randomly at each division, including the first one, whereas spa2Δ/spa2Δ and bud6Δ/bud6Δ mutants still bud at the distal pole at the first division (although not always immediately at the tip) but bud randomly in later divisions (Snyder, 1989; Zahner et al., 1996; Sheu et al., 2000).
One of the interesting classes of proteins is the cell wall components. Bud8 and Bud9 are predicted to be transmembrane proteins; anchoring in the cell wall may be important for forming permanent tags. Because Fks1 and Gas1 are important for only the bipolar pattern, they are likely to be important for anchoring the underlying cytoskeleton or for anchoring the cell wall tags (e.g., Bud8 and Bud9). In contrast, Cwh8 is important for both axial and bipolar budding; it may also serve to anchor Bud8 and Bud9, and perhaps Axl2, a transmembrane protein that has been proposed to be part of the axial tag (Halme et al., 1996; Roemer et al., 1996a).
Another interesting set of proteins is the membrane-trafficking components. Several of the proteins affecting the bipolar pattern are important for exocytosis, including Ypt31, Sec22, and Snc2 (Woodman et al., 1996; Jedd et al., 1997; Spang and Schekman, 1998; Gerst, 1999). We expect the putative membrane tags, such as Bud8, and components that might anchor the tag, such as cell wall components, would be delivered to the incipient bud site with the use of the secretory machinery. The finding that Bud8-GFP localizes diffusely (i.e., as a broader patch) in Ypt31 mutants suggests that Bud8 targeting is defective in this mutant.
Unexpectedly, several mutants that affect the bipolar pattern play a role in endocytosis, including the clathrin light chain and end3 (Benedetti et al., 1994; Chu et al., 1996). This raises the possibility that endocytosis might play an important role in bud site selection. Perhaps recycling distal tags from the surface, especially those that are diffuse, and then redirecting them to distal sites during apical growth helps focus tags at the distal tip. Alternatively, some of these proteins might also play a role in exocytosis. Consistent with this latter possibility for Clc1, little Bud8-GFP is detected at polarized sites or the cell periphery in clc1Δ/clc1Δ mutants, suggesting that it does not get transported properly to the surface. Presumably, clathrin-mediated secretion is needed for targeting of bud site selection components. For end3 mutants, Bud8-GFP may be more concentrated at the tip (data not shown); this result is expected for an endocytosis mutant but does not explain the random budding defect observed in this mutant. Presumably, other components of the tag or those factors that interact with or affect the regulation of tags are defective in these cells.
We also identified a number of components involved in lipid metabolism. These might function in one of several ways. As noted above, several of these mutants might affect the secretory pathway and thus affect the targeting of molecules important for bud site selection. Because deletion of SUR4 affects both haploid and diploids, the Sur4 protein may be involved in delivery or anchoring of both axial and bipolar tags; Erg3 and Erg4 are involved in only the bipolar pattern and presumably only affect the bipolar tags. Interestingly, deletion of Fen1 (Oh et al., 1997), which affects lipid elongation at an earlier step in the lipid elongation than Sur4, affects only the bipolar pattern. Thus, the different lipids have different effects on the distinct budding patterns.
We also identified a number of components important for protein modification, including those involved in N- and O-linked glycosylation (Alg, Mnn2, Pmt2, Ost3), lipid modification (Las2), acetylation (Nat3), initiator methionine processing (Map1), and protein turnover (Rad6). Presumably, these different components function on the tags or components that transport or anchor the tags. Consistent with this interpretation, we note that Axl2 (Roemer et al., 1996a), a component involved in axial bud site selection that may be part of the axial tag, is modified by O-linked glycosylation; pmt4Δ mutants fail in the initial step of O-linked modification and are defective in Axl2 modification and localization (Sanders et al., 1999). Thus, bipolar, as well as distal, tags might require N-linked and O-linked glycosylation.
We identified several transcription factors in our screen. We propose that they are required for the proper expression of either tags (e.g., Bud8) or components responsible for the proper transport or modification of the tag. Several of the RNA-processing components (e.g., Loc1, Rai1, Npl3, Lsm1, Nsr1, Lsm6, and Zuo1; Kondo and Inouye, 1992; Kadowaki et al., 1994; Yan et al., 1998; Costanzo et al., 2000; Tharun et al., 2000; Xue et al., 2000) might be important for the proper expression or transport of RNAs of such components as well. Those nuclear factors whose loss results in random budding are likely to be involved in the expression of multiple tags or their localization; they might affect apical growth or the actin cytoskeleton. Indeed, many of the mutants that exhibit random budding have a round cell shape, consistent with an apical growth defect.
Finally, we identified a number of ribosomal proteins that are important for bud site selection. The random budding of ribosomal protein mutants may be due to the inefficient translation of components important for the bipolar pattern; alternatively, it may be due to an overall growth defect. Consistent with the latter possibility, many of the mutants that exhibit a strong budding pattern defect also exhibit a strong growth defect. Because significant growth control occurs during G1, the coupling of translational efficiency and the onset of budding might be linked.
Novel Components Important for the Bipolar Pattern
In addition to known proteins, we also found a number of previously uncharacterized proteins that participate in bud site selection. Bud13p/Ygl174w and Ist3 localize to the nucleus and have a slightly reduced Bud8-GFP signal, suggesting that they might be important for the proper expression of Bud8 or other components that help localize Bud8. Bud14p/Yar014c and Bud15p/Ybl047c localize to sites of polarized cell growth. For Bud15p/Ybl047c the localization is more diffuse. Recently, another group identified Bud15/Ybl047c as an Eps homology-containing protein that is similar to Pan1 and End3, which are involved in endocytosis and actin organization (Gagny et al., 2000). Perhaps Bud15/Ybl047c is important in localizing Bud8 and other proteins to their proper sites. Bud14p/Yar014c exhibits a highly polarized localization, similar to Spa2, Pea2, and Bni1. However, unlike spa2Δ/spa2Δ, pea2Δ/pea2Δ, and bni1Δ/bni1Δ, bud14Δ/bud14Δ cells are elongated, indicating that they do not have an apical growth defect. We speculate that Bud14p/Yar014c may be important for the placement or recognition of proximal and distal bipolar tags.
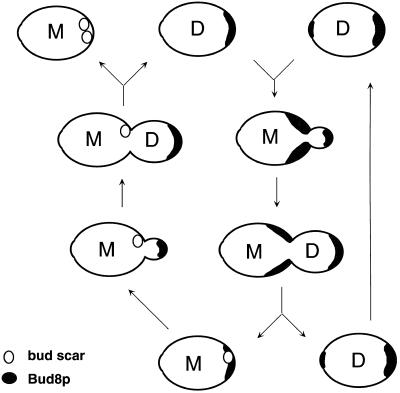
The Unipolar, Random, and Axial-like Budding Patterns
The different mutants affecting the bipolar pattern generally exhibit three types of patterns: unipolar, random, or axial-like budding. As postulated previously, the unipolar patterns are most readily explained by a loss of a functional tag at the appropriate pole (Zahner et al., 1996). Loss of functional Bud8, a component of the distal tag, yields a proximal budding pattern; loss of Bud9, a potential component of the proximal tag, produces a distal pattern. Random budding is most readily explained as either loss or mistargeting of functional tags for both poles.
The new category of mutants described in this study is the axial-like mutants. Previously this budding pattern has only been described for haploid cells (Fujita et al., 1994; Chant and Pringle, 1995; Chant et al., 1995; Halme et al., 1996; Roemer et al., 1996a; Sanders and Herskowitz, 1996). The fact that a significant number of our mutants exhibit this pattern indicates that the axial tags exist but are normally not recognized or are not functional in diploid cells. Consistent with this interpretation, most of the components required for axial budding (Bud3, Bud4, and Axl2/Bud10) are present in diploids (Chant et al., 1995; Halme et al., 1996; Sanders and Herskowitz, 1996). We speculate that in haploids these tags become the preferred site of budding either because the tags are modified to enhance their use or specific machinery that targets the axial sites is present in those cells. One candidate for an axial targeting or modification protein is Axl1 (Fujita et al., 1994); Axl1 is specifically expressed in haploids and can help convert the bipolar pattern to an axial pattern in diploids.
Why do several of the diploid mutants bud axially whereas others bud randomly? We speculate that the random budding mutants either fail to localize bipolar and axial tags or do not recognize these tags. In contrast, the axially budding diploid mutants are specifically defective in forming or recognizing the bipolar tags.
Several Genes Important for Bipolar Budding Are Also Involved in the Axial Pattern
Most mutants that we identified were specific to the bipolar pattern. This includes those affecting the cytoskeleton and polarity, which are important for apical growth. Others components such as transcription factors and cell wall components might be important for helping express, transport, or anchor bipolar tags at their proper sites.
However six of the mutants affect the haploid axial pattern. Bud1 and Bud2 were known previously and are likely to be involved in directing polarity establishment components to the chosen site. As noted above, the coding sequence of Ygr151c partially overlaps with Bud1, and thus mutations in this ORF are likely to affect Bud1. The identification of three new mutants important for bipolar and axial budding doubles the number of gene products thought to be generally important for bud site selection in yeast. Bem4 interacts with several of polarity establishment components (e.g., Rho1 proteins) and may be one of the earliest components delivered to the tag; further characterization of this protein will be reported elsewhere. Cwh8 and Sur4, which have not been well characterized, might participate in anchoring, modifying, and/or transporting both axial and bipolar tags (see above).
It is interesting to note which mutants did not affect haploid axial budding. Several of the cell wall components, Fks1 and Gas1, and Erg3 and Erg4, which are involved in ergosterol biosynthesis, are imporant for only bipolar budding and not axial, suggesting that these constituents are specific for the bipolar tags. Likewise, the Pmt2 and Mnn2 proteins, which act later in the carbohydrate modification pathway, are involved in bipolar budding and not axial budding; whereas Pmt4, which acts at the initial step, does affect axial budding. These results indicate that early chain modification is required for the Axl2 tag and the axial pattern, whereas late chain modification is required for the bipolar pattern.
Bud8 as a Distal Tag in Both New Mother and Daughter Cells
The observation that Bud8 is found both at the tip of the bud and at the neck in new mother-daughter provides an improved model for bud site selection in diploids (Figure 8). As suggested previously (Zahner et al., 1996; Taheri et al., 2000), Bud8 at the tip might serve as part of a distal tag for marking bud formation at that site (alternatively, it might help establish the tag). The Bud8 that resides at the neck of the new mother might also serve as a labile tag and help mark the distal site for the first several divisions. This would account for the distal site bias of new mothers. Consistent with this hypothesis, an increase of Bud8p activity by overexpression of a Bud8-GFP construct results in an increase in distal budding through several generations. Thus, Bud8 may be part of the distal tag, as suggested previously (Zahner et al., 1996; Taheri et al., 2000).
Figure 8.
An improved model of Bud8 localization. Bud8, indicated by the gray-shaded areas, might help direct new growth sites at the distal tip during both the first and subsequent divisions. See DISCUSSION for more details. M, mother; D, daughter.
Genome-wide Mutant Screens
This screen and that of Page, Beaulieu, Maynard, Azuma, Dijkgraaf, Li, Marcoux, Nguyen, Sdicu, and Bussey (unpublished results) are the first genome-wide screen of a defined null mutant collection. Previously, the largest collection of defined mutant alleles screened in yeast contained ∼2000 genes mutated by transposon insertion; these transposon-mutagenized strains were screened for disruption phenotypes with the use of 20 different growth conditions (Ross-Macdonald et al., 1999). The advantage of screening a bank of deletion strains is that nearly every nonessential gene may be investigated for its role in a given cellular process. Our screen for genes involved in bipolar bud site selection identified both previously characterized and novel genes.
At the time we carried out this screen, deletion strains for 4168 genes were available, which accounts for mutations in most annotated genes. We identified nearly all known nonessential genes (16 of 19) affecting bipolar bud site selection, including RSR1/BUD1, BUD2, BUD6, BUD7, BUD8, BUD9, SPA2, BNI1, and SUR4 (Park et al., 1993; Michelitch and Chant, 1996; Zahner et al., 1996; Amberg et al., 1997; Evangelista et al., 1997; Sheu et al., 2000; Taheri et al., 2000). The only known mutants in nonessential genes that were not identified were pea2Δ/pea2Δ, which was not available at the time of the screen, and bud5Δ/bud5Δ, which is not in the deletion collection for technical reasons. In addition, a large number of new genes (113) important for the bipolar pattern were identified. Because many of these mutants (91) corresponded to previously characterized genes, it is possible to identify pathways important for the bipolar pattern. Such models have been valuable in dissecting the secretory pathway(Kaiser et al., 1996). Indeed, based on the types of mutants identified in our screen we can present broad models for bipolar budding in yeast. Thus, one important advantage of screening a defined bank of mutants for which the function of many of the mutated genes is known is that it is possible to obtain a picture of the types of cellular processes that participate in the process of interest.
It is important to note that this collection does not identify all genes involved in a biological process such as bipolar budding. Mutations in essential genes and those that have not been annotated (either because they are <100 codons in length or because they overlap known genes) have not been screened. In addition, redundant genes for bipolar budding would not be identified in this mutation collection, because only one copy has been mutated.
In addition to previously known genes, we found that 22 of our mutants correspond to previously uncharacterized genes. One powerful advantage of screening a large defined collection of strains is that one can determine with certainty which mutants have been screened; thus, in addition to identifying genes that are important for a process, one can determine with certainty what genes are not required for a particular process. In addition, by screening a complete collection of mutants it is possible in principle to achieve saturation (or near saturation) from such an approach.
ACKNOWLEDGMENTS
We thank Greg Michuad, Yi-Jun Sheu and Anuj Kumar for critical comments on the manuscript. Laura Schenkman and John Pringle kindly provided the Bud8-GFP fusion, and Jun-Yi Leu provided the GFP construct for tagging the previously uncharacterized ORFs. This research was supported by National Institutes of Health grants GM-36494, CA-77808, and HG-01627.
REFERENCES
- Adams A, Pringle J. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Pringle JR. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- Adams AEM, Botstein D, Drubin DG. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature. 1991;354:404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Amatruda JF, Cooper JA. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Urdaci M, Aigle M, Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993;13:5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle C, Ho MN, Lindorfer MA, Stevens TH. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H(+)-ATPase membrane sector. J Biol Chem. 1993;268:12749–12757. [PubMed] [Google Scholar]
- Bender A, Pringle JR. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene, RSR1. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H, Silver P. A split zinc-finger protein is required for normal yeast growth. Gene. 1991;107:101–110. doi: 10.1016/0378-1119(91)90302-r. [DOI] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bryant NJ, Piper RC, Gerrard SR, Stevens TH. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45- independent, endocytic route. Eur J Cell Biol. 1998;76:43–52. doi: 10.1016/S0171-9335(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M. The ALG10 locus of Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Corrado K, Pringle JR, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud-site selection in yeast by a set of gene products that comprise a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, Tanaka S, Chant J, Fujita A. Multigenerational cortical inheritance of the rax2 protein in orienting polarity and division in yeast [in process citation] Science. 2000;290:1975–1978. doi: 10.1126/science.290.5498.1975. [DOI] [PubMed] [Google Scholar]
- Chu DS, Pishvaee B, Payne GS. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J Biol Chem. 1996;271:33123–33130. doi: 10.1074/jbc.271.51.33123. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Cyert MS. Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol Biol Cell. 2000;11:2429–2443. doi: 10.1091/mbc.11.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Hogan JD, Cusick ME, Davis BP, Fancher AM, Hodges PE, Kondu P, Lengieza C, Lew-Smith JE, Lingner C, Roberg-Perez KJ, Tillberg M, Brooks JE, Garrels JI. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Snyder M. Cell polarity in the budding yeast, Saccharomyces cerevisiae. Ad Mol Cell Biol. 1998;26:1–66. [Google Scholar]
- Crouzet M, Urdaci M, Dulau L, Aigle M. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast. 1991;7:727–743. doi: 10.1002/yea.320070708. [DOI] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- David D, Sundarababu S, Gerst JE. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- Dix I, Russell C, Yehuda SB, Kupiec M, Beggs JD. The identification and characterization of a novel splicing protein, Isy1p, of Saccharomyces cerevisiae. RNA. 1999;5:360–368. doi: 10.1017/s1355838299981396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG. Actin and actin binding proteins in yeast. Cell Motil Cytoskeleton. 1990;15:7–11. doi: 10.1002/cm.970150103. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Mulholland J, Zhu S, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson MJ, Kaiser CA. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, Schuster T, Hegemann JH, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg CP, Jansen G, Kramer W, Klein S, Kotter P, Kricke J, Launhardt H, Mannhaupt G, Maierl A, Meyer P, Mewes W, Munder T, Niedenthal RK, Ramezani Rad M, Rohmer A, Romer A, Hinnen A, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Finger FP, Novick P. Spatial regulation of exocytosis: lessons from yeast. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G, Lundelius JW. The developmental genetics of dextrality and sinistrality in the gastropod Lymnaea peregra. Wilhelm Rouxs Arch. 1982;191:69–83. doi: 10.1007/BF00848443. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Bud position in Saccharomyces cerevisiae. J Bacteriol. 1960;124:511–523. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugita A, Oka C, Arikawa Y, Katagal T, Tonouchi A, Kuhara S, Misumi Y. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature. 1994;372:567–569. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- Fujita A, Misumi Y, Ikehara Y, Kobayashi H. The yeast SFL2 gene may be necessary for mating-type control. Gene. 1992;112:85–90. doi: 10.1016/0378-1119(92)90306-a. [DOI] [PubMed] [Google Scholar]
- Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature. 1994;372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, ede1p, involved in endocytosis [in process citation] J Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell Mol Life Sci. 1999;55:707–734. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Fink GR. The logic of cell division in the life cycle of yeast. Science. 1992;257:626. doi: 10.1126/science.1496375. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [Google Scholar]
- Halme A, Michelitch M, Mitchell EL, Chant J. Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr Biol. 1996;6:570–579. doi: 10.1016/s0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- Harkins, H.A., Page, N., Schenkman, L.R., Virgilio, C.D., Shaw, S., Bussey, H., and Pringle, J.R. (2001). Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- Hayashibe M, Katohda S. Initiation of budding and chitin-ring. J Gen Appl Microbiol. 1973;19:23–39. [Google Scholar]
- Heesen S, Lehle L, Weissmann A, Aebi M. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Park H-O, Sanders S, Valtz N, Peter M. Programming of cell polarity in budding yeast by endogenous and exogenous signals. Cold Spring Harbor Symp Quant Biol. 1995;60:717–727. doi: 10.1101/sqb.1995.060.01.078. [DOI] [PubMed] [Google Scholar]
- Hirano H, Tanaka K, Ozaki K, Imamura H, Kohno H, Hihara T, Kameyama T, Hotta T, Arisawa M, Watanabe T, Qadota H, Ohya Y. ROM7/BEM4 encodes a novel protein that interacts with the Rho1p small GTP-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;16:4396–4403. doi: 10.1128/mcb.16.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Maeda Y, Watanabe R, Ohishi K, Mishkind M, Riezman H, Kinoshita T. Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J Biol Chem. 1999;274:35099–35106. doi: 10.1074/jbc.274.49.35099. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants [published erratum appears in J. Cell Biol. 1994 Sep;126(6):1627] J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Gimeno RE, Shaywitz DA. Protein secretion, membrane biogenesis, and endocytosis. In: Pringle J R, Broach J R, Jones E W, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 91–228. [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- Knauer R, Lehle L. The oligosaccharyltransferase complex from Saccharomyces cerevisiae: isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J Biol Chem. 1999;274:17249–17256. doi: 10.1074/jbc.274.24.17249. [DOI] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Kobayashi R, Brill SJ. Characterization of a high mobility group 1/2 homolog in yeast. J Biol Chem. 1996;271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- Lussier M, Gentzsch M, Sdicu AM, Bussey H, Tanner W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1- encoded activity. J Biol Chem. 1995;270:2770–2775. doi: 10.1074/jbc.270.6.2770. [DOI] [PubMed] [Google Scholar]
- Mack D, Nishimura K, Dennehey BK, Arbogast T, Parkinson J, Toh EA, Pringle JR, Bender A, Matsui Y. Identification of the bud emergence gene B.E.M4 and its interactions with rho-type G.T.Pases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4387–4395. doi: 10.1128/mcb.16.8.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Costigan C, Snyder M. Cell polarity and morphogenesis in Saccharomyces cerevisiae. Trends Cell Biol. 1992;2:22–29. doi: 10.1016/0962-8924(92)90140-i. [DOI] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and moprhogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Manning BD, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitch M, Chant J. A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr Biol. 1996;6:446–454. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J Bacteriol. 1999;181:3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- Nelson H, Mandiyan S, Nelson N. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase [published erratum appears in J. Biol. Chem. 1989 Mar 25;264(9):5313] J Biol Chem. 1989;264:1775–1778. [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Oberholzer U, Collart MA. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene. 1998;207:61–69. doi: 10.1016/s0378-1119(97)00605-7. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Garrett S. Cdc1 and the vacuole coordinately regulate Mn2+ homeostasis in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1787–1798. doi: 10.1093/genetics/148.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17:6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Park HO, Sanson A, Herskowitz I. Localization of bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 1999;13:1912–1917. doi: 10.1101/gad.13.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Norbeck J, Takakura H, Blomberg A, Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston RA, Manolson MF, Becherer K, Weidenhammer E, Kirkpatrick D, Wright R, Jones EW. Isolation and characterization of PEP3, a gene required for vacuolar biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5801–5812. doi: 10.1128/mcb.11.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J, Bi E, Harkins H, Zahner J, Devirgilio C, Chant J, Corado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem. 1998;273:26836–26843. doi: 10.1074/jbc.273.41.26836. [DOI] [PubMed] [Google Scholar]
- Reiss G, te Heesen S, Zimmerman J, Robbins PW, Aebi M. Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology. 1996;6:493–498. doi: 10.1093/glycob/6.5.493. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci USA. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Madden K, Chang J, Snyder M. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 1996a;10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- Roemer T, Vallier L, Snyder M. Selection of polarized growth sites in yeast. Trends Cell Biol. 1996b;6:434–441. doi: 10.1016/s0962-8924(96)10039-8. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung KH, Sheehan A, Symoniatis D, Umansky L, Heidtman M, Nelson FK, Iwasaki H, Hager K, Gerstein M, Miller P, Roeder GS, Snyder M. Large-scale analysis of the yeast genome by transposon tagging and gene disruption [see comments] Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Ruggieri R, Bender A, Matsui Y, Powers S, Takai Y, Pringle JR, Matsumoto K. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ID, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Gentzsch M, Tanner W, Herskowitz I. O-Glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J Cell Biol. 1999;145:1177–1188. doi: 10.1083/jcb.145.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother-bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of PCR epitope tagging for protein tagging in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Shapiro L. Protein localization and asymmetry in the bacterial cell. Cell. 1993;73:841–855. doi: 10.1016/0092-8674(93)90266-s. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Cold Spring Harbor Laboratories: Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1986. [Google Scholar]
- Sheu YJ, Barral Y, Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y-J, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Smith RL, Johnson AD. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Schekman R. Reconstitution of retrograde transport from the Golgi to the ER in vitro. J Cell Biol. 1998;143:589–599. doi: 10.1083/jcb.143.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, te Heesen S, Aebi M. New phenotype of mutations deficient in glucosylation of the lipid- linked oligosaccharide: cloning of the ALG8 locus. Proc Natl Acad Sci USA. 1994;91:5977–5981. doi: 10.1073/pnas.91.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri N, Kohler T, Braus GH, Mosch HU. Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. EMBO J. 2000;19:6686–6696. doi: 10.1093/emboj/19.24.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping, and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae [see comments] Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ungermann C, von Mollard GF, Jensen ON, Margolis N, Stevens TH, Wickner W. Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J Cell Biol. 1999;145:1435–1442. doi: 10.1083/jcb.145.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vai M, Gatti E, Lacana E, Popolo L, Alberghina L. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J Biol Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel MA, Rieger M, te Heesen S, Ram A F, van den Ende H, Aebi M, Klis F M. The Saccharomyces cerevisiae CWH8 gene is required for full levels of dolichol-linked oligosaccharides in the endoplasmic reticulum and for efficient N-glycosylation. Glycobiology. 1999;9:243–253. doi: 10.1093/glycob/9.3.243. [DOI] [PubMed] [Google Scholar]
- Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A, Haarer B, Field J, Gerst J, Pollard TD, Brown S, Wigler M. Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell. 1991;66:497–505. doi: 10.1016/0092-8674(81)90013-1. [DOI] [PubMed] [Google Scholar]
- Walker KW, Bradshaw RA. Yeast methionine aminopeptidase I: alteration of substrate specificity by site-directed mutagenesis [published erratum appears in J. Biol. Chem. 1999 Sep 17;274(38):27338] J Biol Chem. 1999;274:13403–13409. doi: 10.1074/jbc.274.19.13403. [DOI] [PubMed] [Google Scholar]
- Williams FE, Varanasi U, Trumbly RJ. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Davis RW, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Woodman PG, Rodriguez L, Stirling CJ. Functional conservation of cytosolic proteins required for endosomal vesicle fusion. Yeast. 1996;12:1251–1262. doi: 10.1002/(SICI)1097-0061(19960930)12:12%3C1251::AID-YEA19%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]