Abstract
Nkx2-5 is a homeobox-containing transcriptional regulator that serves as one of the earliest markers of cardiac lineage commitment. To study the role of Nkx2-5-expressing progenitors at specific time points in cardiac development, we have generated a novel and inducible NKX2-5 mouse line by knocking in a CreER cassette into the Nkx2-5 genomic locus, while preserving the endogenous Nkx2-5 gene to avoid haplo-insufficiency. We evaluated the specificity and efficiency of CreER activity after 4-OHT injection by crossing Nkx2-5CreER/+mice with a Rosa26tdT/+ reporter strain. Our immunohistochemistry results confirmed Cre-induced tdTomato expression specifically in cells expressing Nkx2-5. These cells were mainly cardiomyocytes and were observed in the embryonic heart as early as day 9.5. Additionally, quantitative PCR on postnatal hearts showed enriched expression of Nkx2-5 in isolated tdTomato-expressing cells. No tdTomato expression was observed in Nkx2-5CreER/+;Rosa26tdT/+ mice in the absence of 4-OHT, confirming the inducible nature of CreER activity. The Nkx2-5/CreER mouse model described in this paper will serve as an invaluable tool to trace myocardial lineage and to temporally induce genetic manipulation in a selective population of cardiac progenitors during embryonic development and in the adult heart.
Keywords: Nkx2-5, inducible CreER recombinase, cardiac development
INTRODUCTION
The cardiovascular system plays a vital role in embryogenesis and is the first functional organ within the developing embryo. Heart formation occurs around embryonic day (E) 7.5 in the mouse, when the anterior lateral plate mesoderm commits to a cardiogenic fate in response to endodermal signals (Nascone and Mercola, 1996). These precursors converge along the ventral midline to eventually form the linear heart tube around E8.0. The heart then initiates contractions and regular rhythm is generally established by E9.0.
This process is highly regulated by a complex network of signaling pathways and transcription factors. An important transcription factor that serves as one of the earliest markers of cells committed to a cardiac fate is Nkx2-5 (Lyons et al., 1995). It was originally discovered as a homologue of tinman in Drosopholia, a required gene in heart development (Bodmer et al., 1990). This homeobox-containing transcription factor binds to DNA and regulates the transcription of genes such as myosin light chain 2v (MLC2v), MEF2C, and atrial natriuretic peptide (ANP), which are essential for the embryonic heart to develop beyond the looping morphogenesis stage (Akazawa and Komuro, 2005). Nkx2-5 works in conjunction with other cofactors, such as GATA-4, Tbx5, and Srf to promote the expression of cardiac differentiation and maturation genes (Chen and Schwartz, 1996; Sepulveda et al., 1998). Disruption of these regulatory processes contribute to congenital heart diseases, which holds a significant burden in newborns (van der Bom et al., 2011; van der Linde et al., 2011). Mice that completely lack Srf demonstrate restricted mesoderm formation (Niu et al., 2005) and even heterozygous mutations of the Nkx2-5 gene can result in defects in the cardiac conduction system. In homozygous Nkx2-5 null mice, cardiomyocyte differentiation and linear heart tube formation occurs normally, however, looping morphogenesis is disrupted (Lyons et al., 1995). Further studies on Nkx2-5 found that the gene was expressed in the heart starting at E7.5 and in the spleen, tongue, stomach, and thyroid later during embryonic development, however sustained expression to adulthood occurs only in the heart (Kasahara et al., 1998).
To elucidate the role of Nkx2-5 in cardiac development, a cardiac-specific Nkx2-5-driven Cre-loxp mouse system has been developed (Moses et al., 2001). The Cre-loxp system has been a revolutionary tool for developmental research. This system allows for the targeting of genes through promotor-driven Cre recombinase excision of loxp-flanked regions within the genome, allowing for tissue- or cell-specific gene alterations (Orban et al., 1992). The currently available Nkx2-5 Cre mice have proven to be extremely useful in targeting the majority of Nkx2-5 expressing cells, however, there exist limitations to the types of studies that can be performed using these mice. The Cre-loxp system lacks the ability to temporally control the expression of Cre, thus hindering the ability to study a finite number of Nkx2-5-expressing cells at a given time point in cardiac development.
To overcome these limitations, we have generated a novel Nkx2-5 mouse model utilizing the inducible Cre-ER system. The Cre-ER system uses a Cre that is fused with a mutated ligand-binding domain of a 4-hydroxytamoxifen (4-OHT)-responsive estrogen receptor (Feil et al., 1996; Feil et al., 1997; Metzger et al., 1995; Zhang et al., 1996). Cre recombination occurs only in the presence of 4-OHT which then enables the excision of the loxp-flanked sites. This system provides an additional level of versatility, allowing temporal control of recombinase activity. Herein, we provide a technology report characterizing the inducible expression of Cre-recombinase protein specifically in Nkx2-5 cells at various stages of cardiac development.
RESULTS
Generation of Nkx2-5/CreER mice
The targeting vector Nkx2-5IRESCre was a generous gift from Dr. Richard P. Harvey’s laboratory. It consisted of a gene cassette (IRES-Cre) carrying an internal ribosome entry site linked to the gene encoding a nuclear-localizing recombinase, which was targeted to integrate into the 3′ untranslated region (utr) of the endogenous Nkx2-5 gene. This vector was further modified by replacing the Cre sequence with the CreERT2 downstream of the IRES site. The resulting construction vector included a hygromycin resistance gene cassette (pgk-HYGRO-pA) flanked by yeast flp recombinase target (frt) sites. This entire selection cassette was inserted downstream of IRES-CreERT2 on the targeting vector. The correct targeting occurred at a frequency of ~ 1 in 7 (22/152) hygromycin-resistant ES cell clones. Blastocyst injection of a single correctly targeted clone produced chimeric animals that passed the modified allele through the germline, generating the strain Nkx2-5-IRES-CreERT2-HYGRO. To remove the pgk-HYGRO-pA cassette, founders were crossed with transgenic mice expressing the Flp recombinase gene (FLP1) in germ cells (Figure 1a). Founders of this new strain (Nkx2-5IRESCreERT2) were backcrossed onto C57BL/6 mice. Validation and genotyping of mice were performed by Southern blot analysis and PCR. The resulting transgenic mice were fully viable, healthy, and fertile over several generations. In addition, this strategy resulted in preservation of the endogenous Nkx2-5 gene to avoid haplo-insufficiency, which may inadvertently influence developmental studies. Mice were genotyped by PCR using Cre-for (5′-TGCAGGTTTTGAGCCCTAAC-3′) and Cre-rev (5′-CGAGAATGACTTCCCTGTCC-3′) (Figure 1b).
FIG. 1.
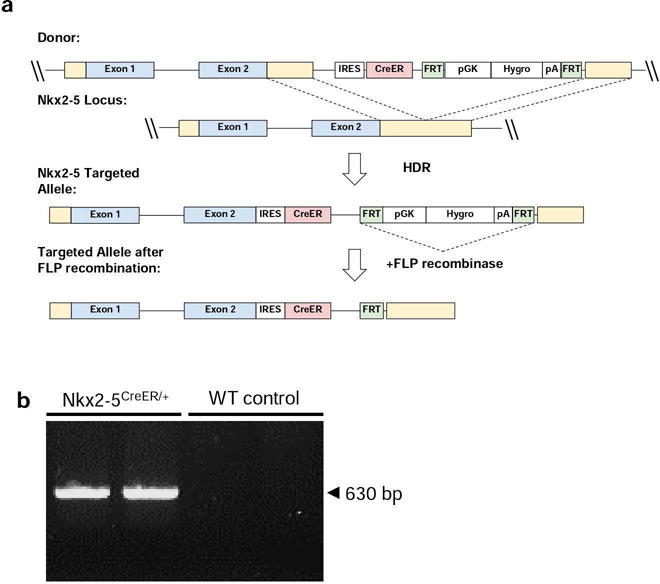
Generation of the Nkx2-5/CreER model. (a) Schematic of the Nkx2-5 wild-type allele, gene targeting construct and resultant mutant alleles before (Nkx2-5IRES-CreER-pgkHYGRO) and after (Nkx2-5IRES-CreER) flp-mediated deletion. (b) DNA gel electrophoresis confirming the presence of the 650 bp Nkx2-5CreER band. Lanes 1 and 2: Nkx2-5CreER/+, Lanes 3 and 4: WT control.
Characterization of Nkx2-5/CreER activity
In order to follow and confirm Nkx2-5/CreER expression, Nkx2-5CreER/+ mice were crossed with Rosa26tdT/+ reporter mice, which contain a loxp-flanked stop cassette preceding a CAG-promoter driven tdTomato gene (Figure 2a). Mice were crossed for timed pregnancies and, at E9.5, dams were injected intraperitoneally with 4-OHT to induce Cre expression in embryos (Figure 2b). Nkx2-5CreER/+;Rosa26tdT/+ embryos demonstrate Cre-induced tdTomato expression as early as E9.5 days. Nkx2-5CreER/+;Rosa26tdT/+ embryos (E16.5) or postnatal (P3) mice that had not been injected with 4-OHT demonstrated no tdTomato expression, confirming the inducible nature of Cre activity (Figure 2c). To confirm specificity of Cre activity, immunohistochemistry for Nkx2-5 on E16.5 embryos was conducted after E9.5 4-OHT injection (Supplementary Figure 1a and Figure 2d). Z-stacking confirmed the co-localization of Nkx2-5 and tdTomato (Supplementary Figure 1b and c), demonstrating that labeled cells do express Nkx2-5. However, we also observed tdTomato+ cells that did not stain positive for Nkx2-5, likely as a result of temporal expression of Nkx2-5. The genetic labeling that occurs from Cre-recombinase activity is a permanent lesion, resulting in the expression of the fluorescent protein in the progeny of the initially labeled cell, even in the absence of Nkx2-5 expression. Additionally, tdTomato+ cells were sorted by fluorescence activated cell sorting (FACS) from P2 pups and quantitative-PCR (qPCR) was conducted for expression of Nkx2-5. Compared to blood samples, tdTomato+ cells exhibited significantly higher expression of Nkx2-5 (Figure 2e). Overall, these results validate the specificity of Nkx2-5/CreER activity through administration of 4-OHT.
FIG. 2.
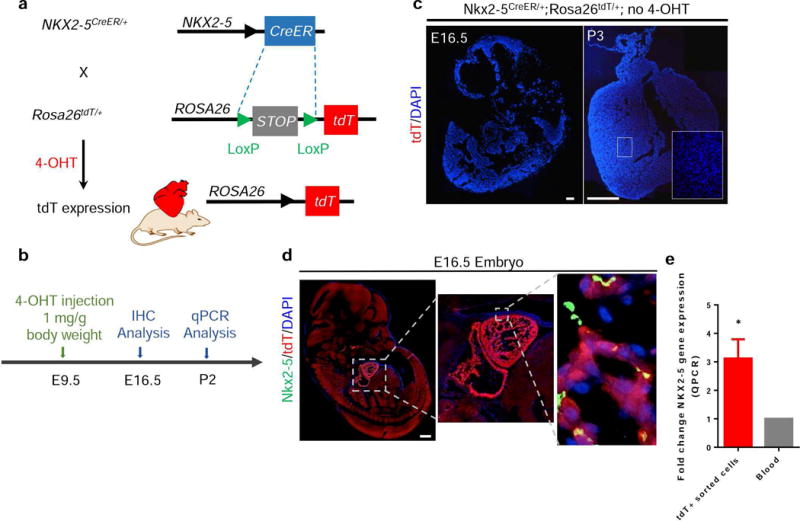
Characterization of Nkx2-5/CreER expression. (a) Schematic of cross-breeding of Nkx2-5CreER/+ with Rosa26tdT/+ mice. (b) Timeline of 4-OHT injection in E9.5 Nkx2-5CreER/+;Rosa26tdT/+ for IHC analysis at E16.5 and qPCR at P2. (c) E16.5 hearts and P3 hearts were analyzed for tdTomato expression in the absence of 4-OHT administration. There was no visible CreER activity. (d) High tdTomato expression is observed in the embryonic heart of mice labeled at E9.5 and analyzed at E16.5. Inset: shows co-localization of Nkx2-5 staining with tdTomato+ cells. (e) qPCR analysis was performed on tdTomato+ sorted cells from P2 hearts showing almost a 3-fold increase in Nkx2-5 expression compared to blood cells. Values are means ± SEM; *P < 0.05, Scale bar = 500 μm.
Tissue Specificity of Nkx2-5/CreER activity
To confirm that Nkx2-5/CreER activity is mainly confined in cardiac tissue during embryonic development, Nkx2-5CreER/+;Rosa26tdT/+ embryos that had been exposed to 4-OHT on E9.5 were isolated at E16.5. Fluorescence microscopy shows that tdTomato expression is seen in the heart, but not in the lung, kidney, liver, spleen, or brain (Figure 3a). Next, we performed a comprehensive time-course analysis of Nkx2-5CreER/+;Rosa26tdT/+ embryos at different embryonic stages (E11.5, E13.5 and E16.5) after 4-OHT injection at E9.5 (Figure 3b). As shown in Figure 3c, tdTomato+ cells mainly contributed to α-sarcomeric actinin+ (α-actinin+) cardiomyocytes within the embryonic hearts, regardless of the time point for analysis. This was further confirmed by qPCR on tdTomato+ cells sorted from P2 neonates, which showed significantly higher levels of α-myosin heavy chain (Myh6) expression, which is specific to cardiomyocytes, compared to blood samples (Supplementary Figure 2a). Quantitative analysis of heart sections over the course of embryonic development showed a significant increase in the percentage of tdTomato+ cardiomyocytes over time, indicating the proliferative nature of tdTomato+ cells during embryonic heart development (Figure 3d). These results show that cells labeled by tdTomato exhibit traits that would be expected of cells expressing Nkx2-5 or progeny of cells that had at one point expressed Nkx2-5.
FIG. 3.
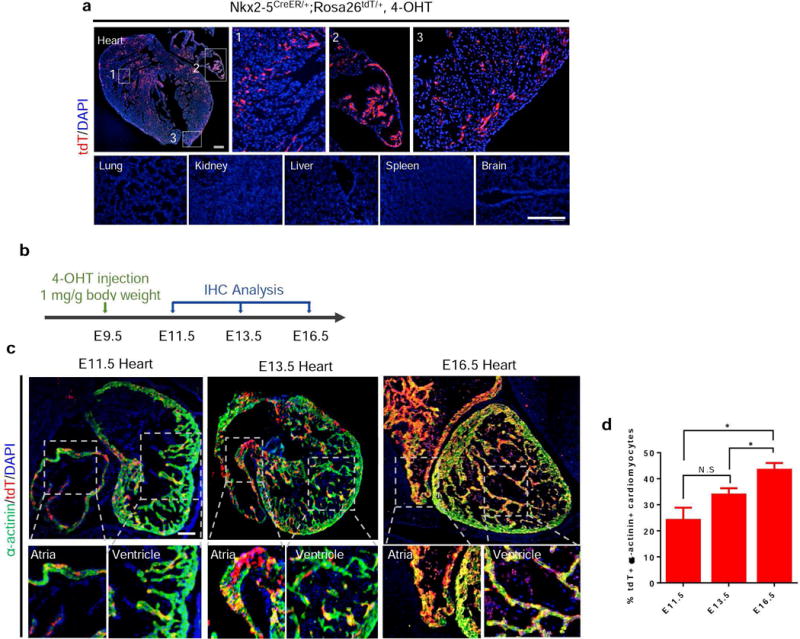
Characterization of Nkx2-5/CreER tissue specificity. (a) In embryos labeled at E9.5 and analyzed at E16.5, expression of tdTomato was observed in the atria, ventricles, and septal regions of the developing heart. No tdTomato expression was observed in the lung, kidney, liver, spleen, or brain. (b) Timeline of 4-OHT injection in E9.5 Nkx2-5CreER/+;Rosa26tdT/+ embryos for time course analysis. (c) Immunohistochemistry of E11.5, E13.5, and E16.5 hearts labeled at E9.5 with α-actinin shows co-localization of tdTomato+ cells with α-actinin. α-actinin+ tdTomato+ cardiomyocytes are found in both the atria and ventricles at each time point of analysis. (d) Quantification of the percentage of α-actinin+ tdTomato+ cardiomyocytes within the tdTomato+ cell population demonstrates a significant increase over the course of embryonic development. Values are means ± SEM; *P < 0.05, NS = non-significant; Scale bar: 500 μm.
To test whether our Nkx2-5/CreER driver is still active at later stages of embryonic development, we injected 4-OHT intraperitoneally at E13.5 and analyzed the heart sections at E16.5 (Supplementary Figure 2b). As shown in Supplementary Figure 2c, tdTomato labeled cells were detected within the heart. We also verified whether recombination that occurs embryonically can be detected in postnatal mice, Nkx2-5CreER/+ mice were crossed with mTmG reporter mice and the dams were injected at E19.5 for postnatal analysis (Supplementary Figure 2d). mTmG is a double fluorescent Cre reporter mouse that expresses membrane-bound tdTomato fluorescent protein prior to Cre mediated excision and, upon Cre recombination, cells permanently express membrane-bound GFP. Nkx2-5CreER/+;Rosa26tdT/+ embryos that were exposed to 4-OHT on E19.5 showed GFP+ recombined cells within the heart at P2 (Supplementary Figure 2e). These results also confirmed that our model is not restricted for use in Rosa26tdT/+ reporter mice. Furthermore, these data indicate that cardiac cells continue to express Nkx2-5 during late stages of embryonic development, which can be labeled and subsequently traced postnatally.
Discussion
In summary, we have shown that the Nkx2-5/CreER mouse model is an effective tool for studying Nkx2-5-expressing cells in an inducible manner. By crossing Nkx2-5CreER/+ mice with Rosa26tdt/+ reporter mice, we have demonstrated the ability to use 4-OHT to induce CreER activity in Nkx-expressing cells during specific time points in development. Upon 4-OHT injection at E9.5, Cre recombinase activity was found to be mainly in the heart and tdTomato+ cells were found to mainly contribute to cardiomyocytes. Patterns of tdTomato expression correlated with what we would expect of Nkx2-5 expression (Kasahara et al., 1998).
The Nkx2-5/CreER mouse model will serve as an invaluable tool for studying cardiogenesis at different time points of development. The inducible nature of this transgenic mouse model allows for targeting of early progenitor cells committed to the cardiac lineage, and can thus be used in multiple applications. As demonstrated by our results, when crossed with a reporter mouse strain, there is effective labeling of Nkx2-5-expressing cells whose progeny can be traced. Use of this mouse model for selective labeling and lineage tracing of cardiac cells at different time points will provide valuable information on the fate of cells originating from Nkx2-5-expressing progenitors. Alternatively, this mouse model can be used to manipulate gene targets in a cell- and time-specific manner. Certain genes are necessary at the beginning stages of embryogenesis and thus full-body knockouts may cause unwarranted phenotypes and lethality. The Nkx2-5/CreER mouse model bypasses these obstacles and allows for a more controlled system for genetic targeting. With this model, it is possible to manipulate the expression of a variety of cardiac-related gene targets such as cardiac-specific transcription factors in order to evaluate their role at specific stages in cardiogenesis. This facilitates a more detailed mapping of the complex transcriptional network that regulates the formation of the embryonic heart. The Nkx2-5/CreER mouse model will serve as an invaluable tool to investigate cells of myocardial lineage during embryonic development.
METHODS
Mice
All procedures were carried out with the approval of the University of California, Los Angeles (UCLA). Rosa26tdT/+ (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) and Rosa26mTmG/+ (Gt (ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J) reporter mice were both obtained from the Jackson Laboratory and have been described previously. Upon publication, Nkx2-5/CreER mice will be available to the academic research community upon request to the Ardehali lab.
4-OHT Preparation and Injection
4-OHT (Sigma, H7904) was first dissolved in absolute ethanol to a concentration of 100 mg/ml by sonication, followed by dilution with corn oil to a final stock concentration of 10 mg/ml. Stock solutions were used within 3 days of preparation. To achieve minimum recombination, one dose of 4-OHT was given through intraperitoneal injection.
Immunohistochemistry
For frozen sections, isolated tissues from murine embryos were fixed in 4% paraformaldehyde for 4 hours and then placed in 30% sucrose overnight in 4°C. Afterwards, the tissues were embedded in OCT (Fisher Healthcare™, catalog # 23-730-571), stored at −80°C, and then cut by a cryostat to 7μm wide sections. These sections were directly used for immunohistochemistry. For paraffin sections, embryos were fixed in 4% paraformaldehyde for 4 hours and then embedded in paraffin. Once cooled to room temperature, the blocks were sectioned 7μm thick. Prior to immunohistochemical staining, sections were deparaffinized by several xylene and ethanol washes and then rehydrated using tap water. Next, slides underwent an antigen retrieval step, where they were boiled (95°C–100°C) in sodium citrate buffer (pH 6) for 10 minutes. For frozen sections, slides were incubated in 0.2% TritonX-100 for 10 minutes at room temperature prior to blocking step. 10% goat serum in PBS with 1% BSA and 0.1% saponin was used for both frozen and paraffin embedded sections for a minimum of an hour in room temperature. The following primary antibodies were added at their respective dilutions: Nkx2-5 (R&D Systems, MAB2444-SP, 1:20), tdTomato (Abcam, ab62341, 1:100), α-actinin (Sigma, A7811, 1:400) in blocking buffer overnight in 4°C. Slides were incubated with the following secondary antibodies for 1 hour at room temperature: Goat Anti-Mouse Alexa Fluor 488 (Invitrogen, A11037, 1:200) and Goat Anti-Rabbit Alexa Fluor 594 (Invitrogen, A11001, 1:200). Washes with PBS-Tween were performed before and after the addition of secondary antibody. Coverslips were then mounted onto slides using DAPI-containing mounting medium (Vector laboratories, catalog #H-1200), and imaged by a Leica inverted fluorescence microscope.
Fluorescence Activated Cell Sorting
Hearts from postnatal mice (day2) were isolated and digested into single cells by incubation with collagenase type II (0.125 U/mL) (Worthington, cat. LS004176) in 37°C for 30 minutes. The cells were pipetted every 10 minutes to ensure complete digestion. The cells were then centrifuged at 300×g for 10 minutes, resuspended in FACS buffer (3% FBS in PBS), and sorted immediately by using a BD FACSAria cell sorter.
cDNA synthesis and Quantitative-PCR
Cells were sorted by FACS directly into Trizol LS (Invitrogen, cat. 10296028) and RNA was extracted following the manufacturer’s protocol. RNA was then quantified by Nanodrop and cDNA was synthesized using the BioRad Reverse Transcriptase kit (BioRad, cat. 1708840). For Q-PCR, SYBR Green mix (BioRad, cat. 1725270) and primers (IDT DNA) targeting respective genes (sequences outlined in Table 1) were added to synthesized cDNA and run through a BioRad cell cycler (BioRad, cat. 1855195).
Table 1.
qPCR Primers
| Gene | Sequence |
|---|---|
| Gapdh | F: AGGTCGGTGTGAACGGATTTG |
| R: TGTAGACCATGTAGTTGAGGTCA | |
| Nkx2-5 | F: ACCTTTAGGAGAAGGGCGATG |
| R: GAGGGTGGGTGTGAAATCTGA | |
| Myh6 | F: ACATGAAGGAGGAGTTTGGG |
| R: GCACTTGGAGCTGTAGGTCA |
Statistical Analysis
Statistical testing was performed with GraphPad Prism version 6. Results are presented as mean ± SEM and were compared using a 2-tailed Student t test (significance was assigned for P < 0.05 and P < 0.0001).
Supplementary Material
SUPPLEMENTARY FIG. 1. Confirmation of Nkx2-5 expression in tdTomato+ cells. (a) Timeline of 4-OHT injection for E16.5 IHC analysis in Nkx2-5CreER/+;Rosa26tdT/+ embryos. (b) Confocal microscopy image of an Nkx2-5CreER/+;Rosa26tdT/+ E16.5 heart stained for Nkx2-5. Z-stacking confirms co-localization (indicated by the yellow due to the green and red overlap) of the cell within the white box. (c) Separate channels showing Nkx2-5 staining, tdTomato fluorescence and DAPI that make up the merged image. The box in the merged image indicates a nucleus that is Nkx2-5+ within a tdTomato+ cell. Scale bar: 10μm.
SUPPLEMENTARY FIG. 2. Nkx2-5/CreER activity in late embryonic development. (a) qPCR comparing fold-expression changes in Myh6 in tdTomato+ cells sorted from P2 Nkx2-5CreER/+;Rosa26tdT/+ neonates compared to blood cells. (b) Timeline of 4OH-T injection in E13.5 Nkx2-5CreER/+;Rosa26tdT/+ embryos for E16.5 analysis. (c) IHC of E16.5 hearts after labeling at E13.5 show significant tdTomato expression throughout the heart. (d) Timeline of 4OH-T injection in E19.5 Nkx2-5CreER/+;Rosa26mTmG/+ embryos for P2 analysis. (e) Analysis of P2 hearts in Nkx2-5CreER/+;Rosa26mTmG/+ mice showed Nkx2-5+ cells labeled GFP within the tdTomato+ myocardium. Values are means ± SEM; **P < 0.0001; Scale bar: 250 μm.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) Director’s New Innovator’s Award (DP2HL127728) (R.A.) Sara Ranjbarvaziri is supported by UCLA Graduate Programs in Bioscience (GPB) and Shuin Park is supported by the Ruth L. Kirschstein National Research Service Award T32HL69766. Ngoc B. Nguyen is supported by the Sarnoff Cardiovascular Research Foundation fellowship.
Grant sponsor: National Institutes of Health (NIH) DP2 HL127728 (R.A.)
Footnotes
Author Information: The authors declare no competing financial interests.
LITERATURE CITED
- Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacology & therapeutics. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan LY, Jan YN. A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development. 1990;110:661. doi: 10.1242/dev.110.3.661. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Molecular and Cellular Biology. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre Recombinase Activity by Mutated Estrogen Receptor Ligand-Binding Domains. Biochemical and Biophysical Research Communications. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and Extracardiac Expression of Csx/Nkx2-5 Homeodomain Protein. Circulation Research. 1998;82:936. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes & Development. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Nascone N, Mercola M. Endoderm and Cardiogenesis. Trends in Cardiovascular Medicine. 1996;6:211–216. doi: 10.1016/S1050-1738(96)00086-2. [DOI] [PubMed] [Google Scholar]
- Niu Z, Yu W, Zhang SX, Barron M, Belaguli NS, Schneider MD, Parmacek M, Nordheim A, Schwartz RJ. Conditional Mutagenesis of the Murine Serum Response Factor Gene Blocks Cardiogenesis and the Transcription of Downstream Gene Targets. Journal of Biological Chemistry. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda JL, Belaguli N, Nigam V, Chen C-Y, Nemer M, Schwartz RJ. GATA-4 and Nkx-2.5 Coactivate Nkx-2 DNA Binding Targets: Role for Regulating Early Cardiac Gene Expression. Molecular and Cellular Biology. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bom T, Zomer AC, Zwinderman AH, Meijboom FJ, Bouma BJ, Mulder BJM. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- van der Linde D, Konings EEM, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJM, Roos-Hesselink JW. Birth Prevalence of Congenital Heart Disease Worldwide: A Systematic Review and Meta-Analysis. Journal of the American College of Cardiology. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Research. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIG. 1. Confirmation of Nkx2-5 expression in tdTomato+ cells. (a) Timeline of 4-OHT injection for E16.5 IHC analysis in Nkx2-5CreER/+;Rosa26tdT/+ embryos. (b) Confocal microscopy image of an Nkx2-5CreER/+;Rosa26tdT/+ E16.5 heart stained for Nkx2-5. Z-stacking confirms co-localization (indicated by the yellow due to the green and red overlap) of the cell within the white box. (c) Separate channels showing Nkx2-5 staining, tdTomato fluorescence and DAPI that make up the merged image. The box in the merged image indicates a nucleus that is Nkx2-5+ within a tdTomato+ cell. Scale bar: 10μm.
SUPPLEMENTARY FIG. 2. Nkx2-5/CreER activity in late embryonic development. (a) qPCR comparing fold-expression changes in Myh6 in tdTomato+ cells sorted from P2 Nkx2-5CreER/+;Rosa26tdT/+ neonates compared to blood cells. (b) Timeline of 4OH-T injection in E13.5 Nkx2-5CreER/+;Rosa26tdT/+ embryos for E16.5 analysis. (c) IHC of E16.5 hearts after labeling at E13.5 show significant tdTomato expression throughout the heart. (d) Timeline of 4OH-T injection in E19.5 Nkx2-5CreER/+;Rosa26mTmG/+ embryos for P2 analysis. (e) Analysis of P2 hearts in Nkx2-5CreER/+;Rosa26mTmG/+ mice showed Nkx2-5+ cells labeled GFP within the tdTomato+ myocardium. Values are means ± SEM; **P < 0.0001; Scale bar: 250 μm.


