Abstract
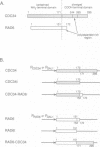
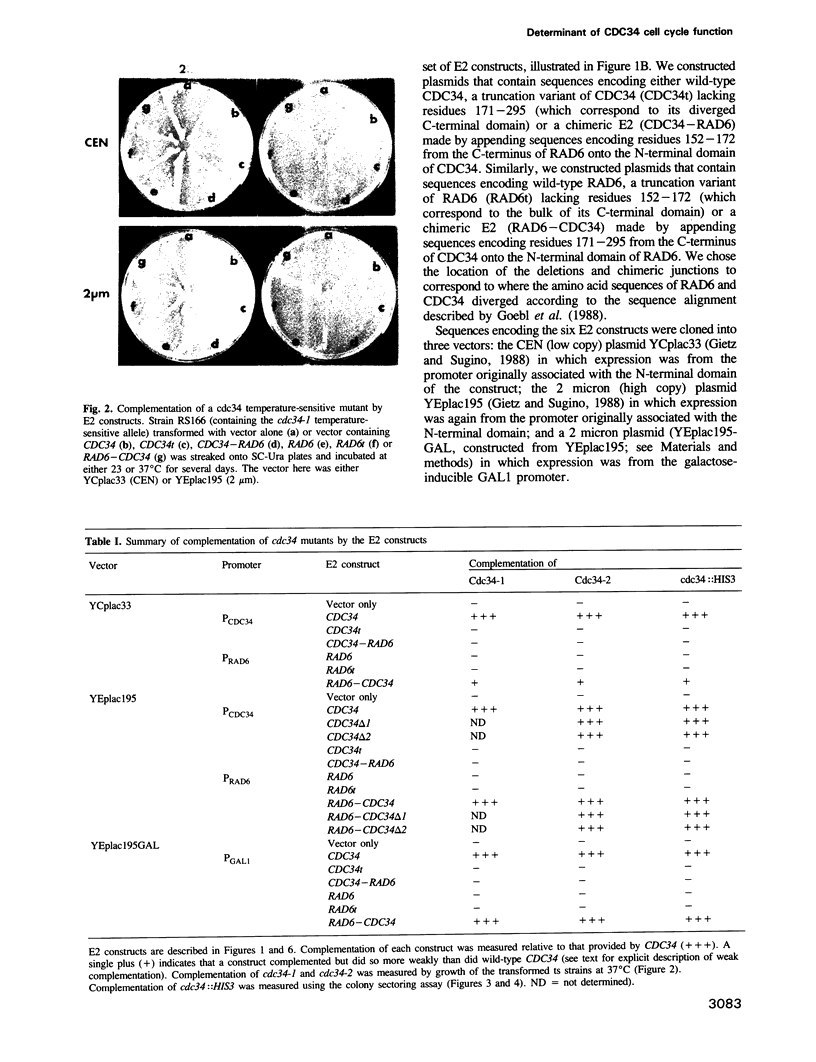
The ubiquitin conjugating (E2) enzyme encoded by CDC34 (UBC3) in Saccharomyces cerevisiae is required for the G1 to S transition of the cell cycle. CDC34 consists of a 170 residue amino-terminal domain that is homologous to that found in other E2s, followed by a 125 residue carboxyl-terminal domain that is specific to CDC34. We found that a truncation mutant of CDC34 which lacked the CDC34 carboxyl-terminal domain could not support the essential function of CDC34 in the cell cycle in vivo. To explore further the role of the carboxyl-terminal domain in determining the cell cycle function of CDC34, we constructed and characterized genes encoding chimeric E2s incorporating sequences from CDC34 and the related but functionally distinct E2 RAD6 (UBC2). We found that a construct encoding a chimeric RAD6-CDC34 ubiquitin conjugating enzyme, in which the 21 residue acidic carboxyl-terminal domain of RAD6 has been replaced with the 125 residue carboxyl-terminal domain of CDC34, performed the essential functions of CDC34 in vivo. This chimeric E2 also complemented the growth deficiency, UV sensitivity and sporulation deficiency of rad6 mutant strains. Deletion analysis of the CDC34 carboxyl-terminal domain in both CDC34 and the RAD6-CDC34 chimeric E2 identified a region comprising residues 171-244 of CDC34 that was sufficient to confer CDC34 function on the amino-terminal domains of CDC34 and RAD6. We suggest that this region interacts with substrates of CDC34 or with trans-acting factors (such as CDC34-specific ubiquitin protein ligases) that govern the substrate selectivity of CDC34. Congruent results demonstrating a positive role for the carboxyl-terminal domain of CDC34 in the essential function of CDC34 have also been obtained by Silver et al. (1992) and are reported in the accompanying paper.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J., Madura K., Bartel B., Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Ellison K. S., Gwozd T., Prendergast J. A., Paterson M. C., Ellison M. J. A site-directed approach for constructing temperature-sensitive ubiquitin-conjugating enzymes reveals a cell cycle function and growth function for RAD6. J Biol Chem. 1991 Dec 15;266(35):24116–24120. [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988 Sep 9;241(4871):1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Haas A. L., Bright P. M., Jackson V. E. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J Biol Chem. 1988 Sep 15;263(26):13268–13275. [PubMed] [Google Scholar]
- Haas A. L., Bright P. M. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J Biol Chem. 1988 Sep 15;263(26):13258–13267. [PubMed] [Google Scholar]
- Haas A. L., Reback P. B., Chau V. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit homologs, E2(20K) AND E2(32K). J Biol Chem. 1991 Mar 15;266(8):5104–5112. [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983 Jul 10;258(13):8206–8214. [PubMed] [Google Scholar]
- Hershko A., Heller H., Eytan E., Reiss Y. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986 Sep 15;261(26):11992–11999. [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Seufert W., Hauser H. P. Genetic analysis of the ubiquitin system. Biochim Biophys Acta. 1991 Jun 13;1089(2):127–139. doi: 10.1016/0167-4781(91)90001-3. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Seufert W., Sommer T., Reins H. A. Ubiquitin-conjugating enzymes: novel regulators of eukaryotic cells. Trends Biochem Sci. 1990 May;15(5):195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- Morrison A., Miller E. J., Prakash L. Domain structure and functional analysis of the carboxyl-terminal polyacidic sequence of the RAD6 protein of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Mar;8(3):1179–1185. doi: 10.1128/mcb.8.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Rose I. A. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985 Feb 10;260(3):1573–1581. [PubMed] [Google Scholar]
- Pickart C. M., Vella A. T. Ubiquitin carrier protein-catalyzed ubiquitin transfer to histones. Mechanism and specificity. J Biol Chem. 1988 Oct 15;263(29):15076–15082. [PubMed] [Google Scholar]
- Qin S., Nakajima B., Nomura M., Arfin S. M. Cloning and characterization of a Saccharomyces cerevisiae gene encoding a new member of the ubiquitin-conjugating protein family. J Biol Chem. 1991 Aug 15;266(23):15549–15554. [PubMed] [Google Scholar]
- Reiss Y., Kaim D., Hershko A. Specificity of binding of NH2-terminal residue of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988 Feb 25;263(6):2693–2698. [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Prakash S. Interactions of the RAD7 and RAD23 excision repair genes of Saccharomyces cerevisiae with DNA repair genes in different epistasis groups. Curr Genet. 1989 Oct;16(4):219–223. doi: 10.1007/BF00422107. [DOI] [PubMed] [Google Scholar]
- Seufert W., McGrath J. P., Jentsch S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990 Dec;9(13):4535–4541. doi: 10.1002/j.1460-2075.1990.tb07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Raboy B., Parag H. A., Dimitrovsky D., Kulka R. G. RAD6 gene product of Saccharomyces cerevisiae requires a putative ubiquitin protein ligase (E3) for the ubiquitination of certain proteins. J Biol Chem. 1991 Aug 25;266(24):15890–15894. [PubMed] [Google Scholar]
- Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992 Aug;11(8):3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. L., Vierstra R. D. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991 Dec 15;266(35):23878–23885. [PubMed] [Google Scholar]
- Sung P., Berleth E., Pickart C., Prakash S., Prakash L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 1991 Aug;10(8):2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988 Nov;2(11):1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]