Abstract
Neuregulin and the neuregulin receptor ERBB4 have been genetically and functionally implicated in schizophrenia. In this study, we used the yeast two-hybrid system to identify proteins that interact with ERBB4, to identify genes and pathways that might contribute to schizophrenia susceptibility. We identified the MAGI scaffolding proteins as ERBB4-binding proteins. After validating the interaction of MAGI proteins with ERBB4 in mammalian cells, we demonstrated that ERBB4 expression, alone or in combination with ERBB2 or ERBB3, led to the tyrosine phosphorylation of MAGI proteins, and that this could be further enhanced with receptor activation by neuregulin. As MAGI proteins were previously shown to interact with receptor phosphotyrosine phosphatase β/ζ (RPTPβ), we postulated that simultaneous binding of MAGI proteins to RPTPβ and ERBB4 forms a phosphotyrosine kinase/phosphotyrosine phosphatase complex. Studies in cultured cells confirmed both a spatial and functional association between ERBB4, MAGI and RPTPβ. Given the evidence for this functional association, we examined the genes coding for MAGI and RPTPβ for genetic association with schizophrenia in a Caucasian United Kingdom case–control cohort (n=~1400). PTPRZ1, which codes for RPTPβ, showed significant, gene-wide and hypothesis-wide association with schizophrenia in our study (best individual single-nucleotide polymorphism allelic P= 0.0003; gene-wide P= 0.0064; hypothesiswide P= 0.026). The data provide evidence for a role of PTPRZ1, and for RPTPβ signaling abnormalities, in the etiology of schizophrenia. Furthermore, the data indicate a role for RPTPβ in the modulation of ERBB4 signaling that may in turn provide further support for an important role of neuregulin/ERBB4 signaling in the molecular basis of schizophrenia.
Keywords: case–control studies, association, protein-tyrosine-phosphatase, receptor proteintyrosine kinases, neuregulins, ERBB4
Introduction
There is now a large body of evidence that the neuregulin 1 (NRG1) gene influences risk for schizophrenia, with an initial linkage and association study in the Icelandic population followed by more than 10 case–control studies and more than five family-based association studies providing positive evidence for association.1–4 NRG1 spans a 1.4-megabase region of chromosome 8p13, and is comprised of at least 20 exons. The gene undergoes complex alternative splicing, generating at least 15 protein isoforms that fall into six families (identified as I–VI).5 These gene products include soluble isoforms, as well as isoforms with one or two transmembrane domains. Gene products of NRG1 include heregulin, neu differentiating factor, acetylcholine receptor-inducing activity, glial growth factor 2 and sensory and motor neuronderived factor. Much of the positive association for NRG1 in schizophrenia centers around the first two exons of NRG1 (E187 and E1006, using the nomenclature of Steinthorsdottir et al.5), around the region of the Icelandic ‘core at-risk haplotype’ first demonstrated to be associated with the disorder. These two alternatively spliced exons are involved in specifying the type IV and type II isoforms of neuregulin 1.5
Expression studies of neuregulin 1 in postmortem samples from subjects with schizophrenia provide support for neuregulin 1 abnormalities in the disorder. Expression of type I-containing isoforms were elevated in schizophrenia.6,7 Moreover, a single-nucleotide polymorphism (SNP; SNP8NRG243177) and a four-marker haplotype (both derived from the Icelandic haplotype) were significantly associated with the expression of type IV neuregulin isoforms in the brain, with the risk allele or haplotype associated with increased expression of these isoforms.7
NRG1 gene products can activate intracellular signaling pathways through interactions with members of the ERB family of receptor tyrosine kinases (ERBB2, ERBB3 and ERBB4).8–11 Of the three ERBB receptors that are involved in NRG1 signaling, ERBB2 lacks an active ligand-binding domain and ERBB3 lacks an active catalytic domain, so both of these receptors must form heterodimers for signal transduction. 8–11 In contrast, ERBB4 can form functional homodimers as well as heterodimers. Recently, it has been shown that ERBB4 is a substrate for γ-secretase cleavage, which results in the liberation of the ERBB4 intracellular/cytoplasmic domain (EICD), which may then be involved in cytoplasmic and/or nuclear signaling.8–11 ERBB4 plays a critical role in the development of the brain, and has been shown to directly influence the proliferation, organization and migration of cells in the rostral migratory stream.12–14
There is now emerging evidence that the gene for ERBB4 also influences risk for schizophrenia,15,16 especially when considered together with NRG1.16 Moreover, there is evidence from postmortem mRNA studies for alterations in the levels of ERBB415 (as well as the ERBB3 coreceptor17–21) in schizophrenia. A recent study that investigated neuregulin stimulation of ERBB4 in slices of prefrontal cortex from control and schizophrenic subjects demonstrated enhanced activation of ERBB4 by neuregulin in schizophrenia.22 Finally, mouse models with disruptions in NRG1-ERBB4 signaling appear to capture some of the behavioral phenotypes associated with schizophrenia, and in certain cases these altered behaviors can be ameliorated by antipsychotics.1,23–25
Altogether, the data support a role for abnormal NRG1-ERB signaling in schizophrenia. Dissecting the pathways of NRG1-ERB signaling in the central nervous system may therefore contribute to the elucidation of pathways of schizophrenia susceptibility and may also reveal targets for interventions. The functional and genetic evidence implicating ERBB4 in schizophrenia suggests that this is a particularly interesting protein for further study.
In the current study we carried out a yeast two-hybrid analysis,26 with the cytoplasmic domain of ERBB4 as bait. We identified two proteins that interacted with ERBB4, one of which was the scaffolding protein MAGI-2.27 MAGI proteins have been shown to bind to receptor phosphotyrosine phosphatase β/ζ (RPTPβ),28,29 indicating the possible existence of a kinase–phosphatase complex brought together by a MAGI scaffold. In support of this, tyrosine phosphorylation ofMAGI proteins was increased in the presence of ERBB4 and decreased in the presence of RPTPβ. In light of these data, we examined PTPRZ1, the gene coding for RPTPβ, and MAGI-1-3, for evidence of association with schizophrenia. We obtained hypo thesis-wide evidence for genetic association between PTPRZ1 and schizophrenia, consistent with the general hypothesis of an etiological role for NRG1-ERBB4 signaling in schizophrenia.
Materials and methods
Yeast two-hybrid screening
The cytoplasmic domain of the cyt1 isoform of ERBB4 (i.e., the isoform which includes the cytoplasmic exon containing a docking site for phosphatidylinositol 3-kinase (PI3K)30) was cloned into the DUAL-hybrid bait vector in frame with the LexA DNA-binding domain, and the resultant construct confirmed by sequencing. After validating that the 96 kDa fusion protein was expressed in yeast and that it did not, by itself, activate reporter gene expression, 1.3×107 colonies from an adult mouse brain cDNA library (complexity 1×106; Clontech, Mountain View, CA, USA) were screened. Seventy clones were identified and subsequently reassayed, and 12 of these clones remained positive after an X-Gal filter test. These 12 clones were analyzed by retransformation with either the ERBB4 bait or with the control bait. All 12 clones showed bait dependency and were considered specific interactors in the yeast assay. Restriction analysis and sequencing of all clones indicated the existence of four unique clones among the 12. Yeast two-hybrid screening was carried out together with Dualsystems Biotech (Zurich, Switzerland).
cDNA constructs
Full-length MAGI-1 and MAGI-2 were cloned into pcDNA3 (Invitrogen, Carlsbad, CA, USA), with the addition of an hemagglutinin (HA) tag at the N terminal. In addition, the ERBB4 EICD, with an initiator methionine and a Kozak31 consensus sequence, was cloned into pcDNA3. Similarly, full-length receptor RPTPβ (short form) was cloned into pcDNA3. Expression vectors for the JMa splice variants of ERBB4 has been described previously.32 All ERBB4 constructs were of the cyt130 splice variant.
Transfection of mammalian cells
H4 human neuroglioma cells, maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in 5% CO2, were transfected with the indicated plasmids, using Lipofectamine (Invitrogen), following the manufacturer’s protocol.
Co-immunoprecipitation
For co-immunoprecipitation from cell lysates, cells were harvested in lysis buffer containing 1 × phosphate-buffered saline (PBS)/0.5% Triton X-100/protease inhibitor cocktail (Roche, Indianapolis, IN, USA). The lysate was sonicated using a Misonix microson sonicator (Misonix Inc., Farmingdale, NY, USA) and centrifuged at 16 000 g for 1 h to clear cellular debris. Cleared lysate (5 μg) was immunoprecipitated using antibodies as indicated, followed by the addition of 20 μl of protein A agarose (Invitrogen). The immunoprecipitate was washed three times in lysis buffer and eluted by boiling in sodium dodecyl sulfate (SDS) sample loading buffer containing 1M Tris–HCl (pH6.8)/10% SDS/50% glycerol/200mM β-mercaptoethanol/bromophenol blue. Samples were run on a 10% NOVEX SDS–polyacrylamide gel (Invitrogen) and transferred to membranes. Immunoblotting was carried out using primary antibodies as indicated, followed by secondary horseradish peroxidase-labeled anti-mouse antibody, and detection using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rochford, IL, USA).
Colocalization
Cells were fixed in 4% formaldehyde in PBS prewarmed to 37°C for 15 min and permeabilized in 0.1% Triton X-100 for 2 min. Cells were incubated with PBS/2% BSA for 1 h at room temperature, and then with antibodies as indicated. After three washes with 1 × PBS/0.2% BSA, cells were incubated with secondary antibody. Coverslips with adherent cells were mounted onto slides with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA). Fluorescent images were captured using a Leica TCSSP (UV) confocal microscope in the Mount Sinai Microscopy Shared Research Facility.
Tyrosine phosphorylation
To characterize tyrosine phosphorylation, MAGI-1 or MAGI-2 were transfected into 293 cells, together with ERBB2/3/4 constructs and/or RPTPβ. Twenty-four hours after transfection, cells were split into smaller wells, and allowed to adhere for an additional 8h. For neuregulin treatment, cells were then washed in serum-free medium and serum-starved for 12 h, followed by treatment for 15min with either 100 ng/ml of recombinant human neuregulin (NRG1-beta EGF domain, R&D systems, Minneapolis, MN, USA) or vehicle. Cells were subsequently lysed in phosphorylation lysis buffer (50mM Hepes (pH 7.5), 100mM NaCl, 10% glycerol, 1mM EGTA, 1.5mM MgCl2, 100mM NaF, 0.6mM Na3VO4 (activated) and Complete Protease EDTA-free Inhibitor Tablets (Roche)), with 2% SDS. After boiling for 2 min, lysates were spun to remove residual debris, and an equal volume of phosphorylation lysis buffer, with 10% Triton X-100, was added. MAGI proteins were then recovered by immunoprecipitation using anti-HA antibody. After electrophoresis and transfer to nitrocellulose, total MAGI was determined by immunoblotting with the anti-HA antibody, whereas tyrosine-phosphorylated MAGI was determined by immunoblotting with an anti-phosphotyrosine antibody (4G10, Upstate, Boston, MA, USA).
Genetic association analysis
The case sample consisted of 673 unrelated subjects with schizophrenia (455 males). All were white and born in the UK or Ireland, and had a consensus diagnosis of schizophrenia according to DSM-IV criteria made by two independent raters following a semistructured interview by trained psychiatrists or psychologists. Full details concerning diagnostic practices and demographics are reported elsewhere.33 The controls for the core sample were 716 individuals (482 males) matched to cases for age, sex and ethnicity, chosen from more than 1400 blood donors recruited from the National Blood Transfusion Service. They were not specifically screened for psychiatric illness but unless there is a major ascertainment bias towards patient inclusion, the use of unscreened controls does not affect power for disorder with the population prevalence of schizophrenia.34 Multicentre and Local Research Ethics Committee approval were obtained, and all subjects, cases and controls, gave written informed consent to participate. Markers that were significantly associated in the primary sample were genotyped in an additional set of controls (n = 617; 233 males) that was drawn from the same source. Although matched for ethnicity, they were not matched for gender. For none of the markers in which this additional sample was typed did the allele frequencies in controls vary by gender (Pmin = 0.12 for marker rs10278079) or between the two sets of controls (Pmin = 0.34 for marker rs1147502).
The SNaPshot (Applied Biosystems, Foster City, CA, USA) protocol for pooled DNA genotyping has been described in detail.35 Positive data in the pooled analyses were followed up by individual genotyping, thereby excluding false positives arising from measurement error. Because pooled analysis usually overestimates evidence for association, and because we genotyped a large number of markers in each gene, to strike a balance between sensitivity and follow-up genotyping burden, individual genotyping was undertaken where the estimated P-value for one marker was ≤0.01, or where more than one marker gave an estimated P-value from pooled analysis of ≤0.05. Individual genotyping was performed with the Sequenom MassARRAY using HME and iPlex chemistries (Sequenom, San Diego, CA, USA, http://www.sequenom.com) according to the recommendations of the manufacturers. All assays were optimized in 30 reference CEU trios (Utah residents with ancestry from Northern and Western Europe) from HapMap (http://www.hapmap.org). All plates contained a mixture of cases, controls, blanks and CEU samples. Genotypes were called in duplicate by a rater blind to sample identity and blind to the other rater. Genotypes of CEU samples were compared to those available on the HapMap to provide a measure of genotyping accuracy. Genotyping assays were only considered suitable for analysis if our own data from CEU individuals were the same as those in the HapMap during optimization and in sample plates.
For statistical analyses, pooled data were tested for association using contingency tables created by multiplying double the number of individuals represented in each pool by the estimated allele frequencies. Contingency tables were also used for single-marker case–control analysis. Haplotypes were analyzed using UNPHASED.36 Genotypes were tested for Hardy–Weinberg equilibrium using a χ2 goodness-of-fit test. Analyses of linkage disequilibrium (LD) between markers (r2 and D′) were performed using Haploview (http://www.broad.mit.edu/mpg/haploview/index.php). The number of effective independent SNPs assayed was estimated by the spectral decomposition method of Nyholt.37 The truncated product of P-values to derive overall gene-wide significance was calculated by multiplying all P-values reaching a threshold of P < 0.05 to achieve a product P-value.38 The significance of this product was established by permuting case–control status for all genotyped markers over 100 000 replicates and counting the number of times this product was exceeded.
Results
Identification of MAGIs as ERBB4-interacting proteins
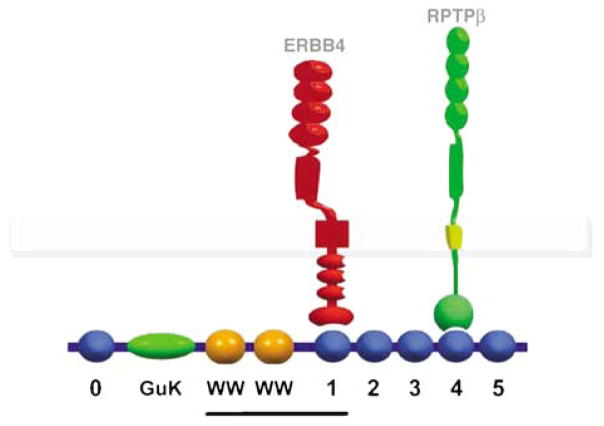
The cytoplasmic domain of ERBB4 was used to screen a mouse brain library for interacting proteins. We identified two classes of interacting clones, one including domains of the MAGI-2 scaffolding protein, and one including domains of the transcription factor PLZF. MAGI proteins include three highly related proteins (MAGI-1, MAGI-2 and MAGI-3), which are comprised of six PDZ domains (blue spheres, Figure 1), a guanylate kinase domain (oval) and WW domains (orange spheres). In the yeast two-hybrid screen, the MAGI-2 clones included the two WW domains and the downstream PDZ domain (PDZ1), corresponding to exons 5–9 of the gene (underlined in Figure 1). Glutathione S-transferase-pulldown experiments using recombinant proteins corresponding to the various domains of MAGI-2 confirmed that PDZ1 was able to bind the C terminus of ERBB4 (data not shown).
Figure 1.
Scheme of ERBB4-MAGI-RPTPβ interactions. PDZ domains of MAGI are numbered 0–5. ERBB4 binds to PDZ1, whereas RPTPβ binds to PDZ4. The line indicates the MAGI-2 sequence recovered in the yeast two-hybrid screen.
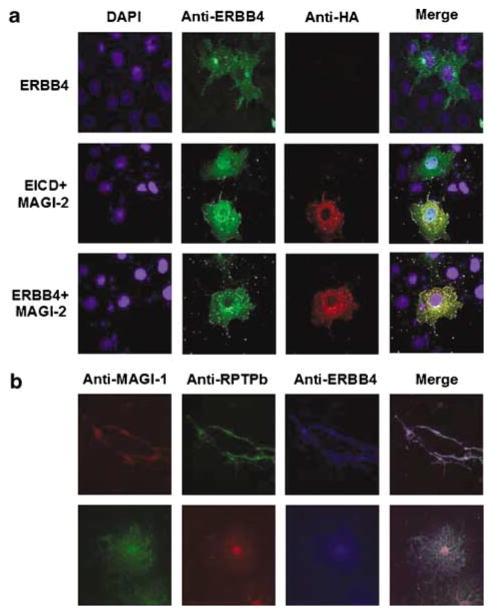
After demonstrating specificity of the MAGI-2/ERBB4 interaction in yeast (data not shown), we validated the interactions between MAGI and ERB4 in mammalian cells (Figure 2). We transfected human neuroglioma H4 cells with either control plasmid (vector alone) or MAGI-2, in the presence or absence of either full-length ERBB4 or EICD. Immunoprecipitation of MAGI-2 with anti-HA antibodies resulted in the co-precipitation of full-length ERBB4, the cytoplasmic fragment (CTF) of ERBB4 resulting from normal processing, and the EICD fragment (Figure 2).
Figure 2.
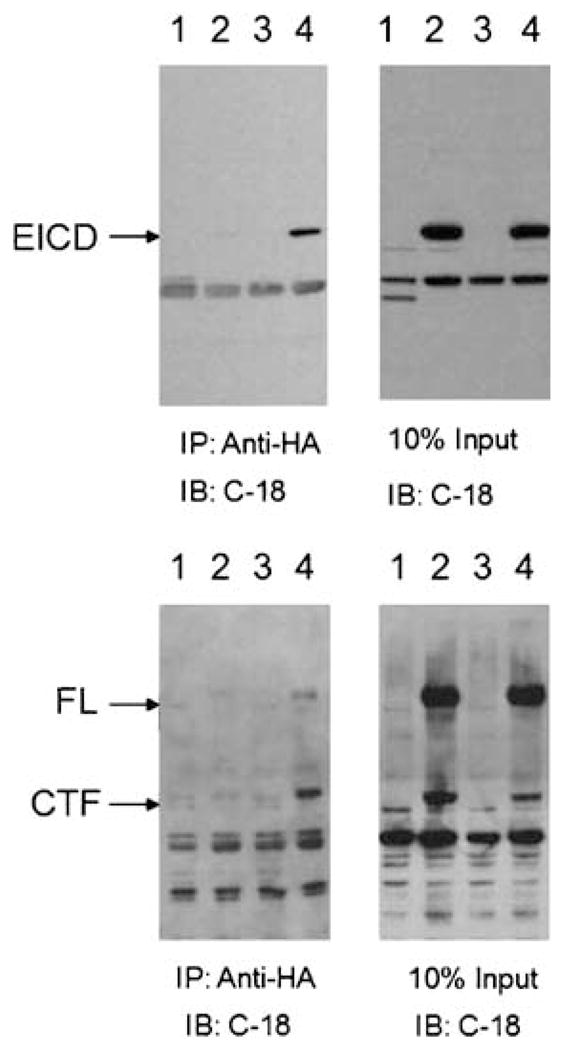
Co-precipitation of MAGI-2 and ERBB4. Cells were transfected with: (1) MAGI-2; (2) EICD (top panels) or ERBB4 (bottom panels); (3) vector; or (4) MAGI-2 and either EICD (top panels) or ERBB4 (bottom panels). Cells were lysed and subjected to immunoprecipitation (IP) with anti-HA antibody (to precipitate HA-tagged MAGI-2), and the amounts of EICD, ERBB4, or cleaved, C-terminal fragment of ERBB4 (CTF), were determined by immunoblotting (IB) with an antibody (C-18) against the C terminal of ERBB4 (left panels). In parallel, an aliquot of cell lysate from transfected cells (10% of the amounts used for the immunopreciptation) was immunoblotted to confirm expression of transfected proteins (right panels).
Expression of exogenous ERBB4 demonstrated primarily perinuclear localization, consistent with a localization of ERBB4 to membrane compartments (Figure 3a). Expression of exogenous MAGI-2 alone resulted in a more diffuse staining of the cytoplasm, whereas expression of EICD alone resulted in diffuse cytoplasmic and nuclear localization (data not shown). When we co-transfected MAGI-2 with either EICD or ERBB4, we observed colocalization of MAGI-2 with the other proteins, particularly in cytoplasmic compartments (Figure 3a). Interestingly, ERBB4 showed a more pronounced reticular staining in the presence of MAGI-2, as compared to in its absence (Figure 3a), indicating that MAGI-2 altered the steadystate distribution of ERBB4.
Figure 3.
Co-localization of MAGI with ERBB4 and RPTPβ. (a) Colocalization of MAGI-2 with ERBB4. H4 cells were transfected with ERBB4, ERBB4 and HA-tagged MAGI-2, or ERBB4 EICD and HA-tagged MAGI-2 and the proteins visualized by immunocytochemistry with an antibody directed against the C-terminus of ERBB4 and with an anti-HA antibody. (b) Colocalization of endogenous MAGI-1 with ERBB4 and RPTPβ. ERBB4, MAGI and RPTPβ were visualized in rat oligodendrocyte progenitor cells (top) and premyelinating oligodendrocytes (bottom) using anti-ERBB4, anti-MAGI-1 and/or anti-RPTPβ antibodies.
We then used purified rat oligodendrocytes, in which we had confirmed that ERBB4 and MAGI (predominantly MAGI-1) were expressed, to demonstrate colocalization of endogenous proteins (Figure 3b). Double labeling for ERBB4 and MAGI demonstrated colocalization of the two proteins.
Modulation of MAGI phosphorylation on tryrosine
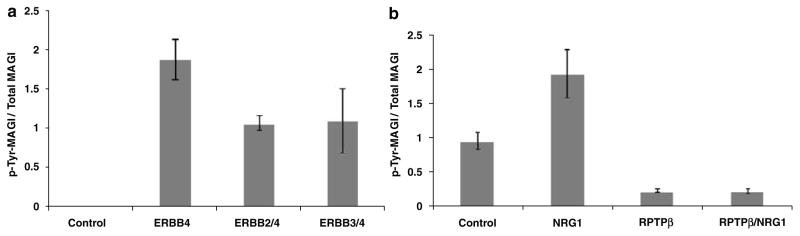
ERBB4 is a receptor tyrosine kinase, found either as a homodimer, or, more typically, as a heterodimer with ERBB2 or ERBB3. As MAGI proteins interact with ERBB4, we investigated whether MAGI proteins are substrates for tyrosine phosphorylation in the presence of ERBB4. When MAGI-1 (Figure 4a) or MAGI-2 (not shown) were co-transfected with ERBB4 (in the absence or presence of ERBB2 or ERBB3), we observed increased tyrosine phosphorylation of the MAGI proteins, when compared to cells transfected with MAGI proteins alone. We then transfected cells with ERBB4, ERBB2 and MAGI-1, treated them with vehicle or with neuregulin 1, and examined tyrosine phosphorylation of MAGI proteins. In the presence of neuregulin 1, we observed an approximately twofold increase in phosphorylation of MAGI-1 on tyrosine (Figure 4b). Similar results were observed when cells were transfected with ERBB3 and ERBB4 in place of ERBB2 and ERBB4 (data not shown).
Figure 4.
Tyrosine-phosphorylation of MAGI proteins. (a) Phosphorylation of MAGI-1 by ERB-receptor kinases. Cells were transfected with MAGI-1, in the presence or absence of ERBB2, ERBB3 and ERBB4 and tyrosine phosphorylation of MAGI-1 determined. (b) Phosphorylation/dephosphorylation of MAGI-1. Cells were transfected with MAGI-1, ERBB2 and ERBB4, in the presence or absence of RPTPβ. Transfected cells were treated with vehicle or NRG1, and tyrosine phosphorylation of MAGI-1 determined. Similar results were observed with MAGI-2 (data not shown).
MAGI proteins, including MAGI-1 and MAGI-3, have previously been shown to bind to RPTPβ (through PDZ4, SH, unpublished).28,29 Furthermore, RPTPβ has been shown to dephosphorylate tyrosinephosphorylated MAGI-1.29,39 As our data indicated that MAGI proteins could bind and be phosphorylated by a tyrosine kinase (ERBB4), we sought to determine whether MAGI proteins could represent a functional (and possibly physical) link between a phosphotyrosine kinase and a phosphatase activity. We therefore transfected cells with ERBB2, ERBB4 and MAGI-1, in the presence or absence of RPTPβ, and assayed the tyrosine phosphorylation of MAGI-1. Addition of RPTPβ resulted in a significant decrease in the phosphorylation of MAGI-1 (Figure 4b), supporting a functional relationship between ERBB receptors, MAGI proteins and RPTPβ. Similar results were observed with MAGI-2 (data not shown). Colocalization of ERBB4, MAGI and RPTPβ in oligodendrocytes indicated that such a complex could form endogenously (Figure 3b).
Genetic association analysis of PTPRZ1 with schizophrenia
Our interest in dissecting ERBB4 signaling was to contribute to the understanding of the etiology of schizophrenia. As RPTPβ appears to modulate ERBB4 (and ERBB2/ERBB4, ERBB3/ERBB4) signaling, we next asked whether sequence variants in PTPRZ1, which encodes RPTPβ, or any of the genes encoding MAGI proteins, were genetically associated with schizophrenia. We carried out association analysis using a two-staged approach. In the first stage, we looked for evidence of association by genotyping pools of DNA of cases and controls, followed by individual genotyping, if indicated. We required gene-wide significance to go on to the second stage, where we genotyped additional SNPs within the gene.
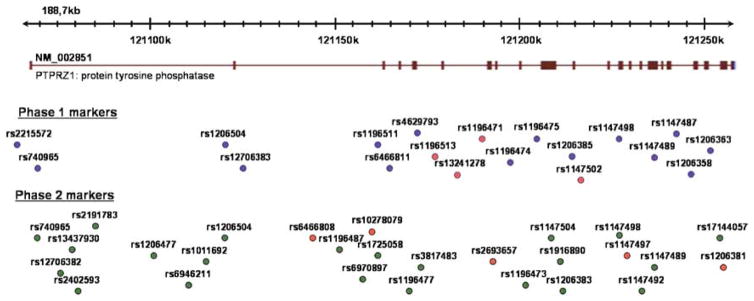
In stage 1 of the analysis of PTPRZ1, we genotyped in pools 19 polymorphic SNPs selected from dbSNP and HapMap (www.hapmap.org) with a focus on exonic sequences based on reference sequence NM_002851. Most of the exons were within 2 kb of a polymorphic SNP, the exceptions being exons 10 (3 kb from the nearest SNP), exon 16 (3.6 kb), exon 17 (3.8 kb), exon 28 (3.3 kb), exon 29 (4.3 kb) and exon 30 (5.6 kb) where the nearest markers were not polymorphic. The positions of the markers are indicated in Figure 5 and the results of the pooled analyses are presented in Table 1. Four markers (rs1196513, rs13241278, rs1196471 and rs1147502) yielded nominally significant evidence (P≤0.05) for association. Assays were designed for three of the markers (we dropped rs1196513 as this was highly correlated (r2 = 0.91) with rs1196471 in the CEU trios). However, the designed assays did not yield reliable genotypes at rs1196471 (but see below). Both rs13241278 and rs1147502 yielded nominally significant evidence for allelic association (P= 0.016 and P = 0.05, respectively) in the case–control sample. To determine if this evidence for association was due to sampling variation in the controls, we typed approximately 600 additional blood donor controls for the two markers. The combined results are given in Table 2. The evidence for rs1147502 remained effectively unchanged (P= 0.02), whereas that for rs13241278 improved somewhat (P= 0.003). To correct for multiple testing, we estimated the effective number of independent SNPs using the method of Nyholt37 based on the Caucasian European Utah Resident (CEU) dataset in HapMap. The number of independent SNPs (MeffLi) was estimated at eight. However, five of the 19 SNPs were not included in HapMap and therefore these were added to give a total of 13 independent SNPs, a conservative estimate because it assumes no LD between any of the additional five markers and any other marker. Thus, for rs13241278, the phase 1 genewide corrected P-value is 0.039. All genotypes were in Hardy–Weinberg equilibrium for cases and controls.
Figure 5.
Position of the markers analyzed across the PTPRZ1 gene. Markers analyzed in phase 1 and phase 2 are shown, with markers with a nominal P > 0.05 indicated in red.
Table 1.
Summarized pooled genotyping for PTPRZ1
| No. | SNP ID | Position | Distance to next SNP (bases) | Nucleotide changeb | Minor allele frequencya | Difference | χ2 | P | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Cases | Controls | ||||||||
| 1 | rs2215572 | 121064040 | G/A | 0.466 | 0.485 | −0.019 | 0.84 | 0.36 | |
| 2 | rs740965 | 121068064 | 4024 | G/T | 0.165 | 0.143 | 0.022 | 2.53 | 0.11 |
| 3 | rs1206504 | 121121853 | 53789 | G/A | 0.446 | 0.425 | 0.021 | 1.92 | 0.17 |
| 4 | rs12706383 | 121124481 | 2628 | C/T | 0.089 | 0.099 | −0.010 | 1.34 | 0.25 |
| 5 | rs1196511 | 121161514 | 37033 | T/C | 0.359 | 0.337 | 0.021 | 1.36 | 0.24 |
| 6 | rs6466811 | 121165045 | 3531 | A/G | 0.129 | 0.125 | 0.003 | 0.07 | 0.80 |
| 7 | rs4629793 | 121170753 | 5708 | A/G | 0.383 | 0.357 | 0.026 | 1.99 | 0.16 |
| 8 | rs1196513 | 121176314 | 5561 | C/T | 0.399 | 0.449 | −0.050 | 6.97 | 0.008 |
| 9 | rs13241278 | 121181151 | 4837 | C/T | 0.478 | 0.510 | −0.041 | 4.60 | 0.03 |
| 10 | rs1196471 | 121189416 | 8265 | C/T | 0.328 | 0.392 | −0.064 | 13.46 | 0.0002 |
| 11 | rs1196474 | 121196863 | 7447 | C/T | 0.117 | 0.120 | −0.003 | 0.00 | 0.97 |
| 12 | rs1196475 | 121203726 | 6863 | T/C | 0.364 | 0.393 | −0.029 | 2.17 | 0.14 |
| 13 | rs1206385 | 121213098 | 9372 | A/G | 0.317 | 0.292 | 0.025 | 1.93 | 0.17 |
| 14 | rs1147502 | 121215433 | 2335 | T/G | 0.463 | 0.501 | −0.038 | 3.84 | 0.05 |
| 15 | rs1147498 | 121225025 | 9592 | C/A | 0.167 | 0.183 | −0.016 | 2.68 | 0.10 |
| 16 | rs1147489 | 121233636 | 8611 | G/A | 0.250 | 0.274 | −0.023 | 1.91 | 0.17 |
| 17 | rs1147487 | 121239221 | 5585 | C/T | 0.400 | 0.424 | −0.024 | 1.58 | 0.21 |
| 18 | rs1206358 | 121244523 | 5302 | T/A | 0.422 | 0.396 | 0.026 | 1.90 | 0.17 |
| 19 | rs1206363 | 121250046 | 5523 | A/G | 0.349 | 0.329 | 0.020 | 1.59 | 0.21 |
Abbreviation: SNP, single-nucleotide polymorphism.
Minor allele frequencies in pools of 648 subjects with schizophrenia and 712 controls.
Minor allele presented first.
Table 2.
Individual genotyping of significant phase 1 markers
| No. | SNP ID | Location | Nucleotide changea | Number of cases | Number of controls | Minor allele frequency | χ2 | Allelic P-value | χ2 | Genotypic P-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Cases | Controls | ||||||||||
| 1 | rs13241278 | 121181151 | C/T | 594 | 1267 | 0.478 | 0.531 | 8.85 | 0.003 | 9.43 | 0.009 |
| 2 | rs1147502 | 121215433 | T/G | 661 | 1317 | 0.463 | 0.502 | 5.45 | 0.02 | 6.27 | 0.04 |
Abbreviation: SNP, single-nucleotide polymorphism.
Minor allele presented first.
Because the pooled analysis was biased towards exons (Figure 5) and also inaccuracies in pooling may result in type II error,34 to follow up our findings, we undertook a more thorough and effectively independent analytic approach based on HapMap. We selected markers using the program Tagger40 based on all markers available at the time in HapMap (Hapmap Data Ref # 19/phase II, Oct 05, on NCBI B34 assembly dbSNP b124) to derive a set of tagSNPs such that each common allele (minor allele frequency (MAF) ≥0.1) was captured with r2 >0.8 by a single marker. This suggested 27 tagSNP markers (Figure 5), which we genotyped in our core sample, in addition to rs1196471, which we had been unsuccessful in genotyping in phase 1. The results from the individual genotyping of the tagSNPs are presented in Table 3. Five of the markers gave nominally significant evidence for association (P≤0.05). These were rs6466808 (P=0.004), rs2693657 (P= 0.009), rs10278079 (P= 0.011), rs1147497 (P=0.032) and rs1206381 (P=0.042). Additionally, rs1196471, a phase 1 marker, yielded nominally significant evidence for association (P= 0.01) but this was not as strong as estimated by the pooled analysis (Table 1). As before, we typed approximately 600 additional blood donor controls for the markers showing strongest evidence: rs6466808, rs2693657, rs1196471 and rs10278079. The combined results are given in Table 3. The evidence for rs2693657 remained unchanged (P=0.01), whereas that for rs6466808 and rs10278079 became stronger (P= 0.0002 and 0.001, respectively). Again, we could not obtain satisfactory genotypes with rs1196471 so these were excluded. All genotypes were in Hardy–Weinberg equilibrium for cases and controls.
Table 3.
Individual genotyping of phase 2 tagSNPs
| No. | SNP ID | Location | Distance to next SNP (bases) | Nucleotide changea | Number of cases | Number of controls | Minor allele frequencya | χ2 | Allelic P-value | χ2 | Genotypic P-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Cases | Controls | |||||||||||
| 1 | rs740965b | 121068064 | G/T | 660 | 707 | 0.165 | 0.147 | 1.69 | 0.19 | 2.11 | 0.35 | |
| 2 | rs12706382 | 121074224 | 6160 | T/C | 659 | 698 | 0.118 | 0.125 | 0.31 | 0.58 | 0.40 | 0.82 |
| 3 | rs13437930 | 121076328 | 2104 | C/G | 650 | 706 | 0.099 | 0.105 | 0.29 | 0.59 | 0.45 | 0.80 |
| 4 | rs2402593 | 121079103 | 2775 | G/A | 665 | 709 | 0.232 | 0.236 | 0.08 | 0.77 | 3.28 | 0.19 |
| 5 | rs2191783 | 121079987 | 884 | T/C | 665 | 710 | 0.165 | 0.151 | 1.01 | 0.32 | 2.23 | 0.33 |
| 6 | rs1206477 | 121101096 | 21109 | C/G | 663 | 707 | 0.452 | 0.455 | 0.01 | 0.91 | 0.03 | 0.99 |
| 7 | rs6946211 | 121109294 | 8198 | T/C | 662 | 710 | 0.157 | 0.138 | 1.99 | 0.16 | 3.32 | 0.19 |
| 8 | rs1011692 | 121114514 | 5220 | C/T | 650 | 709 | 0.143 | 0.126 | 1.80 | 0.18 | 2.90 | 0.23 |
| 9 | rs1206504b | 121121853 | 7339 | G/A | 662 | 708 | 0.446 | 0.424 | 1.25 | 0.26 | 1.43 | 0.49 |
| 10 | rs6466808 | 121142483 | 20630 | A/G | 662 | 1121 | 0.323 | 0.380 | 13.72 | 0.0002 | 15.77 | 0.0004 |
| 11 | rs1196487 | 121146390 | 3907 | C/G | 662 | 705 | 0.477 | 0.443 | 3.19 | 0.074 | 3.60 | 0.17 |
| 12 | rs6970897 | 121156410 | 10020 | G/A | 652 | 705 | 0.102 | 0.084 | 2.71 | 0.10 | 5.02 | 0.081 |
| 13 | rs10278079 | 121159221 | 2811 | G/A | 657 | 1287 | 0.302 | 0.355 | 10.45 | 0.001 | 12.00 | 0.002 |
| 14 | rs1725058 | 121159315 | 94 | A/G | 663 | 705 | 0.167 | 0.158 | 0.66 | 0.42 | 0.72 | 0.70 |
| 15 | rs1196477 | 121168118 | 8803 | A/G | 654 | 700 | 0.386 | 0.396 | 0.30 | 0.58 | 0.41 | 0.81 |
| 16 | rs3817483 | 121171520 | 3402 | G/A | 665 | 710 | 0.116 | 0.133 | 1.89 | 0.17 | 2.32 | 0.31 |
| 17 | rs13241278c | 121181151 | 9631 | C/T | 594 | 1267 | 0.478 | 0.531 | 8.85 | 0.003 | 9.43 | 0.009 |
| 18 | rs1196471c | 121189416 | 8265 | C/T | 594 | 673 | 0.328 | 0.375 | 6.65 | 0.01 | 6.72 | 0.03 |
| 19 | rs2693657 | 121191768 | 2352 | A/G | 596 | 1281 | 0.327 | 0.369 | 6.93 | 0.01 | 6.84 | 0.03 |
| 20 | rs1196473 | 121201610 | 9842 | G/A | 665 | 709 | 0.130 | 0.133 | 0.04 | 0.85 | 0.07 | 0.97 |
| 21 | rs1147504 | 121207901 | 6291 | A/G | 665 | 710 | 0.464 | 0.493 | 2.32 | 0.13 | 2.45 | 0.29 |
| 22 | rs1916890 | 121210293 | 2392 | G/A | 656 | 703 | 0.111 | 0.127 | 1.68 | 0.19 | 1.79 | 0.41 |
| 23 | rs1206383 | 121210312 | 19 | A/G | 652 | 702 | 0.003 | 0.001 | 0.83 | 0.36 | - | - |
| 24 | rs1147502c | 121215433 | 5121 | T/G | 661 | 1317 | 0.463 | 0.502 | 5.45 | 0.02 | 6.27 | 0.04 |
| 25 | rs1147498b | 121225025 | 9592 | C/A | 666 | 708 | 0.167 | 0.18 | 0.86 | 0.35 | 1.28 | 0.53 |
| 26 | rs1147497 | 121225630 | 605 | G/T | 649 | 703 | 0.351 | 0.391 | 4.61 | 0.03 | 5.57 | 0.06 |
| 27 | rs1147492 | 121231427 | 5797 | T/C | 643 | 701 | 0.488 | 0.458 | 2.37 | 0.12 | 2.56 | 0.28 |
| 28 | rs1147489b | 121233636 | 2209 | G/A | 643 | 701 | 0.239 | 0.269 | 3.12 | 0.08 | 4.92 | 0.09 |
| 29 | rs17144057 | 121253030 | 19394 | G/T | 662 | 709 | 0.119 | 0.126 | 0.31 | 0.58 | 0.70 | 0.71 |
| 30 | rs1206381 | 121253330 | 300 | C/A | 653 | 702 | 0.374 | 0.413 | 4.14 | 0.04 | 5.96 | 0.05 |
Abbreviation: SNP, single-nucleotide polymorphism.
Minor allele presented first. Its frequencies in cases and controls are presented in columns 8 and 9. All markers were in Hardy–Weinberg equilibrium (HWE) in cases and controls.
Overlapping markers from pooling and tagSNPs analysis.
Markers significant from pooling experiment.
To evaluate the evidence for overall association in the context of testing multiple markers, we applied the truncated product of P-values method.38 This is a gene-wide test which produces a single significance level for evidence of overall association at a gene, allowing for independent association information from multiple SNPs, multiple testing, and the degree to which the markers are in LD.38 We multiplied together all P-values reaching a threshold of P≤0.05 to achieve a product P-value. The significance of this product was established by permuting case–control status over 100 000 replicates and counting the number of times this product was exceeded. This yielded a gene-wide significance of P= 0.0080, which is stronger to that obtained in phase 1. We also determined the significance, in the context of multiple testing, of the single best marker rs6466808 by running 100 000 permutations of case–control status with all genotyped markers. This gave a similar gene-wide level of significance to that of the other method (P=0.0064).
We also performed sliding window haplotype analyses based on all the markers we had genotyped. Although several haplotypes yielded nominally significant evidence for association (Pmin =0.006), none was more significantly associated than the individual markers.
We analyzed in the DNA pools 18 polymorphic SNPs in MAGI-1, 24 SNPs in MAGI-2 and 21 SNPs in MAGI-3. Several markers yielded nominally significant association in the pools and were individually genotyped (Supplementary Tables). Nominally significant signals were confirmed by individual genotyping for markers in MAGI-1 (rs9880851, P = 0.033), MAGI-2 (rs2868865, P = 0.011) and MAGI-3 (rs1230661, P = 0.028), but none reached gene-wide significance following correction for multiple testing, so these genes were not further pursued using the more thorough HapMap-based approach.
Discussion
NRG1-ERBB4 signaling has been genetically and functionally implicated in schizophrenia, and downstream molecules involved in this pathway and molecules that modulate NRG1-ERBB4 signaling might also play important roles in schizophrenia pathogenesis. Using the cytoplasmic domain of ERBB4, we have identified MAGI proteins as ERBB4-interacting proteins. We further showed that MAGI proteins are tyrosine phosphorylated when ERBB4 is present, either alone or in combination with ERBB2 or ERBB3. MAGI proteins serve as scaffolding proteins underlying the cell membrane, recruiting signaling molecules (such as phosphatase and tensin homolog (PTEN)), channels, receptors and cell adhesion molecules to specific domains in the cell membrane.27,41 Some of these interactions can be modulated by threonine-serine phosphorylation of MAGI.42 It would be of interest to determine whether tyrosine phosphorylation of MAGI proteins might also modulate their interactions with other molecules, modulating the activating different signaling components. Nonetheless, our results indicate that MAGI proteins are apparent downstream molecules in NRG1-ERBB4 signaling that could serve as a site to integrate or modulate signaling events in the cells.
MAGI proteins have previously been shown to interact with RPTPβ.28,29,39 We and others,29,39 have observed that RPTPβ can dephosphorylate MAGI proteins. In addition, we found that RPTPβ, ERBB4 and MAGI proteins can colocalize in cells. Our data suggest that MAGIs can bring ERBB4 tyrosine kinase and RPTPβ tyrosine phosphatase to domains in the cell membrane, modulating the balance among sig naling pathways downstream of these molecules. Given that NRG1-ERBB4 signaling has been etiologically linked with schizophrenia, we carried out a genetic association analysis of PTPRZ1. Several SNPs yielded significant evidence for association, and the evidence in support of association remained significant when corrected for multiple testing at the genewide level (P = 0.0064), and also at the experimentwide level (correcting for all genes examined, n=4, P = 0.026). As the associated SNPs are in introns, it will require further studies to determine how this pattern of association might affect PTPRZ1 gene function. However, if it can be demonstrated that RPTPβ antagonizes NRG1-ERBB4 signaling in vivo, then we would predict that reduced functioning of RPTPβ would result in a phenotype like that of enhanced NRG1-ERBB4 signaling.22 This would provide a potential mechanism by which PTPRZ1/RPTPβ could be involved in the molecular basis of schizophrenia.
PTPRZ1 gene products are expressed mainly in the nervous system.43 There are four splicing isoforms of PTPRZ1, two of which are transmembrane receptor forms (RPTPβ, short and long) and the other two are soluble forms that lack both a transmembrane region and tyrosine phosphatase activity (phosphacan, short and long).44–50 The extracellular region of PTPRZ1 consists of several modular structural domains, including a carbonic anhydrase domain, a fibronectin type III repeat, a spacer domain (all of which are present in all four isoforms), and a region containing ~860 amino acids (that is missing in the RPTPβ and phosphacan short forms) where several glycosaminoglycan chains can be attached.44 The carbonic anhydrase domain interacts with the cell recognition molecule contactin,51 whereas the spacer region can interact with several other cell adhesion molecules including NrCAM, L1CAM, NCAM, contactin-1 and contactin-2/TAG-1.52–56
PTPRZ1 gene products are expressed from early developmental to adulthood in glial cells47,57–59 as well as in certain population of neurons60–63 and are implicated in neuron–glial cell interactions that involve on bi-directional signaling, including myelination and node of Ranvier formation, through interactions with neuronal cell adhesion molecules. 55,56 Interestingly, in situ hybridization analysis has shown that PTPRZ1 is expressed in the subventricular zone of the lateral ventricles in early embryos and in adults, in the subgranular zone in the dentate gyrus of the hippocampus of adults,57,64 and in adult glial progenitor cells,65 suggesting that PTPRZ1 also has a function in adult stem/progenitor cells. In addition, it has been shown that inhibition of PTPRZ1 expression or inhibition of RPTPβ activity in adult glial progenitors leads to their differentiation into mature oligodendrocytes,65 supporting a role for RPTPβ activity in the maintenance of glial progenitors in an undifferentiated state. Using experimental allergic encephalomyelitis (EAE), an inducible mouse model for multiple sclerosis (MS), PTPRZ1-deficient mice could not repair EAE lesions, although PTPRZ1 expression is increased in human remyelinating oligodendrocytes of MS biopsies.66 Taken together, these data suggest that PTPRZ1 could be implicated in adult cell renewal, repair of the nervous system and/or oligodendrocyte development. It should be noted that NRG1-ERBB4 signaling has been shown to play crucial roles in these processes, supporting the possibility that PTPRZ1 might function partly through modulation of the NRG1-ERBB4 signaling pathway.
A role for PTPRZ1 in oligodendendrocyte development is intriguing in light of evidence for altered oligodendrocyte functioning in schizophrenia. Several postmortem studies in schizophrenia have demonstrated reduced expression of oligodendrocyte-related genes,17–21,67–72 some of which may be susceptibility genes for schizophrenia.33,72–75 Moreover, several studies have evidence for deficits in oligodendrocytes in the disorder.76–79 These oligodendrocyte abnormalities may in turn contribute to the loss of coherence of axon tracts, and reduced connectivity, in schizophrenia.80,81 Alterations in NRG1-ERBB4 and/or PTPRZ1 signaling may cause these oligodendrocyte abnormalities, in addition to potential neuronal effects.
In summary, using proteomic and cell–biological approaches, we identified a signaling complex comprised of ERBB4, MAGI and RPTPβ. Moreover, genetic evidence suggests that PTPRZ1 might influence susceptibility for schizophrenia. Further studies delineating the role of PTPRZ1 in the brain will elucidate the role of this gene in ERBB4 signaling, and may contribute to a better understanding of schizophrenia.
Supplementary Material
Acknowledgments
This work was funded by an NIMH Silvio O Conte Centre For The Neuroscience of Mental Disorders Grant (MH063392 to JDB and KLD, PI and co-PI) and in part by grants from the MRC, UK (MRC and MO). VM is funded by a RCUK Fellowship and TS is a Fellow of the Seaver Foundation and is further supported by the Stanley Medical Research Institute (grant 06R-1427) and by a New York State Spinal Cord Injury Grant (SCIRB04-27). We thank Mihaela Gazdoiu and Kate Liberman for excellent technical assistance and Dr Andrew Chan for advice and guidance on the study of MAGI proteins.
Footnotes
Supplementary Information accompanies the paper on theMolecular Psychiatry web site (http://www.nature.com/mp)
References
- 1.Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton N, Williams HJ, Owen MJ. An update on the genetics of schizophrenia. Curr Opin Psychiatry. 2006;19:158–164. doi: 10.1097/01.yco.0000214341.52249.59. [DOI] [PubMed] [Google Scholar]
- 3.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15:1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 4.Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 5.Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, et al. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 7.Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr Opin Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 10.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 11.Jones FE, Golding JP, Gassmann M. ErbB4 signaling during breast and neural development: novel genetic models reveal unique ErbB4 activities. Cell Cycle. 2003;2:555–559. [PubMed] [Google Scholar]
- 12.Anton ES, Ghashghaei HT, Weber JL, McCann C, Fischer TM, Cheung ID, et al. Receptor tyrosine kinase ErbB4 modulates neuroblast migration and placement in the adult forebrain. Nat Neurosci. 2004;7:1319–1328. doi: 10.1038/nn1345. [DOI] [PubMed] [Google Scholar]
- 13.Ghashghaei HT, Weber J, Pevny L, Schmid R, Schwab MH, Lloyd KC, et al. The role of neuregulin-ErbB4 interactions on the proliferation and organization of cells in the subventricular zone. Proc Natl Acad Sci USA. 2006;103:1930–1935. doi: 10.1073/pnas.0510410103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiner H, Blum M, Kitai ST, Fedi P. Differential expression of ErbB3 and ErbB4 neuregulin receptors in dopamine neurons and forebrain areas of the adult rat. Exp Neurol. 1999;159:494–503. doi: 10.1006/exnr.1999.7163. [DOI] [PubMed] [Google Scholar]
- 15.Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 16.Norton N, Moskvina V, Morris DW, Bray NJ, Zammit S, Williams NM, et al. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141:96–101. doi: 10.1002/ajmg.b.30236. [DOI] [PubMed] [Google Scholar]
- 17.Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 18.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- 20.Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr Res. 2005;79:157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Katsel PL, Davis KL, Haroutunian V. Large-scale microarray studies of gene expression in multiple regions of the brain in schizophrenia and Alzheimer’s disease. Int Rev Neurobiol. 2005;63:41–82. doi: 10.1016/S0074-7742(05)63003-6. [DOI] [PubMed] [Google Scholar]
- 22.Hahn CG, Wang HY, Cho DS, Talbot K, Gur RE, Berrettini WH, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA> receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 23.Rimer M, Barrett DW, Maldonado MA, Vock VM, Gonzalez-Lima F. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16:271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- 24.O’Tuathaigh CM, O’Sullivan GJ, Kinsella A, Harvey RP, Tighe O, Croke DT, et al. Sexually dimorphic changes in the exploratory and habituation profiles of heterozygous neuregulin-1 knockout mice. Neuroreport. 2006;17:79–83. doi: 10.1097/01.wnr.0000192738.31029.0a. [DOI] [PubMed] [Google Scholar]
- 25.O’Tuathaigh CM, Babovic D, O’Meara G, Clifford JJ, Croke DT, Waddington JL. Susceptibility genes for schizophrenia: Characterisation of mutant mouse models at the level of phenotypic behaviour. Neurosci Biobehav Rev. 2007;31:60–78. doi: 10.1016/j.neubiorev.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery JM, Zamorano PL, Garner CC. MAGUKs in synapse assembly and function: an emerging view. Cell Mol Life Sci. 2004;61:911–929. doi: 10.1007/s00018-003-3364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adamsky K, Arnold K, Sabanay H, Peles E. Junctional protein MAGI-3 interacts with receptor tyrosine phosphatase beta (RPTP beta) and tyrosine-phosphorylated proteins. J Cell Sci. 2003;116:1279–1289. doi: 10.1242/jcs.00302. [DOI] [PubMed] [Google Scholar]
- 29.Fukada M, Kawachi H, Fujikawa A, Noda M. Yeast substrate-trapping system for isolating substrates of protein tyrosine phosphatases: Isolation of substrates for protein tyrosine phosphatase receptor type z. Methods. 2005;35:54–63. doi: 10.1016/j.ymeth.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Kainulainen V, Sundvall M, Maatta JA, Santiestevan E, Klagsbrun M, Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J Biol Chem. 2000;275:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- 33.Peirce TR, Bray NJ, Williams NM, Norton N, Moskvina V, Preece A, et al. Convergent evidence for 2′,3′-cyclic nucleotide 3′-phosphodiesterase as a possible susceptibility gene for schizophrenia. Arch Gen Psychiatry. 2006;63:18–24. doi: 10.1001/archpsyc.63.1.18. [DOI] [PubMed] [Google Scholar]
- 34.Moskvina V, Holmans P, Schmidt KM, Craddock N. Design of case-controls studies with unscreened controls. Ann Hum Genet. 2005;69:566–576. doi: 10.1111/j.1529-8817.2005.00175.x. [DOI] [PubMed] [Google Scholar]
- 35.Norton N, Williams NM, Williams HJ, Spurlock G, Kirov G, Morris DW, et al. Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum Genet. 2002;110:471–478. doi: 10.1007/s00439-002-0706-6. [DOI] [PubMed] [Google Scholar]
- 36.Dudbridge F. Pedigree disequilibrium tests for multilocus haplo-types. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 37.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- 39.Fukada M, Fujikawa A, Chow JP, Ikematsu S, Sakuma S, Noda M. Protein tyrosine phosphatase receptor type Z is inactivated by ligand-induced oligomerization. FEBS Lett. 2006;580:4051–4056. doi: 10.1016/j.febslet.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 40.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, Hepner K, Castelino-Prabhu S, Do D, Kaye MB, Yuan XJ, et al. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci USA. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolkacheva T, Boddapati M, Sanfiz A, Tsuchida K, Kimmelman AC, Chan AM. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001;61:4985–4989. [PubMed] [Google Scholar]
- 43.Paul S, Lombroso PJ. Receptor and nonreceptor protein tyrosine phosphatases in the nervous system. Cell Mol Life Sci. 2003;60:2465–2482. doi: 10.1007/s00018-003-3123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnea G, Grumet M, Milev P, Silvennoinen O, Levy JB, Sap J, et al. Receptor tyrosine phosphatase beta is expressed in the form of proteoglycan and binds to the extracellular matrix protein tenascin. J Biol Chem. 1994;269:14349–14352. [PubMed] [Google Scholar]
- 45.Barnea G, Grumet M, Sap J, Margolis RU, Schlessinger J. Close similarity between receptor-linked tyrosine phosphatase and rat brain proteoglycan. Cell. 1994;76:205. doi: 10.1016/0092-8674(94)90328-x. [DOI] [PubMed] [Google Scholar]
- 46.Maurel P, Rauch U, Flad M, Margolis RK, Margolis RU. Phosphacan, a chondroitin sulfate proteoglycan of brain that interacts with neurons and neural cell-adhesion molecules, is an extracellular variant of a receptor-type protein tyrosine phosphatase. Proc Natl Acad Sci USA. 1994;91:2512–2516. doi: 10.1073/pnas.91.7.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakurai T, Friedlander DR, Grumet M. Expression of polypeptide variants of receptor-type protein tyrosine phosphatase beta: the secreted form, phosphacan, increases dramatically during embryonic development and modulates glial cell behavior in vitro. J Neurosci Res. 1996;43:694–706. doi: 10.1002/(SICI)1097-4547(19960315)43:6<694::AID-JNR6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Nishiwaki T, Maeda N, Noda M. Characterization and developmental regulation of proteoglycan-type protein tyrosine phosphatase zeta/RPTPbeta isoforms. J Biochem (Tokyo) 1998;123:458–467. doi: 10.1093/oxfordjournals.jbchem.a021959. [DOI] [PubMed] [Google Scholar]
- 49.Garwood J, Heck N, Reichardt F, Faissner A. Phosphacan short isoform, a novel non-proteoglycan variant of phosphacan/receptor protein tyrosine phosphatase-beta, interacts with neuronal receptors and promotes neurite outgrowth. J Biol Chem. 2003;278:24164–24173. doi: 10.1074/jbc.M211721200. [DOI] [PubMed] [Google Scholar]
- 50.Heck N, Klausmeyer A, Faissner A, Garwood J. Cortical neurons express PSI, a novel isoform of phosphacan/RPTPbeta. Cell Tissue Res. 2005;321:323–333. doi: 10.1007/s00441-005-1135-3. [DOI] [PubMed] [Google Scholar]
- 51.Peles E, Nativ M, Campbell PL, Sakurai T, Martinez R, Lev S, et al. The carbonic anhydrase domain of receptor tyrosine phosphatase beta is a functional ligand for the axonal cell recognition molecule contactin. Cell. 1995;82:251–260. doi: 10.1016/0092-8674(95)90312-7. [DOI] [PubMed] [Google Scholar]
- 52.Milev P, Friedlander DR, Sakurai T, Karthikeyan L, Flad M, Margolis RK, et al. Interactions of the chondroitin sulfate proteoglycan phosphacan, the extracellular domain of a receptor-type protein tyrosine phosphatase, with neurons, glia, and neural cell adhesion molecules. J Cell Biol. 1994;127:1703–1715. doi: 10.1083/jcb.127.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milev P, Meyer-Puttlitz B, Margolis RK, Margolis RU. Complex-type asparagine-linked oligosaccharides on phosphacan and protein-tyrosine phosphatase-zeta/beta mediate their binding to neural cell adhesion molecules and tenascin. J Biol Chem. 1995;270:24650–24653. doi: 10.1074/jbc.270.42.24650. [DOI] [PubMed] [Google Scholar]
- 54.Milev P, Maurel P, Haring M, Margolis RK, Margolis RU. TAG-1/axonin-1 is a high-affinity ligand of neurocan, phosphacan/protein-tyrosine phosphatase-zeta/beta, and N-CAM. J Biol Chem. 1996;271:15716–15723. doi: 10.1074/jbc.271.26.15716. [DOI] [PubMed] [Google Scholar]
- 55.Sakurai T, Lustig M, Nativ M, Hemperly JJ, Schlessinger J, Peles E, et al. Induction of neurite outgrowth through contactin and Nr-CAM by extracellular regions of glial receptor tyrosine phosphatase beta. J Cell Biol. 1997;136:907–918. doi: 10.1083/jcb.136.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Revest JM, Faivre-Sarrailh C, Maeda N, Noda M, Schachner M, Rougon G. The interaction between F3 immunoglobulin domains and protein tyrosine phosphatases zeta/beta triggers bidirectional signalling between neurons and glial cells. Eur J Neurosci. 1999;11:1134–1147. doi: 10.1046/j.1460-9568.1999.00521.x. [DOI] [PubMed] [Google Scholar]
- 57.Canoll PD, Barnea G, Levy JB, Sap J, Ehrlich M, Silvennoinen O, et al. The expression of a novel receptor-type tyrosine phosphatase suggests a role in morphogenesis and plasticity of the nervous system. Brain Res Dev Brain Res. 1993;75:293–298. doi: 10.1016/0165-3806(93)90035-9. [DOI] [PubMed] [Google Scholar]
- 58.Engel M, Maurel P, Margolis RU, Margolis RK. Chondroitin sulfate proteoglycans in the developing central nervous system. I. Cellular sites of synthesis of neurocan and phosphacan. J Comp Neurol. 1996;366:34–43. doi: 10.1002/(SICI)1096-9861(19960226)366:1<34::AID-CNE3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 59.Snyder SE, Li J, Schauwecker PE, McNeill TH, Salton SR. Comparison of RPTP zeta/beta, phosphacan, and trkB mRNA expression in the developing and adult rat nervous system and induction of RPTP zeta/beta and phosphacan mRNA following brain injury. Brain Res Mol Brain Res. 1996;40:79–96. doi: 10.1016/0169-328x(96)00039-3. [DOI] [PubMed] [Google Scholar]
- 60.Maeda N, Hamanaka H, Oohira A, Noda M. Purification, characterization and developmental expression of a brain-specific chondroitin sulfate proteoglycan, 6B4 proteoglycan/phosphacan. Neuroscience. 1995;67:23–35. doi: 10.1016/0306-4522(94)00069-h. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Tullai JW, Yu WH, Salton SR. Regulated expression during development and following sciatic nerve injury of mRNAs encoding the receptor tyrosine phosphatase HPTPzeta/RPTPbeta. Brain Res Mol Brain Res. 1998;60:77–88. doi: 10.1016/s0169-328x(98)00175-2. [DOI] [PubMed] [Google Scholar]
- 62.Ohyama K, Kawano H, Asou H, Fukuda T, Oohira A, Uyemura K, et al. Coordinate expression of L1 and 6B4 proteoglycan/phosphacan is correlated with the migration of mesencephalic dopaminergic neurons in mice. Brain Res Dev Brain Res. 1998;107:219–226. doi: 10.1016/s0165-3806(97)00220-4. [DOI] [PubMed] [Google Scholar]
- 63.Hayashi N, Miyata S, Yamada M, Kamei K, Oohira A. Neuronal expression of the chondroitin sulfate proteoglycans receptor-type protein-tyrosine phosphatase beta and phosphacan. Neuroscience. 2005;131:331–348. doi: 10.1016/j.neuroscience.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 64.Levy JB, Canoll PD, Silvennoinen O, Barnea G, Morse B, Honegger AM, et al. The cloning of a receptor-type protein tyrosine phosphatase expressed in the central nervous system. J Biol Chem. 1993;268:10573–10581. [PubMed] [Google Scholar]
- 65.Sim FJ, Lang JK, Waldau B, Roy NS, Schwartz TE, Pilcher WH, et al. Complementary patterns of gene expression by human oligodendrocyte progenitors and their environment predict determinants of progenitor maintenance and differentiation. Ann Neurol. 2006;59:763–779. doi: 10.1002/ana.20812. [DOI] [PubMed] [Google Scholar]
- 66.Harroch S, Furtado GC, Brueck W, Rosenbluth J, Lafaille J, Chao M, et al. A critical role for the protein tyrosine phosphatase receptor type Z in functional recovery from demyelinating lesions. Nat Genet. 2002;32:411–414. doi: 10.1038/ng1004. [DOI] [PubMed] [Google Scholar]
- 67.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 68.Flynn SW, Lang DJ, Mackay AL, Goghari V, Vavasour IM, Whittall KP, et al. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol Psychiatry. 2003;8:811–820. doi: 10.1038/sj.mp.4001337. [DOI] [PubMed] [Google Scholar]
- 69.Sugai T, Kawamura M, Iritani S, Araki K, Makifuchi T, Imai C, et al. Prefrontal abnormality of schizophrenia revealed by DNA microarray: impact on glial and neurotrophic gene expression. Ann NY Acad Sci. 2004;1025:84–91. doi: 10.1196/annals.1316.011. [DOI] [PubMed] [Google Scholar]
- 70.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama Y, Yoshikawa T, et al. A family-based and case-control association study of SOX10 in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141:477–481. doi: 10.1002/ajmg.b.30304. [DOI] [PubMed] [Google Scholar]
- 71.Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci USA. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, et al. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am J Med Genet B Neuropsychiatr Genet. 2006;141:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- 73.Georgieva L, Moskvina V, Peirce T, Norton N, Bray NJ, Jones L, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2006;103:12469–12474. [Google Scholar]
- 74.Wan C, Yang Y, Feng G, Gu N, Liu H, Zhu S, et al. Polymorphisms of myelin-associated glycoprotein gene are associated with schizophrenia in the Chinese Han population. Neurosci Lett. 2005;388:126–131. doi: 10.1016/j.neulet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 75.Yang YF, Qin W, Shugart YY, He G, Liu XM, Zhou J, et al. Possible association of the MAG locus with schizophrenia in a Chinese Han cohort of family trios. Schizophr Res. 2005;75:11–19. doi: 10.1016/j.schres.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 76.Hof PR, Haroutunian V, Friedrich VL, Jr, Byne W, Buitron C, Perl DP, et al. Loss and altered spatial distribution of oligodendrocytes in the superior frontal gyrus in schizophrenia. Biol Psychiatry. 2003;53:1075–1085. doi: 10.1016/s0006-3223(03)00237-3. [DOI] [PubMed] [Google Scholar]
- 77.Byne W, Kidkardnee S, Tatusov A, Yiannoulos G, Buchsbaum MS, Haroutunian V. Schizophrenia-associated reduction of neuronal and oligodendrocyte numbers in the anterior principal thalamic nucleus. Schizophr Res. 2006;85:245–253. doi: 10.1016/j.schres.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 78.Uranova N, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, Vostrikov V, et al. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 79.Uranova NA, Vostrikov VM, Orlovskaya DD, Rachmanova VI. Oligodendroglial density in the prefrontal cortex in schizophrenia and mood disorders: a study from the Stanley Neuropathology Consortium. Schizophr Res. 2004;67:269–275. doi: 10.1016/S0920-9964(03)00181-6. [DOI] [PubMed] [Google Scholar]
- 80.Kubicki M, McCarley R, Westin CF, Park HJ, Maier S, Kikinis R, et al. A review of diffusion tensor imaging studies in schizophrenia. J Psychiatr Res. 2007;41:15–30. doi: 10.1016/j.jpsychires.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buchsbaum MS, Friedman J, Buchsbaum BR, Chu KW, Hazlett EA, Newmark R, et al. Diffusion tensor imaging in schizophrenia. Biol Psychiatry. 2006;60:1181–1187. doi: 10.1016/j.biopsych.2005.11.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.