Summary
Combinatorial inhibition of effector and feedback pathways is a promising treatment strategy for KRAS mutant cancers. However, the particular pathways that should be targeted to optimize therapeutic responses are unclear. Using CRISPR/Cas9, we systematically mapped the pathways whose inhibition cooperates with drugs targeting the KRAS effectors MEK, ERK, and PI3K. By performing 70 screens in models of KRAS mutant colorectal, lung, ovarian, and pancreas cancers, we uncovered universal and tissue-specific sensitizing combinations involving inhibitors of cell cycle, metabolism, growth signaling, chromatin regulation, and transcription. Further, these screens revealed secondary genetic modifiers of sensitivity, yielding a SRC inhibitor-based combination therapy for KRAS/PIK3CA double mutant colorectal cancers with clinical potential. Surprisingly, acquired resistance to combinations of growth signaling pathway inhibitors develops rapidly following treatment, but by targeting signaling feedback or apoptotic priming, it is possible to construct three-drug combinations that greatly delay its emergence.
Graphical abstract
Introduction
In 1982, the discovery of activating RAS mutations in human cancer cells launched an intensive effort to both understand this potent family of driver oncogenes and develop selective treatment strategies (Cox and Der, 2014). Although more recent large-scale sequencing efforts have catalogued an extensive and growing list of driver mutations in human cancers, mutations in RAS family oncogenes, and in particular KRAS, remain among the most common oncogenic alterations in many cancers (Berger et al., 2016; Cox et al., 2014; Vogelstein et al., 2013). After neoplastic initiation, the continued reliance on mutant RAS signaling for tumor maintenance (Lim and Counter, 2005; Brummelkamp et al., 2002) provides a strong rationale to design therapies aimed at ablating its signaling activity. The literature to date recognizes at least four strategies to interfere with mutant RAS signaling: (i) direct pharmacological inhibition of RAS, (ii) interfering with RAS membrane association, (iii) exploiting RAS synthetic lethal interactions, and (iv) inhibiting key RAS effector pathways (Cox et al., 2014). Although several groups have reported recent progress towards strategy (i) above (Athuluri-Divakar et al., 2016; Lito et al., 2016), whether these can be advanced towards clinically active and selective inhibitors remains to be determined. Similarly, while excitement surrounded the initial development of farnesyltransferase inhibitors that block the post-translational farnesyl lipid modification controlling RAS localization, the unexpected compensatory activity of a related enzyme rendered KRAS refractory to inhibition, explaining the disappointing clinical outcomes in KRAS mutant cancers (Berndt et al., 2011). The strategy of selectively targeting RAS-driven cells by inhibiting mutation-induced synthetic lethal dependencies is a promising concept that has yielded actionable candidates, but one whose progress has been slowed by concerns about model system and reagent fidelity (Downward, 2015). Finally, as a result of promising early findings, the strategy of targeting key RAS effector pathways has emerged as the focus of most translational research efforts (Cox et al., 2014).
The activity of single agent therapies targeting individual RAS effector pathways is limited; however, combinations of inhibitors targeting multiple effector and/or feedback pathways have shown impressive activity in KRAS mutant model systems (Al-Lazikani et al., 2012; Samatar and Poulikakos, 2014). Specifically, the combined inhibition of MEK and the PI3K/AKT/mTOR pathway has been widely described, as have approaches combining MEK inhibitors with inhibitors of ERK feedback reactivation, including the receptor tyrosine kinases (RTKs) ERBB3, IGF1R, and FGFR1. More recent studies have also identified combinations targeting MEK alongside YAP1 and BCL-XL survival pathways as promising strategies for these tumors. (Corcoran et al., 2013; Ebi et al., 2011; Engelman et al., 2008; Lamba et al., 2014; Lin et al., 2015; Manchado et al., 2016; Sun et al., 2014). To date, however, these approaches have shown only limited clinical activity in patients with KRAS mutant cancers (Singh et al., 2015). Therapies targeting MEK alongside the PI3K/AKT/mTOR pathway have been limited by toxicities, making it difficult to achieve complete pathway inhibition in patients (Ebi et al., 2014). Ongoing clinical studies involving strategies targeting MEK alongside feedback or alternative survival pathways have thus far revealed activity in only subsets of patients, a fact which is likely attributed to the intrinsic molecular, genetic, and clinical heterogeneity of KRAS mutant tumors (Lamba et al., 2014). For example, recent integrative genomic analyses have revealed substantial diversity across, and even within, KRAS mutant tumors from distinct tissue types, a finding consistent with recent evidence that certain combination therapies may only have activity in defined tumor subsets (Kitai et al., 2016; Skoulidis et al., 2015).
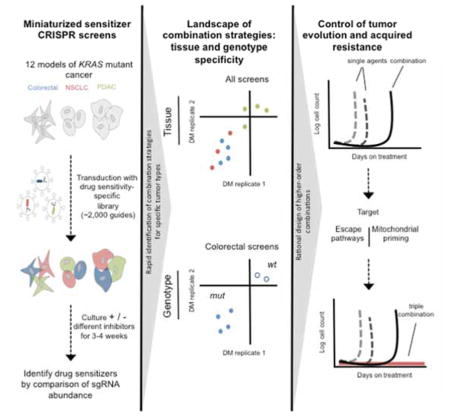
Together, these findings highlight three key unanswered questions. First, what is the landscape of druggable signaling pathways whose inhibition cooperates with inhibitors of KRAS effector signaling? Second, how do the activities of combination therapies targeting these pathways vary both across and within KRAS mutant tumors from distinct tissues? Finally, might a landscape view of drug target interactions shed light on strategies to build more effective combination therapies? To date, no studies have been powered to address these questions. Pooled RNA interference (RNAi) sensitizer screens are in principle suitable for the challenge, but historically they have only been performed with drugs targeting a single KRAS effector node in a single cell line (Corcoran et al., 2013; Lamba et al., 2014; Manchado et al., 2016; Sun et al., 2014). Further, the deficiencies of RNAi-based methods in the areas of reagent fidelity and scalability make it difficult to use this approach to accurately map drug target interactions across a large matrix of cell lines and drug treatments (Shalem et al., 2015). CRISPR/Cas9 screening methods have the potential to overcome these limitations by enabling the construction of high fidelity screening libraries that enable complete loss-of-function with minimal off-target effects (Shalem et al., 2014; Wang et al., 2014). In this study, we sought to use a custom CRISPR/Cas9-based screening library to map the landscape of druggable pathways that cooperate with inhibitors of the key KRAS effectors MEK, ERK, and PI3K in KRAS mutant lung, colon, pancreas, and ovarian cancer models based on the hypothesis that such an effort may reveal potent and durable strategies to target these tumors.
Results
Development of a CRISPR/Cas9-based method to identify drug sensitizers
Recent work has demonstrated the ability of CRISPR/Cas9-based loss-of-function screening to efficiently identify essential genes in mammalian cells using one- (Wang et al., 2014; 2015) and two-plasmid lentiviral systems (Shalem et al., 2014). We chose to clone optimized short guide RNAs (sgRNAs) (Wang et al., 2014) into a second generation single plasmid lentiviral system in which both an sgRNA and Cas9 are expressed from the same vector (Sanjana et al., 2014; Shalem et al., 2014) to create a custom screening library targeting 378 genes (5 guides per gene) alongside 50 non-targeting controls. These genes represent hand selected, key nodes in major oncogenic growth, survival, and metabolic pathways, RTKs, key druggable kinases and epigenetic modifiers, and frequently amplified or mutated oncogenes, genes that were judged to be both likely to modify drug sensitivity and are often druggable (Figure 1A and Supplemental Tables 1–2). We intentionally limited the size of this library to ~2,000 sgRNAs to enable parallel screening across many adherent cell line/drug combinations, as this limited library size enables negative selection screening using 50-fold fewer cells than genome-wide libraries (e.g., 2×106 vs. 1×108 cells per condition). To identify genes whose inhibition sensitized cells to drug treatments, cells were first transduced with the pooled library at a low multiplicity of infection (MOI = 0.2) and then cultured in the presence of sub-lethal drug doses for 3–4 weeks followed by deconvolution with next generation sequencing (Figure 1A).
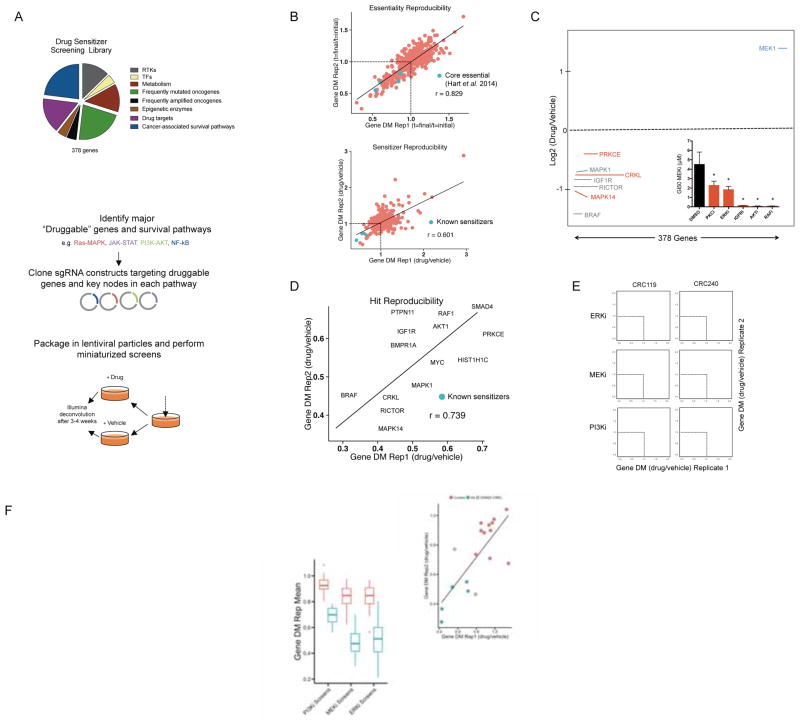
Figure 1. Construction and validation of miniaturized CRISPR/Cas9 library for drug sensitizer screening.
A) Breakdown of the 378 genes included (top) and schematic depicting the construction and implementation of the 1,940 sgRNA library (bottom). B) Replicate comparison of gene-level essentiality phenotypes in the colorectal cancer cell line HCT116 pilot screen. The mean depletion metric (DM; t = 4 weeks/t = initial) for all 5 constructs targeted to each gene in the library is plotted and fit to a linear model. Previously identified “core essential” genes (Hart et al., 2014) RPL5, SF3B1, PPP2R1A, SMC3, and U2AF1 are noted in blue as positive controls (top). Replicate comparison of sensitizer phenotypes from HCT116 pilot screen. Cells were cultured in the presence of 0.1 μM AZD6244 or vehicle for 4 weeks. The mean depletion metric (DM; t = 4 weeks, drug/vehicle) for all 5 constructs targeted to each gene in the library is plotted and fit to a linear model. Select sensitizers identified in previous studies (BRAF, IGF1R, AKT1, and MAPK1) are noted in blue as positive controls. C) Log2(DM) for each gene in a representative HCT116 MEKi sensitizer screen. The 50 control sgRNAs are randomly assigned to 10 control genes and are labeled in yellow. Hits in red are genes that scored reproducibly in the bottom 10% of genes in the library for both replicates (grey text, known sensitizer; red text, previously unknown sensitizer) Inset: Pharmacologic validation of sensitizers to the MEKi AZD6244 in HCT116 cells identified by the pilot screen (ERKi, SCH772984 0.02μM; IGFRi, GSK1838705A 1μM; PKCi LY317615 2μM; AKTi MK2206 10μM; RAFi LY3009120, 0.1μM, right). Data are GI50 values (mean ± SD of three replicates) in the presence of DMSO or the indicated sensitizer drugs. D) Replicate comparison of the DM (most active 3 sgRNAs per gene) for hits in the pilot screen (known sensitizers, blue; previously unknown sensitizers, orange). E) Replicate comparison of screens performed in two primary patient-derived CRC cell lines across the three inhibitors tested (SCH772984, ERKi; GDC-0623, MEKi; BKM-120, PI3Ki). Data are the DM for the 3 most active sgRNAs per gene. F) Relationship of control guides to hits in replicate 1 across all CRC screens (DM is the average of the 3 most active sgRNAs). Boxplots for the mean DM score of each hit and control in every CRC cell line for each drug screen (bottom right). G) Screen results for ControlA1 (orange) and guides targeting CRKL (blue) are plotted and fit to a linear model (adj R2= 0.54; top). Cells expressing sgCRKL constructs were treated with the MEK inhibitor AZD6244 and normalized to cells expressing sgControl constructs. Data are fold change in GI50 values (mean ± SD of three replicates; bottom). *p<0.05 by Student’s t test. See also Figure S1 and Tables S1, S2, and S3.
To validate drug sensitizer screening with CRISPR/Cas9, we first tested the approach in a well-defined model system. Specifically, we screened KRAS mutant HCT116 colorectal cancer (CRC) cells, a well-validated KRAS-dependent cell line, in the presence of the MEK inhibitor (MEKi) selumetinib (AZD6244), as several MEKi sensitizers have been previously identified in this cell line. Although our library lacked extensive representation of generally essential genes, the reproducibility of essentiality phenotypes in replicate screens confirmed consistent reagent behavior and identified several known “core essential” genes identified in a recent study (Figure 1B, top) (Hart et al., 2014). Drug screens were performed using a dose of 0.1 μM AZD6244, a concentration yielding on-target MEK inhibition and approximately 25% growth inhibition (Supplemental Figure 1a). After calculating the depletion metric (DM) for each sgRNA (its relative abundance in the presence of drug normalized to the same quantity in the presence of vehicle), we converted sgRNA-level scores to gene-level scores by calculating the cumulative sum of the rank ordered scores derived using five common metrics: the mean and median scores for the five sgRNAs targeting each gene alongside the average score of the three best and two best sgRNAs per gene and the score of the second best sgRNA alone (Supplemental Table 3; for details, see Methods (Gilbert et al., 2014; Marcotte et al., 2012)). This multi-metric gene scoring method was used to minimize false positive hits by balancing the relative strengths and weaknesses of each metric. Using this approach, replicates were highly correlated for sensitizer phenotypes, and known sensitizers behaved as expected (Figure 1B, bottom). Nominally, we considered genes to be sensitizer hits when they scored in the bottom 10% of all genes in both replicates of a given screen (Figure 1C, hits indicated with red dots). All hits scoring at this threshold had multiply corrected p-values less than 0.05 (Benjamini-Hochberg correction), suggesting that they are likely to be true positives. Further, sensitizers from this screen could be validated in an eight-point growth inhibition-50% (GI50) assay (See Methods) using selective small molecule kinase inhibitors targeting sensitizer gene products (Figure 1C, inset; Supplemental Figure 1b). Importantly, non-targeting control sgRNAs, randomly binned into 10 control genes, failed to score (Figure 1C, in yellow). The sensitizer phenotypes for hit genes were highly reproducible across replicate screens (Figure 1D), and included the previously identified sensitizers AKT1, RAF1 (C-RAF), BRAF, and IGF1R.
Having validated the sensitizer screening method, we next performed screens to identify sensitizers to the MEKi GDC-0623, the ERKi SCH772984, and pan-PI3Ki BKM120 (buparlisib). GDC-0623 and SCH772984 were chosen because of their abilities to selectively and potently inhibit both the kinase activities and feedback-mediated phosphorylation of their targets MEK and ERK, respectively. Similarly, we chose a pan- rather than isoform-selective PI3K inhibitor BKM120 because isoform-selective inhibitors can be overcome by compensation by other isoforms (Costa et al., 2015; Schwartz et al., 2015). Screens were performed in a panel of four KRAS mutant CRC models – two established cell lines (HCT116 and SW620) and two primary patient-derived xenograft (PDX)-established cell lines (CRC119 and CRC240) (Uronis et al., 2012) – at doses yielding on-target kinase inhibition and partial cell growth inhibition in each cell line (Supplemental Table 3 and Supplemental Figure 1c and d). Correlation plots from replicate screens demonstrate that while screens with MEKi and ERKi identified numerous reproducible sensitizers, screens with PI3Ki yielded fewer and weaker sensitizers, perhaps reflecting the limited driver role for PI3K in KRAS-dependent effector signaling in these cancers (Figure 1E, where sensitizers are found in the lower left hand quadrant of each plot, and 1F and Supplemental Figure 1e) (Cox et al., 2014). Additionally, the low passage PDX-derived cell lines behaved similarly to established cell lines, and both MEKi and ERKi screens produced broadly similar sensitizer profiles across all cell lines (Supplemental Figure 1f) Finally, as a demonstration of the reliability of the screening approach, we generated knockout lines for the adaptor protein CRKL. CRKL validation assays in all screened cell lines yielded consistent sensitizing phenotypes with the exception of SW620 (grey dots and bars), whose variable efficacy in validation assays mimicked that seen in the screen results for this cell line (Figure 1G, Supplemental Figure 1g). Taken together, these findings suggest that CRISPR/Cas9-based screens can reproducibly identify drug sensitizers with low false positive hit rates, suggest consistency between screens performed in primary patient-derived and established cellular models, and underscore the extensive cooperation between inhibitors of the MEK/ERK pathway and other cellular targets in KRAS mutant CRCs.
Mapping a landscape of combination therapies in KRAS mutant cancers
To broaden our analysis to additional KRAS mutant tumor types, we next performed replicate screens in cellular models of KRAS mutant non-small cell lung adenocarcinoma (NSCLC) (Calu-6, A549, Calu-1), pancreatic ductal adenocarcinoma (PDAC) (Capan-1, Panc 02.03, Panc 03.27, and CFPAC-1), and ovarian mucinous carcinoma (MCAS) treated with GDC-0623, SCH772984, and BKM120 as above. By integrating the results of these screens with those performed in KRAS mutant CRC cell lines – a total of 70 screens (35 screens in duplicate) in 12 cellular models – we identified 51 genes scoring as sensitizers to either MEKi, ERKi, or PI3Ki (top 10% in both replicate screens in ≥2 cell lines), with eight sensitizers scoring for PI3Ki and 47 sensitizers scoring for either MEKi or ERKi. When analyses were restricted by tissue type, a similar pattern was observed, with 3, 3, and 2 PI3Ki sensitizers observed in colon-, lung-, and pancreas-derived models, respectively, and 24, 22, and 12 MEKi and/or ERKi sensitizers observed in the same models (Figure 2A, Supplemental Figure 1c, d).
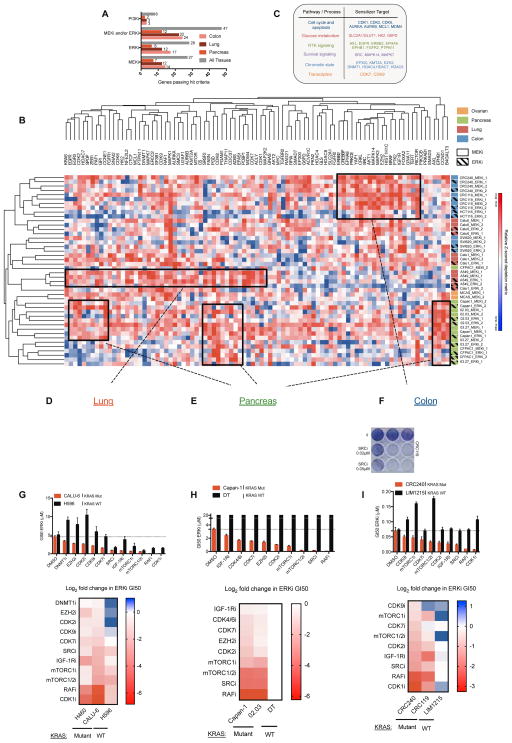
Figure 2. Landscape view and validation of sensitizers to MEK/ERK inhibitors across KRAS mutant cancers.
A) Comparison of hit frequency across tissues and drugs. A gene is considered a hit if it scores reproducibly in two cell lines per tissue. B) Hierarchical clustering of the Z-scored DM for the 3 most active sgRNAs per gene in each replicate for GDC-0623 (MEKi) and SCH772984 (ERKi) screens. For each condition, cells were grown either in vehicle or low doses of the indicated inhibitor (see Supplemental Table 3 for doses) for 3–4 weeks and then results were de-convoluted by deep sequencing (boxes highlight representative areas of heat, indicating groups of possible tissue-specific sensitizers). C) Table with representative processes and corresponding target genes that modulate sensitivity to MEK/ERK inhibition uncovered by the screens. D–F) Crystal violet staining of 7 day colony growth in cell lines treated with the indicated, candidate sensitizers. Cells were treated with the indicated inhibitors in combination with AZD6244 (MEKi) or ERKi (SCH772984) at the listed concentrations (ERK5i, XMD8-92; MDM2/4i, MI-773; EGFRi, Gefitinib; mTORC 1/2, Torin1; SRCi, dasatinib; CDK1i RO-3306). Data are a representative image of each experiment performed in duplicate. (G–I, top) Pharmacologic validation of 12 sensitizers. Mutant and wild type cells were tested in 8-point GI50 assays with either SCH772984 (ERKi) alone or in the presence of a constant background concentration of the indicated drugs. Relative viability was measured at 72 hours post-treatment using Cell Titer Glo. Dotted line indicates ERKi GI50 value for DMSO treated KRAS mutant cells. Data are mean ± SEM of three replicate experiments. (G–I, bottom) Similar to top, log2 transformed GI50 values for two KRAS mutant cell lines and one KRAS WT cell line per tissue. Data are normalized to DMSO treated samples for each cell line. (DNMT1i, Azacitadine 0.5μM; EZH2i UNC1999, 0.5μM; CDK2i Roscovitine, 5μM; CDK9i LDC000067, 2μM; CDK7i BS-181, 2μM; SRCi dasatinib, 0.2μM; IGFRi, GSK1838705A 1μM; mTORC1 Rapamycin, 0.1μM; mTORC 1/2, Torin1, 0.2μM; RAFi LY3009120, 1μM; CDK1i RO-336 5μM; CDK4/6i PD0332991, 2μM). *p<0.05. See also Figure S1, S2 and Tables S3, S4.
To visualize the landscape of MEKi and ERKi sensitizers across tissues, we performed unsupervised hierarchical clustering of all sensitizers identified in at least two pairs of replicate MEKi or ERKi screens (n = 89 genes). This analysis revealed several overarching patterns. First, sensitizer profiles from replicate screens tended to cluster together, as did profiles from MEKi and ERKi screens in a given cell line. Second, sensitizer profiles generally clustered by tissue of origin, with pancreas and ovarian models clustering separately from colorectal and lung models, which were partially intermingled (Figure 2B). Our screens identified previously described MEKi sensitizers, including those that function through the PI3K/AKT/mTOR pathway (AKT1, AKT2, RICTOR, MTOR, and IGF1R), through suppression of ERK pathway feedback reactivation (or incomplete pathway inhibition) (BRAF, RAF1, MAPK1, MAPK3, KRAS, and FGFR1), and through the YAP1 pathway (Ebi et al., 2011; Engelman et al., 2008; Lamba et al., 2014; Lin et al., 2015; Manchado et al., 2016). Importantly, these screens also identified a myriad of cellular processes not previously implicated as therapeutic co-targets with MEKi or ERKi in KRAS mutant cancers. These druggable sensitizers included regulators of cell cycle and apoptosis (CDK1, CDK2, AURKA, AURKB, MCL1, and MDM4), glucose metabolism (SLC2A1/GLUT1, HK2, G6PD), RTK signaling (AXL, ERBB2, EPHA8, EPHB1, FGFR2, and PTPN11), survival signaling (SRC, MAPK14, MAPK7), chromatin state (EP300, KMT2A, EZH2, DNMT1, HDAC4, HDAC7, and HDAC8), and transcription (CDK7 and CDK9), as well as the currently undruggable oncogenic transcription factors MYC, NFE2L2, ZEB1, and SNAI2 (Figure 2B and C, Supplemental Table 3).
To functionally validate the druggable sensitizers identified in primary screens, we began by testing a selection of hits identified in lung, pancreas, and colorectal cancer screens using pharmacological inhibitors of each sensitizer. Seven-day clonogenic growth assays confirmed greater than additive growth inhibition resulting from MEKi or ERKi plus sensitizer drug co-treatments in lung, pancreas, and colon cancer models (Figure 2D–F, Supplemental Figure 2a). To better understand the strength, spectrum of activity, and KRAS mutation selectivity of these combination therapies, we tested a broader panel of candidate sensitizers using validated small molecule inhibitors of each sensitizer. Inhibitors of known sensitizers mTORC1, B- and C-RAF (pan RAFi), and IGF-1R served as positive controls. Specifically, we assessed sensitization to ERKi mediated by each candidate sensitizer in three cell line models for each tissue type (two KRAS mutant and one wild-type (WT)) using an eight-point GI50 growth assay. For pancreatic cancer, we modeled KRAS WT disease, which occurs in <5% of patients and is not well-represented by established cell lines, using hTERT-immortalized pancreatic ductal epithelial (DT) cells (Campbell et al., 2007). We note that BxPC3 cells, which are sometimes used as models of KRAS WT disease, actually harbor a MAPK pathway dependency owing to a recently characterized, activating deletion in BRAF, and thus were not used in this setting (Chen et al., 2016; Foster et al., 2016). Overall, we pharmacologically validated 44/46 (96%) of the sensitizers examined in distinct tissues, in the process finding that while the strength of sensitization varied by tissue type and drug, each combination was more effective in the KRAS mutant setting (Figure 2G–I, Supplemental Figure 2b). For an expanded and complete list of the 44 sensitizing combinations validated in secondary assays, their method(s) of validation, and the degree of sensitization observed by GI50 assays, refer to Supplemental Table 4.
Lung and colon tissue specificity of p38α
An obvious advantage of screening many cell line models is the ability to rapidly credential the tissues where a combination will have the greatest effect. For instance, knockout of MAPK14 (encoding the p38α MAPK) scored strongly as a MEKi/ERKi sensitizer in lung and colorectal models, but less so in pancreas models (Supplemental Figure 2c). Subsequent validation of this interaction with sgRNAs and a small molecule inhibitor against p38α confirmed this tissue-specific interaction (Supplemental Figure 2d–f). Inhibition of p38 as a monotherapy has not demonstrated significant anti-tumor activity in RAS-mutant cancer cell lines in vitro (Campbell et al., 2014). Our findings suggest that the combination of a clinical candidate p38α/β inhibitor (LY2228820) with MEK/ERK inhibition may be an actionable strategy for KRAS mutant lung and colorectal cancer patients due to its ability to block compensatory, treatment-induced p38 pathway activation (Supplemental Figure 2g) and induce greater than additive levels of apoptosis (Supplemental Figure 2h).
SRC inhibitors cooperate with MEK/ERK inhibitors in a PIK3CA mutation-dependent manner
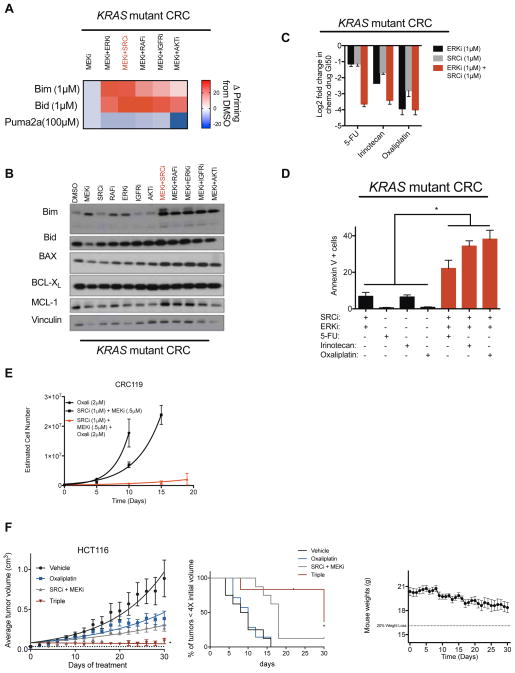
In several instances, we observed genes whose knockout conferred differential sensitization across cell lines from a single tissue, suggesting the presence of secondary modifiers of sensitivity and leading us to hypothesize that some of these secondary modifiers may be mutationally encoded. For example, MDM4 knockout strongly sensitized lung cancer cells with wild-type (WT) TP53, but not those with mutant TP53, to MEKi/ERKi, a result that could be phenocopied using a pharmacological MDM2/4 inhibitor (MI-773) and that is consistent with MDM4’s established role as a negative regulator of p53 activity (Supplemental Figure 3a). Similarly, we observed that SRC knockout sensitized the CRC cell lines CRC119, CRC240, and HCT 116 to MEKi/ERKi, but not the SW620 cell line, differential sensitization that correlated with the presence of canonical activating PIK3CAH1047R mutations in the former lines, but not in the latter (Figure 3A, Supplemental Figure 3b). We confirmed this sensitization in eight-point GI50 assays that revealed that pharmacological ERK inhibition conferred a highly synergistic, >100-fold sensitization to SRC inhibition in CRC240 cells (CI < 1.0 by Chou-Talalay method, Figure 3B (Chou, 2010)).
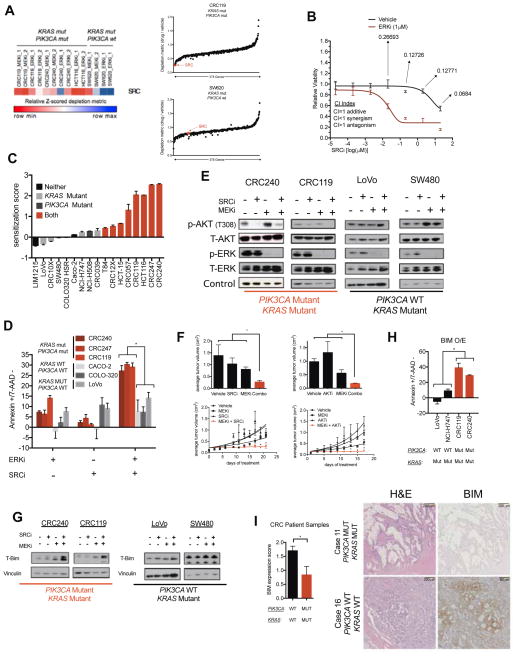
Figure 3. Co-inhibition of the MEK/ERK pathway plus SRC induces synergistic apoptosis in KRAS/PIK3CA double mutant colorectal cancers (CRCs) through induction of BIM.
A) At right, relative depletion of SRC across CRC screens. At left, rank ordered relative depletion scores (threescore) plotted for all 378 genes in a KRAS/PIK3CA double mutant and a KRAS mutant/PIK3CA wild-type cell line. B) GI50 for a SRC inhibitor (dasatinib) in the presence of either vehicle or a constant background dose of ERK inhibitor (VX-11e) in CRC240 cells. CI values are calculated for each dose on the curve. C) SRC inhibitor (dasatinib) sensitization score across a panel of CRC cell lines with indicated alterations in KRAS and PIK3CA. Sensitization score is calculated as the log10 ratio of the GI50 values for SRCi (dasatinib) relative to the same quantity in the presence of a constant background dose of 1 μM ERKi (VX-11e). Additive effects center at zero, antagonistic effects are negative, and sensitization effects are positive. D) Apoptosis measurements, reported as the percentage of annexin v+/7-AAD- cells in six CRC cell lines representing various mutational backgrounds treated with vehicle, a SRC inhibitor (dasatinib, 200nM), an ERK inhibitor (VX-11e, 1μM), or the combination of both. E) Immunoblots of P-AKT, T-AKT, P-ERK, T-ERK, and a loading control in four CRC cell lines representing different mutational backgrounds treated with vehicle, a SRC inhibitor (dasatinib, 1μM), a MEK inhibitor (AZD6244, 0.5μM), or the combination of both for 6 hrs. Loading control for CRC240 and LoVo is Histone H3 and control for CRC240 and SW480 is vinculin. Blots are cropped for clarity. F) HCT116 xenografts treated with vehicle, SRCi (dasatinib 15 mg/kg, daily) or AKTi (MK2206 60 mg/kg, daily), MEKi (AZD6244 10 mg/kg, twice daily), or the combination of a MEKi with either SRCi or AKTi for 21 days, shown as tumor size at endpoint (top) or growth curve (bottom). G) Immunoblot of T-BIM and vinculin in four CRC cell lines representing different mutational backgrounds treated with vehicle, a SRC inhibitor (dasatinib, 1μM), a MEK inhibitor (AZD6244, 0.5μM), or the combination of both for 6 hrs. Blots are cropped for clarity. H) Apoptosis (annexin V+/7-AAD- percentage) following ectopic overexpression of BIM in the absence of drug in indicated CRC cell lines. I) Quantification by immunohistochemistry (IHC) for T-BIM in CRC patient samples stratified into WT/WT or KRAS/PIK3CA mutant groups. To the right are representative images of each case, also showing H&E staining. Error bars show data ± SEM. *p<0.05. See also Figure S3 and Table S5.
Given the dramatic synergy and unclear mechanism(s) of action associated with SRC plus MEK/ERK pathway inhibition, we examined this combination in further detail. First, we confirmed the on-target MEK/ERK pathway and SRC specificity of the combination by performing cell proliferation assays using a MEKi (AZD6244), an additional SRCi (saracatinib), and two independent shRNAs targeting SRC (Supplemental Figure 3c). Next, we tested the combination in a panel of 17 cellular models of CRC, 7 of which were PDX-derived primary cell lines. In this panel, 8 cell lines with mutations in both KRAS and PIK3CA (heretofore “double mutant”) exhibited pronounced sensitization in growth inhibition assays, while cell lines with either mutation alone, or WT for both, failed to respond (Figure 3C, Supplemental Figure 3d). Similarly, double mutant cell lines exhibited greater than additive apoptosis induction following treatment with SRCi plus ERKi, while single mutant and WT/WT lines failed to respond (Figure 3D). Mechanistically, SRC inhibition blocked AKT phosphorylation and blunted MEKi-driven AKT feedback activation, but only in double mutant cells (Figure 3E, Supplemental Figure 3e). Importantly, AKT inhibition by SRC inhibitors was required for the activity of the combination therapy, as ectopic expression of a constitutively active, myristoylated AKT1 (myr-AKT1) blocked the combination’s activity in growth and apoptosis assays (Supplemental Figure 3f, g). Further, combined SRCi plus MEKi therapy in mice bearing double mutant xenograft tumors led to greater than additive tumor growth inhibition, a result that could be phenocopied using the combination of a MEKi and an allosteric AKTi (Figure 3F).
To better understand the mechanism of apoptosis induction following MEK/ERK pathway plus SRC inhibition, we used BH3 profiling, which identified an increase in overall apoptotic priming in KRAS/PIK3CA mutant cells treated with the drug combination as evidenced by increased mitochondrial depolarization following treatment with BIM- or BID-derived peptides (Supplemental Figure 3h). Given that increased priming implies a change in the stoichiometry of BH3-only proteins associated with the mitochondrial membrane (Montero et al., 2015; Winter et al., 2014), we probed lysates of KRAS mutant/PIK3CA mutant and KRAS mutant/PIK3CA WT cells for alterations in the levels of BIM. Indeed, combination treatment at 6 hr and 24 hr increased levels of the pro-apoptotic protein BIM only in KRAS/PIK3CA double mutant cells, with BIM increases being predominantly driven by ERK pathway inhibition (Figure 3G, Supplemental Figure 3i). Further, SRC/AKT signaling also regulates BAD, which cooperates with BIM upstream through BCL-2/BCL-XL (Supplemental Figure 3j). This led us to hypothesize that BIM activation may be necessary and sufficient for the activity of the combination, a hypothesis we confirmed experimentally (Figure 3H and Supplemental Figure 3k). Interestingly, during the course of these experiments we noticed that ectopic BIM expression only induced marginal apoptosis in single mutant cell lines despite the established importance of this protein as a key activator of the intrinsic apoptotic pathway (Figure 3H) (Letai, 2008). Consistent with this observation, BIM protein was found to be expressed at significantly lower steady state levels in double mutant cell lines and human patient tumors than in corresponding tumors having WT KRAS or PIK3CA alleles (Figure 3I, Supplemental Figure 3l, and Supplemental Table 5). Collectively, these results demonstrate that KRAS/PIK3CA double mutant CRCs undergo apoptosis and tumor growth inhibition following treatment with inhibitors of SRC and the MEK/ERK pathway. Mechanistically, SRC inhibition appears to function by suppressing AKT phosphorylation only in double mutant tumors, an event that drives BIM activation when combined with MEK/ERK inhibition through both direct up-regulation of BIM protein levels and through suppression of BAD phosphorylation. Surprisingly, BIM induction only leads to apoptosis in double mutant tumors, an observation that may be explained by the fact that tumors lacking double mutations have evolved in the presence of high steady state BIM protein levels, and therefore are insensitive its death-inducing effects. Because SRC inhibition leads to AKT inhibition only in double mutant cells, and because these cells are particularly sensitive to BIM induction, this therapy may have a substantial therapeutic window that exceeds that of combined, direct MEK and AKT inhibition, which has failed clinically because of the toxicity associated with systemic inhibition of these pathways (Singh et al., 2015).
Leveraging the landscape of sensitivity modifiers to suppress resistance
As demonstrated above, large scale CRISPR/Cas9 screening can uncover potent combination therapies; however, it is clear from clinical experience that resistance can emerge even in the context of combination therapies to which patients initially respond (Robert et al., 2015). To better understand the potential for acquired resistance to combination therapies in KRAS mutant cancers, and to credential the kinetics of resistance acquisition, we used time-to-progression (TTP) modeling (Misale et al., 2015). In this assay, KRAS/PIK3CA double mutation CRC cells developed resistance to single agent ERKi (as measured by the acquisition of exponential growth kinetics) on the time scale of three to four weeks, while the combination of SRCi plus ERKi delayed resistance by an additional two weeks (Figure 4A). To extend these studies, we performed TTP assays on a panel of additional, combination therapies arising from our screens. These assays focused specifically on inhibitor combinations targeting growth signaling pathways, as these agents may have near-term clinical potential. Interestingly, as in the case of the SRCi + ERKi combination, we observed delayed resistance emergence for each assayed combination therapy (Figure 4B). This observation implies that resistance to two drug combination therapies, while delayed relative to monotherapies, is likely to eventually arise, thereby placing an emphasis on defining strategies to more durably block resistance.
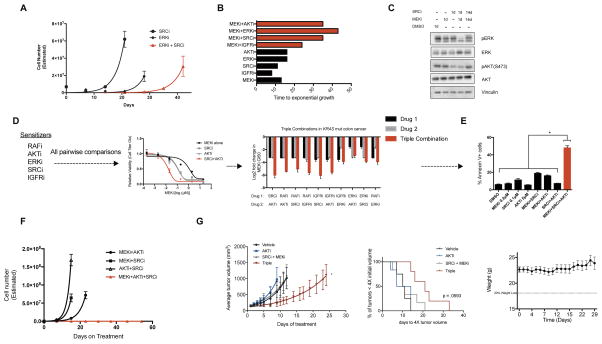
Figure 4. Leveraging the landscape of sensitizers to suppress resistance.
A) Time to progression (TTP) assay in CRC240 cells treated with the ERKi (VX-11e, 1μM) + SRCi (dasatinib, 1μM) combination. Data are mean ± SEM of three replicates. B) TTP for several candidate CRC combinations tested in CRC119 cells. MEKi, AZD6244 1μM; IGFRi, GSK1838705A 1μM; SRCi, dasatinib 0.5μM; ERKi, SCH772984 0.1μM; AKTi, MK2206 5μM. C) Immunoblots of indicated targets in CRC119 cells treated with DMSO, MEKi (AZD6244, 1μM), SRCi (dasatinib, 0.5μM), or the combination for 14 days and probed at the indicated times. Blots are cropped for clarity. D) Pairwise combinations of sensitizers in CRC119 cells to identify triple combinations (left). Triple combinations (red) were tested for their ability to shift the GI50 curves to a greater extent than either of the two body combinations (grey; center). Log2 fold shifts from baseline are shown for all combinations tested, where negative values indicate leftward shift of the curve as in the center plot. Data are mean ± SD of three replicate experiments. Drug identities as above except: RAFi, LY3009120 0.05μM; ERKi 0.05μM; SRCi 1μM. E) CRC119 annexin V+ cells after 48 hours of treatment with the indicated combinations (MEKi, SRCi, and AKTi identity same as above). F) TTP assay for a candidate triple combination (MEKi AZD6244, 0.2μM; SRCi dasatinib, 1μM; AKT MK2206, 10μM). G) HCT116 xenograft treated with vehicle, MK-2206 (15 mg/kg, daily), dasatinib (15 mg/kg, daily) and AZD6244 (15 mg/kg, daily), or the triple combination. For average tumor volumes, each arm only plots the data up to the point at which the first mouse in the group reached the humane endpoint. To the right, a survival curve showing percent of mice with tumors less than 4X the starting volume at a given time. To the right of survival curve are the mouse weights for the triple combination group over the course of the study. Data are mean ± SEM. *p>0.05. See also Figure S4.
Feedback and bypass signaling mechanisms have been well documented to limit the activity of MEK/ERK pathway targeted monotherapies in KRAS mutant cancers (Bernards, 2012), and it is reasonable to hypothesize that similar mechanisms may also limit the activity of combination therapies, as has been demonstrated when combined BRAFi and MEKi therapies are used in BRAF mutant melanomas (Moriceau et al., 2015). Indeed, in KRAS/PIK3CA double mutant CRC cells treated for two weeks with SRCi plus MEKi, we observed rebound of both ERK and AKT phosphorylation (Figure 4C). Reasoning that multiple mechanisms of feedback and bypass signaling may exist, and that these mechanisms are likely to be driven by the sensitizer hits identified in our primary screens, we hypothesized that these hits may be enriched for drug targets that can be combined to yield triple drug therapies that cause greater than additive cell death at low doses and delay TTP in vitro. Indeed, effective triple drug therapies have been previously described, but efficiently navigating the space of potential three drug combinations to identify those capable of suppressing resistance is technically challenging (Ahronian et al., 2015). To test this concept, we assembled panels of drugs targeting sensitizers that cooperate with MEKi in KRAS mutant colorectal and lung cancers (AKT, IGFR, RAF, ERK, SRC, MAPK7, and ERK5). First, we performed GI50 assays to quantify the degree to which each individual sensitizer, and all combinations of two sensitizers, potentiated the activity of MEKi. In most cases, combinations of two sensitizers potentiated the activity of MEKi more than either drug alone (Figure 4D and Supplemental Figure 4a), a result that translated to greater than additive apoptosis induction using low dose triple combinations (Figure 4E and Supplemental Figure 4b). Importantly, low dose triple drug combinations also suppressed the emergence of resistance over time in multiple TTP models (Figure 4F and Supplemental Figure 4c). Additionally, low, tolerable doses of this triple combination could suppress resistance in vivo for a short period, however, the tumors ultimately rebounded on treatment (Figure 4G). Collectively, these data suggest that the landscape of MEKi and ERKi sensitizers identified by CRISPR/Cas9 screening identifies multiple, parallel mechanisms of resistance, and that by targeting these mechanisms in combination, it is possible to derive higher order drug combinations that suppress resistance in vitro. However, the eventual outgrowth of resistance to these triple combinations in vivo suggests that alternative approaches to delay resistance will be needed.
Suppressing resistance by exploiting drug-induced apoptotic priming
Although targeting signaling feedback-based resistance is a potentially viable approach to suppress resistance, an alternative approach is to take advantage of underlying properties shared by drug combinations. In agreement with our SRCi plus MEKi/ERKi findings, recent work has demonstrated that targeted therapies can tilt the balance of pro- versus anti-apoptotic signals in the mitochondria, “priming” cells for death (Supplemental Figure 3h (Montero et al., 2015)). Indeed, BH3 profiling revealed that each pair of a selection of the most potent combination therapies targeting growth signaling pathways also forced KRAS mutant cells into a more primed state (Figure 5A, Supplemental Figure 5a) likely through the induction of BIM at the protein level, as only modest changes were observed in other candidate BCL-2 family proteins (Figure 5B, Supplemental Figure 5a). Drug-induced priming was not observed in KRAS WT cells (Supplemental Figure 5b). Given recent evidence that cancer cells in a primed state are more sensitive to treatment with cytotoxic chemotherapies (Chonghaile et al., 2011), we reasoned that combination therapies may sensitize tumors to standard of care chemotherapy. Indeed, treatment of KRAS/PIK3CA double mutant colorectal cancer cells with ERKi plus SRCi sensitized these cells to 5-fluorouracil (5-FU), irinotecan, and oxaliplatin, the cytotoxic agents used in the standard-of-care regimens FOLFOX, FOLFIRI, and FOLFIRINOX, by over tenfold in GI50 assays (Gustavsson et al., 2015). Further, treatment with low doses of ERKi, SRCi, and cytotoxics caused greater than additive apoptosis induction, delayed TTP in vitro, and suppressed the growth of a xenograft tumor model in vivo (Figures 5D–F).
Figure 5. Leveraging the priming ability of two-body combinations to design triple combination therapies involving cytotoxic chemotherapies.
A) BH3 profiling in the KRAS mutant CRC cell line HCT116 treated with the indicated combinations (MEKi AZD6244, 1μM; RAFi LY3009120, 0.5μM; IGFRi GSK1838705A, 3μM; SRCi dasatinib, 0.5μM; ERKi SCH772984, 0.25μM; AKTi MK2206, 5μM). B) Immunoblot of indicated proteins in CRC119 CRC cell line treated with the indicated combinations (Drug identities same as above with following doses: MEKi 1μM; RAFi 0.2μM; IGFRi 1μM; SRCi 0.5μM; ERKi 0.1; AKTi 5μM). Blots are cropped for clarity. C) Log2 transformed GI50 values for three separate cytotoxic chemotherapeutic drugs treated with either vehicle or a constant background dose of an ERKi (VX-11e), a SRCi (dasatinib), or the combination of both. D) Apoptosis measurements reported as percent annexin V+ cells treated with the indicated drugs for 48hrs in CRC240 cells. SRCi (dasatinib 100nM), ERKi (VX-11e 500nM), 5-FU (5μM), irinotecan (5μM), oxaliplatin (5μM). E) TTP in CRC119 cells treated with the indicated drugs. SRCi (dasatinib), MEKi (AZD6244). F) HCT116 xenograft treated with vehicle, Oxaliplatin (7.5 mg/kg once every 4 days), dasatinib (15 mg/kg, daily) and AZD6244 (10 mg/kg, twice daily), or the triple combination. To the right, a survival curve showing percent of mice with tumors less than 4X the starting volume at a given time. To the right of survival curves, mouse weights for the triple combination group over the course of the study. Error bars show data ± SEM. *p<0.05. See also Figure S5.
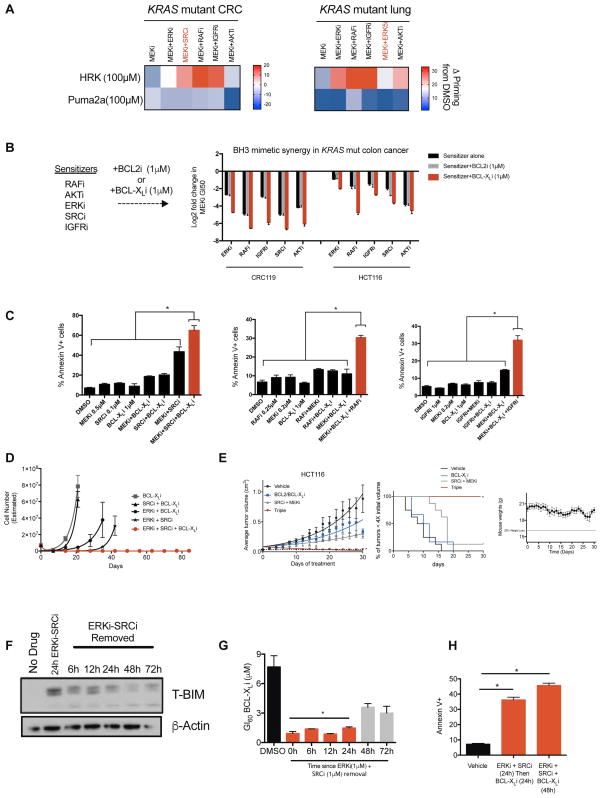
Along with increasing overall apoptotic priming, BH3 profiling of cells treated with various combinations also revealed that drug treatment induced a newfound dependence on the anti-apoptotic protein BCL-XL, as indicated by an increased sensitivity to the BCL-XL-neutralizing HRK peptide (Figure 6A). Indeed, across combination therapies spanning KRAS mutant colorectal, lung, pancreas, and ovarian models, inhibition of BCL-XL (but not the related anti-apoptotic protein BCL-2) conferred greater than additive growth inhibition and apoptosis induction, suggesting that BCL-XL inhibitors may be a common method to increase the therapeutic window for drug combinations targeting KRAS effector and feedback pathways (Figure 6B–C, Supplemental Figure 6a). Interestingly, we note that the combination of BCL-XL and MEK inhibitors was previously proposed as a strategy to treat KRAS driven cancers (Corcoran et al., 2013). However, we found that this combination yielded more modest apoptosis induction than corresponding triple therapies, and further, that cells were able to rapidly develop resistance to this treatment, even when administered at high doses (Figures 6C–D and Supplemental Figure 6b), a feature which may partially explain the heterogeneous clinical responses observed in patients treated with this combination (Singh et al., 2015). In contrast, when administered as part of a triple combination that maximally induces BIM expression, this approach blocked acquired resistance in the TTP assay performed using multiple cellular models (Figure 6D, Supplemental Figures 6c–f). The candidate MEKi, SRCi, BCL-XLi triple combination was also well tolerated and efficacious in a KRAS/PIK3CA mutant CRC xenograft model in vivo (Figure 6E).
Figure 6. Targeting the unmasked BCL-XL dependency to design triple combination therapies.
A) BH3 profiling in KRAS mutant CRC (HCT116) and lung cancer (Calu6) cell lines treated with indicated combinations (Drug doses and identities are the same as in Figure 5A with the addition of ERK5i XMD8-92, 5μM). B) Log2 transformed GI50 values for MEKi (AZD6244) in the presence of background treatment containing one of the sensitizers indicated (RAFi LY3009120 0.1μM; AKTi MK2206, 5μM; ERKi SCH772984, 0.05μM; SRCi dasatinib, 0.5μM; or IGFRi GSK1838705A, 1μM) and either a BCL-XL inhibitor (WEHI-539, 1μM) or a BCL2 inhibitor (ABT-199, 1μM) in two CRC cell lines. C) Apoptosis measurements reported as percent annexin V+ in CRC119 cells treated with the indicated combinations. Each graph represents a different sensitizer in combination with a BCL-XL inhibitor and a MEK inhibitor (Drug identities same as in B). D) TTP assay in CRC240 cells treated with the indicated combinations (BCL-XLi WEHI-539, 1μM; SRCi dasatinib, 1μM; ERKi VX-11e, 1μM). E) HCT116 xenograft treated with the indicated drugs. ABT-737 (BCL-2/BCL-XLi, 25 mg/kg, daily), Dasatinib (SRCi, 15 mg/kg, daily), AZD6244 (MEKi, 10 mg/kg, twice daily). To the right, a survival curve showing percent of mice with tumors less than 4X the starting volume at a given time. To the right of survival curves, mouse weights for the triple combination group over the course of the study. F) Immunoblot of total BIM in CRC240 cells treated with an ERKi (VX-11e, 1μM) and a SRCi (dasatinib, 1μM) for 24hrs. Drugs were removed, then lysates were probed at the indicated time points. Blots are cropped for clarity. G) GI60 value for a BCL-XL inhibitor (WEHI-539) in CRC240 cells. Each of the bars on the graph represents the time at which the BCL-XL inhibitor was added to the cells after background dose of ERKi+SRCi (drug identities as above) was removed. The DMSO bar is the average of the DMSO values for each of the time points (0h, 6h, 12h, 24h, 48h, 72h). H) Apoptosis measurements are reported as percent annexin V+ cells in CRC240 cells treated with each of the indicated drugs for each of the indicated times. BCL-XLi (WEHI-539, 1μM), ERKi (VX-11e, 500nM), SRCi (dasatinib, 100nM). Error bars show data ± SEM. *p<0.05. See also Figure S6.
Finally, a potential clinical limitation of BCL-XLi is the fact that these inhibitors can cause on-target, reversible thrombocytopenia (Zhang et al., 2007). To determine whether drug combinations that potentiate the activity of BCL-XLi in KRAS mutant tumors also exacerbate thrombocytopenia, we isolated platelets from fresh blood obtained from two healthy human donors, then treated them with BCL-XLi in the presence or absence of MEKi-based combination therapies. Importantly, these assays revealed that short-term treatment with combination therapies failed to influence the sensitivity of platelets to BCL-XLi (Supplemental Figure 6g). Further, the reversible nature of toxicities associated with these agents also suggests that serial or intermittent dosing regimens may be advantageous. Given the fact that BIM induction sensitizes cells to BCL-XLi, we measured BIM protein levels over time following treatment with MEKi/ERKi-based combinations. After ERKi+SRCi removal, BIM levels in KRAS mutant cells remain elevated for at least 48 h (Figure 6F), suggesting that there could be an opportunity to temporally separate BIM-inducing combination treatment from BCL-XL inhibition. Consistent with this hypothesis, cells remained highly sensitive to BCL-XLi treatment for up to 48 h after the removal of ERKi+SRCi (Figure 6G–H). Collectively, the findings described in Figures 5–6 demonstrate that MEKi/ERKi-based combination therapies sensitize cells to apoptosis, unmasking a dependency on the pro-survival BCL-2 family protein BCL-XL. Because of these properties, combination therapies sensitize KRAS mutant tumors to both cytotoxic chemotherapies and BCL-XLi. Finally, owing to the temporal nature of BIM induction following treatment with combination therapies, it may be possible to schedule triple therapies by first administering a combination that drives BIM induction, then sequentially administering an agent such as a BCL-XLi that exploits that induction to drive apoptosis, an approach that may reduce systemic toxicities while retaining anti-tumor efficacy.
Discussion
CRISPR/Cas9-based loss-of-function screening has recently been shown to be a powerful approach in functional genomics owing to its ability to generate complete genetic loss-of-function mutations with minimal off-target effects relative to RNAi (Barrangou and Doudna, 2016). Here, we applied this technology to design a strategy for drug sensitizer screening with low false positive hit rates, as evidenced by the fact that nearly all of the hits identified in our screens could be validated using small molecule inhibitors of their encoded proteins. Coupling CRISPR’s high reagent fidelity with careful selection of a miniaturized, sub-genomic library, our pan-cell line analysis revealed the first working landscape of drug targets and pathways that sensitize KRAS mutant tumors to MEK, ERK, or PI3K inhibition. This analysis revealed both universal and tissue specific sensitizers that corroborate the known sensitizer literature while also uncovering numerous previously unknown sensitizer interactions targeting diverse cellular processes. Indeed, the literature to date has implicated combinations that largely target MAPK pathway feedback re-activation and RTK-induced PI3K pathway activation. Our screens revealed the unanticipated finding that diverse cellular processes can alter the sensitivity of KRAS mutant tumors to MEK/ERK pathway inhibition (Figure 2C), often in a manner that appears to be KRAS mutation-specific (Figure 2G–I). These findings warrant future studies to more deeply credential the mechanistic basis for, and translational potential of, these combination therapies, as well as to characterize the degree to which these combinations exhibit selectivity for KRAS mutant tumors relative to tissues with wild-type KRAS. Additionally, the breadth of these screens allowed for the identification of sensitizers with activity only in tissue- or mutationally-defined subsets of KRAS mutant tumors. Thus, this strategy may be well suited for mapping similar sensitivity landscapes in other tumor types.
We identified PIK3CA mutations as secondary modifiers of sensitivity to combined SRCi plus MEKi/ERKi therapy in KRAS mutant CRC. Interestingly, this combination’s selectivity for KRAS/PIK3CA double mutant tumors, which represent ~10–11% of all CRCs (Network, 2012), owes to two features: (1) SRC inhibits AKT phosphorylation, leading to BIM induction, only in double mutant tumors; and (2) double mutant tumors have significantly lower levels of BIM at steady state, and are therefore highly sensitive to BIM-induced apoptosis, a finding that is corroborated in cellular models, animal models, and primary human tumors. Previous studies have shown that high levels of pretreatment BIM can predict the apoptotic response of oncogene-addicted cancers to their cognate kinase inhibitors (Faber et al., 2011). Our data provide a contrasting case wherein double mutant CRC tumors maintain low levels of BIM and are therefore highly sensitive to its induction, whereas WT cells, which have presumably adapted to the presence of higher steady-state BIM levels, are insensitive to further increases. Importantly, these distinctions provide a therapeutic window, as combined SRC plus MEK/ERK inhibition leads to tumor growth inhibition in vivo. Several open questions remain, including the mechanisms by which SRC regulates AKT phosphorylation in PIK3CA mutant, but not WT, tumors, and how SRC/AKT inhibition cooperates with ERK pathway inhibition to induce BIM in this setting. Nevertheless, this strategy may be particularly promising clinically, as dual MEK/AKT inhibition, while promising in model systems, has failed clinically owing to toxicities caused by direct pathway inhibition in patients (Ebi et al., 2014). Our data suggests that selective SRC inhibitors may make it possible to inhibit the AKT arm of this pathway selectively in tumors, thereby reducing systemic toxicity. However, one limitation of this work is that dasatinib, the compound used to inhibit SRC in most of our assays, is a notoriously promiscuous compound with many potential targets (Greuber et al., 2013). Although SRC was validated as a target in our assays using multiple genetic and pharmacological methods, we cannot rule out the possibility that some of the effects of dasatinib inhibition may owe to the inhibition of other complementary targets. Nonetheless, because drugs targeting SRC and MEK/ERK are clinically approved, the potential utility of SRC plus MEK/ERK inhibition is straightforward to test clinically.
Finally, and surprisingly, we found that it was possible to rapidly acquire resistance to a selection of the most potent combination therapies uncovered in our screens, in particular those combinations targeting growth signaling pathways. By taking advantage of common features of the sensitizer landscape, we identified several strategies to potentially combat resistance. Specifically, by targeting feedback reactivation or drug-induced apoptotic priming – shared features of many of the combination therapies identified here - it is possible to derive triple drug therapies that suppress resistance evolution, using drug doses at which we observed no evidence of substantial toxicities in in vivo mouse models. To further address potential toxicity issues associated with these strategies, we suggest that it may be possible to schedule apoptosis targeting treatments in series by taking advantage of a treatment-induced therapeutic window, a finding that is the focus of ongoing studies. Additionally, it will be critical to determine through future studies whether the emergence of resistance can be more effectively delayed using combination therapies targeting alternative cellular processes such as metabolism, chromatin state, and transcription. Together, the findings described here reveal a rich and biologically diverse landscape of cooperating drug therapies with activity in KRAS mutant cancers, providing a starting point for the design of next-generation treatment strategies.
Experimental Procedures
Cell lines and reagents
All cell lines were grown at 37°C in 5% CO2. Colon cell lines were grown in RPMI 10% FBS 1% penicillin/streptomycin, pancreas lines were grown in DMEM/F12 10% FBS 1% penicillin/streptomycin, Lung lines were grown in RPMI 10% FBS 1% penicillin/streptomycin, Ovarian were grown in MEM 10% FBS 1% penicillin/streptomycin. CRC240 and CRC119 were generated by David Hsu (Duke University). All other cell lines were purchased from American Type Culture Collection (ATCC) or Duke University Cell Culture Facility (CCF). All cell lines were authenticated using Promega PowerPlex 18D kit or were purchased within 6 months from Duke CCF. Drugs were purchased from Selleck Chemicals, ChemieTek, MedChemExpress, Ontario Chemicals, Sigma-Aldrich, or APExBIO.
Cloning CRISPR drug-sensitivity library
Our sub-genomic CRISPR library was cloned following previous methods (Shalem et al., 2014) using previously characterized sgRNAs (Wang et al., 2014). Five unique sgRNA inserts along with 50 non-targeting controls were synthesized by Custom Array of the form:
GGAAAGGACGAAACACCGXXXXXXXXXXXXXXXXXXXXGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC
“X” denotes unique 20mer sgRNA sequence
The oligo pool was diluted 1:10 in water and amplified using NEB Phusion Hotstart Flex enzyme master mix and the following primers:
ArrayF: TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG
ArrayR: ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC
PCR Protocol: 98°C/30s, 18x[98°C/10s, 63°C/10s, 72°C/15s], 72°C/3min
Inserts were cleaned with Axygen PCR clean-up beads (1.8x; Fisher Scientific) and resuspended in molecular biology grade water. lentiCRISPRv2 (Addgene ID# 52961) was digested with BsmBI (Thermo Fisher) for 2 hours at 37°C. The large ~ 13 kB band was gel extracted after size-selection on a 1% agarose gel. Using 100ng of cut lentiCRISPRv2 and 40ng of sgRNA library inserts, a 20μL Gibson assembly reaction was performed (30min, 50°C). After Gibson assembly, 1μL of the reaction was transformed into electrocompetent Lucigen cells and spread on LB-ampicillin plates and incubated overnight. After counting dilution plates to ensure library coverage, colonies were scraped and combined for plasmid extraction using a Plasmid Maxiprep kit (Qiagen).
Lentivirus production, viral tittering and transduction
Letiviral production was performed as previously described with slight modification (Martz et al., 2014). HEK 293T cells were grown in 15cm to ~50% confluence. For each plate, transfection was performed using Fugene6 (Promega), 5.6ug of psPAX2, 0.625ug pVSVg, 6.25ug of library plasmid. After 30min of incubation at room temperature, the mixture was added to the cells and incubated overnight. The next day harvest media was added (DMEM 30% FBS). After 48 hours harvested virus was passed through a 0.45 μm filter. Viral titers and transductions were performed as previously described (Martz et al., 2014).
Pooled screening using CRISPR drug sensitizer library
The desired cell line was seeded at 500,000 cells per well in 6-well plates and the next day transduced at an MOI of 0.2. After puromycin selection, a day two sample is taken to check library representation. Transduced cells were then maintained at 1000x coverage of the library in puro for 10 days to allow for the generation of knockout cells. After 10 days in puromycin, cells were split into vehicle and drug treatment conditions and maintained at 1000x library coverage, in a low dose of drug (~25% growth inhibition). After 3–4 weeks, DNA was extracted (DNeasy Blood & Tissue Kit, Qiagen) and prepared for sequencing as previously described (Shalem et al., 2014). To determine essentiality phenotypes, the fractional representation (FR) for a given guide in the t=final condition is compared to its FR in the t=initial condition giving the depletion metric (DM) for each guide. The 5 construct DMs per gene are then collapsed to gene level scores corresponding to their means and medians for all 5 guides, three score (best performing 3 guides), GARP score (best two performing guides) (Marcotte et al., 2012), and second best performing guide. The genes are then rank ordered by each metric and the sum of the ranks by each metric gives the cumulative score and determines the overall rank ordered list. The same analysis is carried out for the sensitizer phenotypes except the final time point is compared for both conditions, FR in the drug treated sample normalized to its FR in the vehicle treated sample. Thus, for a gene to score as a hit, it must be depleted specifically in the drug-treated condition.
Short-term growth-inhibition assay (GI50)
Cells were seeded into 96-well plates at 5,000 cells/well. To generate GI50 curves, cells were treated with vehicle (DMSO) or an eight-log serial dilution of drug. Each treatment condition was represented by at least three replicates. Three days after drug addition, cell viability was measured using Cell Titer Glo® (Promega). Relative viability was then calculated by normalizing luminescence values for each treatment condition to control treated wells. To generate GI50 curves for drug combinations, slight modifications are made. Primary drug was applied and diluted as above while the second drug was kept at a constant concentration across all wells except the DMSO-only condition. Viability for all primary drug dilutions was then calculated relative to luminescence values from the secondary drug-only condition. We plot the viability versus concentration curve for drug A (normalized appropriately to the viability of cells treated with DMSO in media control). Next, we plot the viability versus concentration curve for drug A in the presence of a fixed dose of drug B (this time normalizing to the viability of cells treated with drug B alone). Sensitization of cells to drug A by drug B is evidenced by a leftward shift in the curve. Dose-response curves were fit using Graph pad/Prism 6 software.
Western blotting and antibodies
Immunoblotting was performed as previously described (Wood et al., 2012) and membranes were probed with primary antibodies (1:1,000 dilution) recognizing vinculin (CST#4650), H3 (CST#4499), BIM (CST#2933), p-AKT (S473, T308) (CST#4058, CST#13038), AKT (CST#4691), p-ERK (CST#9101), T-ERK (CST#4695), p-SRC (Tyr416) (CST#6943), T-SRC (CST#2123), Na, K-ATPase (CST#3010), T-BID (CST#2002), T-BAX (CST#5023), T-BCL-XL (CST#2764), T-MCL-1 (5453), B-Actin (CST#4970), T-p38 (CST#9212), p-p38 (CST#9211), T-p38α (CST#9218), p-HSP27 (Ser82) (CST#2401), T-HSP27 (CST#95357).
Quantification of apoptosis by annexin-V
Cells were seeded in six-well plates and treated the next day with either the indicated amount of drug, vehicle (DMSO), or combination. Cells were incubated for two days, washed twice with ice-cold PBS, and resuspended in 1X annexin V binding buffer (10mM HEPES, 140mM NaCl, 2.5mM CaCl2; BD Biosciences). Surface exposure of phosphatidylserine was measured using APC-conjugated annexin V (BD Biosciences). 7-AAD (BD Biosciences) was used as a viability probe. Experiments were analyzed at 20,000 counts/sample using BD FACSVantage SE. Gatings were defined using untreated/unstained cells as appropriate.
shRNA and ORF constructs
TRC shRNA clones were obtained from the Duke RNAi Facility as glycerol stocks. Constructs were prepared in lentiviral form and used to infect target cells as previously described. X2 (HcRed) and myr-AKT1 sequence information can be found in Martz et al. (2014). BCL2L11 (Bim) ORF was obtained from transomic technologies (clone ID# TOLH-1508630).
shSRC (1) TRCN0000038150 GACAGACCTGTCCTTCAAGAA
shSRC (2) TRCN0000195339 CATCCTCAGGAACCAACAATT
shBIM (1) TRCN0000001051 ATGGTTATCTTACGACTGTTA
shBIM (2) TRCN0000001052 GTCTCGATCCTCCAGTGGGTA
Clonogenic growth assay
Cells were seeded at 2,000 cells per well. The next day cells were drugged at the indicated doses. Seven to ten days following drug addition, plates were rinsed with PBS and fixed and stained with 0.5% (wt/vol) crystal violet in 6.0% (vol/vol) gluteraldehyde solution (ThermoFisher Scientifics) for 30 min at room temperature. Plates were rinsed in distilled H2O and photographed the following day.
BH3 profiling
BH3 profiling was performed as previously described (Sarosiek et al., 2013). Briefly, cells are resuspended in sample buffer containing JC-1 (a mitochondrial dye) and plated into a 384-well plate containing individual peptides of the BCL-2 family of proteins. Fluorescence is measured over time in order to capture the percentage of depolarization caused by each peptide. Slight modifications are made for drug incubations; cells are treated with drug for 16 hours prior to BH3 profiling as previously described (Montero et al., 2015; Winter et al., 2014).
Platelet isolation and growth inhibition studies
Approximately 50 milliliters of fresh, whole blood was obtained from Texas Gulf Coast Medical. Five milliliters of a sodium citrate solution (3.8% w/vol in sterile water) was added to the blood and the sample was centrifuged for 10 minutes at 1,000 RPM. The platelet rich plasma (the top layer) was transferred to a new tube, centrifuged an additional 10 minutes at 1,000 RPM and the resulting plasma was transferred to a new 50 mL tube. The sample was analyzed for red or white blood cell contamination under a microscope, and if pure was diluted with 3 volumes (relative to the final plasma volume) of RPMI 1640 media containing 10% FBS and 1% Pen. Strep. Platelets were then seeded in equal parts into 96-well plates. Drug was added 30 minutes later and after 24 hours of drug incubation, platelet viability was measure by Cell Titer Glo® (Promega).
Xenograft tumor studies
Animal studies were approved by the Duke University Medical Center Institutional Animal Care and Use Committee. HCT116 cells (approximately 1×107 in PBS) were injected subcutaneously into 6–8 week old female athymic NOD/SCID gamma mice or nude mice (AKT triple). Once tumors reached ~100 mm3, mice were randomly assigned to treatment groups with 1) ABT737 Triple: dasatinib (15 mg/kg/day by oral gavage); AZD6244 (10 mg/kg twice daily by oral gavage); ABT737 (25 mg/kg/day by IP injection); the dual combination of dasatinib + AZD6244; or the triple combination of drugs. 2) MK2206 Triple: MK-2206 (15 mg/kg/day by oral gavage); the dual combination of AZD6244 (15 mg/kg/day by oral gavage) + dasatinib (15 mg/kg/day by oral gavage); or the triple combination of drugs. 3) Oxaliplatin Triple: Oxaliplatin (7.5 mg/kg every 4 days by IP injection); or the triple combination of AZD6244, dasatinib and Oxaliplatin. Tumors were measured every other day with calipers and tumor volume was calculated using the formula length/2xwidth^2. Mice were housed under standard conditions and monitored daily for symptoms of morbidity, including weight loss, hunched posture and other humane endpoints.
Mutational analysis of PIK3CA
E542, E545, and H1047 sites were analyzed as previously described (Kalaany and Sabatini, 2009).
Mutational analysis of KRAS
G12, G13, Q61 sites were analyzed as previously described (Berg et al., 2010).
In vitro TTP assay
Cells were seeded at 300,000 per plate in 10cm dishes in duplicate or triplicate. The next day, drugs were added at indicated concentrations. One week later, plates were counted and 200,000 cells were re-plated with drug. This process was carried out until exponential growth rates were observed. Virtual cell counts were calculated based upon the number plated, the growth rate, and the counts each week.
Patient Samples
Colorectal patient samples (paraffin embedded slices) were obtained from UNC-CH (Autumn McRee) and were then sent to the Duke Pathology Research Immunohistology Lab for T-BIM staining. Samples were then sent to a pathologist (Shannon McCall) to be scored for T-BIM staining. Scoring was performed blinded to the mutational status of the samples.
Statistical analysis
Unless otherwise specified, student’s t tests, or for grouped analyses, one-way ANOVA with Tukey’s post-hoc test, were performed and p values < 0.05 were considered significant. Results are presented as means ±SEM.
Supplementary Material
Supplemental Figures S1–S6.
Genes included in the CRISPR screening library, Related to Figure 1.
Guide RNAs in the miniaturized CRISPR library, Related to Figure 2.
Screen results for each drug and cell line, Related to Figure 2.
List of validated sensitizers to MEK/ERK pathway inhibition across KRAS mutant tissue types, Related to Figure 2.
Colorectal patient samples stained for BIM levels, Related to Figure 3.
Acknowledgments
We would like to thank to the members of the Wood, Counter, and Der labs for helpful discussions and research support, and David Hsu for providing primary patient-derived CRC cell lines. We would also like to thank Joseph Herbert and Greg Palmer of the Duke Preclinical Translational Research Unit for their assistance and expertise with the in vivo data. This work was supported by start-up funds from the Duke University School of Medicine and the Duke Cancer Institute (K.C.W.), a scholar award from the NIH Building Interdisciplinary Research Careers in Women’s Health Program (K12HD043446, to K.C.W.), a Golfers Against Cancer Research Award (K.C.W.), a Stewart Trust Fellowship (K.C.W.), a V Scholar Award from the V Foundation for Cancer Research (K.C.W.), NIH/NCI awards (R01CA207083 to K.C.W., R01CA123031 to C.M.C., and U01CA199235 to C.J.D.), an AACR-Pancreatic Cancer Action Network Research Acceleration Network Grant (C.J.D.), National Science Foundation Graduate Research Fellowship awards (DGE-1106401 to G.R.A. and DGF-1106401 to L.A.C.), a Schlumberger Foundation Faculty for the Future Fellowship (M.C.), NIH National Research Service Awards (F31CA195967 to P.S.W., F30CA206348 to K.H.L., F32CA180569 to D.P.N., and F32CA206234 to R.S.S.), and the Duke NIH CTSA award (UL1TR001117). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
Footnotes
Author Contributions
Conceptualization, K.C.W., C.M.C., C.J.D., P.S.W, and G.R.A; Methodology, K.C.W., P.S.W, and G.R.A; Validation, G.R.A., P.S.W., D.P.N, M.C., E.M.S, R.S.S, M.A., J.C.L., R.N., P.S., C.Y., J.P.T., and M.X.; Formal Analysis, K.H.L., L.C., S.E.W., P.S.W., and G.R.A.; Investigation, G.R.A., P.S.W., D.P.N, M.C., E.M.S, R.S.S, M.A., J.C.L., R.N., P.S., C.Y., J.P.T., and M.X.; Resources, K.C.W., C.M.C., C.J.D., S.J.M., and A.J.M.; Writing-Original Draft, K.C.W, P.S.W., and G.R.A., Writing-Review and Editing, All authors; Visualization, G.R.A., P.S.W., K.H.L., and K.C.W.; Supervision, K.C.W.; Funding Acquisition, K.C.W., C.M.C., C.J.D., P.S.W., G.R.A., L.A.C., M.C., K.H.L., D.P.N., and R.S.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, Nishimura K, Lynch KD, Mermel CH, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discovery. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- Athuluri-Divakar SK, Carpio RVD, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MVR, Ueno L, Hart JR, et al. A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling. Cell. 2016;165:643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I, et al. DNA Sequence Profiles of the Colorectal Cancer Critical Gene Set KRAS-BRAF-PIK3CA-PTEN-TP53 Related to Age at Disease Onset. PLoS ONE. 2010;5:e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Brooks AN, Wu X, Shrestha Y, Chouinard C, Piccioni F, Bagul M, Kamburov A, Imielinski M, Hogstrom L, et al. High-throughput Phenotyping of Lung Cancer Somatic Mutations. Cancer Cell. 2016;30:214–228. doi: 10.1016/j.ccell.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R. A Missing Link in Genotype-Directed Cancer Therapy. Cell. 2012;151:465–468. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras Promotes Growth Transformation and Invasion of Immortalized Human Pancreatic Cells by Raf and Phosphatidylinositol 3-Kinase Signaling. Cancer Research. 2007;67:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- Campbell RM, Anderson BD, Brooks NA, Brooks HB, Chan EM, De Dios A, Gilmour R, Graff JR, Jambrina E, Mader M, et al. Characterization of LY2228820 Dimesylate, a Potent and Selective Inhibitor of p38 MAPK with Antitumor Activity. Molecular Cancer Therapeutics. 2014;13:364–374. doi: 10.1158/1535-7163.MCT-13-0513. [DOI] [PubMed] [Google Scholar]
- Chen SH, Zhang Y, Van Horn RD, Yin T, Buchanan S, Yadav V, Mochalkin I, Wong SS, Yue YG, Huber L, et al. Oncogenic BRAF Deletions That Function as Homodimers and Are Sensitive to Inhibition by RAF Dimer Inhibitor LY3009120. Cancer Discovery. 2016;6:300–315. doi: 10.1158/2159-8290.CD-15-0896. [DOI] [PubMed] [Google Scholar]
- Chonghaile TN, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore VDG, Deng J, Anderson KC, Richardson P, Tai YT, et al. Pretreatment Mitochondrial Priming Correlates with Clinical Response to Cytotoxic Chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Research. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. Synthetic Lethal Interaction of Combined BCL-XL and MEK Inhibition Promotes Tumor Regressions in KRAS Mutant Cancer Models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C, Ebi H, Martini M, Beausoleil SA, Faber AC, Jakubik CT, Huang A, Wang Y, Nishtala M, Hall B, et al. Measurement of PIP3 Levels Reveals an Unexpected Role for p110β in Early Adaptive Responses to p110α-Specific Inhibitors in Luminal Breast Cancer. Cancer Cell. 2015;27:97–108. doi: 10.1016/j.ccell.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Ras history. Small GTPases. 2014;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission Possible? Nat Rev Drug Discov. 2014;3:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clinical Cancer Research. 2015;21:1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. Journal of Clinical Investigation. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Faber AC, Engelman JA, Yano S. Not just gRASping at flaws: Finding vulnerabilities to develop novel therapies for treating KRASmutant cancers. Cancer Sci. 2014;105:499–505. doi: 10.1111/cas.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, et al. BIM Expression in Treatment-Naive Cancers Predicts Responsiveness to Kinase Inhibitors. Cancer Discovery. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Whalen DM, Özen A, Wongchenko MJ, Yin J, Yen I, Schaefer G, Mayfield JD, Chmielecki J, Stephens PJ, et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer Cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Controlof Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer:from leukaemia to solid tumours. Nat Rev Cancer. 2013;13:559–571. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll HJ, Tveit KM, Gibson F. A Review of the Evolution of Systemic Chemotherapy in the Management of Colorectal Cancer. Clinical Colorectal Cancer. 2015;14:1–10. doi: 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, Oizumi S, Nishimura M, Faber AC, Yano S. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discovery. 2016;6:754–769. doi: 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, Lieftink C, Bernards R, Di Nicolantonio F, Bardelli A. RAF Suppression Synergizes with MEK Inhibition in KRAS Mutant Cancer Cells. Cell Reports. 2014;8:1475–1483. doi: 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8:381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, Asthana S, Neel D, Yan JJ, Lu X, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47:250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JP, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn Y, Koh JLY, et al. Essential Gene Profiles in Breast, Pancreatic, and Ovarian Cancer Cells. Cancer Discovery. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, Anderson GR, Winter PS, Wang T, Alley HM, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Science Signaling. 2014;7:ra121–ra121. doi: 10.1126/scisignal.aaa1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, Lin KH, Truini M, Trusolino L, Bertotti A, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nature Communications. 2015;6:1–9. doi: 10.1038/ncomms9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, Piao H, Horowitz NS, Berkowitz RS, Matulonis U, et al. Drug-Induced Death Signaling Strategy Rapidly Predicts Cancer Response to Chemotherapy. Cell. 2015;160:977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, Koya RC, Samatar AA, Khanlou N, Braun J, et al. Tunable-Combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Cotargeting but Result in Melanoma Drug Addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, Lichinitser M, Dummer R, Grange F, Mortier L, et al. Improved Overall Survival in Melanoma with Combined Dabrafenib and Trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- Samatar AA, Poulikakos PI. Targeting RAS–ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Meth. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A. BID Preferentially Activates BAKwhile BIM Preferentially Activates BAX, Affecting Chemotherapy Response. Molecular Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, Will M, Yellen P, de Stanchina E, Baselga J, et al. Feedback Suppression of PI3Kα Signaling in PTEN-Mutated Tumors Is Relieved by Selective Inhibition of PI3Kβ. Cancer Cell. 2015;27:109–122. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomicsusing CRISPR–Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Longo DL, Chabner BA. Improving Prospects for Targeting RAS. Journal of Clinical Oncology. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discovery. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, et al. Intrinsic Resistance to MEK Inhibitionin KRAS Mutant Lung and Colon Cancer through Transcriptional Induction of ERBB3. Cell Reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- Uronis JM, Osada T, McCall S, Yang XY, Mantyh C, Morse MA, Lyerly HK, Clary BM, Hsu DS. Histological and Molecular Evaluation of Patient-Derived Colorectal Cancer Explants. PLoS ONE. 2012;7:e38422. doi: 10.1371/journal.pone.0038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter PS, Sarosiek KA, Lin KH, Meggendorfer M, Schnittger S, Letai A, Wood KC. RAS signaling promotes resistance to JAK inhibitors by suppressing BAD-mediated apoptosis. Science Signaling. 2014;7:ra122–ra122. doi: 10.1126/scisignal.2005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KC, Konieczkowski DJ, Johannessen CM, Boehm JS, Tamayo P, Botvinnik OB, Mesirov JP, Hahn WC, Root DE, Garraway LA, et al. MicroSCALE Screening Reveals Genetic Modifiers of Therapeutic Response in Melanoma. Science Signaling. 2012;5:rs4–rs4. doi: 10.1126/scisignal.2002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Nimmer PM, Tahir SK, Chen J, Fryer RM, Hahn KR, Iciek LA, Morgan SJ, Nasarre MC, Nelson R, et al. Bcl-2 family proteins are essential for platelet survival. Cell Death Differ. 2007;14:943–51. doi: 10.1038/sj.cdd.4402081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures S1–S6.
Genes included in the CRISPR screening library, Related to Figure 1.
Guide RNAs in the miniaturized CRISPR library, Related to Figure 2.
Screen results for each drug and cell line, Related to Figure 2.
List of validated sensitizers to MEK/ERK pathway inhibition across KRAS mutant tissue types, Related to Figure 2.
Colorectal patient samples stained for BIM levels, Related to Figure 3.