Abstract
The RNA interference pathway is an evolutionary conserved post-transcriptional gene regulation mechanism that is exclusively triggered by double-stranded RNA inducers. RNAi-based methods and technologies have facilitated the discovery of many basic science findings and spurred the development of novel RNA therapeutics. Transient induction of RNAi via transfection of synthetic small interfering RNAs can trigger the selective knockdown of a target mRNA. For durable silencing of gene expression, either artificial short hairpin RNA or microRNA encoding transgene constructs were developed. These miRNAs are based on the molecules that induce the natural RNAi pathway in mammals and humans: the endogenously expressed miRNAs. Significant efforts focused on the construction and delivery of miRNA cassettes in order to solve basic biology questions or to design new therapy strategies. Several viral vectors have been developed, which are particularly useful for the delivery of miRNA expression cassettes to specific target cells. Each vector system has its own unique set of distinct properties. Thus, depending on the specific application, a particular vector may be most suitable. This field was previously reviewed for different viral vector systems, and now the recent progress in the field of miRNA-based gene-silencing approaches using lentiviral vectors is reported. The focus is on the unique properties and respective limitations of the available vector systems for miRNA delivery.
Keywords: : miRNA cassettes, gene therapy, viral vectors, vector tropism
Introduction
Non-coding RNA molecules play an essential role in gene regulation of the cell via a mechanism called RNA interference (RNAi). The RNAi mechanism is evolutionary conserved in eukaryotes and triggers the sequence-specific inhibition or degradation of a single or a unique set of complementary mRNAs. Three small RNA classes have been described to participate in mammalian RNAi mechanisms: microRNAs (miRNAs),1,2 endogenous small interfering RNAs (endo-siRNAs),3 and PIWI-associated RNAs (piRNAs).4 The endo-siRNAs and piRNAs are involved in suppression of transposable elements. The miRNAs are broadly involved in the regulated expression of multiple cellular genes and execute their effect at the posttranscriptional level.5,6
MiRNA-mediated regulation of gene expression plays a significant role in cell metabolism and cellular developmental and differentiation processes in mammals. More than a thousand human miRNAs have already been identified that are involved in the regulated expression of around 30% of all genes, which is a conservative estimate.7 Because of the miRNA abundance and their important regulatory functions, these molecules have received much attention from the scientific community over the last decade. For instance, efforts have been made to explain the biological function of the natural miRNAs by identifying the target mRNA and the role in cellular physiology. On the other hand, the design of man-made miRNA mimics to impose control over gene expression became of interest for the control of newly introduced transgenes in research and therapeutic applications. The development of vector-mediated RNAi allowed the establishment of durable gene silencing approaches, in particular for vector systems that are stably inherited by insertion into the host cell genome.8–10 Especially attractive is the delivery of RNAi-inducing gene cassettes with viral vector systems. The use of different viral vector systems for miRNA delivery and their advantages and disadvantages will be discussed, followed by a more detailed discussion of the lentiviral vector system.
RNAi Strategies
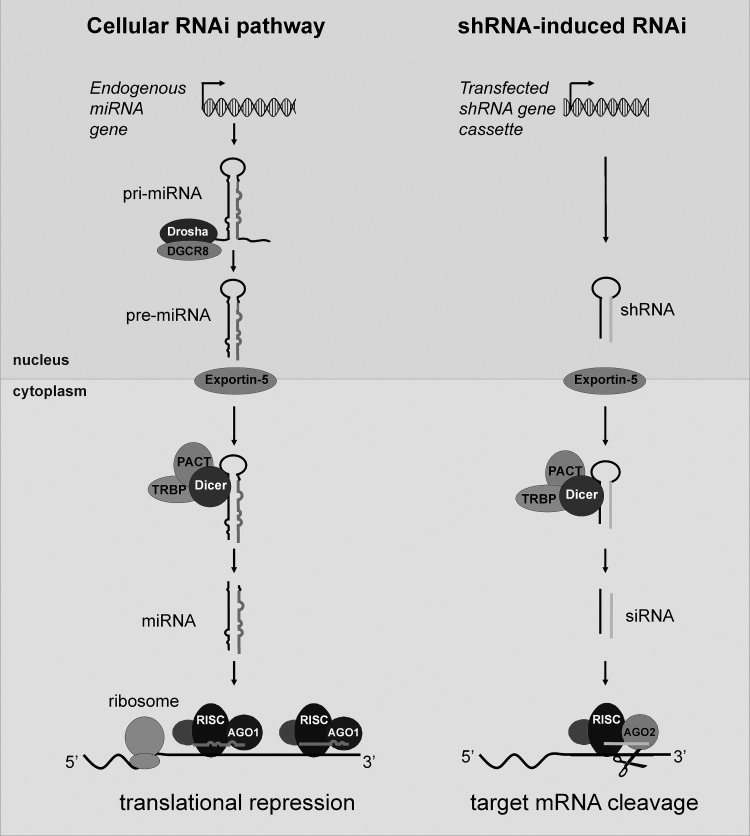
Since its discovery, RNAi was used frequently to knockdown specific genes for fundamental research, biotechnology applications, and therapeutic strategies.11,12 The cellular RNAi pathway uses the nuclear Drosha and cytoplasmic Dicer enzymes to produce mature miRNA (Fig. 1, left pathway). The RNAi mechanism can also be induced to mediate transient gene silencing via transfected synthetic siRNA duplexes that are ∼21 nucleotides complementary RNA strands with two-nucleotide 3′-overhangs. These artificial siRNA molecules are modeled after the natural miRNA duplexes formed upon Drosha/Dicer cleavage.13–15 Man-made siRNAs are usually fully complementary to the mRNA target by their design, causing site-specific mRNA cleavage via interaction with the argonaute 2 (Ago2) protein. Nevertheless, the use of siRNAs has several disadvantages. First, degradation by RNase A-like nucleases causes a relatively short intracellular half-life. Second, it may also be difficult to deliver the siRNAs efficiently to specific target cells, especially primary cell types.16–19 Perhaps most importantly, siRNAs can only cause a transient silencing effect because their intracellular concentration drops upon cell division.
Figure 1.
The RNA interference (RNAi) pathway. Illustrated are the endogenous miRNA pathway (left side) and the man-made exogenous short hairpin RNA (shRNA) pathway (right side). The cellular gene and the transgene, respectively, are transcribed into the pri-miRNA transcript and the shRNA precursor. Pri-miRNAs are processed by DGCR8/Drosha into pre-miRNAs and subsequently by Dicer/TRBP/PACT into miRNA duplexes with imperfect ∼22 base pairs. Exogenous shRNAs are directly processed by Dicer/TRBP/PACT. The miRNA and siRNA duplexes are subsequently incorporated into RISC, and one strand (thick-lined RNA strand) of the duplex directs this complex toward complementary mRNA targets. A typical miRNA causes translational repression by binding to multiple partially complementary target sites in the mRNA. In exceptional cases, a near perfect base pairing complementarity of the miRNA and mRNA will result in cleavage of the latter. Similarly, siRNAs are designed to be fully complementary to the target mRNA, which is inactivated by cleavage.
This led to the development of gene cassettes for short hairpin RNA (shRNA) molecules (Fig. 1, right pathway) that are processed into siRNAs. Such constructs encode short transcripts that typically fold ∼19–29 base pairs shRNAs, which mimic the pre-miRNA molecules with a base-paired stem, small loop and 3′-terminal UU overhang.8–10 RNA polymerase III promoters (H1, U6, or tRNA) usually control the expression of such shRNA constructs.8,10,20,21 RNA polymerase III is specialized in the production of high levels of small cellular transcripts and uses exact initiation and termination signals (four to six consecutive U residues) that result in synthesis of a defined shRNA molecule.
Optimization of the original shRNA design was achieved by incorporation of certain miRNA-derived characteristics.22–24 Compared to the perfectly base-paired shRNAs (Fig. 1, right pathway), modified structures were designed that closely resemble the natural miRNAs by introducing typical pri-miRNA specifics such as imperfect hairpin structures with bulges and mismatches, larger loops, flanking sequences, and mismatches. The mature miRNA strands are usually designed with perfect complementarity to the target, thereby inducing mRNA cleavage. Artificial miRNAs, like their natural counterparts, are frequently transcribed from a promoter for RNA polymerase II such as the strong immediate early promoter of the cytomegalovirus (CMV).25 However, RNA polymerase III promoters were also used.26 RNA polymerase II promoters have certain benefits over RNA polymerase III systems, including regulated and tissue-specific expression,27,28 and inducible RNA polymerase III systems have also been designed.29,30 Regulated shRNA expression is crucial, as high-level constitutive shRNA synthesis can trigger serious adverse effects. Artificial siRNA/shRNA molecules can target unwanted mRNAs (off-targeting)31 or induce immunological responses that cause toxicity.32–34 High level shRNA expression can cause morbidity and mortality in mouse models,35,36 but these adverse effects are usually dose-dependent. These results suggest that one or more key components of the RNAi pathway in mammalian cells may become saturated. Two key factors of the RNAi pathway, Ago2 and Exportin-5 (Fig. 1, left pathway), have indeed been shown to become rate-limiting upon shRNA overexpression, resulting in considerable disruption of the cellular miRNA pathway.35,37
Viral Vector Systems
Viral vectors have emerged as attractive vehicles for the delivery of transgenes to particular target cell types. In the simplest version, the pathogenic genes are removed from these viruses and replaced by the therapeutic gene(s). One vector may be more suitable than other systems, depending on the specifics of the transgene cassette, the therapeutic purpose (acute or chronic), and the cell type that is targeted. The properties of four popular viral vector systems that can facilitate high level transgene and miRNA expression are briefly presented: adenovirus and adeno-associated virus, retrovirus, and the subclass lentivirus (Table 1).
Table 1.
Viral vector systems
| Viral vector | Advantages | Shortcomings |
|---|---|---|
| Adenovirus | High vector titers, high efficiency | No integration, short-term expression |
| Efficient uptake in dividing and nondividing cells | Requires repeated administrationa | |
| Insert capacity (max 8 kb, extended to 37 kb in “gutless” vectors) | High immunogenicity | |
| Adeno-associated virus | High vector titers, high efficiency | Requires helper virus for replication |
| Uptake in dividing and nondividing cells | Time-consuming production protocol | |
| Relatively low toxicity, non-pathogenic | Limited insert capacity (max 3–5 kb) | |
| Remains predominantly episomal | ||
| Low risk of insertional mutagenesis | ||
| Retrovirus | Low immune response in host | Low vector titers |
| Modest insert capacity (max 8 kb) | Incorporates into dividing cells only | |
| Integrates into genome | Restricted tropism (expanded by pseudotyping) | |
| High risk of insertional mutagenesis | ||
| Lentivirus | Uptake in dividing and nondividing cells | Low vector titers |
| Modest insert capacity (max 8 kb) | Restricted tropism (expanded by pseudotyping) | |
| Integrates into genome | Risk of insertional mutagenesis | |
| New generation is self-inactivating for safety |
Episomal DNA is rapidly lost from dividing cells but may be retained by nonmitotic cells.
Vectors Based on Adenovirus
Members of the Adenoviridae family are non-enveloped viruses from with a linear dsDNA genome that is characterized by terminal inverted repeats of 37 kb.38 Adenoviruses cause 5–10% of upper respiratory tract infections in children. Initial binding to the cell occurs by interaction with the ubiquitously expressed coxsackievirus B receptor (CAR), and cell entry is triggered by binding to integrins αvβ3 and αvβ5.39,40 Adenoviral vectors are attractive delivery tools due to the relatively ease of manipulation and the ability to transduce a broad variety of quiescent and proliferating cells without integrating its cargo into the host genome.41 Serotypes 2 and 5 are frequently used for vaccination and gene delivery purposes. The initial adenoviral vectors were deficient for the E1 gene and provided a large packaging capacity of 8 kb. To reduce the chance of evoking immunostimulatory responses further, second-generation adenoviral vectors were designed by omitting the E2, E3, and E4 genes. Next-generation “gutless” adenoviral vectors are completely stripped of all viral proteins and encode only the packaging signal and regulatory elements, which results in an increased packaging capacity of ∼37 kb.42,43 The major complication in vector production is that a helper virus is needed for vector propagation. Reduced stimulation of the immune response by gutless adenoviral vectors can lead to reduced vector toxicity and prolonged transgene expression.44,45 The adenoviral gene therapy vector is ideal for short-term use because recombinant adenoviral vectors are replication deficient and do not integrate into the host genome.46,47 The major shortcoming these vectors is the potent induction of immune responses (adaptive and innate), which may impose cell toxicity and thereby limit the gene transfer efficiency, thus hampering repeated use.48–50
Adenoviral vectors can be used for delivery of shRNA/miRNA expression cassettes to specific cells in vitro51–54 and in vivo.55–58 The first in vivo study using adenoviral vectors demonstrated reduced expression of the target gene in the liver and brain,57 arguing that adenoviral-mediated RNAi approaches may be of special interest for neurodegenerative disorders.59 Adenoviral vectors can deliver shRNA cassettes to liver cells and induce gene silencing for five weeks without perturbing or saturating the endogenous miRNA silencing machinery.60,61 Due to its large cargo capacity, adenoviral vectors are especially suited for the simultaneous delivery of protein and sh/miRNA cassettes. Several publications used of adenoviral vectors for transgene expression of shRNA or miRNA molecules,62–66 and these studies are covered in excellent recent review articles.67,68
Vectors Based on the Adeno-Associated Virus
Adeno-associated virus (AAV) represents a non-enveloped member from the Parvoviridae family with icosahedral capsids. AAV contain a single-stranded DNA (ssDNA) genome of 4.7 kb flanked by inverted terminal repeats that are required for replication of the genome, its packaging, and integration. AAV vectors can transduce both dividing and nondividing cells. Wild-type AAV is dependent for its replication on a helper virus such as adenovirus or herpes simplex virus. In the absence of a helper virus, the AAV particle releases its genome that persists in episomal forms, and sometimes the virus remains latent by integrating its DNA into the AAVS1 region of chromosome 19. Infection of the cell by a helper virus can rescue the latent state.69 The AAV Rep proteins are essential for targeted integration. In recombinant AAV (rAAV), the Rep protein is deleted from the viral genome but can be supplied in trans. Consequently, integration of rAAV is less efficient than wild-type AAV and not targeted to chromosome 19.
High-level and durable gene expression was reported in the mouse for diverse organs, including the lung, muscle, liver, brain, and eyes.70–74 As with other integrating vectors, insertional mutagenesis remains a concern, especially for rAAV that lacks the specificity for chromosome 19. Wild-type AAV and rAAV integration has been linked to hepatocellular carcinoma in mice due to insertional oncogene activation,75–77 but other long-term studies did not describe such a correlation.78–82 Genome integration seems to occur preferentially at breaks in the host cell genome, including regions sensitive to endonuclease attack,83 but other motifs have also been implicated, such as CpG islands,84,85 sites of active transcription84–86 and palindromic chromosome sequences.87
The 11 known AAV serotypes exhibit distinct host cell tropisms as well as varying immunological properties, such that one can pick the best match for a certain therapeutic application in a particular target cell type.88 Pseudotyping of AAV allows vector retargeting to non-natural target cells or tissues.89 An extensive set of capsid variants has been engineered to avoid neutralization by antibodies that are directed against a particular capsid, as this can significantly affect the in vivo transduction efficiency.90,91
The absence of AAV pathogenicity in humans makes them an attractive therapeutic vehicle. In contrast to other viral vectors, AAV is the only vector system for which the wild-type virus is not associated with human malignancies. AAV has a relatively small packaging capacity, which commonly suffices for sh/miRNA cassettes that are limited in size. Accordingly, many in vivo studies were performed with AAV-RNAi vectors for human cancers, muscular dystrophies, neurodegenerative disorders, and cardiac, retinal, metabolic, and infectious diseases (see Grim,92 McCown,93 and Borel et al.94 for reviews).
RNAi-related toxicity and mortality were first observed in AAV-injected mice when the shRNA was overexpressed in the liver.35 Toxicity was found to be dose-dependent and due to the constitutive high shRNA expression, leading to saturation of Exportin-5. Toxicity was also observed in the mouse brain for shRNAs against the Huntington gene that were delivered with an AAV-vector.95 Toxicity was avoided by inserting the siRNA sequences in a backbone of a miRNA instead of a shRNA. The miRNA backbone caused reduced steady-state levels of the mature miRNA and prevented saturation of the RNAi machinery.96 Injection of this AAV-miRNA vector caused effective gene silencing of the target in Purkinje cells, suggesting that miRNA approaches are suitable for therapy applications in the brain. A second study confirmed that artificial miRNAs outperform shRNAs in suppressing a photoreceptor gene in the retina both in vitro and in vivo.97 These and other studies indicate the superiority of miRNA-based AAV approaches for some therapeutic applications with respect to the efficiency and safety.98–101
Vectors Based on Retroviruses
Retroviruses belong to the Retroviridae family and the enveloped virion particles contain a ssRNA genome of ∼10 kb. Retroviral vector (RV) systems are usually derived from Moloney murine leukemia virus, with a simple genome encoding the Gag, Pol, and Env proteins flanked by long terminal repeats (LTR).102 The RNA genome is copied upon cell entry by the virus-encoded reverse transcriptase enzyme into dsDNA that subsequently integrates randomly into one of the host chromosomes. This retrovirus-specific property is obviously favorable for the durable expression of inserted transgenes, but also raises a specific risk. RNA packaging sequences and other regulatory elements are relatively well defined to allow the design of RV systems. All protein-encoding sequences can be replaced by foreign sequences, as the required viral proteins can be supplied in trans during vector production.
Despite RV integration, persistent transgene expression is not warranted because the transgene may be transcriptionally silenced over time.103,104 A significant safety concern is RV integration in or near unwanted genome sites. In fact, several patients treated with a RV developed T-cell leukemia in a gene therapy trial for X-linked inherited immunodeficiency in which hematopoietic stem cells (HSCs) are modified.105–107 The leukemic event was linked to proto-oncogene LMO2 activation by adjacent RV integration. Dysregulation of host cell gene expression can occur because retroviruses were found to integrate preferentially near the start sites of transcription.108–111 The presence of transcriptional enhancers in the RV can result in enhanced expression of cellular (proto-) oncogenes.
Self-inactivating (SIN) vector systems were designed to improve the safety profile.112 SIN vectors carry an inactivating 3′ LTR deletion that is transferred into the 5′ LTR during vector amplification, which results in inactivation of this promoter. The vector requires an internal promoter to drive transgene expression. In addition, transcriptional enhancers that can affect cellular gene expression were removed. A significant limitation of RVs is the inability to infect quiescent cells due to an inability of the infecting particle to enter the nucleus, as the membrane is disassembled exclusively during mitosis.
Despite early disappointments, two recent Phase I clinical trials with RVs showed that ex vivo transduction of CD4+ T cells as well as CD34+ HSCs, followed by re-infusion of the transduced cells, is feasible and safe.113,114 A Phase II trial confirmed safety of such a gene therapy, but also demonstrated a lack of efficacy of RVs that encode antiviral ribozymes for HIV-1 infected individuals.115 Despite other RV-miRNA applications,116 most recent studies have shifted toward the use of lentiviral vectors.
Vectors Based on Lentiviruses
Lentiviruses represent a subgroup of the Retroviridae family that includes the human immunodeficiency virus type 1 (HIV-1). HIV-1 encodes the standard RV proteins Gag, Pol, and Env, but also the regulatory Tat and Rev proteins and the accessory Vif, Vpr, Vpu, and Nef proteins. Tat is the viral trans-activator that induces HIV-1 transcription from the LTR promoter. The Rev protein is responsible for export of singly spliced and unspliced HIV-1 transcripts from the nucleus.117–120 All protein-coding information is deleted in the HIV-based lentiviral vector (LV), leaving only the necessary regulatory sequences. LVs have been developed based on a variety of immunodeficiency viruses of human (HIV-2), simian (SIV), horse (EIAV), bovine (BIV), and feline (FIV) origin.121–125
A main benefit of LV over RV vectors is that the former are able to transduce dividing and nondividing cells. LVs allow persistent transgene expression by stably integrating into the host DNA, although transcriptional silencing may occur over time.126 LVs have a better safety profile than RV vectors because they favor integration within active transcriptional units, thereby reducing the risk of insertional oncogenesis.127–129 In addition, safer SIN-based LV variants were designed. Both RV and LV are amenable to pseudotyping, meaning that heterologous envelope proteins can be accommodated. Pseudotyping with pantropic envelopes such as the vesicular stomatitis virus G protein can mediate viral entry into a wide variety of cells that otherwise would be refractory to infection, including hematopoietic and embryonic stem cells.130
The CD4+ T cells of HIV-infected patients were treated ex vivo in a clinical trial with a LV that encodes an anti-HIV antisense transcript and reinfused into the patients after ex vivo expansion. The integration sites of the vectors were mapped in CD4+ T cells, and no adverse events were reported.131,132 These results confirmed the previously described HIV-1 integration profile and thus further backed up the safety profile of LVs.131,133 Several Phase I gene therapy trials were performed with LV to modify CD34+ HSC. Patients received a LV expressing a TAR RNA decoy, a ribozyme against CCR5 mRNA and an anti-tat/rev shRNA. This treatment was both feasible and safe, although no therapeutic effect could be documented.134,135 The other trial concerned two children with adrenoleukodystrophy (ALD), a lipid-storage disease of the brain. Disease progression was stabilized, and transduced stem cells showed polyclonal reconstitution of diverse cell subsets, including monocytes that migrate to the brain to become astrocytes that will express the therapeutic protein.136 The ALD trial also raised safety concerns due to detection of common insertion sites (CIS) of the LV. CIS are typically associated with insertional mutagenesis in mouse models and humans.137–141 A similar LV integration profile was observed in xenotransplanted immunodeficient mice.142 However, these CIS differed from the previously detected genotoxic CIS by clustering in mega-base-wide chromosomal regions, whereas genotoxic CIS were confined in contracted genome regions, always targeting a single gene. Recent LV successes in HSC gene transfer include trials in patients with metachromatic leukodystrophy and Wiskott–Aldrich syndrome.143,144 The combined data indicate that LV-mediated gene therapy is safe and effective, but this should be confirmed in larger patient groups and with extended follow-up.
LV-induced shRNA expression can cause cytotoxicity in cell lines, as was described for AAV vectors. This effect depends on the shRNA sequence and dose.145 Several basic and applied studies tested different LV–miRNA combinations.146–149 For instance, Bcr-Abl oncogene expression was effectively controlled by LVs encoding three artificial miRNAs, thus preventing outgrowth of leukemic cells in vitro and in vivo.150 Furthermore, LV-delivered anti-osteopontin miRNAs could inhibit cell proliferation in vitro and in vivo tumor growth of hepatocellular carcinoma.151
Vector Production Issues
When using viral vectors for miRNA or shRNA delivery, it is crucial that production of the vector is not influenced by insertion of the RNAi-inducing elements. Each vector system will exhibit some unique advantages and disadvantages, but a universal issue that may hamper vector production is genomic instability, although the details will differ among vector systems. DNA packaging in optimized adenoviral vectors demonstrates a strict size limit (27–37.8 kb).152,153 RNAi cassettes are normally small, such that DNA stuffers could be added to prevent destabilization of the capsid. LV titers were detectable, even with a viral RNA genome size of 18 kb that greatly exceeds that of the wild-type virus.154 However, the titer was much reduced, suggesting that increasing LV size hampers RNA genome encapsidation. This may not be a serious problem because RNAi cassettes are relatively small, and even multiple RNAi cassettes can be accommodated based on size arguments in the LV system. The recently described fourth generation of LV may facilitate longer inserts.155,156
Repeated-sequence motifs can also have a negative impact on the genetic stability of RVs and LVs. Retroviruses are known to be recombination prone, and repeats in RVs can thus trigger sequence duplication or deletion during transduction of target cells.157–160 For instance, it was demonstrated that combinatorial RNAi cassettes with repeat H1 promoter sequences are not stable, resulting in deletion of one or multiple shRNA cassettes by recombination.161,162 Removal of such repeat sequences, for example by use of multiple different promoter elements, did indeed increase the vector genome stability,163 although repeat-associated problems were not reported for DNA virus vectors, but homologous recombination on repeat sequences can in theory occur during production of DNA vectors.
Mammalian cells sense a viral infection by the innate immune system and respond by induction of the interferon (IFN) response.164–168 Accumulating evidence indicates that other antiviral defense mechanisms are also triggered, including the RNAi pathway.169–176 To counteract such cellular responses, mammalian viruses frequently encode IFN antagonists.177 Some of these IFN antagonists also act as potent RNA-silencing suppressors.169,171,172,178 Vector production can be regulated by innate antiviral mechanisms in mammalian cells, especially when the viral counter defense mechanism is absent from these vector systems compared to the fully equipped wild-type virus. Provision of RNAi-silencing suppressors in trans can indeed improve the production of LV, adenoviral vectors, and especially Sindbis virus vectors.179
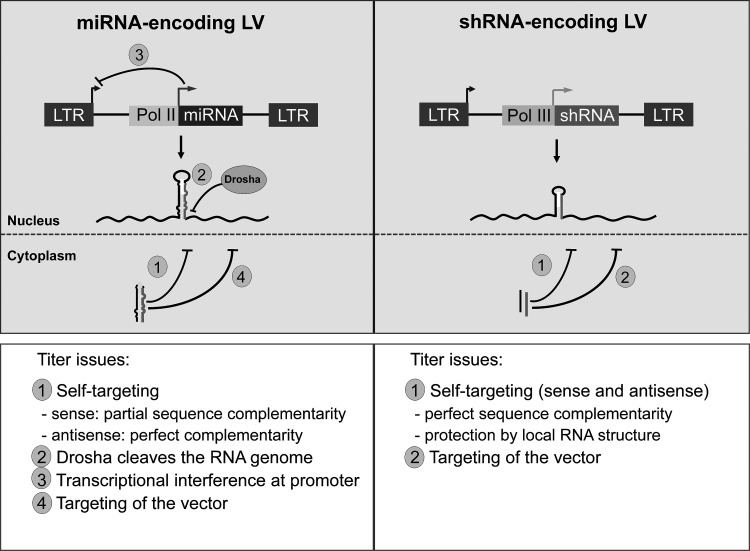
Several RNAi-specific problems may be encountered when viral vectors encode shRNAs or miRNAs. First, high transgene expression levels may impact on viability of the vector-producing cell and the eventual amount of vector that is produced. Overexpression of the vector-encoded shRNA or miRNA can cause activation of the IFN response and related toxicity, saturation of the endogenous miRNA pathway, or off-target effects. Such effects can easily be missed, as producer cells are usually thrown away after vector production. Second, specific RV and LV problems may arise when a shRNA/miRNA is introduced in the viral RNA genome. LV production requires transient co-transfection of the shRNA/miRNA-LV and packaging constructs that yield the structural proteins.180 The full-length viral RNA genome encoding the RNAi cassette will be expressed by RNA polymerase II during vector production. Besides the RNA genome, the shRNA/miRNA will be simultaneously transcribed from its own promoter and processed into a mature siRNA/miRNA that can target the complementary sequences in the LV RNA genome (Fig. 2, left and right panels: self-targeting). Self-targeting of the vector RNA genome can theoretically cause a drop in the yield of produced vector.
Figure 2.
Titer issues in lentiviral vectors (LV) expressing miRNA or shRNA. LV encoding miRNAs can potentially cause self-targeting due to expression of the mature miRNA that can target the homologous sequences in the miRNA encoding LV genome during vector production (mechanism 1, self-targeting) If the miRNA cassette is cloned in the sense orientation in respect to the vector RNA genome, then the target will be partially complementary to the miRNA, whereas the antisense miRNA cassette in the LV will have a perfect complementarity with the expressed miRNA. The LV genome encoding the miRNA can be recognized and cleaved by the Drosha during vector production (mechanism 2, Drosha cleavage). As the LV genome and the miRNA are both transcribed from an RNA polymerase II promoter, promoter interference could possibly reduce the titer (mechanism 3, promoter interference). In case of lentiviral RNAi vectors that target HIV-1, the mature anti-HIV-1 miRNA can target sequences from the HIV-1 based LV genome (mechanism 4, vector targeting). LV encoding shRNA (right side) could also potentially target their own complementary sequences as part of the LV genome (mechanism 1, self-targeting). However, this is not likely to occur in shRNA expressing LV due to occlusion of the target in a tight hairpin structure. Anti-HIV-1 shRNAs can possibly target similar sequences present in the LV genome.
Contradictory reports are present in literature on the impact of this “self-targeting” by LV-encoded shRNAs. It was suggested that self-targeting does not take place, as the target sequence is masked in a stable RNA structure and thus is shielded from the RISC complex.181,182 There is indeed good evidence in the literature for a major suppressive effect of stable target RNA structure on RNAi efficiency.183,184 However, another study showed dramatic titer reductions when shRNAs were produced by LV.185 The cause of these differences has not yet been resolved, but likely relates to the different LV systems and/or cell types used in these studies. Dramatically low titers of dsRNA-encoding LV were reported due to RNAi-mediated cleavage of the RNA genome during vector production.182 For miRNA-encoding LVs, it remains to be investigated whether self-targeting is excluded, since miRNAs are typically less tightly structured and thus likely more vulnerable to such attack.
RV and LV with a miRNA cassette face yet another problem. The vector RNA can be cleaved by Drosha (Fig. 2, left panel only: Drosha cleavage of vector genome). It was demonstrated that knockdown of Drosha can indeed increase the titer of such vectors.186,187 When comparing a large series of LV constructs, a dramatically low titer was measured for LVs that express one or multiple artificial miRNA(s), but this reduction was largely due to the internal Pol II promoter element.186 The titer could be restored almost completely by deletion or replacement of this promoter. It is possible that transgene expression from the CMV promoter is favored over RSV promoter-driven expression of the vector RNA genome (Fig. 2, left panel only: promoter interference).188,189 Such effects have indeed been described for RV vectors.190
In special cases, the RNAi reagents could attack the vector backbone to cause a reduction in titer. Because the LV system is based on the HIV-1 genome, anti-HIV shRNA and miRNA molecules can theoretically also target HIV-derived sequences in the vector (Fig. 2, left and right panels: vector targeting). It was shown that anti-HIV shRNAs indeed cause a severely titer reduction when the LV or packaging construct is targeted.181,186 Restoration of the vector production titer can be obtained via inhibition of the RNAi pathway, either by production of an excess of shRNAs to saturate the RNAi pathway or by expression of a siRNA/shRNA against Dicer or Drosha. To prevent vector targeting, one could also select shRNAs that target HIV-1 sequences that are absent from the LV genome and the Gag-Pol vector that is required for vector production. To prevent targeting of the packaging construct, a Gag-Pol construct optimized for human codons can be utilised.191 At last, the usage of an alternative LV system can be considered, for example based on HIV-2, SIV, FIV, or BIV.
Controlled Transgene Expression Via Mirnas
The focus of this review will now be expanded by discussing novel technologies to regulate the expression of transgenes or viral vectors by means of introduced miRNA target sequences. Because most miRNAs have a distinct cell type or tissue expression profile, this approach allows one to control the cellular tropism of genes, vectors, oncolytic viruses, and viral vaccines. By incorporating miRNA targets, mRNA expression becomes vulnerable to RNAi control in the cells programmed to express this particular miRNA.
Brown et al. tried to confine transgene expression to hepatocytes by the use of tissue-specific promoter elements because transgene activity in antigen-presenting cells (APCs) could induce an immune response.192,193 This approach could not block an immune response, and transgene-expressing hepatocytes were eventually cleared. Next, one tried the insertion of four fully complementary targets downstream of the transgene for miR-142-3p that is specific for the hematopoietic lineage. Inclusion of the miRNA targets caused profound suppression of transgene expression, but exclusively in hematopoietic cells, including APCs.194 By successfully avoiding immune-mediated clearance of the vector, stable gene transfer was achieved in this mouse model.195,196 Soon thereafter, target sequences of different cell-type-specific miRNAs were tested and promising results were reported, indicating that this is a broadly applicable approach.197 The strategy was used to separate transgene expression in neurons versus astrocytes (miR-124), undifferentiated versus differentiated embryonic stem cells (miR-302), immature versus mature dendritic cells (miR-155), and lymphoid versus myeloid lineage cells (miR-223). Two different miRNA targets can be combined to restrict transgene expression to a specific cell type. Notably, these vectors did not perturb the endogenous miRNA pathway.
This research team also used miR-126 targets in a gene therapy for treatment of globoid cell leukodystrophy.198 HSCs were treated ex vivo with LV that encodes the therapeutic galactosylceramidase (GALC) gene. This approach is toxic without miRNA targets because of GALC overexpression in HSCs and early progenitor cells. GALC expression was blocked by the inserted miR-126 targets in HSCs and early progenitors, but vigorous expression was preserved in the mature hematopoietic cell lineages. Elevated GALC levels were obtained in multiple tissues upon transduced HSC transplantation in mice, causing reduced malignancies and prolonged survival.
The same strategy has been applied to control a stem cell–based gene therapy. Several miR-181 targets were inserted in the LV that encodes tumor antigen receptors.199 This miRNA is expressed in developing thymocytes, but its expression ceases in mature thymocytes. This property was used to restrict the LV to developing thymocytes in mice upon re-engraftment with modified bone-marrow cells. One could prevent the clonal deletion of autoreactive cells by avoiding early transgene exposure in developing thymocytes. The pioneering Naldini team published a review on this topic.200
Conclusions
The mechanism of RNAi allows the researcher to silence genes in a sequence-specific manner. This major breakthrough in molecular biology prompted the development of several technologies for basic research and therapeutic applications. It was realized that these technologies need further refinement when artificially induced RNAi were shown to cause serious side effects, for example off-target effects on unrelated genes or saturation of the RNAi pathway, thus affecting normal cellular physiology. Early RNAi experiments focused on siRNA inducers in transient systems and shRNA inducers in stably transduced cells. Improvements were reported for enhancement of the safety and efficacy by the use of artificial miRNAs. An important safety precaution concerns the implementation of inducible or tissue-specific promoters for miRNA or shRNA expression.
Future research may lead to the development of novel classes of more specific shRNA/miRNA reagents. One recent example started with the discovery of Dicer-independent miRNAs such as miR-451.201,202 This lack of Dicer recognition is due to the too-short base-paired duplex.203,204 Surprisingly, this class of miRNA is not destroyed but processed by other nucleases and is shown to be active in the RNAi pathway. In fact, the Ago2 enzyme as part of the RISC complex processes these short duplexes to activate the mature miRNA strand.205,206 Independent studies led to the description of a strikingly similar phenomenon for shRNAs that are active, despite being too short for Dicer processing.207 It was demonstrated that Ago2 is key in this alternative shRNA processing route.208,209 Compared to conventional shRNA designs, this alternative Ago2 route activates the “opposite” RNA strand. This design was called “AgoshRNA”, as Ago2 is required for both its processing and subsequent silencing action.209 Most importantly, whereas siRNA and shRNA reagents have the potential to generate two active strands, the AgoshRNA produces only a single active strand, thus minimizing off-target effects. The AgoshRNA design may have additional advantages, including full activity in Dicer-minus cells.210
Viral vectors represent smart vehicles to transport RNAi-inducing gene cassettes into cells. The characteristics of different vector systems were discussed, in particular with regard to the expression of miRNA payloads. After several setbacks in clinical gene therapy trials using viral vectors, significant modifications were introduced to augment the safety profile of these vectors. Recent clinical studies indicate that these optimized approaches can be used in a safe manner to achieve a clinical benefit. Thus, the future also seems bright for miRNA-based therapeutic interventions using a variety of viral vector systems.
Progressive knowledge of RNAi pathways inside the cell has resulted in optimized RNAi inducers that combine enhanced safety and efficiency. Concurrently, the viral vectors were improved to exhibit augmented efficacy and better safety profiles. Further studies should reveal the potential for RNAi-based therapeutics to be applied in the clinic.
Acknowledgments
RNAi research in the Berkhout laboratory is sponsored by ZonMw (Translational Gene Therapy program) and NWO-CW (TOP grant).
Author Disclosure
There are no conflicts of interest.
References
- 1.Ambros V, Lee RC, Lavanway A, et al. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 2003;13:807–818 [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature 2004;431:350–355 [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science 2002;297:1831. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007;318:761–764 [DOI] [PubMed] [Google Scholar]
- 5.Lau NC. Small RNAs in the animal gonad: guarding genomes and guiding development. Int J Biochem Cell Biol 2010;42:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126–139 [DOI] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500 [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550–553 [DOI] [PubMed] [Google Scholar]
- 9.Siolas D, Lerner C, Burchard J, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 2005;23:227–231 [DOI] [PubMed] [Google Scholar]
- 10.Paddison PJ, Caudy AA, Bernstein E, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002;16:948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet 2007;8:173–184 [DOI] [PubMed] [Google Scholar]
- 12.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol 2007;25:1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001;15:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494–498 [DOI] [PubMed] [Google Scholar]
- 15.Caplen NJ, Parrish S, Imani F, et al. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci U S A 2001;98:9742–9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bumcrot D, Manoharan M, Koteliansky V, et al. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol 2006;2:711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuschl T, Borkhardt A. Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol Interv 2002;2:158–167 [DOI] [PubMed] [Google Scholar]
- 18.Holen T, Amarzguioui M, Wiiger MT, et al. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res 2002;30:1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 2009;8:129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res 2003;31:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z, Harwig A, Berkhout B, et al. Mutation of nucleotides around the +1 position of type 3 Polymerase III promoters: the effect on transcriptional activity and start site usage. Transcription 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 2002;9:1327–1333 [DOI] [PubMed] [Google Scholar]
- 23.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods 2006;3:707–714 [DOI] [PubMed] [Google Scholar]
- 24.Koornneef A, van LR, Timmermans E, et al. AAV-mediated in vivo knockdown of luciferase using combinatorial RNAi and U1i. Gene Ther 2011;18:929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006;13:1097–1101 [DOI] [PubMed] [Google Scholar]
- 27.Zhou X, Vink M, Klaver B, et al. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther 2006;13:1382–1390 [DOI] [PubMed] [Google Scholar]
- 28.Toniatti C, Bujard H, Cortese R, et al. Gene therapy progress and prospects: transcription regulatory systems. Gene Ther 2004;11:649–657 [DOI] [PubMed] [Google Scholar]
- 29.van de Wetering M, Oving I, Muncan V, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep 2003;4:609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta S, Schoer RA, Egan JE, et al. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci U S A 2004;101:1927–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003;21:635–637 [DOI] [PubMed] [Google Scholar]
- 32.Bridge AJ, Pebernard S, Ducraux A, et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 2003;34:263–264 [DOI] [PubMed] [Google Scholar]
- 33.Sledz CA, Holko M, de Veer MJ, et al. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 2003;5:834–839 [DOI] [PubMed] [Google Scholar]
- 34.Kleinman ME, Yamada K, Takeda A, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature 2008;452:591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006;441:537–541 [DOI] [PubMed] [Google Scholar]
- 36.Beer S, Bellovin DI, Lee JS, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther 2010;18:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D, Wang L, Lee JS, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest 2010;120:3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douglas JT. Adenoviral vectors for gene therapy. Mol Biotechnol 2007;36:71–80 [DOI] [PubMed] [Google Scholar]
- 39.Bergelson JM, Cunningham JA, Droguett G, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997;275:1320–1323 [DOI] [PubMed] [Google Scholar]
- 40.Wickham TJ, Mathias P, Cheresh DA, et al. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 1993;73:309–319 [DOI] [PubMed] [Google Scholar]
- 41.Cao H, Koehler DR, Hu J. Adenoviral vectors for gene replacement therapy. Viral Immunol 2004;17:327–333 [DOI] [PubMed] [Google Scholar]
- 42.Chen HH, Mack LM, Kelly R, et al. Persistence in muscle of an adenoviral vector that lacks all viral genes. Proc Natl Acad Sci U S A 1997;94:1645–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amalfitano A. Next-generation adenoviral vectors: new and improved. Gene Ther 1999;6:1643–1645 [DOI] [PubMed] [Google Scholar]
- 44.Crettaz J, Otano I, Ochoa L, et al. Treatment of chronic viral hepatitis in woodchucks by prolonged intrahepatic expression of interleukin-12. J Virol 2009;83:2663–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maione D, Della RC, Giannetti P, et al. An improved helper-dependent adenoviral vector allows persistent gene expression after intramuscular delivery and overcomes preexisting immunity to adenovirus. Proc Natl Acad Sci U S A 2001;98:5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer D, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther 2003;8:846–852 [DOI] [PubMed] [Google Scholar]
- 47.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther 2004;11 Suppl 1:S10–S17 [DOI] [PubMed] [Google Scholar]
- 48.Marshall E. Gene therapy death prompts review of adenovirus vector. Science 1999;286:2244–2245 [DOI] [PubMed] [Google Scholar]
- 49.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008;372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 2008;372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SY, Kim KA, Kim CH, et al. CD44-shRNA recombinant adenovirus inhibits cell proliferation, invasion, and migration, and promotes apoptosis in HCT116 colon cancer cells. Int J Oncol 2017;50:329–336 [DOI] [PubMed] [Google Scholar]
- 52.Huang M, Li G, Pan T, et al. A novel multi-target RNAi adenovirus inhibits hepatoma cell proliferation, migration, and induction of angiogenesis. Oncotarget 2016;7:57705–57713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan Y, He B, Chen J, et al. Gene therapy for colorectal cancer by adenovirus-mediated siRNA targeting CD147 based on loss of the IGF2 imprinting system. Int J Oncol 2015;47:1881–1889 [DOI] [PubMed] [Google Scholar]
- 54.Jiang AG, Lu HY, Zhang DG, et al. Short hairpin RNA targeting AKT1 and PI3K/p85 suppresses the proliferation and self-renewal of lung cancer stem cells. Mol Med Rep 2015;12:363–370 [DOI] [PubMed] [Google Scholar]
- 55.Cai CM, Xiao X, Wu BH, et al. Targeting endogenous DLK1 exerts antitumor effect on hepatocellular carcinoma through initiating cell differentiation. Oncotarget 2016;7:71466–71476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin WP, Lin JH, Cai B, et al. Effect of adenovirus-mediated RNA interference of IL-1beta expression on spinal cord injury in rats. Spinal Cord 2016;54:778–784 [DOI] [PubMed] [Google Scholar]
- 57.Xia H, Mao Q, Paulson HL, et al. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002;20:1006–1010 [DOI] [PubMed] [Google Scholar]
- 58.Chao TC, Chan LC, Ju SY, et al. In vivo growth suppression of CT-26 mouse colorectal cancer cells by adenovirus-expressed small hairpin RNA specifically targeting thymosin beta-4 mRNA. Cancer Gene Ther 2014;21:389–396 [DOI] [PubMed] [Google Scholar]
- 59.Wu R, Wang H, Xia X, et al. Nerve injection of viral vectors efficiently transfers transgenes into motor neurons and delivers RNAi therapy against ALS. Antioxid Redox Signal 2009;11:1523–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narvaiza I, Aparicio O, Vera M, et al. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J Virol 2006;80:12236–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mowa MB, Crowther C, Ely A, et al. Inhibition of hepatitis B virus replication by helper dependent adenoviral vectors expressing artificial anti-HBV pri-miRs from a liver-specific promoter. Biomed Res Int 2014;2014:718743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hutcheson R, Terry R, Chaplin J, et al. MicroRNA-145 restores contractile vascular smooth muscle phenotype and coronary collateral growth in the metabolic syndrome. Arterioscler Thromb Vasc Biol 2013;33:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ibrisimovic M, Kneidinger D, Lion T, et al. An adenoviral vector-based expression and delivery system for the inhibition of wild-type adenovirus replication by artificial microRNAs. Antiviral Res 2013;97:10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Donnell JM, Kalichira A, Bi J, et al. In vivo, cardiac-specific knockdown of a target protein, malic enzyme-1, in rat via adenoviral delivery of DNA for non-native miRNA. Curr Gene Ther 2012;12:454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakurai F, Furukawa N, Higuchi M, et al. Suppression of hepatitis C virus replicon by adenovirus vector-mediated expression of tough decoy RNA against miR-122a. Virus Res 2012;165:214–218 [DOI] [PubMed] [Google Scholar]
- 66.Kumar M, Follenzi A, Garforth S, et al. Control of HBV replication by antiviral microRNAs transferred by lentiviral vectors for potential cell and gene therapy approaches. Antivir Ther 2012;17:519–528 [DOI] [PubMed] [Google Scholar]
- 67.Mowa MB, Crowther C, Arbuthnot P. Therapeutic potential of adenoviral vectors for delivery of expressed RNAi activators. Expert Opin Drug Deliv 2010;7:1373–1385 [DOI] [PubMed] [Google Scholar]
- 68.Raoul C, Barker SD, Aebischer P. Viral-based modelling and correction of neurodegenerative diseases by RNA interference. Gene Ther 2006;13:487–495 [DOI] [PubMed] [Google Scholar]
- 69.Schultz BR, Chamberlain JS. Recombinant adeno-associated virus transduction and integration. Mol Ther 2008;16:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakai H, Herzog RW, Hagstrom JN, et al. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood 1998;91:4600–4607 [PubMed] [Google Scholar]
- 71.Flotte TR, Afione SA, Conrad C, et al. Stable in vivo expression of the cystic fibrosis transmembrane conductance regulator with an adeno-associated virus vector. Proc Natl Acad Sci U S A 1993;90:10613–10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaplitt MG, Leone P, Samulski RJ, et al. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet 1994;8:148–154 [DOI] [PubMed] [Google Scholar]
- 73.Grant CA, Ponnazhagan S, Wang XS, et al. Evaluation of recombinant adeno-associated virus as a gene transfer vector for the retina. Curr Eye Res 1997;16:949–956 [DOI] [PubMed] [Google Scholar]
- 74.Back S, Peranen J, Galli E, et al. Gene therapy with AAV2-CDNF provides functional benefits in a rat model of Parkinson's disease. Brain Behav 2013;3:75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Donsante A, Vogler C, Muzyczka N, et al. Observed incidence of tumorigenesis in long-term rodent studies of rAAV vectors. Gene Ther 2001;8:1343–1346 [DOI] [PubMed] [Google Scholar]
- 76.Donsante A, Miller DG, Li Y, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science 2007;317:477. [DOI] [PubMed] [Google Scholar]
- 77.Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015;47:1187–1193 [DOI] [PubMed] [Google Scholar]
- 78.Nichols TC, Dillow AM, Franck HW, et al. Protein replacement therapy and gene transfer in canine models of hemophilia A, hemophilia B, von willebrand disease, and factor VII deficiency. ILAR J 2009;50:144–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Niemeyer GP, Herzog RW, Mount J, et al. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood 2009;113:797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bell P, Moscioni AD, McCarter RJ, et al. Analysis of tumors arising in male B6C3F1 mice with and without AAV vector delivery to liver. Mol Ther 2006;14:34–44 [DOI] [PubMed] [Google Scholar]
- 81.Li H, Malani N, Hamilton SR, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 2011;117:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ghazi NG, Abboud EB, Nowilaty SR, et al. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a Phase I trial. Hum Genet 2016;135:327–343 [DOI] [PubMed] [Google Scholar]
- 83.Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet 2004;36:767–773 [DOI] [PubMed] [Google Scholar]
- 84.Nakai H, Wu X, Fuess S, et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol 2005;79:3606–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller DG, Trobridge GD, Petek LM, et al. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol 2005;79:11434–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakai H, Montini E, Fuess S, et al. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet 2003;34:297–302 [DOI] [PubMed] [Google Scholar]
- 87.Inagaki K, Lewis SM, Wu X, et al. DNA palindromes with a modest arm length of greater, similar 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice. J Virol 2007;81:11290–11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther 2006;14:316–327 [DOI] [PubMed] [Google Scholar]
- 89.Grimm D, Kay MA. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr Gene Ther 2003;3:281–304 [DOI] [PubMed] [Google Scholar]
- 90.Scallan CD, Jiang H, Liu T, et al. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood 2006;107:1810–1817 [DOI] [PubMed] [Google Scholar]
- 91.Lin J, Calcedo R, Vandenberghe LH, et al. Impact of preexisting vector immunity on the efficacy of adeno-associated virus-based HIV-1 Gag vaccines. Hum Gene Ther 2008;19:663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grimm D. Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev 2009;61:672–703 [DOI] [PubMed] [Google Scholar]
- 93.McCown TJ. Adeno-associated virus (AAV) vectors in the CNS. Curr Gene Ther 2005;5:333–338 [DOI] [PubMed] [Google Scholar]
- 94.Borel F, Kay MA, Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol Ther 2014;22:692–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A 2008;105:5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther 2009;17:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Georgiadis A, Tschernutter M, Bainbridge JW, et al. AAV-mediated knockdown of peripherin-2 in vivo using miRNA-based hairpins. Gene Ther 2010;17:486–493 [DOI] [PubMed] [Google Scholar]
- 98.Yang X, Marcucci K, Anguela X, et al. Preclinical evaluation of an anti-HCV miRNA cluster for treatment of HCV infection. Mol Ther 2013;21:588–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pihlmann M, Askou AL, Aagaard L, et al. Adeno-associated virus-delivered polycistronic microRNA-clusters for knockdown of vascular endothelial growth factor in vivo. J Gene Med 2012;14:328–338 [DOI] [PubMed] [Google Scholar]
- 100.Karali M, Manfredi A, Puppo A, et al. MicroRNA-restricted transgene expression in the retina. PLoS One 2011;6:e22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Han Y, Khodr CE, Sapru MK, et al. A microRNA embedded AAV alpha-synuclein gene silencing vector for dopaminergic neurons. Brain Res 2011;1386:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pages JC, Bru T. Toolbox for retrovectorologists. J Gene Med 2004;6 Suppl 1:S67–S82 [DOI] [PubMed] [Google Scholar]
- 103.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther 2005;16:1241–1246 [DOI] [PubMed] [Google Scholar]
- 104.Ellis J, Yao S. Retrovirus silencing and vector design: relevance to normal and cancer stem cells? Curr Gene Ther 2005;5:367–373 [DOI] [PubMed] [Google Scholar]
- 105.Gaspar HB, Thrasher AJ. Gene therapy for severe combined immunodeficiencies. Expert Opin Biol Ther 2005;5:1175–1182 [DOI] [PubMed] [Google Scholar]
- 106.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 2008;118:3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 2010;16:198–204 [DOI] [PubMed] [Google Scholar]
- 108.Wu X, Li Y, Crise B, et al. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003;300:1749–1751 [DOI] [PubMed] [Google Scholar]
- 109.Mitchell RS, Beitzel BF, Schroder AR, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2004;2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lewinski MK, Yamashita M, Emerman M, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog 2006;2:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bushman F, Lewinski M, Ciuffi A, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 2005;3:848–858 [DOI] [PubMed] [Google Scholar]
- 112.Yu SF, von Ruden T, Kantoff PW, et al. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci U S A 1986;83:3194–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Macpherson JL, Boyd MP, Arndt AJ, et al. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J Gene Med 2005;7:552–564 [DOI] [PubMed] [Google Scholar]
- 114.Amado RG, Mitsuyasu RT, Rosenblatt JD, et al. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a Phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum Gene Ther 2004;15:251–262 [DOI] [PubMed] [Google Scholar]
- 115.Mitsuyasu RT, Merigan TC, Carr A, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34(+) cells. Nat Med 2009;15:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rezaeian AH, Gao Y, Lin HK. Cloning, expression, and functional analysis of genomic miRNA using retroviral system in cancer cells. Methods Mol Biol 2013;936:157–172 [DOI] [PubMed] [Google Scholar]
- 117.Jeang KT, Chang Y, Berkhout B, et al. Regulation of HIV expression: mechanisms of action of Tat and Rev. [Review]. AIDS 1991;5:Suppl 2:S3–14 [PubMed] [Google Scholar]
- 118.Gait MJ, Karn J. RNA recognition by the human immunodeficiency virus Tat and Rev proteins. Trends Biochem Sci 1993;18:255–259 [DOI] [PubMed] [Google Scholar]
- 119.Kjems J, Askjaer P. Rev protein and its cellular partners. Adv Pharmacol 2000;48:251–298 [DOI] [PubMed] [Google Scholar]
- 120.Hope TJ. The ins and outs of HIV Rev. Arch Biochem Biophys 1999;365:186–191 [DOI] [PubMed] [Google Scholar]
- 121.Poeschla E, Gilbert J, Li X, et al. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J Virol 1998;72:6527–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mangeot PE, Negre D, Dubois B, et al. Development of minimal lentivirus vectors derived from simian immunodeficiency virus (SIVmac251) and their use for gene transfer into human dendritic cells. J Virol 2000;74:8307–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Poeschla EM, Wong-Staal F, Looney DJ. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med 1998;4:354–357 [DOI] [PubMed] [Google Scholar]
- 124.Berkowitz R, Ilves H, Lin WY, et al. Construction and molecular analysis of gene transfer systems derived from bovine immunodeficiency virus. J Virol 2001;75:3371–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leroux C, Cadore JL, Montelaro RC. Equine infectious anemia virus (EIAV): what has HIV's country cousin got to tell us? Vet Res 2004;35:485–512 [DOI] [PubMed] [Google Scholar]
- 126.Greber UF, Fassati A. Nuclear import of viral DNA genomes. Traffic 2003;4:136–143 [DOI] [PubMed] [Google Scholar]
- 127.Laufs S, Guenechea G, Gonzalez-Murillo A, et al. Lentiviral vector integration sites in human NOD/SCID repopulating cells. J Gene Med 2006;8:1197–1207 [DOI] [PubMed] [Google Scholar]
- 128.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol 2006;24:687–696 [DOI] [PubMed] [Google Scholar]
- 129.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest 2009;119:964–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang AH, Sadelain M. The genetic engineering of hematopoietic stem cells: the rise of lentiviral vectors, the conundrum of the LTR, and the promise of lineage-restricted vectors. Mol Ther 2007;15:445–456 [DOI] [PubMed] [Google Scholar]
- 131.Wang GP, Levine BL, Binder GK, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther 2009;17:844–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McGarrity GJ, Hoyah G, Winemiller A, et al. Patient monitoring and follow-up in lentiviral clinical trials. J Gene Med 2013;15:78–82 [DOI] [PubMed] [Google Scholar]
- 133.Levine BL, Humeau LM, Boyer J, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci U S A 2006;103:17372–17377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li MJ, Kim J, Li S, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 2005;12:900–909 [DOI] [PubMed] [Google Scholar]
- 135.DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med 2010;2:36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009;326:818–823 [DOI] [PubMed] [Google Scholar]
- 137.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer 2009;9:389–399 [DOI] [PubMed] [Google Scholar]
- 138.Akagi K, Suzuki T, Stephens RM, et al. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res 2004;32:D523–D527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 2006;12:401–409 [DOI] [PubMed] [Google Scholar]
- 140.Schwarzwaelder K, Howe SJ, Schmidt M, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest 2007;117:2241–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baum C. Insertional mutagenesis in gene therapy and stem cell biology. Curr Opin Hematol 2007;14:337–342 [DOI] [PubMed] [Google Scholar]
- 142.Biffi A, Bartolomae CC, Cesana D, et al. Lentiviral-vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood 2011;117:5332–5339 [DOI] [PubMed] [Google Scholar]
- 143.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:864. [DOI] [PubMed] [Google Scholar]
- 145.Fish RJ, Kruithof EK. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC Mol Biol 2004;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen L, Zhang R, Li P, et al. P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2. Neurosci Lett 2013;534:327–332 [DOI] [PubMed] [Google Scholar]
- 147.Di Martino MT, Leone E, Amodio N, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res 2012;18:6260–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li YT, Chen SY, Wang CR, et al. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum 2012;64:3240–3245 [DOI] [PubMed] [Google Scholar]
- 149.Feng SY, Dong CG, Wu WK, et al. Lentiviral expression of anti-microRNAs targeting miR-27a inhibits proliferation and invasiveness of U87 glioma cells. Mol Med Rep 2012;6:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McLaughlin J, Cheng D, Singer O, et al. Sustained suppression of Bcr-Abl-driven lymphoid leukemia by microRNA mimics. Proc Natl Acad Sci U S A 2007;104:20501–20506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sun BS, Dong QZ, Ye QH, et al. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology 2008;48:1834–1842 [DOI] [PubMed] [Google Scholar]
- 152.Parks RJ, Graham FL. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol 1997;71:3293–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bett AJ, Prevec L, Graham FL. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol 1993;67:5911–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kumar M, Keller B, Makalou N, et al. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther 2001;12:1893–1905 [DOI] [PubMed] [Google Scholar]
- 155.Vink CA, Counsell JR, Perocheau DP, et al. Eliminating HIV-1 packaging sequences from lentiviral vector proviruses enhances safety and expedites gene transfer for gene therapy. Mol Ther 2017;25:1790–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Berkhout B. A fourth generation lentiviral vector: simplifying genomic gymnastics molecular therapy. Mol Ther 2017;25:1741–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.An W, Telesnitsky A. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 2001;286:475–482 [DOI] [PubMed] [Google Scholar]
- 158.Marzio G, Verhoef K, Vink M, et al. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc Natl Acad Sci U S A 2001;98:6342–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhuang J, Jetzt AE, Sun G, et al. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J Virol 2002;76:11273–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jetst AE, Yu H, Klarmann GJ, et al. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol 2000;74:1234–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ter Brake O, Konstantinova P, Ceylan M, et al. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 2006;14:883–892 [DOI] [PubMed] [Google Scholar]
- 162.McIntyre GJ, Yu YH, Tran A, et al. Cassette deletion in multiple shRNA lentiviral vectors for HIV-1 and its impact on treatment success. Virol J 2009;6:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ter Brake O, Berkhout B. Development of an RNAi-based gene therapy against HIV-1. In: Kurreck J, ed. Therapeutic Oligonucleotides. London: RSC Publishing, 2008:296–311 [Google Scholar]
- 164.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature 2006;442:39–44 [DOI] [PubMed] [Google Scholar]
- 165.Basler CF, Mikulasova A, Martinez-Sobrido L, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 2003;77:7945–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Garcia-Sastre A, Egorov A, Matassov D, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998;252:324–330 [DOI] [PubMed] [Google Scholar]
- 167.Cai R, Carpick B, Chun RF, et al. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 2000;373:361–367 [DOI] [PubMed] [Google Scholar]
- 168.McMillan NA, Chun RF, Siderovski DP, et al. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virol 1995;213:413–424 [DOI] [PubMed] [Google Scholar]
- 169.Bennasser Y, Le SY, Benkirane M, et al. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 2005;22:607–619 [DOI] [PubMed] [Google Scholar]
- 170.Bucher E, Hemmes H, de Haan P, et al. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 2004;85:983–991 [DOI] [PubMed] [Google Scholar]
- 171.Haasnoot J, de Vries W, Geutjes EJ, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 2007;3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li WX, Li H, Lu R, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A 2004;101:1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Triboulet R, Mari B, Lin YL, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007;315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 174.Soifer HS, Zaragoza A, Peyvan M, et al. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res 2005;33:846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol 2007;88:2627–2635 [DOI] [PubMed] [Google Scholar]
- 176.Aparicio O, Razquin N, Zaratiegui M, et al. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol 2006;80:1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Basler CF, Wang X, Muhlberger E, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A 2000;97:12289–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science 2005;308:557–560 [DOI] [PubMed] [Google Scholar]
- 179.de Vries W, Haasnoot J, van der Velden J, et al. Increased virus replication in mammalian cells by blocking intracellular innate defense responses. Gene Ther 2008;15:545–552 [DOI] [PubMed] [Google Scholar]
- 180.Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol 1998;72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ter Brake O, Berkhout B. Lentiviral vectors that carry anti-HIV shRNAs: problems and solutions. J Gene Med 2007;9:743–750 [DOI] [PubMed] [Google Scholar]
- 182.Zhou D, Zhang J, Wang C, et al. A method for detecting and preventing negative RNA interference in preparation of lentiviral vectors for siRNA delivery. RNA 2009;15:732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Westerhout EM, Ooms M, Vink M, et al. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res 2005;33:796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Westerhout EM, Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res 2007;35:4322–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Poluri A, Sutton RE. Titers of HIV-based vectors encoding shRNAs are reduced by a Dicer-dependent mechanism. Mol Ther 2007;16:378–386 [DOI] [PubMed] [Google Scholar]
- 186.Liu YP, Vink MA, Westerink JT, et al. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA 2010;16:1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Brandl A, Wittmann J, Jack HM. A facile method to increase titers of miRNA-encoding retroviruses by inhibition of the RNaseIII enzyme Drosha. Eur J Immunol 2011;41:549–551 [DOI] [PubMed] [Google Scholar]
- 188.Lee AH, Suh YS, Sung JH, et al. Comparison of various expression plasmids for the induction of immune response by DNA immunization. Mol Cells 1997;7:495–501 [PubMed] [Google Scholar]
- 189.Xu ZL, Mizuguchi H, Ishii-Watabe A, et al. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene 2001;272:149–156 [DOI] [PubMed] [Google Scholar]
- 190.Schambach A, Mueller D, Galla M, et al. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther 2006;13:1524–1533 [DOI] [PubMed] [Google Scholar]
- 191.Kotsopoulou E, Kim VN, Kingsman AJ, et al. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J Virol 2000;74:4839–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Brown BD, Shi CX, Powell S, et al. Helper-dependent adenoviral vectors mediate therapeutic factor VIII expression for several months with minimal accompanying toxicity in a canine model of severe hemophilia A. Blood 2004;103:804–810 [DOI] [PubMed] [Google Scholar]
- 193.Follenzi A, Battaglia M, Lombardo A, et al. Targeting lentiviral vector expression to hepatocytes limits transgene-specific immune response and establishes long-term expression of human antihemophilic factor IX in mice. Blood 2004;103:3700–3709 [DOI] [PubMed] [Google Scholar]
- 194.Brown BD, Venneri MA, Zingale A, et al. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med 2006;12:585–591 [DOI] [PubMed] [Google Scholar]
- 195.Brown BD, Cantore A, Annoni A, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood 2007;110:4144–4152 [DOI] [PubMed] [Google Scholar]
- 196.Majowicz A, Maczuga P, Kwikkers KL, et al. Mir-142-3p target sequences reduce transgene-directed immunogenicity following intramuscular adeno-associated virus 1 vector-mediated gene delivery. J Gene Med 2013;15:219–232 [DOI] [PubMed] [Google Scholar]
- 197.Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 2007;25:1457–1467 [DOI] [PubMed] [Google Scholar]
- 198.Gentner B, Visigalli I, Hiramatsu H, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med 2010;2:58ra84. [DOI] [PubMed] [Google Scholar]
- 199.Papapetrou EP, Kovalovsky D, Beloeil L, et al. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J Clin Invest 2009;119:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gentner B, Naldini L. Exploiting microRNA regulation for genetic engineering. Tissue Antigens 2012;80:393–403 [DOI] [PubMed] [Google Scholar]
- 201.Cheloufi S, Dos Santos CO, Chong MM, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010;465:584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010;328:1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA 2012;18:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dueck A, Ziegler C, Eichner A, et al. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 2012;40:9850–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 2010;9:4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Havens MA, Reich AA, Duelli DM, et al. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 2012;40:4626–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dallas A, Ilves H, Ge Q, et al. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res 2012;40:9255–9271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res 2013;41:3723–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Herrera-Carrillo E, Harwig A, Liu YP, et al. Probing the shRNA characteristics that hinder Dicer recognition and consequently allow Ago-mediated processing and AgoshRNA activity. RNA 2014;20:1410–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Herrera-Carrillo E, Harwig A, Berkhout B. Silencing of HIV-1 by AgoshRNA molecules. Gene Ther 2017. [Epub ahead of print]; DOI: 10.1038/gt.2017.44 [DOI] [PubMed] [Google Scholar]