Abstract
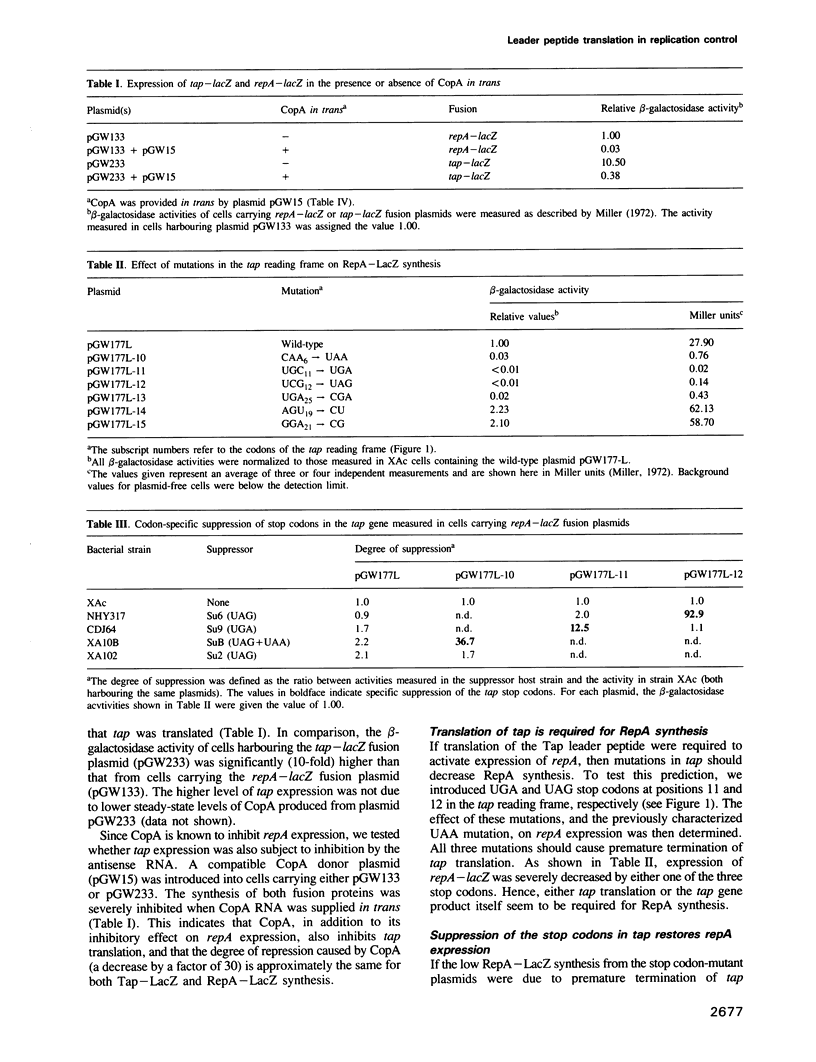
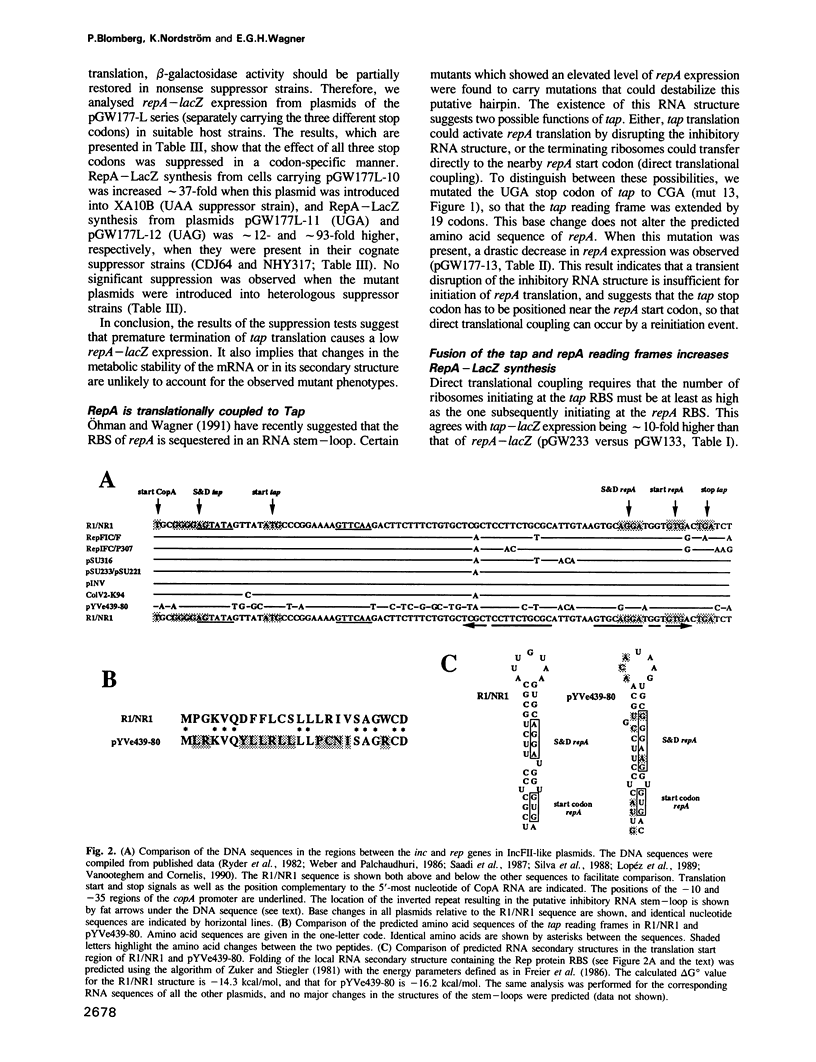
The replication frequency of plasmid R1 is post-transcriptionally controlled by an antisense RNA, CopA, that binds to the leader region in the RepA mRNA, CopT, and ultimately inhibits the synthesis of the replication initiator protein RepA. We present results demonstrating that CopA controls RepA synthesis indirectly. A reading frame for a 24 amino acid leader peptide (Tap, translational activator peptide) is located in the region between the copA and repA genes. A translational fusion between the tap and lacZ genes was used to demonstrate that tap is translated and controlled by CopA. Stop codons (UAA, UAG and UGA) introduced at three different positions within the tap gene led to a severe decrease in repA expression. Specific suppression of the stop codons reversed the effect. This indicates that tap translation is required for RepA synthesis. Phylogenetic comparisons between IncFII-like plasmids, together with previous in vitro and in vivo results (Ohman and Wagner, 1989, 1991), suggest that a stable RNA stem-loop structure sequesters the repA ribosome binding site irrespective of CopA-CopT duplex formation. The results presented here show that ribosomes translating the tap reading frame have to terminate close to the start codon of repA to permit reinitiation (direct translational coupling), and that transient disruption of the inhibitory RNA stem-loop is insufficient for activation of repA translation. The possibility that direct translational coupling is required because of a suboptimal repA RBS cannot be excluded.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asano K., Kato A., Moriwaki H., Hama C., Shiba K., Mizobuchi K. Positive and negative regulations of plasmid CoLIb-P9 repZ gene expression at the translational level. J Biol Chem. 1991 Feb 25;266(6):3774–3781. [PubMed] [Google Scholar]
- Asano K., Moriwaki H., Mizobuchi K. An induced mRNA secondary structure enhances repZ translation in plasmid ColIb-P9. J Biol Chem. 1991 Dec 25;266(36):24549–24556. [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Schmidt B. F., van Strien A., van Boom J., van Westrenen J., van Duin J. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J Mol Biol. 1987 Jun 5;195(3):517–524. doi: 10.1016/0022-2836(87)90180-x. [DOI] [PubMed] [Google Scholar]
- Berzal-Herranz A., Wagner E. G., Díaz-Orejas R. Control of replication of plasmid R1: the intergenic region between copA and repA modulates the level of expression of repA. Mol Microbiol. 1991 Jan;5(1):97–108. [PubMed] [Google Scholar]
- Blomberg P., Wagner E. G., Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990 Jul;9(7):2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer K. J., Ivey M. R., Steege D. A. Translational control of phage f1 gene expression by differential activities of the gene V, VII, IX and VIII initiation sites. J Mol Biol. 1987 Oct 5;197(3):439–451. doi: 10.1016/0022-2836(87)90557-2. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. Transcriptional pausing in a region important for plasmid NR1 replication control. J Bacteriol. 1987 Dec;169(12):5353–5363. doi: 10.1128/jb.169.12.5353-5363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Poulsen L. K., Thisted T., Nielsen A. K., Martinussen J., Andreasen P. H. The hok killer gene family in gram-negative bacteria. New Biol. 1990 Nov;2(11):946–956. [PubMed] [Google Scholar]
- Gerhart E., Wagner H., Nordström K. Structural analysis of an RNA molecule involved in replication control of plasmid R1. Nucleic Acids Res. 1986 Mar 25;14(6):2523–2538. doi: 10.1093/nar/14.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givskov M., Molin S. Copy mutants of plasmid R1: effects of base pair substitutions in the copA gene on the replication control system. Mol Gen Genet. 1984;194(1-2):286–292. doi: 10.1007/BF00383529. [DOI] [PubMed] [Google Scholar]
- Hama C., Takizawa T., Moriwaki H., Mizobuchi K. Role of leader peptide synthesis in repZ gene expression of the ColIb-P9 plasmid. J Biol Chem. 1990 Jun 25;265(18):10666–10673. [PubMed] [Google Scholar]
- Ivey-Hoyle M., Steege D. A. Translation of phage f1 gene VII occurs from an inherently defective initiation site made functional by coupling. J Mol Biol. 1989 Jul 20;208(2):233–244. doi: 10.1016/0022-2836(89)90385-9. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Baer M. F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J Mol Biol. 1988 Dec 20;204(4):879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- López J., Rodríguez J. C., Andrés I., Ortiz J. M. Characterization of the RepFVII replicon of the haemolytic plasmid pSU233: nucleotide sequence of an incFVII determinant. J Gen Microbiol. 1989 Jun;135(6):1763–1768. doi: 10.1099/00221287-135-6-1763. [DOI] [PubMed] [Google Scholar]
- Ma C., Simons R. W. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 1990 Apr;9(4):1267–1274. doi: 10.1002/j.1460-2075.1990.tb08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Projan S. J., Kornblum J., Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989 Oct 20;59(2):395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- Ohman M., Wagner E. G. Regulation of replication of plasmid R1: an analysis of the intergenic region between copA and repA. Mol Gen Genet. 1991 Nov;230(1-2):321–328. doi: 10.1007/BF00290683. [DOI] [PubMed] [Google Scholar]
- Ohman M., Wagner E. G. Secondary structure analysis of the RepA mRNA leader transcript involved in control of replication of plasmid R1. Nucleic Acids Res. 1989 Apr 11;17(7):2557–2579. doi: 10.1093/nar/17.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: formation of an initial transient complex is rate-limiting for antisense RNA--target RNA pairing. EMBO J. 1990 Nov;9(11):3777–3785. doi: 10.1002/j.1460-2075.1990.tb07591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988 Oct;7(10):3279–3288. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 1990 Nov;9(11):3767–3775. doi: 10.1002/j.1460-2075.1990.tb07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- Saadi S., Maas W. K., Hill D. F., Bergquist P. L. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987 May;169(5):1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R. M., Saadi S., Maas W. K. A basic replicon of virulence-associated plasmids of Shigella spp. and enteroinvasive Escherichia coli is homologous with a basic replicon in plasmids of IncF groups. Infect Immun. 1988 Apr;56(4):836–842. doi: 10.1128/iai.56.4.836-842.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Schmidt W., Cosstick R., Okruszek A., Eckstein F. The use of phosphorothioate-modified DNA in restriction enzyme reactions to prepare nicked DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8749–8764. doi: 10.1093/nar/13.24.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanooteghem J. C., Cornelis G. R. Structural and functional similarities between the replication region of the Yersinia virulence plasmid and the RepFIIA replicons. J Bacteriol. 1990 Jul;172(7):3600–3608. doi: 10.1128/jb.172.7.3600-3608.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G., Blomberg P., Nordström K. Replication control in plasmid R1: duplex formation between the antisense RNA, CopA, and its target, CopT, is not required for inhibition of RepA synthesis. EMBO J. 1992 Mar;11(3):1195–1203. doi: 10.1002/j.1460-2075.1992.tb05160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G., von Heijne J., Nordström K. Control of replication of plasmid R1: translation of the 7k reading frame in the RepA mRNA leader region counteracts the interaction between CopA RNA and CopT RNA. EMBO J. 1987 Feb;6(2):515–522. doi: 10.1002/j.1460-2075.1987.tb04783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Palchaudhuri S. Incompatibility repressor in a RepA-like replicon of the IncFI plasmid ColV2-K94. J Bacteriol. 1986 Jun;166(3):1106–1112. doi: 10.1128/jb.166.3.1106-1112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]




