Abstract
MicroRNAs and other small oligonucleotides in biofluids are promising disease biomarkers, yet conventional assays require complex processing steps that are unsuitable for point-of-care testing or for implantable or wearable sensors. Single-walled carbon nanotubes are an ideal material for implantable sensors, owing to their emission in the near-infrared spectral region, photostability and exquisite sensitivity. Here, we report an engineered carbon-nanotube-based sensor capable of real-time optical quantification of hybridization events of microRNA and other oligonucleotides. The mechanism of the sensor arises from competitive effects between displacement of both oligonucleotide charge groups and water from the nanotube surface, which result in a solvatochromism-like response. The sensor, which allows for detection via single-molecule sensor elements and for multiplexing by using multiple nanotube chiralities, can monitor toehold-based strand-displacement events, which reverse the sensor response and regenerate the sensor complex. We also show that the sensor functions in whole urine and serum, and can non-invasively measure DNA and microRNA after implantation in live mice.
Introduction
The detection of oligonucleotide hybridization is important for a broad range of applications, from devices based on dynamic DNA nanotechnology to clinical point-of-care diagnostics. Oligonucleotides in biofluids such as serum, urine, and peritoneal fluid1, 2 are a promising source of biomarkers for a variety of pathologies, including cancer3, 4. Of potential biomarker oligonucleotides5–7, microRNA (miRNA) was found to be highly stable in body fluids, as many studies to date identify specific patterns of miRNA expression indicative of disease states3, 8–13. Devices that can measure changes in biomarker miRNA14 or cell-free DNA15 concentrations in biofluids such as urine, serum, or ascites in patients with risk factors for a disease or its relapse may improve early detection and treatment. Implantable or wearable devices that enable minimally-invasive continuous biomarker monitoring may extend this strategy to detect biomarkers at the earliest possible stages of disease.
The current standard for miRNA measurement, with limits of detection ranging from aM to fM16, is quantitative PCR (qPCR)17, but this method requires purification and amplification of miRNA that can introduce biases and variability18. Commercially available techniques that do not involve amplification, such as microarrays, suffer from poorer sensitivity (pM to nM) and high false positive rates16, 18. Detection strategies that avoid amplification, labeling, and purification from biofluids are under investigation19, 20, but in vivo detection strategies are sparse. The detection of nucleic acid biomarkers in real-time and in situ within living tissues and organisms remains an important challenge.
Of potential materials for detecting nucleic acids, individually-dispersed semiconducting single-walled carbon nanotubes (SWCNTs) exhibit exciting properties for use as optical biomedical sensors21, 22. Semiconducting carbon nanotubes are fluorescent in the near-infrared spectral region23, a wavelength range penetrant to tissue24, and they do not photobleach25. Their emission wavelength26 and intensity27 are sensitive to the local environment, allowing perturbations at the nanotube surface to be transduced via modulation of their emission, with up to single-molecule sensitivity28. Up to 17 distinct nanotube (n,m) species (chiralities) with unique and resolvable emission wavelengths can be measured, potentiating multiplexed detection schemes29.
Herein, we describe a label-free approach to detect hybridization events of miRNA and other oligonucleotides transiently and in vivo. We designed a sensor which transduces the hybridization of small DNA and RNA oligonucleotides into spectral changes of carbon nanotube photoluminescence, and we determined its mechanism of action via experiments and molecular dynamics simulations to be a competitive response to local dielectric and electrostatic factors. Using this understanding, we designed a scheme wherein amphiphilic moieties undergo triggered assembly on the nanotube surface upon binding of target miRNA, resulting in a markedly enhanced spectral response. We showed that the sensor enables multiplexed detection using different nanotube chiralities and real-time monitoring of toehold-mediated DNA-strand displacement, which caused a reversal of the signal response. The sensor was highly resistant to non-specific interactions with biological molecules, allowing for direct detection in urine and serum. Finally, we showed the first in vivo optical detection of target DNA and miRNA by encasing the sensor within an implantable device through which we detected hybridization non-invasively via near-infrared fluorescence in live mice.
Results and Discussion
Development of sensor
We synthesized a DNA-nanotube complex consisting of a single oligonucleotide sequence with two domains—one to impart nanotube colloidal stability (nanotube-binding sequence) and a second with a complementary sequence to a target oligonucleotide (miRNA capture sequence). For the miRNA capture sequence, we chose a specific 23-mer miRNA (miR-19) as a model target due to its role in oncogenesis30. Figure 1a shows the synthesis scheme. A screen of potential nanotube-binding sequences found (GT)15, known to efficiently encapsulate nanotubes31, to provide superior resistance to non-complementary oligonucleotides (Figure S1). The purified DNA-nanotube construct was found to be highly photoluminescent and is herein referred to as the GT15mir19 sensor.
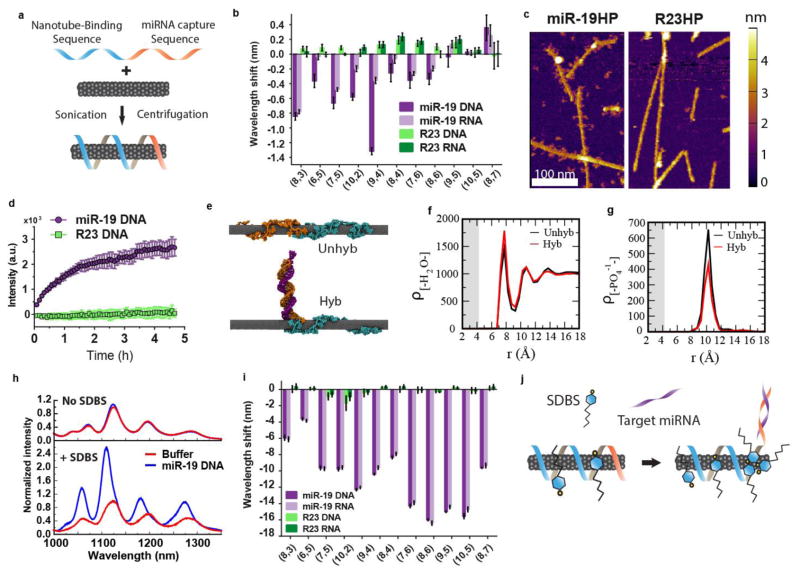
Figure 1. Carbon nanotube sensor for the detection of miRNA hybridization events.
a, Construction scheme of the miRNA sensor complex consisting of a single DNA oligonucleotide containing a nanotube-binding sequence (blue) and miRNA capture sequence (orange) which is non-covalently bound to the carbon nanotube surface. b, Response of the GT15mir19 sensor to analyte DNA or RNA with the miR-19 sequence, or a control sequence (R23). Response of nanotube emission is shown for each nanotube chirality. Positive wavelength shift denotes a red-shift in the emission peak, and negative numbers denote a blue-shift. c. Atomic force microscopy images of the sensor complex upon incubation with non-complementary (R23HP) or complementary (miR-19HP) hairpin DNA. d, Intensity of Cy5 emission from the GT15mir19-Cy5-nanotube complex after introducing miR-19 DNA or R23 DNA. e, Snapshot images of molecular dynamics simulations of the GT15mir19 sensor (Unhyb) and GT15mir19 sensor hybridized with miR-19 (Hyb) after equilibrating for 250 ns. Teal color denotes the (GT)15, nanotube-binding sequence, and orange denotes the miR-19 capture sequence. The purple strand denotes miR-19. f. Density of water as a function of radial distance from the nanotube, calculated for both simulations. g, Density of phosphate groups as a function of radial distance from the nanotube, calculated for the final frame of both simulations. h, Photoluminescence spectra of the GT15mir19 sensor on hybridization with miR-19 in the absence (Top) or presence (Bottom) of SDBS, normalized to the buffer condition i, Response of the sensor complex to analyte DNA or RNA with the miR-19 sequence, or a control sequence (R23), in the presence of SDBS, for each nanotube chirality. j, Cartoon depicting the proposed mechanism of SDBS-mediated enhancement of the blue shifting response upon hybridization. Error bars represent standard deviation for n = 3 technical replicates.
The optical response of the GT15mir19 sensor was then tested using both a DNA-based and RNA-based analyte miR-19 sequence, as well as a length-matched, randomly generated, non-complementary control (R23). After incubation with miR-19 or R23, eleven different nanotube chiralities were measured via two-dimensional excitation/emission photoluminescence spectroscopy32 (“PL plots”, Figure S2). Each nanotube emission peak exhibited a shift in wavelength which was specific to the miR-19 target sequence over the R23 control (Figure 1b). In general, nanotube emission peak wavelengths blue-shifted and intensity increased upon introduction of the target oligonucleotide (Figure S3); excitation peaks (E22 transitions) also blue-shifted (Figure S4).
To verify that hybridization to the GT15mir19 sensor occurred upon introduction of the target, we designed a hairpin oligonucleotide which would make binding of the target more apparent by atomic force microscopy (AFM). The oligonucleotide was composed of the miR-19 or R23 sequence, a short spacer, and a 52-nucleotide hairpin region (diagram in Figure S5). After incubation with the miR-19-hairpin, the average height of the sample increased by ~0.6 nm, as measured by AFM in dry conditions (Figure S5). Upon imaging in aqueous conditions, we observed distinct protrusions from the nanotubes which were absent in the R23 hairpin-treated sample and buffer controls (Figure 1c, Figure S6). Based on this pattern and other AFM studies33, we estimate that the GT15mir19 sensor presents 5–10 binding sites per 100 nm of nanotube. Our preparation method yielded nanotubes with a mean length of 166 nm (SD 149nm)34; thus we calculate that an average single nanotube could potentially bind approximately 8–17 copies of miRNA.
Because the mechanism of nanotube spectral changes induced by oligonucleotide hybridization is poorly understood, we designed a set of experiments to better understand the structural changes of the sensor induced by hybridization. We first investigated whether the hybridized duplex remained near the nanotube surface after the binding of target miRNA. We developed an assay using an organic fluorophore conjugated to the miRNA capture sequence under the premise that the fluorophore intensity would increase upon hybridization if the fluorophore desorbed from the nanotube surface, as organic fluorophores are known to quench upon interaction with the nanotube surface via an energy transfer mechanism35. We suspended nanotubes with the sequence GT6mir19, (shortened due to synthesis constraints) containing the Cy5 dye conjugated to the 3′ end of the miR-19-binding domain (scheme in Figure S7). Upon addition of miR-19 to the modified complex, we found that Cy5 fluorescence increased over time, while the R23 sequence caused no change in Cy5 fluorescence (Figure 1d). To validate GT6mir19-Cy5 as a proxy for the GT15mir19 sequence, we measured the nanotube emission upon introduction of the miR-19 sequence to the fluorophore-labeled complex. Again, we saw blue-shifting upon hybridization with the target oligonucleotide, suggesting the same sensor function despite the shortened nanotube-binding domain (Figure S8). In agreement with the Cy5 fluorescence change, we found nanotube fluorescence emission to blue-shift at a slower rate compared to GT6mir19 without Cy5. (Figure S8). We interpret this relatively slow rate as a result of the affinity of Cy5 for the nanotube surface, based on π stacking interactions between the Cy5 dye, which is rich with π electrons, and the graphitic π electrons of the nanotube. The fluorophore de-quenching and AFM together suggest a final hybridized structure consisting of a partial duplex dissociating from the nanotube surface.
Using all-atom molecular dynamics simulations, we assessed whether the GT15mir19 sequence could remain stable on the nanotube upon partial hybridization. The pre-hybridized sequence was placed in the vicinity of the (9,4) nanotube with explicit water and counterions, and a simulation was run for 250 ns (details in methods). The single-stranded portion of the oligomer bound to the nanotube and the hybridized construct remained stable on the nanotube surface for the remainder of the simulation (Figure 1e “Hyb”, Movie S1). A second simulation was run in absence of the hybridization strand. During the simulation, the entire oligomer bound to the surface and wrapped the nanotube, with the nucleobases orienting closely to the nanotube surface in a parallel orientation (Figure 1e, “Unhyb”, Movie S2).
The simulations allowed the quantification of nucleobase adsorption to the nanotube surface. We measured the radial distance of the nucleobases from the nanotube surface and their stacking angles relative to the nanotube surface (Figure S9). We observed that all bases of the (GT)15 nanotube-binding domain remained adsorbed on the nanotube surface, whereas only 1–2 terminal bases of the double-stranded miR-19/miRNA capture sequence adsorbed to the nanotube surface. In the simulation without the complementary strand, all bases of the (GT)15 nanotube binding domain adsorbed to the nanotube surface, as well as most of the bases of the miR-19 miRNA capture sequence (Figure S9).
We assessed the thermodynamic concerns regarding the stability of the hybridized duplex in the presence of the nanotube. Molecular dynamics simulations of hybridized miR-19, without the (GT)15 nanotube binding domain, in the presence of the nanotube were run using several different initial conditions (Figure S10, Movie S3). In all simulations, no de-hybridization of the duplex was observed, suggesting that the nanotube would not destabilize the hybridized duplex. To understand how the partial hybridized state of the DNA is stable on the nanotube (or preferred over single strand adsorption on the nanotube), we conducted a free energy analysis (Supplementary Text, Figure S11). The analysis suggests that hybridization of the dsDNA is favored if the analyte strand is not initially adsorbed on the nanotube surface, as is the case in our experiments.
We also analyzed the molecular dynamics simulations to gain a quantitative understanding of the carbon nanotube spectral response upon hybridization. On comparing the water density as a function of distance at the end of the two simulations, we found a slight increase in the water concentration near the nanotube in the hybridized structure (Figure 1f). In addition, we found that the density of phosphate ions as a function of distance from the nanotube decreased upon hybridization (Figure 1g). While an increase in local water density is known to cause red-shifting36 of the nanotube emission wavelength, a decrease in local anionic charge density in the local environment of the nanotube was found to cause a blue-shifting response, according to recent work34. As the nanotube emission exhibited a net blue-shift upon hybridization, we thus conclude that the effect of the removal of phosphate charges from the nanotube surface out-competed the effects of increased local water density.
As the simulations showed an increase in available nanotube surface area upon hybridization, we hypothesized that additional small amphipathic molecules might assemble on this newly exposed nanotube surface to enhance the optical response. Low concentrations of several candidate surfactants (Table S1) were tested to determine whether they changed the optical response of the GT15mir19 sensor (Supplementary Text, Figure S12, S13). The study found that a low concentration (0.2% wt/vol, or 5.7 mM) of sodium dodecylbenzenesulfonate (SDBS), a mild surfactant known to associate with nanotubes37, resulted in an increase in the degree of hybridization-dependent blue-shifting and intensity enhancement by an order of magnitude (Figure 1h). In the SDBS-supplemented buffer-only condition and in the presence of the R23 control, the emission bands broadened slightly but did not increase or shift appreciably. Upon hybridization in the presence of SDBS, all nanotube chiralities exhibited a greatly enhanced blue-shift (Figure 1i, S14–17), even those that that did not blue-shift in the absence of SDBS (Figure 1b). A significant blue-shift in the excitation wavelength was also observed (S15). The magnitude of blue-shifting and intensity enhancement (Figure S16) upon hybridization of DNA and RNA were identical. In the absence of the target oligonucleotide, the GT15mir19 sensor emission remained stable over a wide-range of SDBS concentrations (Figure S17). We present our model of SDBS-mediated hybridization-dependent signal enhancement in Figure 1j, wherein hybridization triggers SDBS assembly on the newly-exposed nanotube surface. For a more detailed analysis of the observed spectroscopic changes induced by SDBS, see the Supplementary Text.
To further assess the specificity of the sensor response, we introduced an ensemble of randomly generated oligonucleotides. A random library of 23 nt oligonucleotides, with a diversity of approximately 423 different sequences, was introduced to the GT15mir19 sensor, resulting in no response (Figure S19). In the presence of the random library, the GT15mir19 sensor maintained sensitivity to miR-19.
Detection limit, kinetics, and breadth of applicability
Given the variety of potential miRNA biomarkers, we sought to assess the modularly of the sensor. The miRNA capture sequence was substituted with several sequences specific to 9 different serum or urine miRNA biomarkers, as well as a sequence not found in humans (C. elegans miR-39) used for standardization in clinical applications38 (Table 1). Each GT15mirX sensor was treated with SDBS and interrogated with its respective miRNA target sequence, resulting in a wavelength shift which was comparable to that of the original miR-19 sensor, with slight sequence-to-sequence variations (Figure 2a and Figure S20). Intensity was similarly enhanced (Figure S21). In all of the sensors, no appreciable responses from the control sequence (R23) were observed.
Table 1.
Disease-relevant miRNA biomarkers
| Name | Disease relevance | Biofluid | Ref |
|---|---|---|---|
| miR-21 | Diffuse large B-cell lymphoma | serum | 9 |
| miR-96 | Urothelial carcinoma | urine sediment | 10 |
| miR-183 | Urothelial carcinoma | urine sediment | 10 |
| miR-126 | Urinary bladder cancer | voided urine | 11 |
| miR-182 | Urinary bladder cancer | voided urine | 12 |
| miR-152 | Healthy control | voided urine | 11 |
| miR-494 | Acute kidney injury | voided urine | 13 |
| miR-509 | Healthy control, highly expressed | voided urine | 8 |
| miR-39 | Found only in C. elegans; spike-in control | N/A | 3 |
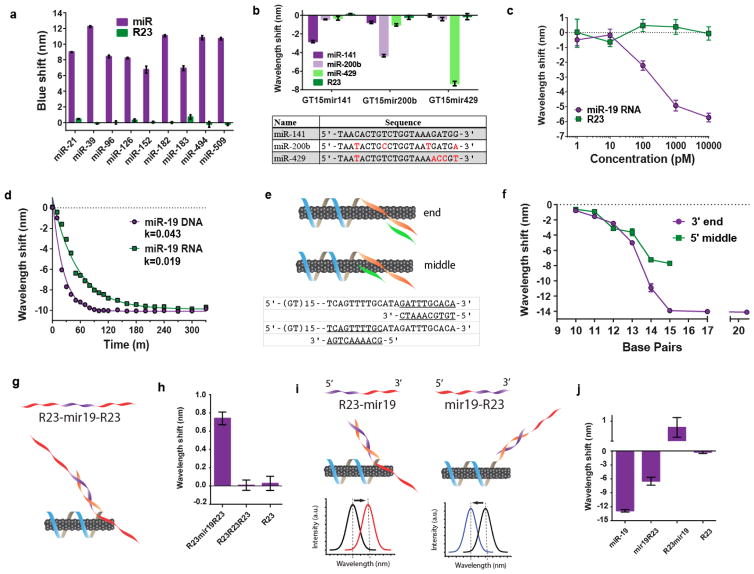
Figure 2. Characterization of miRNA detection limit, kinetics, and functionality.
a, Optical responses of the sensor composed of different capture sequences to recognize the specified miRNA sequences (DNA analogues), shown for the (7,5) chirality. b, Spectral responses (blue-shifts), of the sensor composed of the specified capture sequences, to related miR-200 family sequences (shown in the table; differences shown in red). c, Dose-response curve of the GT15mir19 sensor, shown for the (7,6) nanotube species. d, Kinetics of the sensor response upon interrogating with miR-19 RNA or the DNA analogue, fitted with the exponential function, y = y0 e−kt. R2=0.97 and 0.99 for DNA and RNA, respectively. e, Cartoon illustrating truncated analyte sequences complementary to the middle or 3′ end of the capture sequence. f, Emission response of the sensor to a series of truncated sequences (length specified in the x axis) designed to hybridize to either the middle or 3′ end of the capture sequence. g, Cartoon illustrating a modified analyte sequence and expected configuration upon binding to the GT15mir19 sensor. h, Spectral response of the (8,6) nanotube species upon introduction of long analyte sequences to the sensor. i, Cartoons depicting experiment designed to assess orientation of partial complementary sequences, as well as predicted sensor responses. j, Response of the (8,6) nanotube species upon interrogating the GT15mir19 sensor sensor with the partial complementary oligonucleotides. All error bars represent standard deviation for n = 3 technical replicates.
To determine if the SDBS-GT15mirX sensor could discriminate among similar sequences, three related sequences from the miR-200 family were selected. The miR-200 family plays an essential role in the epithelial-to-mesenchymal transition (EMT)39 in cancer. Focusing on the wavelength response of the (9,4) nanotube chirality, we observed a high degree of discrimination between the three sequences after one hour of incubation (Figure 2b). Complete time-course data for both the (9,4) and (8,6) nanotubes (Figure S22 and S23, respectively) revealed that the intensity increase provided near-perfect discrimination in most cases. Although the SDBS-GT15mirX sensor responded to target miRNA via both wavelength shifting and intensity changes, we assessed detection limits, kinetics, and other sensor characteristics using the wavelength response, due to the inherent quantifiability and internal standard provided by this mode.
To determine the limit and range of detection, a dose-response curve of the sensor was constructed over several orders of magnitude of miR-19 concentrations. At a minimal sensor concentration of 0.02 mg/L, the limit of detection of miRNA was between 10 and 100 pM (500 attomoles to 5 femtomoles) (Figure 2c). Signal saturation occurred at a high concentration between 1 and 10 nM. The dynamic range was tuned by adjusting the concentration of the GT15mir19 sensor to cover at least 5 orders of magnitude, from 10 pM to 1 uM (Figure S24). We calculated the number of binding sites using the mass of DNA used to suspend 1 mg of nanotubes (see Methods). We estimated that 2.117 nM of miR-19 binding sites were available in a solution of 0.02 mg/L of the GT15mir19 sensor, consistent with the observed saturating range of concentrations (between 1 and 10 nM).
The kinetics of both DNA and miRNA detection were assessed via transient measurements. The kinetics of eleven different nanotube chiralities were measured by excitation/emission spectroscopy (Figure S25). Within 10 minutes of introducing the analyte to the sensor, significant blue-shifting was discerned. The rate of blue-shifting behaved with pseudo-first order kinetics and showed no obvious dependence on nanotube structure (Figure S26). The sensor kinetics were consistently faster for DNA (1.8x on average), as compared to RNA (Figure 2d). We ascribe this difference to the longer persistence length and higher rigidity of single-stranded RNA as compared to single-strand DNA40.
To test if the composition of the miRNA capture sequence influenced sensor kinetics, we compared the response rates for the sensor using 8 different miRNA capture sequences (Figure S27). On comparing the sensor kinetics as a function of guanine content, we found a significant correlation, with Pearson coefficients of −0.74195 (p=0.035) for the (9,4) nanotube and −0.77215 (p=0.0248) for the (8,6) nanotube (Figure S28). This result may be explained by the high affinity of guanine for the nanotube surface, which was determined previously via both molecular dynamics and ab initio calculations41 to fall in the order: G > A > T > C. Thus, the affinity of guanine to the nanotube surface may slow the hybridization process. The content of other nucleotide bases, as well as the overall ΔG of hybridization, did not show any statistically significant correlations (Figure S29 and S30).
To better understand how the length and thermodynamics of hybridization relate to the optical response of the nanotube, we conducted several experiments using modified analyte oligonucleotides. We interrogated the G15mir19 sensor using analyte sequences between 10 and 23 nucleotides long which were complementary to either the 3′ terminal end of the miRNA capture sequence, or the middle of the sequence, as depicted in Figure 2e. We found that, in general, a shorter analyte sequence resulted in a smaller blue-shifting response of the nanotube, down to approximately 10 nucleotides, where there was virtually no response (Figure 2f). Additionally, the magnitude of the blue-shifting response was consistently smaller when the analyte sequence was designed to hybridize to the middle of the capture sequence. This difference may be explained by the affinity of the capture sequence to the nanotube, as suggested by the results of the fluorophore quenching experiment, MD simulations, and free energy analyses41.
To assess its broad applicability for the detection of different nucleic acid types, we assessed whether the sensor could detect oligonucleotides longer than miRNA sequences. First, we assessed how the GT15mir19 sensor would respond to a long oligonucleotide designed to contain a complementary sequence flanked by non-complementary sequences (Figure 2g). On interrogating the sensor with R23-mir19-R23, a 69-bp oligonucleotide with 23 complementary bases in the middle of the sequence, the sensor emission red-shifted—opposite of the expected blue-shifting response—even in the presence of SDBS (Figure 2h, shown for the (8,6) chirality). We therefore hypothesized that, when R23-mir19-R23 hybridizes to the recognition sequence, the R23 portion at the 5′ end may disrupt the sensor function by increasing the phosphate content near the nanotube surface to cause a red-shift of the nanotube emission. To test this hypothesis, we designed two long oligonucleotide sequences, R23-mir19 and mir19-R23, placing the R23 portion at either the 3′ end or 5′ end. The proposed orientation upon binding and predicted spectral shifts are shown in Figure 2i—the two sensors were hypothesized to give the opposite spectral responses. Upon interrogating the sensor, the R23-mir19 oligonucleotide produced a red-shifting response, and the mir19-R23 oligonucleotide produced a blue-shift, as predicted (Figure 2j). The magnitude of the blue-shift in response to mir19-R23 was smaller than that produced by the miR-19 control, suggesting that the unhybridized single-stranded nucleotides may bind to the nanotube surface, diminishing the response. This hypothesis warrants further study. Regardless, we conclude that sensors based on the GT15 nanotube binding domain and a general capture sequence can be extended to detect longer nucleic acid sequences, but the orientation of the oligonucleotide is critical for eliciting a desired spectral response.
Measurements of single sensor complexes
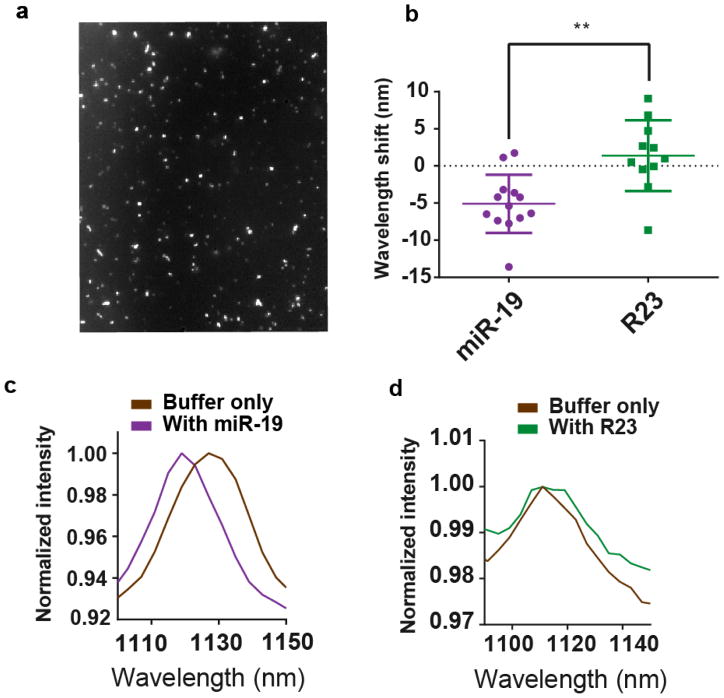
We assessed the sensor function on the single-nanotube level via spectral imaging. The sensor was deposited on a lysine-coated glass surface with sodium dodecyl sulfate (SDS). We then used hyperspectral microscopy29 to spectroscopically image the (9,4) nanotube (Figure 3a). The blue-shifting of single nanotubes was apparent upon interrogating the sensor with miR-19 RNA, but not upon introducing R23 RNA (Figure 3b–d, Figure S31). Using the number of binding sites per nanotube length determined from AFM measurements (Figure 1d), we attempted to estimate the number of copies of miRNA detected per nanotube. Because a diffraction-limited spot could contain a nanotube up to ~600nm long42, we estimate a range of detection between 1–60 miRNA molecules.
Figure 3. Single nanotube response to miRNA hybridization.
a, Broadband near-infrared image of the GT15mir19 sensor adsorbed to a glass surface. b, Change in emission wavelength of individual (9,4) nanotubes after incubation of the surface-bound complexes with miR-19 RNA or R23 RNA at room temperature (unpaired, two-tailed t test, p=0.0014, n = 24). c, Representative spectra of the same sensor complex in the buffer-only condition and 50 min after introducing miR-19. d, Representative spectra of the same sensor complex in the buffer-only condition and 50 min after introducing the R23 control sequence.
Sensor multiplexing
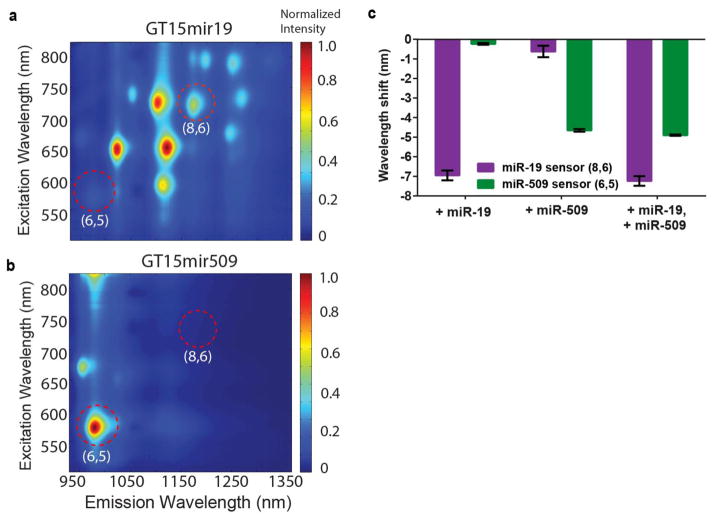
We assessed the potential for the multiplexed detection of several miRNA sequences via the use of different nanotube chiralities. Two nanotube preparations enriched for different nanotube chiralities were suspended with binding sequences for either miR-19 or miR-509. A preparation enriched in large diameter species, (Nano-C APT-200) was suspended by the GT15mir19 sequence, and a CoMoCAT preparation enriched in small diameter species was suspended using the GT15mir509 sequence. Excitation/emission plots found that the GT15mir19 sensor, encapsulating the APT-200 nanotubes, effectively lacked the (6,5) species (Figure 4a), while the GT15mir509 sensor, encapsulating the narrow-diameter enriched CoMoCAT preparation, lacked the (8,6) species (Figure 4b). Absorbance spectra verified the differential enrichment of nanotube chiralites between these samples (Figure S32). After mixing the two nanotube preparations, each miRNA sequence added individually was recognized by the appropriate nanotube chirality (Figure 4c). When miR-19 and miR-509 were added together, the spectral shift almost identically recapitulated the shifts seen when either miRNA was added alone.
Figure 4. Sensor multiplexing.
a, Photoluminescence excitation/emission plot of the GT15mir19 sensor synthesized using a large-diameter nanotube preparation. b, Photoluminescence excitation/emission plot of the GT15mir509 sensor synthesized using a small-diameter preparation of carbon nanotubes. Red circles highlight the differences in chiralities present. c, Responses of the two sensors mixed together, upon introducing miR-19 alone, miR-509 alone, or both analytes simultaneously. Error bars represent standard deviation for n = 3 technical replicates.
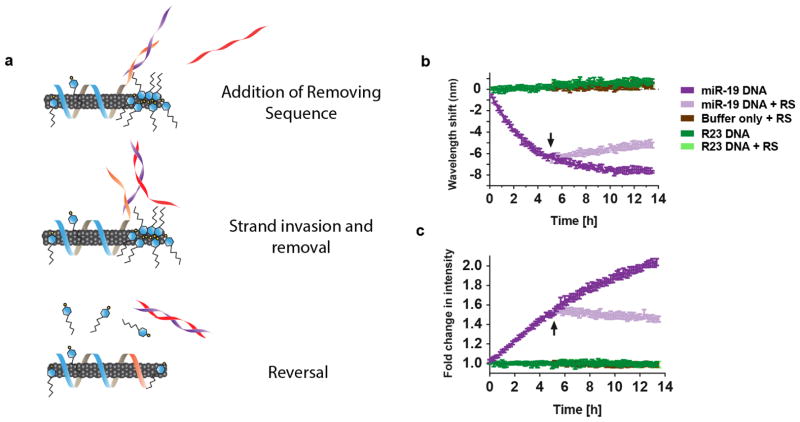
Toehold-mediated strand displacement
We asked whether the spectral response of the sensor could be reversed via toehold-mediated strand displacement. Strand displacement reactions occur through the use of “toeholds,” single-strand overhangs on duplexed DNA that facilitate binding of a complementary strand, which is thermodynamically favored due to complete complementarity, and is thus able to displace the shorter bound strand43. We truncated the miRNA capture sequence of the GT15mir19 sensor to leave a 6 nucleotide overhang44 after hybridization with the target strand to test whether the addition of a removing strand (RS) to bind the toehold and displace the target would reverse the spectral shift, according to the proposed scheme in Figure 5a. Upon addition of miR-19 to the modified GT15mir19 sensor, the nanotube emission blue-shifted and the intensity increased as expected (Figure 5b and c). After 5 hours, the removing strand was added, at which point the blue-shifting ceased and the emission began to undergo a steady red-shift (Figure 5b). The emission intensity exhibited a similar reversal (Figure 5c). We note that the signal reversal was slower than detection in the forward direction, which is likely due to the energetic barrier for the truncated capture sequence to displace SDBS from the nanotube surface.
Figure 5. Monitoring toehold-mediated strand displacement.
a, Cartoon depicting the experiment, wherein the GT15mir19 sensor contains a truncated miRNA-capture sequence (orange domain). Upon hybridization with miR-19, a 6-nucleotide toehold remains on the miRNA sequence (purple strand). The removing strand (red), which is complementary to the entire miRNA sequence, is introduced to remove miR-19. b, Kinetic response of the GT15mir19-sensor wavelength upon addition of the target miRNA sequence, and after introducing the removing strand (RS), at the time denoted by the arrow. c, Kinetic response of the GT15mir19 sensor intensity, measured during the same experiment. Error bars represent standard deviation for n = 3 technical replicates.
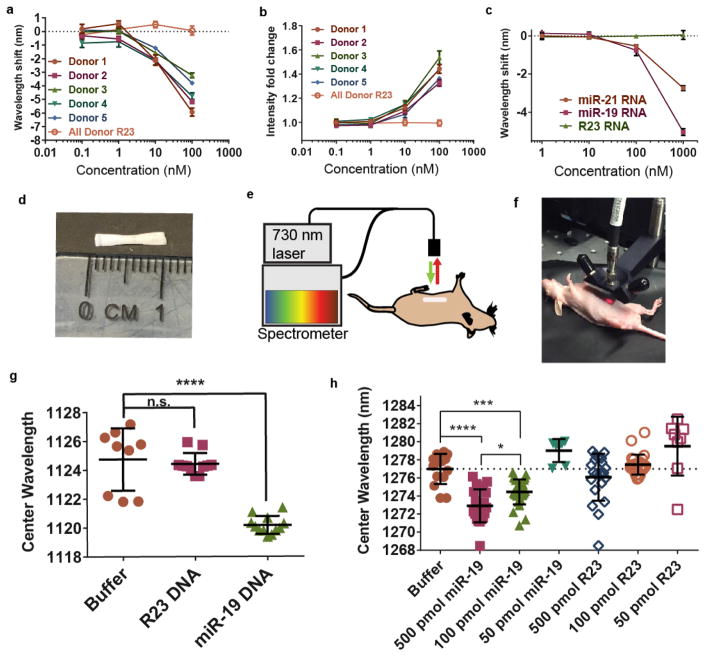
Detection of miRNA in biofluids
We sought to assess the ability of the GT15mir19 sensor to detect miRNA binding events in common biofluids—urine and serum—due to their clinical value as sources of microRNA biomarkers8. The GT15mir19 sensor and SDBS were introduced concomitantly to whole urine from 5 healthy donors before interrogating with miR-19 RNA. The wavelength shifting response was clearly detectable against controls down to 1 nM of miRNA, and intensity enhancement gave a similar sensitivity, between 1 and 10 nM (Figure 6a–b, Figure S33). Variation from sample-to-sample was minimal. In whole serum, we found that target miR-19 DNA was similarly detectable in the presence of SDBS (Fig S34). When target miR-19 RNA was introduced to the sensor, we found only a small response at the highest tested concentration (Fig S34). We hypothesized that the RNA detection was complicated by RNases in the serum which might degrade the analyte sequence, as reported elsewhere for synthetic RNA sequences45. We therefore introduced proteinase K, a detergent-stable protease used to deactivate RNase, into the serum. Introduction of proteinase K allowed the detection of miR-19 RNA with the same sensitivity as for the DNA analogue (Figure 6c, Figure S34). When proteinase K was introduced 12 h after mixing miR-19 with serum, the sensitivity of the response to miR-19 RNA was not improved, suggesting that the RNA had been destroyed. To verify broad applicability with this method, we also used miR-21 as a target, due to its significance as a serum colorectal cancer biomarker46. We similarly tested GT15mir21 sensor in whole serum treated with proteinase K and found that miR-21 RNA could be detected directly in minimally-treated serum via both blue-shifting and intensity enhancement (Figure 6c, Figure S35).
Figure 6. Detection of miRNA in biofluids and non-invasively within live mice.
a, Response of the GT15mir19 sensor emission wavelength to miR-19 spiked into urine from 5 healthy donors. The (7,6) nanotube chirality was measured. Error bars represent standard deviation of technical triplicates. b, Intensity response of the sensor in urine. Error bars represent standard deviation of technical triplicates. c, Wavelength response of the nanotube sensor complex to miR-21 and miR-19 miRNA in fetal bovine serum (FBS). Error bars represent standard deviation of technical triplicates. d, Semi-permeable membrane encapsulating the GT15mir19 sensor for implantation. e, Diagram of NIR probe apparatus for illuminating and measuring the sensor response in vivo. f, Image of NIR probe system measuring the nanotube response within a live mouse. g, Response of the implanted sensor device to miR-19 DNA within the live mouse (3–4 measurements per mouse; 3 mice per group). The (9,4) nanotube species was measured (p<0.0001, Dunnet’s multiple comparison test, ordinary one-way ANOVA). h, Response of the implanted sensor device to 500, 100, and 50 pmol of miR-19 RNA or R23 RNA injected into mice intraperitoneally (3–4 measurements per animal; 3 animals per group), shown for the (8,7) nanotube species. 50 pmol R23 was slightly red-shifted compared to buffer control. Error bars represent standard deviation. Statistical significance was calculated with Dunnet’s multiple comparison test. Ordinary ANOVA was used to compare the mean of each group to the mean of the buffer control. Sidak’s multiple comparison test with an alpha of 0.05 was used to compare miR-19 groups.
Detection of miRNA detection in vivo
We further explored the potential of the sensor to detect miRNA in vivo via a minimally-invasive implantable device. We loaded the SDBS-treated GT15mir19 sensor into a semipermeable membrane capillary with a MWCO of 500 kDa (Figure 6d). To determine whether this cutoff would to prevent the diffusion of the GT15mir19 sensor complexes outside of the membrane, we calculated the molecular weight of the GT15mir19 sensor. We estimated that the sensor complexes composed of the narrowest diameter (0.8 nm) and average length of 166nm fall within the range of 701 kDa to 839 kDa (see Supplementary Text). We surmised that the miR-19 miRNA, with a molecular weight of 7.055 kDa, would pass through the membrane. We also assessed the likelihood that the enhanced signal response provided by SDBS would continue after device implantation. We thus filled the semi-permeable capillary with SDBS-pretreated GT15mir19 sensor and placed it in buffer dialysate for 6 hours. The buffer was changed and the sensor response was assessed with miR-19 every 2 hours (Figure S36). We found that the GT15mir19 sensor exhibited a nearly identical blue-shifting response after 6 hours of dialysis, suggesting that the SDBS remained associated with the sensor even under these conditions.
We tested the sensor response in vivo after surgically implanting the membrane into the peritoneal cavity of NU/J (nude) mice. The membrane was placed medially over the intestines and sutured to the parietal peritoneum to immobilize the device. We first tested whether DNA could be detected intraperitoneally by injecting 1 nanomole of miR-19 DNA, R23, or the vehicle control. The mice exhibited no obvious adverse effects or changes in behavior following the implantation or injection. After 90m, the mice were anesthetized using isofluorane. A fiber optic-based probe system was developed to excite an 0.8 cm2 area with a 730 nm CW laser (Figure 6e), collect the emitted near-infrared light through the same fiber bundle, disperse the light with a Czerny-Turner spectrograph, and detect the light via a 1D InGaAs array (Figure 6f). Using the nanotube emission signal collected from the mouse, we found that the target miR-19 DNA exhibited a significant blue-shifting response as compared to controls (Figure 6g). The experiment was repeated using the RNA version of the analyte, resulting in a similar response (Figure S37). We tested the implantable device in vitro by immersing the filled capillary into buffer containing RNA, finding that the threshold of detection was below 10 pmol (Figure S38). To determine the limit of detection in vivo, we injected 500 pmol, 100 pmol, or 50 pmol of miR-19 RNA intraperitoneally into mice implanted with the devices. After 120 min, significant wavelength shifting responses were measured down to 100 pmol (Figure 6h). The devices were removed from the animals and measured ex vivo, resulting in similar results (Figures S39). We suspect the higher limit of detection of the device in vivo as compared to in vitro was due to the degradation of microRNA in the peritoneal fluid as well as fluid exchange out of the peritoneal cavity. The measurement of endogenous microRNA targets, which are highly stable due in part to their association with proteins such as Ago247, 48, may help improve sensor performance in future investigations.
Outlook
Herein, we engineered a label-free, amplification-free optical sensor for the quantitative detection of oligonucleotide hybridization events in vitro and non-invasively in vivo. The sensor mechanism, resulting from competitive effects of the displacement of both electrostatic charge and water from the carbon nanotube surface, has implications for the improvement of carbon nanotube-based optical and electronic sensors. We also gained a clear understanding over the effects of length, mismatches in sequence, and orientation of longer oligonucleotides on the optical response of the carbon nanotube, providing a basis for continued optimization. The GT15mirX sensor enabled detection via single-molecule sensor elements and multiplexing using multiple nanotube chiralities. The monitoring of toehold-based strand displacement events portends use in nucleic acid-based logic circuits49 and also allowed the reversal of the sensor response and regeneration of the sensor complex, which may potentially be exploited for continuous use.
Using the current iteration of the sensor, in vitro applications such as point-of-care diagnostics may provide the most immediate route to clinical use. We found that the sensor can directly detect oligonucleotides in heterogeneous biofluids such as urine and serum with minimal pre-treatment, potentially circumventing biases and variability related to typical pre-analytical steps required for RT-qPCR38. Regarding sensor parameters pertinent to clinical measurements, Weber et al.8 surveyed the microRNA content in 12 body fluids, providing useful quantitative information to estimate the physiological range of microRNA. In urine, the median concentration of a microRNA species is on the scale of tens of pM8, while in plasma and peritoneal fluid it is in the hundreds of pM8. Other quantitative sensors50 found biofluid microRNA concentrations in the high femtomolar range, suggesting that the dynamic range may fall between about 10 fM and 100 pM. The current limit of detection of our sensor in bulk solution is in the picomolar range, although the threshold of detection and dynamic range depends on several factors, including binding site coverage, which have not yet been modulated in the development process. We also demonstrated the ability to measure single-nanotube responses representing 1–60 copies of microRNA binding, suggesting developments that might attain sensitivities down to 10’s of copies of microRNA, potentially rivaling the most sensitive techniques45, 51, 52.
An implantable optical sensor device for the non-invasive detection of biomarkers such as miRNA may potentially be used in conjunction with wearable devices to facilitate the optical readout and data recording. Our sensor implant quantified miRNA down to 100 pmol in vivo, although further optimization of the sensitivity and other parameters is warranted. While in vitro experiments suggest the current version of the sensor to be robust over at least several hours, more testing is needed to determine stability over longer periods. Investigations are also needed to ensure that oligonucleotides can be detected in their physiological states. For example, miRNA is often found associated with the small protein Ago247, which makes it physiologically stable. Functionally, Ago2 binds to microRNA in a conformation to favor hybridization with target sequences, especially over an 8 nucleotide section called the seed sequence53, 54, but steric hindrance or charge interactions of the protein with miRNA could slow access to the protein-bound sections of the strand. Future iterations of a sensor may include locked nucleic acids55 or peptide nucleic acids56 to enhance binding affinities to short, unbound sections of miRNA. Future studies are also needed to investigate device form factors for implantation and data collection strategies, such as sensor interrogation using wearable devices.
Methods
DNA-suspension of carbon nanotubes
Carbon nanotubes produced by the HiPco process (Unidym, Sunnyvale, CA), CoMoCAT process (SG65i grade, Sigma-Aldrich, St. Louis, MO, US), or a combustion process (APT-200, Nano-C, Westwood, MA) were mixed with DNA oligonucleotides (IDT DNA, Coralville, IA) at a 2:1 mass ratio in 1 mL of saline-sodium citrate (SSC) buffer and ultrasonicated for 30 minutes at 40% amplitude (Sonics & Materials, Inc.). The complete list of DNA sequences used for suspension can be found in Supplementary Methods. Following ultrasonication, the dispersions were ultracentrifuged (Sorvall Discovery 90SE) for 30 minutes at 280,000 × g. The top 80% of the supernatant was collected. Absorbance spectra were acquired using a UV/Vis/nIR spectrophotometer (Jasco V-670, Tokyo, Japan). The concentration was calculated using the extinction coefficient Abs910 = 0.02554 L mg−1c−1. To remove free DNA, 100 kDa Amicon centrifuge filters (Millipore) were used. The DNA-nanotube complexes were re-suspended in saline-sodium citrate buffer (G Biosciences, St. Louis, MO).
Fluorescence spectroscopy of carbon nanotubes in solution
Fluorescence emission spectra from aqueous nanotube solutions were acquired using a home-built apparatus consisting of a tunable white light laser source, inverted microscope, and InGaAs nIR detector. The SuperK EXTREME supercontinuum white light laser source (NKT Photonics) was used with a VARIA variable bandpass filter accessory capable of tuning the output 500 – 825 nm with a bandwidth of 20 nm. The light path was shaped and fed into the back of an inverted IX-71 microscope (Olympus) where it passed through a 20x nIR objective (Olympus) and illuminated a 50–100 μL nanotube sample in a 96-well plate (Corning). The emission from the nanotube sample was collected through the 20x objective and passed through a dichroic mirror (875 nm cutoff, Semrock). The light was f/# matched to the spectrometer using several lenses and injected into an Isoplane spectrograph (Princeton Instruments) with a slit width of 410 μm which dispersed the emission using a 86 g/mm grating with 950 nm blaze wavelength. The spectral range was 930 – 1369 nm with a resolution of ~0.7 nm. The light was collected by a PIoNIR InGaAs 640 × 512 pixel array (Princeton Instruments). A HL-3-CAL-EXT halogen calibration light source (Ocean Optics) was used to correct for wavelength-dependent features in the emission intensity arising from the spectrometer, detector, and other optics. A Hg/Ne pencil style calibration lamp (Newport) was used to calibrate the spectrometer wavelength. Background subtraction was conducted using a well in a 96-well plate filled with DI H2O. Following acquisition, the data was processed with custom code written in Matlab which applied the aforementioned spectral corrections, background subtraction, and was used to fit the data with Lorentzian functions.
Atomic force microscopy
The GT15mir19 sensor was incubated overnight at 20 mg/L with 10 uM of the miR-19-hairpin or 10 uM of the R23-hairpin in saline sodium citrate diluted 20x in 20 mM HEPES + 5 mM MgCl2. The sample was plated on a freshly cleaved mica substrate (SPI) for 4 minutes before washing with 10 mL of dH2O and blowing dry with argon gas. An Asylum Research MFP-3D-Bio instrument was used with an Olympus AC240TS AFM probe in AC mode. Data was captured at 2.93 nm/pixel XY resolution and 15.63 pm Z resolution. For AFM under aqueous conditions, 20 mg/L of the GT15mir19 sensor was incubated with 10 uM of the miR-19-hairpin, R23-hairpin, or buffer overnight. All three conditions were spin-filtered 3x with 100 kDa Amicon centrifuge filters, and resuspended with 5 mM NiCl2, 20 mM HEPES pH 6.7 buffer. The samples were plated onto freshly cleaved mica for 2 minutes before gently washing with the same buffer. Samples were imaged in a droplet of the buffer using an Asylum Research Cypher ES + BlueDrive AFM with an Olympus AC55 probe and imaged using BlueDrive excitation at the ambient temperature of 31°C within the AFM enclosure. All three samples were imaged with the same probe, consecutively, with the same scan settings, starting with the miR-19-hairpin sample, followed by the R23-hairpin control and the buffer control.
Hybridization experiments in buffer conditions and biofluids
Hybridization experiments were conducted with 2 mg/L of the GT15mir19 sensor in saline-sodium citrate buffer at room temperature. Target DNA or RNA was introduced to reach a final concentration of 1 uM. Samples were incubated for 4 hours, unless otherwise noted. Free energy of hybridization was predicted using OligoAnalyzer 3.1 (IDT). Kinetics experiments were measured every 10 minutes using custom LabView code. Hybridization experiments with sodium dodecylbenzenesulfonate (SDBS) were conducted using a final concentration 0.2% wt/v. SDBS was added to the GT15mir19 sensor and allowed to equilibrate overnight at room temperature before target oligonucleotides were added. Toehold-mediated strand displacement experiments were performed with 1 uM of target miR-19 DNA, and 10 uM of the removing strand, composed of an ssDNA oligonucleotide with the complementary sequence to miR-19. Hybridization experiments in urine were conducted in samples from 5 healthy volunteers and stored on ice until the experiment. Concentrated GT15mir19 was added to each sample to final concentration of 0.2 mg/L and SDBS to final concentration of 0.2%. Concentrated DNA and RNA target were added to the indicated concentrations and incubated at room temperature overnight. Serum experiments used fetal bovine serum (Life Sciences) with GT15mir19 added to final concentration 0.2 mg/L and SDBS at 0.2%. Where indicated, proteinase K (New England Biolabs) was added to a final concentration of 0.5 mg/mL. Spectra was acquired after overnight incubation at room temperature.
Single-nanotube measurements
Single-nanotube measurements were performed by incubating SDS-treated GT15mir19 sensor (0.2% SDS in SSC buffer) on a poly-D-lysine coated glass bottom plate (Mattek, Ashland, MA) for 10 minutes before gently washing with 0.2% SDS in SSC buffer. A final volume of 1 mL SDS-buffer was left in the plate during hyperspectral imaging measurements of the surface-bound nanotubes. A small volume (1 uL) of 1 mM solutions of miR-19 RNA or R23 RNA were then mixed with the buffer. Hyperspectral imaging measurements were repeated after 15 minutes and 50 minutes. Single nanotube emission spectra were collected via a custom near-infrared hyperspectral microscope, as described previously29. Data was processed with ImageJ software. Peaks were fit to Voigt functions using custom Matlab code to obtain center wavelength values.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were conducted using the (9,4) nanotube chirality in explicit water. The DNA molecule for GT15mir19 (without complementary strand) was generated as an unstructured single stranded DNA and placed near the (9,4) nanotube, followed by a sufficiently long equilibration MD simulation enhanced with a replica-exchange based method57, 58 to let the entire strand adsorb on (9,4) nanotube surface. Analysis of an additional 250 ns long MD simulation is presented in the results. The DNA molecule for GT15mir19 hybridized with the complementary strand was created in a partially double stranded form. miR-19 was generated in the double stranded form using NAB program of AmberTools59 and was appropriately bonded via phosphodiester bond to the ss(GT)15 segment of the GT15mir19 DNA. The ss(GT)15 nanotube binding portion of the first strand was adsorbed to the nanotube. The entire DNA and nanotube construct was solvated in a 10.65 × 10.65 × 14.7179 nm water-box containing approximately 55,000 water molecules and 74 sodium counter-ions, placed randomly, to balance the negative charges from phosphates on the DNA. The total system was approximately 170,000 atoms. The nanotube extended to the edges of the water box and was kept frozen in place during the entire equilibration and simulation time. The nanotube atoms were modeled as sp2 hybridized carbon. All structures were visualized in VMD60.
The Gromacs 4.6.7 simulation package61, 62 was used with the Charmm36/TIP3P nucleic acid/water model63. Long-range electrostatics were calculated using the particle mesh Ewald method with a 0.9 nm real space cutoff64. For van der Waals interactions, a cutoff value of 1.2 nm was used. The energy minimized simulation box was then subjected to 100 ps equilibration in an NVT (T=300 K) ensemble where the number of water molecules were fine-tuned to make average pressure approximately equivalent to atmospheric pressure. Further equilibration runs were performed for 100–200 ns in NVT (T=300 K) ensemble. The movie and analysis used the 250 ns production run, followed by equilibration. Systems were propagated with stochastic Langevin dynamics with a time step of 2 fs. The trajectories were saved every 10 ps, yielding a total of 25,000 snapshots for production analysis. VMD Movie Maker was used to create the movie of the trajectory. Homemade python scripts calling MDAnalysis module65 were used for all other analysis presented.
Quantification of DNA on the nanotube complex
The GT15mir19 sequence was used to suspend nanotubes as described earlier. After each of 4 centrifugation filter steps using the Amicon centrifuge filter (100 kDa MWCO), the concentration of the filtered DNA was measured using Abs260 on a NanoDrop spectrophotometer (ThermoScientific, Waltham, MA). The pellet from centrifugation was also filtered to measure free DNA. The final mass of DNA from the combined values was calculated from the concentration and subtracted from the initial value. From three suspensions, we found that 3.5 (+/−1.8) mg of DNA suspended 1 mg of nanotube, matching previous reports of 2.5 to 5 mg of DNA per 1 mg of nanotube66.
Device implantation and in vivo spectroscopy
All animal experiments were approved by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center. KrosFlo Implant Membranes (500 kD MWCO) were obtained from Spectrum Labs (Rancho Dominguez, CA). The membrane was cut to about 1 cm in length and filled with approximately 15 uL of 2 mg/L GT15mir19-nanotubes. Each end was heat sealed. A total of 36 NU/J (nude) mice (Jackson Labs) were anesthetized with 2% isoflurane and implanted with the membrane. Nine mice were divided into three cohorts of three mice to receive miR-19 DNA, R23 DNA, or buffer vehicle via an intraperitoneal injection of 1 nanomole in 1 mL sodium saline citrate buffer. An identical experiment was performed with miR-19 RNA, R23 RNA, or buffer vehicle at 1 nanomole, 500 picomole, 100 picomole, or 50 picomole in 1 mL sodium saline citrate buffer The mice were removed from anesthesia and allowed to regain consciousness. After 90 or 120 minutes, mice were anesthetized and measured using a custom-built reflectance probe-based spectroscopy system. The system consisted of a continuous wave 1 watt 730 nm diode laser (Frankfurt). The laser light was injected into a bifurcated fiber optic reflection probe bundle. The sample leg of the bundle included one 200 μm, 0.22 NA fiber optic cable for sample excitation located in the center of six 200 μm, 0.22 NA fiber optic cables for collection of the emitted light. Emission below 1050 nm was filtered using longpass filters, and the light was focused into the slit of a Czerny-Turner spectrograph with 303 mm focal length (Shamrock 303i, Andor). The slit width of the spectrograph was set at 410 μm. The light was dispersed using a 85 g/mm grating with 1350 nm blaze wavelength and collected with an iDus InGaAs camera (Andor). Spectra were fit to Voigt functions using custom Matlab code.
Supplementary Material
Acknowledgments
This work was supported by the NIH Director’s New Innovator Award (DP2-HD075698), NIH/NCI Cancer Center Support Grant (P30-CA008748), the Center for Molecular Imaging and Nanotechnology, the Louis V. Gerstner Jr. Young Investigator’s Fund, the Experimental Therapeutics Center, the Alan and Sandra Gerry Metastasis Research Initiative, Cycle for Survival, the Frank A. Howard Scholars Program, the Honorable Tina Brozman Foundation for Ovarian Cancer Research, the Byrne Research Fund, the Anna Fuller Fund, Mr. William H. Goodwin and Mrs. Alice Goodwin the Commonwealth Foundation for Cancer Research, and the Imaging and Radiation Sciences Program at Memorial Sloan Kettering Cancer Center. Molecular simulation work was performed at the Lehigh University and is supported by the U.S. Department of Energy (DOE) Office of Science, Basic Energy Sciences (BES), and Division of Material Sciences and Engineering, under Award DE-SC0013979. Use of the high-performance computing capabilities of the Extreme Science and Engineering Discovery Environment (XSEDE) was supported by the National Science Foundation (NSF) under grant number TG-MCB-120014. This research also used resources of the National Energy Research Scientific Computing Center, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. P.V.J was supported by the National Cancer Institute (NCI) Grant NIH T32 Training Grant 2T32CA062948-21. H.A.B. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. RMW was supported by the Ovarian Cancer Research Fund Alliance [Anna Schreiber Mentored Investigator Award 370463]. D.R. was supported by an American Cancer Society 2013 Roaring Fork Valley Research Fellowship. We thank the Molecular Cytology Core Facility at Memorial Sloan Kettering Cancer Center and N. Paknejad for atomic force microscopy measurements.
Footnotes
MATLAB code availability. All MATLAB code is available upon request.
Author Contributions
J.D.H., P.V.J., and D.A.H. conceived the research, designed experiments, and analyzed the data. J.D.H. and H.A.B. performed the experiments. Programs to facilitate data analysis were written by P.V.J. and D.R. Molecular dynamics simulations and analysis were designed and conducted by G.H.Z., D.R., and J.M. Assistance with in vivo work was provided by R.M.W. The probe system for in vivo measurements was designed and built by T.V.G. and D.A.H. The manuscript was prepared and written by J.D.H. and D.A.H.; all authors contributed to editing the manuscript. D.A.H. supervised the project.
Competing Financial Interests
The authors have no competiting financial interests to declare.
Data Availability. The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information. Source data for the figures in this study are available in Figshare with the identifier doi:10.6084/m9.figshare. 4567945 (ref. 67).
References
- 1.Tokuhisa M, Ichikawa Y, Kosaka N, et al. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PloS one. 2015;10:e0130472. doi: 10.1371/journal.pone.0130472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrella P, Zangen R, Sidransky D, Nicol T. Molecular analysis of peritoneal fluid in ovarian cancer patients. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2003;16:636–640. doi: 10.1097/01.MP.0000076979.28106.ED. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. The New England journal of medicine. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Science translational medicine. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nature medicine. 2014;20:430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 7.Deras IL, Aubin SM, Blase A, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. The Journal of urology. 2008;179:1587–1592. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clinical chemistry. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. British journal of haematology. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 10.Yamada Y, Enokida H, Kojima S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer science. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanke M, Hoefig K, Merz H, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urologic oncology. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 12.Snowdon J, Boag S, Feilotter H, Izard J, Siemens R. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Cuaj-Can Urol Assoc. 2013;7:28–32. doi: 10.5489/cuaj.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan YF, Chen HH, Lai PF, et al. MicroRNA-494 Reduces ATF3 Expression and Promotes AKI. Journal of the American Society of Nephrology. 2012;23:2012–2023. doi: 10.1681/ASN.2012050438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung YW, Bae HS, Song JY, et al. Detection of MicroRNA as Novel Biomarkers of Epithelial Ovarian Cancer From the Serum of Ovarian Cancer Patient. International Journal of Gynecological Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 15.Pajek J, Kveder R, Gucek A, et al. Cell-free DNA in the Peritoneal Effluent of Peritoneal Dialysis Solutions. Ther Apher Dial. 2010;14:20–26. doi: 10.1111/j.1744-9987.2009.00717.x. [DOI] [PubMed] [Google Scholar]
- 16.Johnson BN, Mutharasan R. Biosensor-based microRNA detection: techniques, design, performance, and challenges. The Analyst. 2014;139:1576–1588. doi: 10.1039/c3an01677c. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic acids research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker M. MicroRNA profiling: separating signal from noise. Nature methods. 2010;7:687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- 19.Hunt EA, Broyles D, Head T, Deo SK. MicroRNA Detection: Current Technology and Research Strategies. Annual review of analytical chemistry. 2015;8:217–237. doi: 10.1146/annurev-anchem-071114-040343. [DOI] [PubMed] [Google Scholar]
- 20.Dong H, Lei J, Ding L, et al. MicroRNA: function, detection, and bioanalysis. Chemical reviews. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 21.Kruss S, Hilmer AJ, Zhang J, et al. Carbon nanotubes as optical biomedical sensors. Advanced drug delivery reviews. 2013;65:1933–1950. doi: 10.1016/j.addr.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Iverson NM, Barone PW, Shandell M, et al. In vivo biosensing via tissue-localizable near-infrared-fluorescent single-walled carbon nanotubes. Nature nanotechnology. 2013;8:873–880. doi: 10.1038/nnano.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connell MJ, Bachilo SM, Huffman CB, et al. Band gap fluorescence from individual single-walled carbon nanotubes. Science. 2002;297:593–596. doi: 10.1126/science.1072631. [DOI] [PubMed] [Google Scholar]
- 24.Cheong WF, Prahl SA, Welch AJ. A review of the optical properties of biological tissues. IEEE Journal of Quantum Electronics. 1990;26:2166–2185. [Google Scholar]
- 25.Wang F, Dukovic G, Brus LE, Heinz TF. The optical resonances in carbon nanotubes arise from excitons. Science. 2005;308:838–841. doi: 10.1126/science.1110265. [DOI] [PubMed] [Google Scholar]
- 26.Heller DA, Jeng ES, Yeung TK, et al. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 27.Barone PW, Baik S, Heller DA, Strano MS. Near-infrared optical sensors based on single-walled carbon nanotubes. Nature materials. 2005;4:86–92. doi: 10.1038/nmat1276. [DOI] [PubMed] [Google Scholar]
- 28.Cognet L, Tsyboulski DA, Rocha JD, et al. Stepwise quenching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science. 2007;316:1465–1468. doi: 10.1126/science.1141316. [DOI] [PubMed] [Google Scholar]
- 29.Roxbury D, Jena PV, Williams RM, et al. Hyperspectral Microscopy of Near-Infrared Fluorescence Enables 17-Chirality Carbon Nanotube Imaging. Scientific reports. 2015;5:14167. doi: 10.1038/srep14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17–92. Genes & development. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng M, Jagota A, Strano MS, et al. Structure-based carbon nanotube sorting by sequence-dependent DNA assembly. Science. 2003;302:1545–1548. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- 32.Bachilo SM, Strano MS, Kittrell C, et al. Structure-assigned optical spectra of single-walled carbon nanotubes. Science. 2002;298:2361–2366. doi: 10.1126/science.1078727. [DOI] [PubMed] [Google Scholar]
- 33.Campbell JF, Tessmer I, Thorp HH, Erie DA. Atomic force microscopy studies of DNA-wrapped carbon nanotube structure and binding to quantum dots. Journal of the American Chemical Society. 2008;130:10648–10655. doi: 10.1021/ja801720c. [DOI] [PubMed] [Google Scholar]
- 34.Roxbury D, Jena PV, Shamay Y, Horoszko CP, Heller DA. Cell Membrane Proteins Modulate the Carbon Nanotube Optical Bandgap via Surface Charge Accumulation. ACS nano. 2016;10:499–506. doi: 10.1021/acsnano.5b05438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang R, Jin J, Chen Y, et al. Carbon Nanotube-Quenched Fluorescent Oligonucleotides: Probes that Fluoresce upon Hybridization. Journal of the American Chemical Society. 2008;130:8351–8358. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 36.Heller DA, Pratt GW, Zhang JQ, et al. Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8544–8549. doi: 10.1073/pnas.1005512108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore VC, Strano MS, Haroz EH, et al. Individually suspended single-walled carbon nanotubes in various surfactants. Nano letters. 2003;3:1379–1382. [Google Scholar]
- 38.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clinical chemistry. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 39.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 40.Landry MP, Vukovic L, Kruss S, et al. Comparative Dynamics and Sequence Dependence of DNA and RNA Binding to Single Walled Carbon Nanotubes. The journal of physical chemistry. C, Nanomaterials and interfaces. 2015;119:10048–10058. doi: 10.1021/jp511448e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson RR, Johnson AT, Klein ML. The nature of DNA-base-carbon-nanotube interactions. Small. 2010;6:31–34. doi: 10.1002/smll.200901481. [DOI] [PubMed] [Google Scholar]
- 42.Cognet L, Tsyboulski DA, Weisman RB. Subdiffraction far-field imaging of luminescent single-walled carbon nanotubes. Nano letters. 2008;8:749–753. doi: 10.1021/nl0725300. [DOI] [PubMed] [Google Scholar]
- 43.Machinek RR, Ouldridge TE, Haley NE, Bath J, Turberfield AJ. Programmable energy landscapes for kinetic control of DNA strand displacement. Nature communications. 2014;5:5324. doi: 10.1038/ncomms6324. [DOI] [PubMed] [Google Scholar]
- 44.Srinivas N, Ouldridge TE, Sulc P, et al. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic acids research. 2013;41:10641–10658. doi: 10.1093/nar/gkt801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson-Buck A, Su X, Giraldez MD, et al. Kinetic fingerprinting to identify and count single nucleic acids. Nature biotechnology. 2015;33:730–732. doi: 10.1038/nbt.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 50.Joshi GK, Deitz-McElyea S, Liyanage T, et al. Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing at Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS nano. 2015;9:11075–11089. doi: 10.1021/acsnano.5b04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wanunu M, Dadosh T, Ray V, et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nature nanotechnology. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 52.Gunnarsson A, Jonsson P, Marie R, Tegenfeldt JO, Hook F. Single-molecule detection and mismatch discrimination of unlabeled DNA targets. Nano letters. 2008;8:183–188. doi: 10.1021/nl072401j. [DOI] [PubMed] [Google Scholar]
- 53.Schirle NT, MacRae IJ. The Crystal Structure of Human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koshkin AA, Singh SK, Nielsen P, et al. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 56.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 57.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314:141–151. [Google Scholar]
- 58.Bonomi M, Parrinello M. Enhanced Sampling in the Well-Tempered Ensemble. Physical review letters. 2010:104. doi: 10.1103/PhysRevLett.104.190601. [DOI] [PubMed] [Google Scholar]
- 59.Case DA, RMB, Botello-Smith W, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, York DM, Kollman PA. AMBER. 2016;2016 [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Berendsen HJC, Vanderspoel D, Vandrunen R. Gromacs - a Message-Passing Parallel Molecular-Dynamics Implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 62.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 63.Hart K, Foloppe N, Baker CM, et al. Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J Chem Theory Comput. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Essmann U, Perera L, Berkowitz ML, et al. A Smooth Particle Mesh Ewald Method. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- 65.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. Software News and Updates MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. Journal of computational chemistry. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng M, Jagota A, Semke ED, et al. DNA-assisted dispersion and separation of carbon nanotubes. Nature materials. 2003;2:338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]
- 67.Harvey J, et al. Dataset for “A Carbon Nanotube Reporter of miRNA Hybridization Events In Vivo.” figshare. 2017 doi: 10.1038/s41551-017-0041. http://dx.doi.org/10.6084/m9.figshare.4567945. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.