Abstract
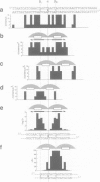
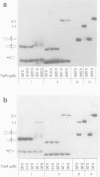
Unexpected features seen by high resolution X-ray crystallography at the interface of the trp repressor and the 'traditional' trp operator provoked the claim that the DNA fragment used in the crystal structure is not the true operator, and therefore that the crystal structure of the trp repressor-operator complex does not portray a specific interaction. An alternative sequence was proposed mainly on the basis of mutational studies and gel retardation analysis of short target duplexes (Staacke et al., 1990a,b). We have reexamined the sequence consensus in trpR-repressible promoters and analyzed the mutagenesis experiments of others including Staacke et al. (1990a) and found them fully consistent with the interactions of the traditional operator sequence seen in the crystal structure, and stereochemically inconsistent with the above referenced alternative model. Moreover, an in vitro trp repressor-DNA binding analysis, employing both novel DNA constructs devised to avoid previously encountered artifacts as well as full-length promoter sequences, indicates that the traditional operator used in the crystal structure is the preferred target of the trp repressor.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrowsmith C. H., Pachter R., Altman R. B., Iyer S. B., Jardetzky O. Sequence-specific 1H NMR assignments and secondary structure in solution of Escherichia coli trp repressor. Biochemistry. 1990 Jul 10;29(27):6332–6341. doi: 10.1021/bi00479a002. [DOI] [PubMed] [Google Scholar]
- Bass S., Sorrells V., Youderian P. Mutant Trp repressors with new DNA-binding specificities. Science. 1988 Oct 14;242(4876):240–245. doi: 10.1126/science.3140377. [DOI] [PubMed] [Google Scholar]
- Bass S., Sugiono P., Arvidson D. N., Gunsalus R. P., Youderian P. DNA specificity determinants of Escherichia coli tryptophan repressor binding. Genes Dev. 1987 Aug;1(6):565–572. doi: 10.1101/gad.1.6.565. [DOI] [PubMed] [Google Scholar]
- Bennett G. N., Yanofsky C. Sequence analysis of operator constitutive mutants of the tryptophan operon of Escherichia coli. J Mol Biol. 1978 May 15;121(2):179–192. doi: 10.1016/s0022-2836(78)80004-7. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci U S A. 1988 Feb;85(4):975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J., Lewis D. E., Lavoie T. A., Yang J. How does trp repressor bind to its operator? J Biol Chem. 1991 Dec 25;266(36):24509–24513. [PubMed] [Google Scholar]
- Carey J. trp repressor arms contribute binding energy without occupying unique locations on DNA. J Biol Chem. 1989 Feb 5;264(4):1941–1945. [PubMed] [Google Scholar]
- Chu B. C., Orgel L. E. Binding of hairpin and dumbbell DNA to transcription factors. Nucleic Acids Res. 1991 Dec 25;19(24):6958–6958. doi: 10.1093/nar/19.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydock P. V., Bogosian G., Brechling K., Somerville R. L. Studies on the interaction of Trp holorepressor with several operators. Evidence that the target need not be palindromic. J Mol Biol. 1983 Nov 15;170(4):1019–1030. doi: 10.1016/s0022-2836(83)80201-0. [DOI] [PubMed] [Google Scholar]
- Heatwole V. M., Somerville R. L. The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in Escherichia coli K-12. J Bacteriol. 1991 Jun;173(11):3601–3604. doi: 10.1128/jb.173.11.3601-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt B. K., Yanofsky C. Enhanced operator binding by trp superrepressors of Escherichia coli. J Biol Chem. 1990 May 15;265(14):7853–7858. [PubMed] [Google Scholar]
- Joachimiak A., Kelley R. L., Gunsalus R. P., Yanofsky C., Sigler P. B. Purification and characterization of trp aporepressor. Proc Natl Acad Sci U S A. 1983 Feb;80(3):668–672. doi: 10.1073/pnas.80.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak A., Marmorstein R. Q., Schevitz R. W., Mandecki W., Fox J. L., Sigler P. B. Crystals of the trp repressor-operator complex suitable for X-ray diffraction analysis. J Biol Chem. 1987 Apr 5;262(10):4917–4921. [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Mutational studies with the trp repressor of Escherichia coli support the helix-turn-helix model of repressor recognition of operator DNA. Proc Natl Acad Sci U S A. 1985 Jan;82(2):483–487. doi: 10.1073/pnas.82.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klig L. S., Crawford I. P., Yanofsky C. Analysis of trp repressor-operator interaction by filter binding. Nucleic Acids Res. 1987 Jul 10;15(13):5339–5351. doi: 10.1093/nar/15.13.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto A. A., Miller W. G., Gunsalus R. P. Escherichia coli tryptophan repressor binds multiple sites within the aroH and trp operators. Genes Dev. 1987 Aug;1(6):556–564. doi: 10.1101/gad.1.6.556. [DOI] [PubMed] [Google Scholar]
- Lawson C. L., Zhang R. G., Schevitz R. W., Otwinowski Z., Joachimiak A., Sigler P. B. Flexibility of the DNA-binding domains of trp repressor. Proteins. 1988;3(1):18–31. doi: 10.1002/prot.340030103. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Sigler P. B. The stereochemistry and biochemistry of the trp repressor-operator complex. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):113–126. doi: 10.1016/0167-4781(90)90047-6. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Q., Sigler P. B. Stereochemical effects of L-tryptophan and its analogues on trp repressor's affinity for operator-DNA. J Biol Chem. 1989 Jun 5;264(16):9149–9154. [PubMed] [Google Scholar]
- Oppenheim D. S., Bennett G. N., Yanofsky C. Escherichia coli RNA polymerase and trp repressor interaction with the promoter-operator region of the tryptophan operon of Salmonella typhimurium. J Mol Biol. 1980 Dec 5;144(2):133–142. doi: 10.1016/0022-2836(80)90029-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Robertson M. Gene regulation: specificity and flexibility. Nature. 1987 Jun 11;327(6122):464–466. doi: 10.1038/327464a0. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Otwinowski Z., Joachimiak A., Lawson C. L., Sigler P. B. The three-dimensional structure of trp repressor. 1985 Oct 31-Nov 6Nature. 317(6040):782–786. doi: 10.1038/317782a0. [DOI] [PubMed] [Google Scholar]
- Staacke D., Walter B., Kisters-Woike B., v Wilcken-Bergman B., Müller-Hill B. How Trp repressor binds to its operator. EMBO J. 1990 Sep;9(9):3023–3023. doi: 10.1002/j.1460-2075.1990.tb07495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staacke D., Walter B., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. How Trp repressor binds to its operator. EMBO J. 1990 Jun;9(6):1963–1967. doi: 10.1002/j.1460-2075.1990.tb08324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P., Vershon A., Bouvier S., Sauer R. T., Susskind M. M. Changing the DNA-binding specificity of a repressor. Cell. 1983 Dec;35(3 Pt 2):777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Berg O. G. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]