SUMMARY
During transcription initiation, RNA polymerase binds to promoter DNA to form an initiation complex containing a DNA bubble and enters into abortive cycles of RNA synthesis before escaping the promoter to transit into the elongation phase for processive RNA synthesis. Here we present the crystal structures of E. coli transcription initiation complexes containing a complete transcription bubble and de novo synthesized RNA oligonucleotides at about 6 Å resolution. The structures show how RNA polymerase recognizes DNA promoters that contain spacers of different lengths and reveal a bridging interaction between the 5’-triphosphate of the nascent RNA and the σ factor that may function to stabilize the short RNA-DNA hybrids during the early stage of transcription initiation. The conformation of the RNA oligonucleotides and the paths of the DNA strands in the complete initiation complexes provide insights into the mechanism that controls both the abortive and productive RNA synthesis.
INTRODUCTION
Transcription initiation is a dynamic process that begins with the sequence-specific binding of the RNA polymerase (RNAP) to a DNA promoter to form a closed complex (RPc), followed by sequential conformational changes in both the DNA and the enzyme to form an open complex (RPo) that is capable of de novo RNA synthesis (Saecker et al., 2011). The DNA strands near the transcription start site remain basepaired in the RPc, but are separated in the RPo and form a transcription bubble. As the nascent RNA extends during transcription initiation, the initiation complex forms a stressed intermediate containing a short RNA-DNA duplex (Straney and Crothers, 1987). Ejection of the short RNA (abortive initiation) or release of the σ factor (promoter escape) relieves the strain energy. Many of the initiation RNA transcripts are aborted with the release of short RNA oligonucleotides, and only when the RNA is extended to about 10 nucleotides (nt) or longer will the transcription initiation survive and progress into the elongation phase for processive RNA synthesis (Goldman et al., 2009; Hansen and McClure, 1979).
During transcription initiation, RNA polymerase remains associated with the upstream promoter DNA while moving downstream for RNA synthesis, causing “DNA scrunching” as the downstream DNA is unwound and pulled into the RNA polymerase (Kapanidis et al., 2006; Revyakin et al., 2006). DNA scrunching is expected to build stress within the enzyme and was suggested to be the driving force for both abortive initiation and promoter escape for productive RNA synthesis (Kapanidis et al., 2006; Revyakin et al., 2006). While the details of DNA scrunching remain obscure, it is known that the σ3.2 loop of a σ factor lies on the pathway of the nascent RNA. The growing RNA is expected to press on the σ3.2 loop and this clashing of RNA with the σ factor was also suggested to play an important role in destabilizing the hybrid for abortive initial RNA synthesis (Murakami et al., 2002b; Samanta and Martin, 2013).
The dynamic feature of transcription initiation makes it challenging for structural studies of the initiation complex as a whole. Previous structural studies of transcription initiation frequently involve DNA fragments that form a partial transcription bubble (Basu et al., 2014; Liu et al., 2011; Murakami et al., 2002a; Sainsbury et al., 2013; Zhang et al., 2014; Zhang et al., 2012), and there are no structural data on transcription complexes containing a complete transcription bubble that is in the process of de novo RNA synthesis. While significant progress has been made in our understanding of transcription initiation based on these studies, the structure of an initiation complex with both the upstream and downstream DNA duplexes and an intact transcription bubble remains essential for understanding both the abortive feature of transcription initiation and the promoter escape for transition into transcription elongation.
We have determined the crystal structures of the E. coli RNA polymerase holoenzyme in complex with DNA promoters containing the consensus -35 and -10 elements, a complete transcription bubble, and de novo synthesized RNA oligonucleotides (Figure 1). The structures of the transcription initiation complexes (TICs) show the specific interactions between a complete transcription bubble and the RNA polymerase, reveal the conformational changes of the RNA polymerase that allow it to accommodate variations in the spacer length of the promoter DNA and an interaction of the 5’-triphosphate of the nascent RNA with the σ3.2 loop, as well as provide insights into the mechanism of abortive RNA synthesis and promoter escape during transcription initiation.
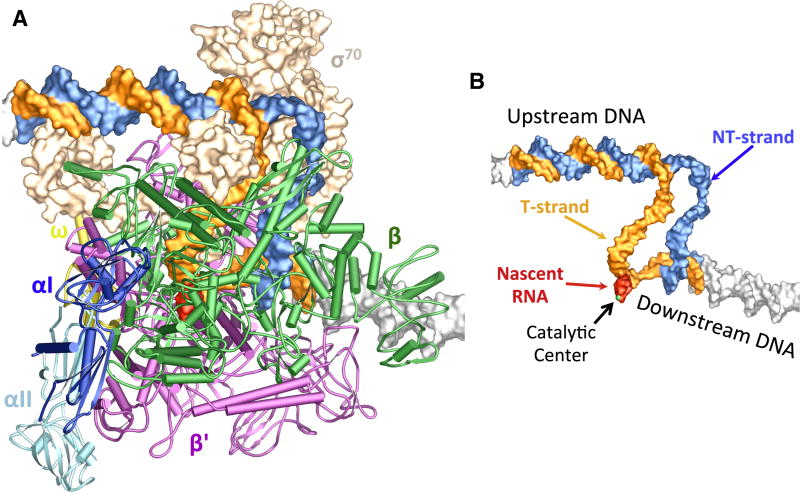
Figure 1. Structure of the E. coli transcription initiation complex.
(A) Overall structure. The E. coli RNAP core enzyme is shown in a tube and arrow cartoon representation: blue – αI subunit; light blue – αII subunit; orange – βsubunit; green – β’ subunit; yellow – ω subunit. The σ70 factor (wheat) is shown in a surface representation. The promoter DNA (blue – NT-strand; orange – T-strand) and the nascent RNA (red) are shown in color, and the DNA duplexes from symmetry-related molecules are shown in grey surface representation.
(B) An overview of the nucleic acids that form the complete transcription initiation bubble. Nucleic acid chains are color-coded as in Figure 1A, and protein subunits are omitted for clarity. A metal ion (green ball) marks the catalytic center.
RESULTS AND DISCUSSION
Overall Structure of the E. coli transcription initiation complexes
To form TICs of the E. coli RNA polymerase, we used synthetic DNA scaffolds corresponding to the promoter region between positions -37 and +13 relative to the transcription start site (Figure S1). Nascent RNA oligonucleotides were synthesized de novo by incubating the synthetic DNA promoters with the E. coli RNAP holoenzyme in the presence of selected nucleotide triphosphates (NTPs). The E. coli transcription initiation complexes crystallize in the monoclinic P21 space group. There are three copies of the complexes per asymmetric unit in the TIC crystals. The structures were solved by molecular replacement using the structure of the E. coli RNAP holoenzyme (Zuo et al., 2013) as a starting model. The final models were refined to 5.5 Å resolution (Table 1).
Table 1.
Data Collection and Refinement Statistics
| TIC1 17 bp spacer 4 nt RNA |
TIC2 16 bp spacer 4 nt RNA |
TIC3 16 bp spacer 5 nt RNA |
|
|---|---|---|---|
| Data Collection* | |||
| Space group | P21 | P21 | P21 |
| Cell Dimensions | |||
| a, b, c (Å) | 237.39, 206.07, 248.70 | 240.89, 208.17, 256.32 | 237.68, 204.99, 248.84 |
| α, β, γ (°) | 90, 116.55, 90 | 90, 119.31, 90 | 90, 116.86, 90 |
| Resolution (Å) | 40-5.50 (5.59-5.50)** | 40-6.00 (6.10-6.00)** | 40-5.50 (5.59-5.50)** |
| Rmeas(%) | 13.6 | 10.3 | 8.4 |
| I/σI | 9.4 (0.71) | 13.4 (0.48) | 12.3 (0.45) |
| Completeness (%) | 99.9 (100) | 99.9 (100) | 99.9 (100) |
| Redundancy | 4.7 (4.6) | 5.1 (5.2) | 3.4 (3.2) |
| Refinement | |||
| Resolution (Å) | 40-5.50 | 40-6.00 | 40-5.50 |
| No. reflections | 69,849 | 55,356 | 69,757 |
| Rwork/Rfree (%) | 25.5/32.9 | 24.1/32.2 | 23.4/30.8 |
| No. atoms | 94,608 | 94,608 | 94,668 |
| Protein | 88,284 | 88,284 | 88,284 |
| DNA/RNA/Ions | 6,324 | 6,324 | 6,384 |
| B-factors | 219.5 | 238.3 | 198.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.011 | 0.014 | 0.011 |
| Bond angles (°) | 1.51 | 1.67 | 1.56 |
Values in parentheses are for highest-resolution shell.
I/σI=2.0 at 6.6Å(TIC1), 6.9Å(TIC2), 6.7Å(TIC3); I/σI=1.0 at 6.0Å(TIC1), 6.5Å(TIC2), 6.1Å(TIC3).
The structures provide a complete picture of the transcription initiation complex, including the upstream DNA interacting sequence-specifically with the RNA polymerase, the downstream duplex DNA, an intact transcription bubble, and a nascent RNA oligonucleotide (Figure 1). The downstream dsDNA of each complex packs head-to-tail against the upstream dsDNA of a symmetry-related molecule to form an extended DNA duplex in the TIC crystals.
Compared to the E. coli RNAP apo holoenzyme structure that displays a relatively closed conformation, the E. coli TICs appear to have both pincers of its cleft moved further toward each other to adopt a tightly closed conformation resembling the one observed for a bacterial elongation complex (Vassylyev et al., 2007). It has been shown that the acidic N-terminal domain of σ70 (σ1.1) in an apo holoenzyme is inserted into the downstream channel and makes contacts with both sides of the RNAP cleft (Figure S2A) (Bae et al., 2013). σ1.1 prevents σ70 from interacting with promoter DNA in the absence of the RNAP core and functions in σ70 auto-regulation (Camarero et al., 2002). In the initiation complexes, the downstream channel is occupied by the downstream DNA duplex (Figure S2B). Although σ1.1 could not be built in the TIC structures, the linker helix that connects σ1.1 to the conserved σ1.2 region folds back and becomes ordered, suggesting that σ1.1 is relocated to the outside of the downstream channel, consistent with a previous study (Mekler et al., 2002). The positively charged end of the “jaw” domain interacts with the DNA backbone of the NT-strand some 15 nt downstream of the active center (Figure S2C).
Promoter Recognition
Bacterial RNA polymerase recognizes the promoter DNA through sequence-specific interactions of the σ factor with the conserved -10 and -35 DNA elements (Campagne et al., 2014; Campbell et al., 2002; Feklistov and Darst, 2011; Feklistov et al., 2014). In the E. coli TICs, the bases of the -11A and -7T residues of the -10 NT-strand are clearly flipped out, and the σ2 domain interacts with the -10 NT-strand in essentially the same manner as that observed with shorter DNA fragments (Feklistov and Darst, 2011; Zhang et al., 2012), whereas the -10 T-strand wraps around the σ3 globular domain and passes through a cleft between σ2 and σ3 (Figure 2A and Figure S3A). It is not clear whether there are any sequence-specific interactions between the -10 T-strand and the σ factor at the current resolution of our map. The TIC structures also confirm that DNA promoters remain basepaired at position -12 upon open complex formation (Guo and Gralla, 1998).
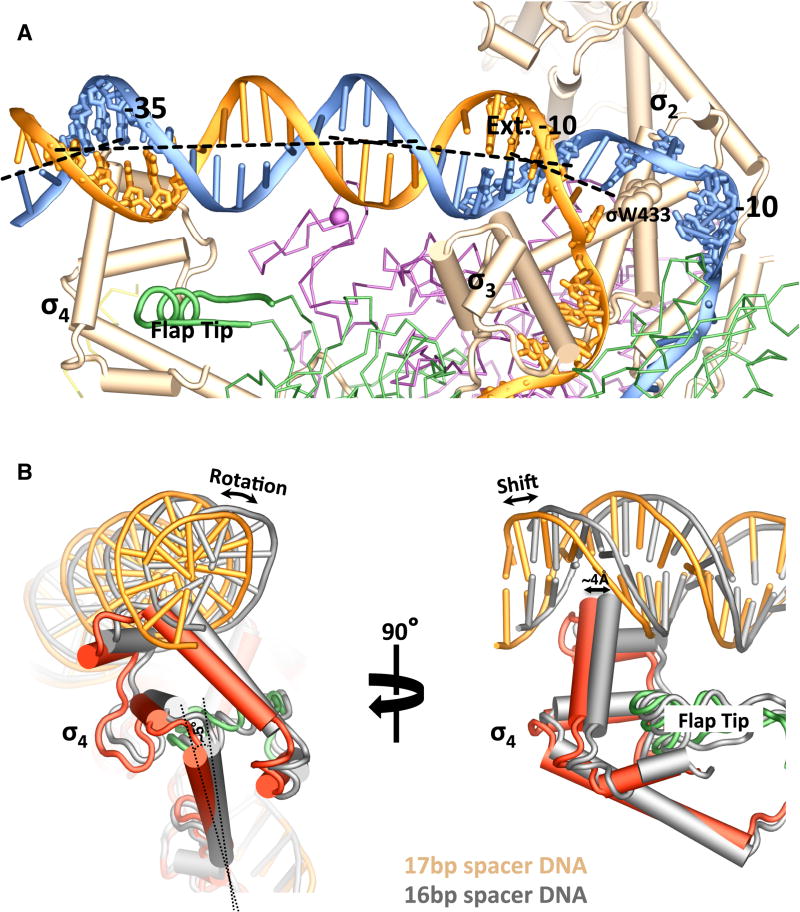
Figure 2. Promoter Recognition.
(A) An overview of the promoter-recognition interactions. Nucleic acid chains (ladders) and protein subunits are color-coded as in Figure 1A. The conserved -10, extended -10 and -35 regions of the promoter DNA are also shown as sticks. The tryptophan residue of σ70 (σ W433) that stacks on the -12 basepair is shown as a space-filled model.
(B) Accommodation of DNA promoters with different spacer length. σ4 domains that interact with a promoter with a 17-bp spacer (colored red – σ4; green –β flap tip; orange – promoter DNA) or a promoter with a 16-bp spacer (grey) are superimposed based on the β’ subunits of the corresponding TICs.
A subclass of σ70 promoters contain an “extended -10” motif, a sequence motif of 5’- (TR)TGn-3’ immediately upstream of the -10 element (Hook-Barnard and Hinton, 2007; Keilty and Rosenberg, 1987). To investigate the potential sequence-specific recognition of the “extended -10” motif by the RNA polymerase, we used synthetic promoters containing a 5’-TATGc-3’ sequence immediately upstream of the -10 hexamer. Although details of the interactions could not be established at the current resolution, two perpendicular helices spanning the σ2 and σ3 domains of σ70 insert into the major groove of the promoter extended -10 region and cause significant bending of the promoter DNA toward the σ3 domain (Figure 2A and Figure S3B). Several conserved residues of the σ3 domain are likely to make sequence-specific interactions with the extended -10 residues.
The highly conserved C-terminal domain of σ70 (σ4) recognizes the -35 element of the DNA promoter. It was shown previously that the -35 element interacts with the σ4 domain primarily through the insertion of a helix-turn-helix (HTH) into the DNA major groove (Campbell et al., 2002; Murakami et al., 2002a). This insertion of a HTH into the major groove of the promoter -35 region also happens in the E. coli TICs (Figure 2A and Figure S3C). The extensive contacts between σ4 and the promoter DNA lead to a significant bending of the promoter DNA axis by about 30°.
For σ70 promoters, the spacer that separates the conserved -35 and -10 hexamers is generally not conserved in sequence, but instead is rather constant in length, with a consensus of 17±2 basepairs (bp) (Harley and Reynolds, 1987; Shimada et al., 2014). The allowance of the spacer length variation results from the flexibility of the σ4 domain that is anchored on the flexible tip helix of the β flap. As shown in Figure 2B, a change of the spacer length from 17 bp to 16 bp causes a rotation of the σ4 domain by about 5°. This rotation translates into a shift of about 4 Å, equivalent to a 1-bp step, at the turn of the HTH that interacts with the -35 element. It is conceivable that the spacer length variation is limited by the amount that the σ4 domain is able to rotate.
σ3.2 loop and de novo synthesized RNA
Association of a σ factor with the RNAP core enzyme involves inserting the σ3.2 loop deep into the RNAP active-site chamber. Immediately after the structure of the RNAP holoenzyme was determined, it was suggested that the σ3.2 loop would play a role in abortive transcription initiation (Murakami et al., 2002b). σ3.2 does not interact with the initiating nucleotides directly but was found to stimulate the binding of initiating NTPs; this led to a suggestion that σ3.2 helps constrain the template strand DNA in the A-form conformation to facilitate transcription initiation(Hansen and McClure, 1979, 1980; Kulbachinskiy and Mustaev, 2006; Zenkin and Severinov, 2004). In the E. coli TIC structures, the σ3.2 loop appears to shift slightly toward the template strand DNA and its tip interacts with the bases of the T-strand residues that lie 4–5 nt upstream of the catalytic center, consistent with a role for σ3.2 in positioning the template DNA for the initial RNA synthesis.
For both DNA promoters used in this study, the TICs initially crystallized contain a 14-nt bubble and an RNA tetranucleotide synthesized de novo from three NTPs (Figure S1). The RNA tetranucleotides in both TICs were found to lie at the pretranslocation position (Figure 3A). The TIC crystals are active in RNA synthesis. Soaking of the crystals with the fourth NTP extends the RNA to 5 nt and expands the bubble to 15 nt as well. All the RNA oligonucleotides in the TICs contain a well-ordered 5’-triphosphate resulted from de novo RNA synthesis (Figure 3A).
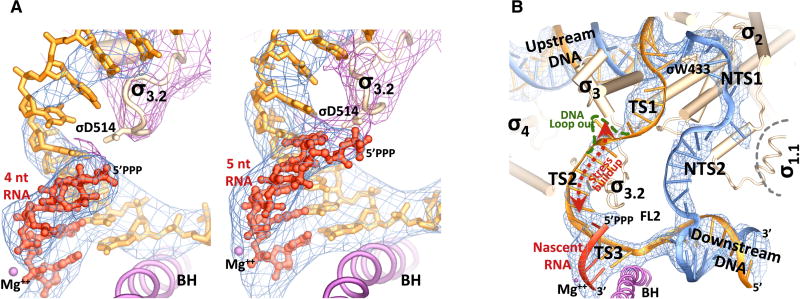
Figure 3. Nascent RNA and the transcription initiation bubble.
(A) (left) An RNA tetranucleotide. (right) An RNA pentanucleotide. Blue meshes show the σA-weighted Fobs-Fcalc electron density map (contoured at 2.0 σ) calculated in the absence of the nucleic acids. Purple meshes show the σA-weighted 2Fobs-Fcalc electron density map (contoured at 1.5 σ) covering the σ70 factor. The bridge helix (BH) and the catalytic metal center (Mg++) are shown for reference.
(B) A close-up view of the 15-nt initiation bubble. The σ70 factor is shown in wheat, and the RNAP core subunits are omitted for clarity. Blue meshes show the σA-weighted Fobs-Fcalc electron density map (contoured at 2.0 σ) calculated for the TIC in the absence of the nucleic acids. The upstream fork of the initiation bubble is maintained through specific interactions of the σ2 domain with the promoter -10 element and a stacking of σ W433 on the -12 basepair, and the downstream fork is maintained with the help of the fork loop 2 (FL2) and the bridge helix (BH).
The observation that in the TIC crystals all the nascent RNA oligonucleotides lie at the pre-translocation position, which is in contrast to the post-translocation position frequently observed in transcription complexes that contain an incomplete transcription bubble (Liu et al., 2015; Sainsbury et al., 2013; Vassylyev et al., 2007; Zhang et al., 2012), suggests that there is stress buildup in the initiation complexes with a complete transcription bubble of 14 or 15 nt. While a pretranslocated RNA tetranucleotide is not long enough to reach the σ3.2 loop, interestingly, when a pretranslocated RNA pentanucleotide reaches the σ3.2 loop, the 5’-triphosphate of the RNA appears to interact with the acidic σ3.2 loop favorably as shown by the well-defined electron density bridging the 5’-triphosphate and the σ3.2 loop (Figure 3A). This suggests that the stress buildup in the TICs could not be the result of repulsion of the RNA 5’- triphosphate by the acidic σ3.2 loop. We speculate that one function of the σ3.2 loop is to stabilize the short nascent hybrids through this bridging interaction during the early stage of transcription initiation.
DNA scrunching during transcription initiation
As the nascent RNA extends to about 9 to 10 nt for promoter escape during transcription initiation, the transcription bubble expands from an initial size of about 13 nt to over 20 nt. Since σ2 makes sequence-specific interactions with the -10 element, it is expected that the upstream portion of the initiation bubble undergoes minimal changes whereas the rest of the bubble DNA scrunches during this initiation process.
The TIC structures shown here contain complete transcription bubbles of 14 or 15 nt respectively (Figure S1). The bubble region of the NT-strand DNA in the TICs can be divided into two parts: a well-defined upstream segment (NTS1) comprised of the single-stranded -10 residues and the first discriminator residue (position -6) that interact specifically with the RNAP, and a downstream segment (NTS2) comprised of the rest of the discriminator and the newly unwound initially-transcribed region that scrunches into the enzyme (Figure 3B). Although individual bases could not be resolved at this resolution, it is plausible to assume that all the discriminator and downstream residues in the TICs shown here remain inside of an internal space between the β’ clamp, β1 and β2 domains of the RNAP, since continuous electron density is observed for the entire ssDNA region of the NT-strand for both the 14-nt and 15-nt bubbles. For large initiation bubbles, the protein internal space will not be spacious enough to enclose all the NT-strand nucleotides. It is currently not known how the extra NT-strand residues are accommodated; significant scrunching of the NT-strand DNA could promote the DNA to loop out at the joint of the upstream and downstream segments to release the stress buildup by the NT-strand DNA scrunching.
For the T-strand DNA, the bubble region might be divided into three segments: an upstream segment of the -10 residues (TS1), a downstream segment that forms the hybrid with the nascent RNA (TS3), and the middle segment of ssDNA residues that resides inside the active site chamber (TS2) (Figure 3B). The TS3-RNA hybrid is well ordered and superimposes well on the DNA-RNA hybrid in a transcription elongation complex (Vassylyev et al., 2007), but there appears to be a change in the pathway for the single-stranded TS2 from that of the A-form hybrid (Figure S4). The TS1 of -10 T-strand residues that wraps around the σ3 globular domain joins TS2 inside a space enclosed by the β1, β flap and σ3 domains. Much weaker electron densities suggest that the TS1-TS2 joint region is conformationally flexible. The T-strand DNA of a 15-nt bubble appears to start looping out at the TS1-TS2 joint region toward the internal space enclosed by the β1, β flap and σ3 domains. If this DNA looping-out at the TS1-TS2 joint is the mechanism for T-strand DNA scrunching, it is conceivable that scrunched DNA residues would press on the σ3 globular domain.
Consistent with the speculation that scrunching of the T-strand DNA presses on σ3, mutations of σ70 residues lying immediately C-terminal to the σ3 globular domain that interact with the RNAP core to help anchor the domain were known to affect abortive RNA synthesis in a promoter-dependent manner and facilitate promoter escape (Cashel et al., 2003). We have not been able to obtain TIC crystals containing bubbles larger than 15 nt in size no matter the size of the RNA oligonucleotides. We suspect that additional T-strand DNA scrunching of an initiation bubble of 16 nt or larger would require a movement of the σ3 globular domain to expand the inter-domain space. This forced movement would eventually cause σ3 to separate from the RNAP core; however, the σ3 globular domain in the TIC crystals is blocked from an outward movement by crystal packing.
Concluding remarks
For a typical σ70 promoter with a discriminator of 6–8 bp (Shimada et al., 2014), the initiation bubble would grow to 16–18 nt and the initiation complex would accumulate significant stress when the nascent RNA grows to 5 nt and reaches the σ3.2 loop. This implies that DNA scrunching, but not the σ3.2 repulsion, might be the primary contributor for the stress buildup that drives abortive synthesis and the σ-core separation for promoter escape. In certain cases, σ3.2 might even be helpful for transition into transcription elongation since the RNA 5’- triphosphate- σ3.2 bridging interaction could serve to drag and guide the nascent RNA into the RNA exit channel. Consistent with this hypothesis, it is known that initial RNA syntheses are mostly aborted as di- and tri-nucleotides and an AMPPCP substitution in the first step of transcription leads to only one-tenth to one-sixth of full transcripts (Fujioka et al., 1991). This might also provide an explanation for the observation that amino acid substitutions and deletions in the σ3.2 loop resulted in disappearance of middle-sized abortive RNAs and a significant increase of trinucleotides (Pupov et al., 2014). In the absence of a complete transcription bubble, the σ3.2 loop, and the related B-reader loop in eukaryotes, would block the growing RNA, as observed in other structural studies (Basu et al., 2014; Sainsbury et al., 2013).
EXPERIMENTAL PROCEDURES
Preparation of α C-terminal domain truncated (ΔαCTD) RNA polymerase
We modified the pVS10 plasmid (Belogurov et al., 2007) using the QuickChange site-directed mutagenesis kit (Agilent Technologies) to generate a new plasmid pVS10D for overexpression of the E. coli RNA polymerase core enzyme that lacks the C-terminal 94 amino acids of the α subunit (Δα236–329, or ΔαCTD). We also replaced the original β’ C-terminal his-tag with an N-terminal his-tag to the truncated α subunit to help the mutant protein purification. The ΔαCTD RNAP core enzyme was overexpressed and purified in essentially the same way as that for the full-length RNAP core enzyme (Zhi et al., 2003). The ΔαCTD RNAP holoenzyme was prepared by mixing the purified mutant core enzyme with purified σ70 protein (about 1:3 molar ratio) at room temperature for 15 minutes followed by size exclusion chromatography to remove the extra σ70 protein. The holoenzyme was concentrated to over 30 mg/ml and stored in small aliquots at -80°C after flash freezing in liquid nitrogen.
Crystallization of the E. coli transcription initiation complexes
The synthetic promoters, which contains the consensus -35 and -10 hexamers and the extended -10 motif, were prepared by annealing the non-template (NT) strand to an equal molar amount of the template (T) strand DNA that is complementary to the NT-strand except for a 6-nucleotide (nt) discriminator region (Figure S1). The transcription initiation complexes were assembled by incubating the synthetic DNA promoters (50 µM) with the mutant RNAP holoenzyme (10 mg/ml) at 37°C for 20 minutes in the presence of ATP, GTP and UTP (1 mM each). The promoters contain a C residue at the -1 position of the template strand, and the in vitro transcription was found to start at the -1 position and generate an RNA tetranucleotide 5’-GAGU-3’ (TIC1 & TIC2, Figure S1). The TICs were used directly for crystallization at room temperature by vapor diffusion with a reservoir containing 7% PEG3350, 150 mM MgCl2 and 100 mM HEPES-NaOH (pH7.0). After the crystals grew for about one week, they were cryo-protected in the mother liquor containing 20% sucrose before flash-freezing in liquid nitrogen. The TIC crystals containing an RNA pentanucleotide (TIC3, Figure S1) was prepared by soaking TIC2 crystals in the mother liquor supplemented with 1 mM CTP at room temperature for 30 minutes before flash-freezing. X-ray data were collected at 100 K at the NSLS X25 beamline. Diffraction data were integrated and scaled with HKL2000 (Otwinowski and Minor, 1997).
Structure determination of the E. coli transcription initiation complexes
The structures were solved by molecular replacement with PHASER (McCoy et al., 2007) using the structure of the E. coli RNAP holoenzyme (Zuo et al., 2013) as a starting model. The initial difference Fourier map that was calculated using the Fobs-Fcalc amplitudes and phases calculated using the structure of the holoenzyme showed unambiguously the difference electron density for the helical dsDNA region. The phases were improved by NCS averaging and the maps were further improved by temperature factor sharpening that allowed fitting the nucleic models into the density. After fitting DNA into the difference density in Coot (Emsley and Cowtan, 2004), ten cycles of TLS (translation libration screw-motion) and restrained refinement were performed using Refmac5 (Murshudov et al., 2011) in the CCP4 suite (Winn et al., 2011). The data collection and structural refinement statistics are summarized in Table 1. All figures were created using PyMOL (Delano, 2002).
Supplementary Material
HIGHLIGHTS.
Crystal structures of transcription initiation complexes with a complete DNA bubble
σ4 domain rotates to accommodate promoter spacer variations
A bridging interaction between the nascent RNA 5’-triphosphate and the σ3.2 loop
Template-strand DNA scrunching presses on the σ3 globular domain
Acknowledgments
We thank the staffs of the National Synchrotron Light Source beamline X25 for help during data collection and the Center for Structural Biology facility at Yale University for computational support. This work was supported by NIH grant GM057510 to T.A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The coordinates and structure factor files have been deposited with the Protein Data Bank under accession codes 4YLN (TIC1 with 17 bp spacer and 4 nt RNA), 4YLO (TIC2 with 16 bp spacer and 4 nt RNA), and 4YLP (TIC3 with 16 bp spacer and 5 nt RNA).
References
- Bae B, Davis E, Brown D, Campbell EA, Wigneshweraraj S, Darst SA. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl Acad. Sci. USA. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernandez-Tornero C, Kulbachinskiy A, Murakami KS. Structural Basis of Transcription Initiation by Bacterial RNA Polymerase holoenzyme. J. Biol. Chem. 2014;289:24549–24559. doi: 10.1074/jbc.M114.584037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial sigma factor explored by using segmental isotopic labeling and NMR. Proc. Natl Acad. Sci. USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne S, Marsh ME, Capitani G, Vorholt JA, Allain FH. Structural basis for -10 promoter element melting by environmentally induced sigma factors. Nat. Struct. Mol. Biol. 2014;21:269–276. doi: 10.1038/nsmb.2777. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Cashel M, Hsu LM, Hernandez VJ. Changes in conserved region 3 of Escherichia coli sigma 70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 2003;278:5539–5547. doi: 10.1074/jbc.M211430200. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PYMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Hirata T, Shimamoto N. Requirement for the beta, gamma-pyrophosphate bond of ATP in a stage between transcription initiation and elongation by Escherichia coli RNA polymerase. Biochemistry. 1991;30:1801–1807. doi: 10.1021/bi00221a011. [DOI] [PubMed] [Google Scholar]
- Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc. Natl Acad. Sci. USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen UM, McClure WR. A noncycling activity assay for the omega subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1979;254:5713–5717. [PubMed] [Google Scholar]
- Hansen UM, McClure WR. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. II. Release of sigma from ternary complexes. J. Biol. Chem. 1980;255:9564–9570. [PubMed] [Google Scholar]
- Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul. Syst. Bio. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- Kulbachinskiy A, Mustaev A. Region 3.2 of the sigma subunit contributes to the binding of the 3'-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- Liu B, Zuo Y, Steitz TA. Structural basis for transcription reactivation by RapA. Mol. Cell. 2015;112:2006–2010. doi: 10.1073/pnas.1417152112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333:633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002a;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002b;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pupov D, Kuzin I, Bass I, Kulbachinskiy A. Distinct functions of the RNA polymerase sigma subunit region 3.2 in RNA priming and promoter escape. Nucleic Acids Res. 2014;42:4494–4504. doi: 10.1093/nar/gkt1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- Samanta S, Martin CT. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J. Biol. Chem. 2013;288:31993–32003. doi: 10.1074/jbc.M113.497669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki Y, Tanaka K, Ishihama A. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PloS one. 2014;9:e90447. doi: 10.1371/journal.pone.0090447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straney DC, Crothers DM. A stressed intermediate in the formation of stably initiated RNA chain at the Escherichia coli lac UV5 promoter. J. Mol. Biol. 1987;193:267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy AA, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkin N, Severinov K. The role of RNA polymerase sigma subunit in promoter-independent initiation of transcription. Proc. Natl Acad. Sci. USA. 2004;101:4396–4400. doi: 10.1073/pnas.0400886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Degen D, Ho MX, Sineva E, Ebright KY, Ebright YW, Mekler V, Vahedian-Movahed H, Feng Y, Yin R, et al. GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides. eLife. 2014;3:e02450. doi: 10.7554/eLife.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi H, Yang W, Jin DJ. Escherichia coli proteins eluted from mono Q chromatography, a final step during RNA polymerase purification procedure. Methods Enzymol. 2003;370:291–300. doi: 10.1016/s0076-6879(03)70026-3. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.