Abstract
The fidelity of protein biosynthesis rests largely on the correct aminoacylation of transfer RNAs by their cognate aminoacyl-tRNA synthetases. Previous studies have demonstrated that the interaction of Escherichia coli tRNA(Gln) with glutaminyl-tRNA synthetase (GlnRS) provides an excellent system for studying the basis of this highly specific recognition process. Correct aminoacylation depends on the set of nucleotides (identity elements) in tRNA(Gln) responsible for correct interaction with GlnRS. Specific contacts between tRNA(Gln) and GlnRS include the 2-amino group of guanosines. Therefore, we made a set of tRNA(Gln) variants in which specific guanosines were replaced by inosine using recombinant RNA technology. This resulted in a set of tRNAs that varied by single deletions of the amino group from guanine residues, thus allowing us to test the functional importance of these contacts. In addition, a number of mutants were made by transcription of mutated tRNA genes with base changes at position 10, 16 or 25. In vitro aminoacylation of these mutants showed decreases in the specificity constant (kcat/KM) of up to 300-fold, with kcat being the parameter most affected. These experiments reveal G10 as a new element of glutamine identity. In addition, the interaction of G2, G3 and G10 with GlnRS via the 2-amino group is significant for tRNA discrimination. Based on these results, and on earlier data, we propose a complete set of bases as identity elements for tRNA(Gln).
Full text
PDF
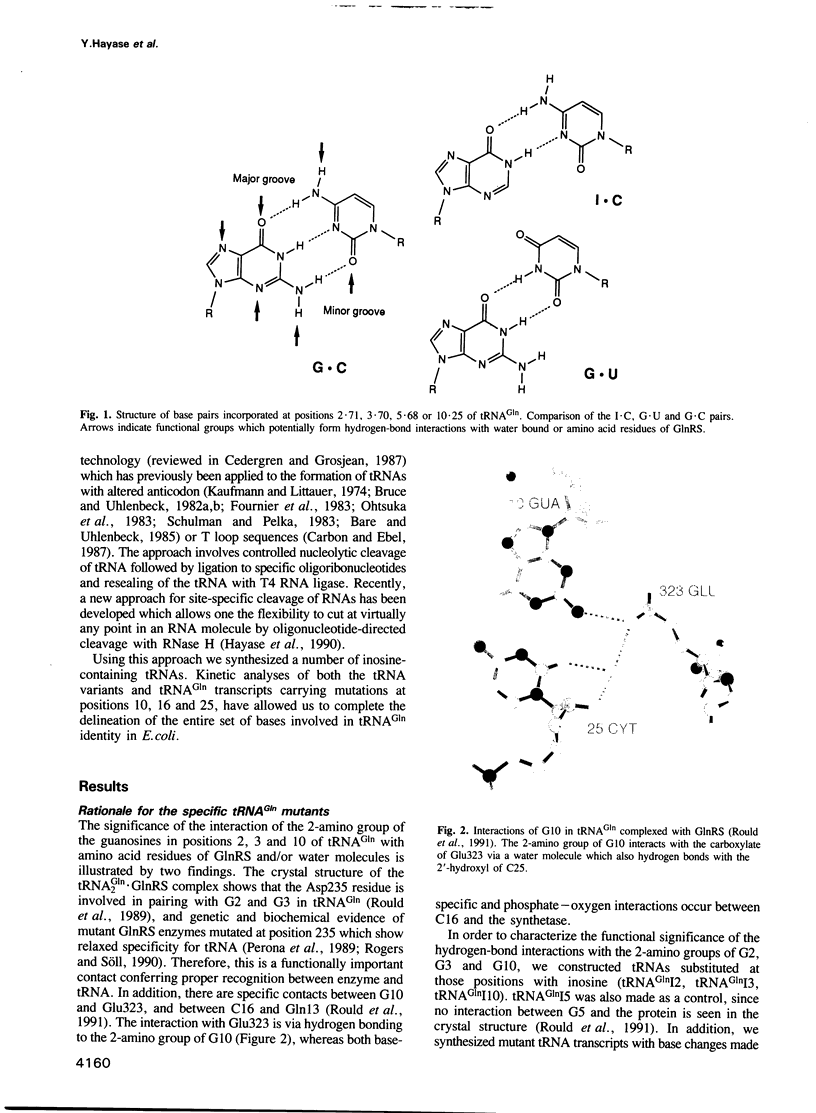
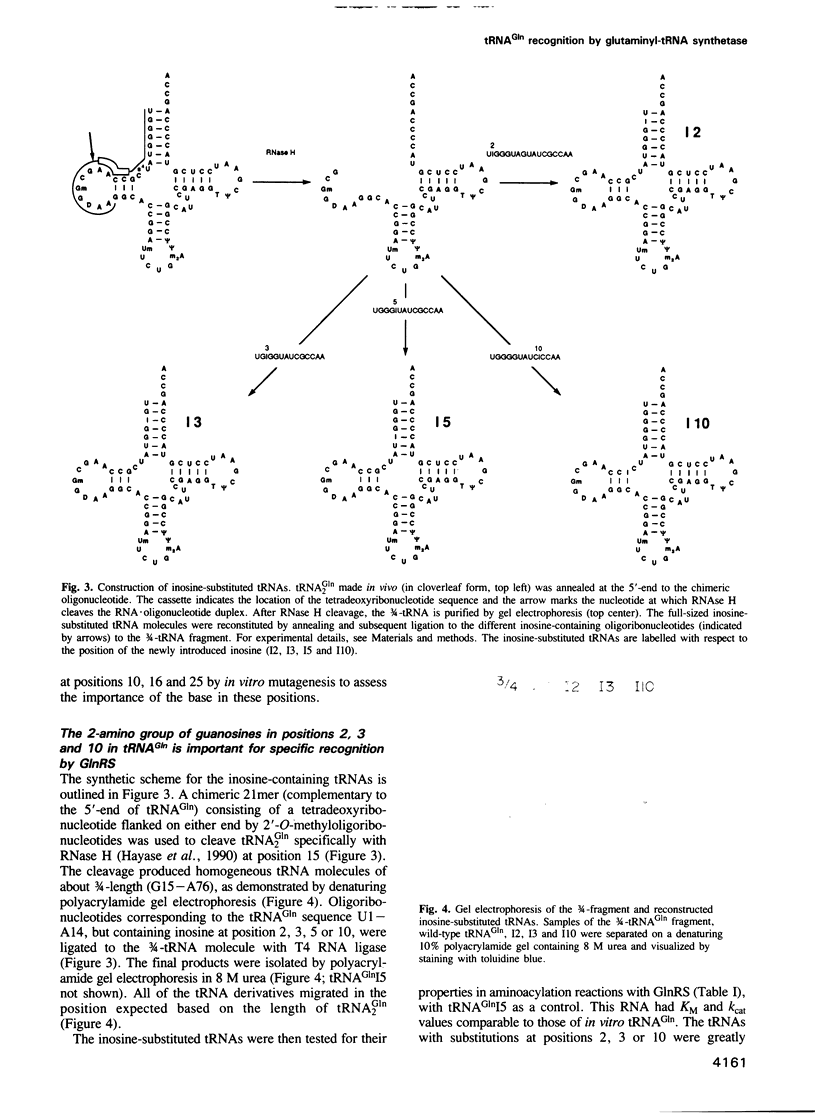

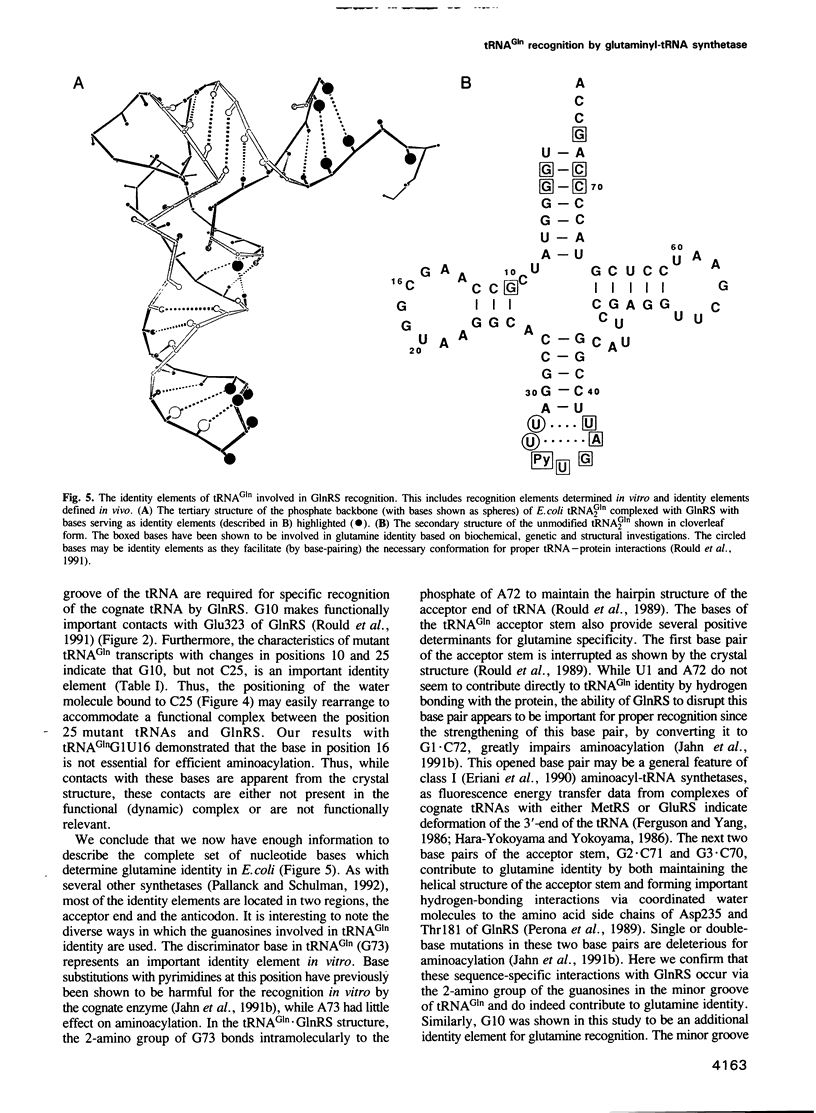


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bare L., Uhlenbeck O. C. Aminoacylation of anticodon loop substituted yeast tyrosine transfer RNA. Biochemistry. 1985 Apr 23;24(9):2354–2360. doi: 10.1021/bi00330a034. [DOI] [PubMed] [Google Scholar]
- Bedouelle H. Recognition of tRNA(Tyr) by tyrosyl-tRNA synthetase. Biochimie. 1990 Aug;72(8):589–598. doi: 10.1016/0300-9084(90)90122-w. [DOI] [PubMed] [Google Scholar]
- Beresten S., Jahn M., Söll D. Aminoacyl-tRNA synthetase-induced cleavage of tRNA. Nucleic Acids Res. 1992 Apr 11;20(7):1523–1530. doi: 10.1093/nar/20.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Specific interaction of anticodon loop residues with yeast phenylalanyl-tRNA synthetase. Biochemistry. 1982 Aug 17;21(17):3921–3926. doi: 10.1021/bi00260a003. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P. In vitro construction of yeast tRNAAsp variants: nucleotide substitutions and additions in T-stem and T-loop. Nucleic Acids Res. 1987 Mar 11;15(5):1933–1950. doi: 10.1093/nar/15.5.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergren R., Grosjean H. RNA design by in vitro RNA recombination and synthesis. Biochem Cell Biol. 1987 Aug;65(8):677–692. doi: 10.1139/o87-090. [DOI] [PubMed] [Google Scholar]
- Chattapadhyay R., Pelka H., Schulman L. H. Initiation of in vivo protein synthesis with non-methionine amino acids. Biochemistry. 1990 May 8;29(18):4263–4268. doi: 10.1021/bi00470a001. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Ferguson B. Q., Yang D. C. Topographic modeling of free and methionyl-tRNA synthetase bound tRNAfMet by singlet-singlet energy transfer: bending of the 3'-terminal arm in tRNAfMet. Biochemistry. 1986 Oct 21;25(21):6572–6578. doi: 10.1021/bi00369a035. [DOI] [PubMed] [Google Scholar]
- Fournier M., Haumont E., de Henau S., Gangloff J., Grosjean H. Post-transcriptional modification of the wobble nucleotide in anticodon-substituted yeast tRNAArgII after microinjection into Xenopus laevis oocytes. Nucleic Acids Res. 1983 Feb 11;11(3):707–718. doi: 10.1093/nar/11.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Fu D. J., McLaughlin L. W. Importance of specific purine amino and hydroxyl groups for efficient cleavage by a hammerhead ribozyme. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3985–3989. doi: 10.1073/pnas.89.9.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Szostak J. W., Benner S. A., Rich A., Usman N. Synthesis of RNA containing inosine: analysis of the sequence requirements for the 5' splice site of the Tetrahymena group I intron. Nucleic Acids Res. 1991 Aug 11;19(15):4161–4166. doi: 10.1093/nar/19.15.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Yokoyama M., Yokoyama S., Miyazawa T. Conformation change of tRNAGlu in the complex with glutamyl-tRNA synthetase is required for the specific binding of L-glutamate. Biochemistry. 1986 Nov 4;25(22):7031–7036. doi: 10.1021/bi00370a041. [DOI] [PubMed] [Google Scholar]
- Hayase Y., Inoue H., Ohtsuka E. Secondary structure in formylmethionine tRNA influences the site-directed cleavage of ribonuclease H using chimeric 2'-O-methyl oligodeoxyribonucleotides. Biochemistry. 1990 Sep 18;29(37):8793–8797. doi: 10.1021/bi00489a041. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Miura K. I. Functional sites in transfer ribonucleic acid. Nature. 1966 Jan 22;209(5021):376–378. doi: 10.1038/209376a0. [DOI] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. Conversion of aminoacylation specificity from tRNA(Tyr) to tRNA(Ser) in vitro. Nucleic Acids Res. 1990 Dec 11;18(23):6815–6819. doi: 10.1093/nar/18.23.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoben P., Söll D. Glutaminyl-tRNA synthetase of Escherichia coli. Methods Enzymol. 1985;113:55–59. doi: 10.1016/s0076-6879(85)13011-9. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Imura N., Weiss G. B., Chambers R. W. Reconstitution of alanine acceptor activity from fragments of yeast tRNA-Ala II. Nature. 1969 Jun 21;222(5199):1147–1148. doi: 10.1038/2221147a0. [DOI] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Imura A., Iwai S., Miura K., Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2'-O-methyl)ribonucleotides. Nucleic Acids Res. 1987 Aug 11;15(15):6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Littauer U. Z. Covalent joining of phenylalanine transfer ribonucleic acid half-molecules by T4 RNA ligase. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3741–3745. doi: 10.1073/pnas.71.9.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R., Caruthers M. H., Longfellow C. E., Swinton D., Turner D. H., Freier S. M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986 Dec 2;25(24):7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Seong B. L., RajBhandary U. L. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J Biol Chem. 1991 Sep 25;266(27):18012–18017. [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a "variable pocket". Science. 1988 Sep 30;241(4874):1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Rapid determination of nucleotides that define tRNA(Gly) acceptor identity. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Nucleotides that contribute to the identity of Escherichia coli tRNA(Phe). J Mol Biol. 1988 Aug 20;202(4):697–709. doi: 10.1016/0022-2836(88)90551-7. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Schimmel P. Functional contacts of a transfer RNA synthetase with 2'-hydroxyl groups in the RNA minor groove. Nature. 1992 Jun 11;357(6378):513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991 Aug 16;253(5021):784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Doi T., Fukumoto R., Matsugi J., Ikehara M. Modification of the anticodon triplet of E.coli tRNAMetf by replacement with trimers complementary to non-sense codons UAG and UAA. Nucleic Acids Res. 1983 Jun 25;11(12):3863–3872. doi: 10.1093/nar/11.12.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona J. J., Swanson R. N., Rould M. A., Steitz T. A., Söll D. Structural basis for misaminoacylation by mutant E. coli glutaminyl-tRNA synthetase enzymes. Science. 1989 Dec 1;246(4934):1152–1154. doi: 10.1126/science.2686030. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Rogers M. J., Adachi T., Inokuchi H., Söll D. Switching tRNA(Gln) identity from glutamine to tryptophan. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3463–3467. doi: 10.1073/pnas.89.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Inaccuracy and the recognition of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:185–208. doi: 10.1016/s0079-6603(08)60627-3. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon loop size and sequence requirements for recognition of formylmethionine tRNA by methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6755–6759. doi: 10.1073/pnas.80.22.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Sherman J. M., Rogers M. J., Söll D. Competition of aminoacyl-tRNA synthetases for tRNA ensures the accuracy of aminoacylation. Nucleic Acids Res. 1992 Jun 11;20(11):2847–2852. doi: 10.1093/nar/20.11.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S., Mukai S., Nishihara T., Inoue H., Ohtsuka E., Morisawa H. Site-directed cleavage of RNA. Nucleic Acids Res. 1987 Jun 11;15(11):4403–4415. doi: 10.1093/nar/15.11.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. B., Hawley D. K. TFIID binds in the minor groove of the TATA box. Cell. 1991 Dec 20;67(6):1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Swanson R., Hoben P., Sumner-Smith M., Uemura H., Watson L., Söll D. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988 Dec 16;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- Söll D. The accuracy of aminoacylation--ensuring the fidelity of the genetic code. Experientia. 1990 Dec 1;46(11-12):1089–1096. doi: 10.1007/BF01936918. [DOI] [PubMed] [Google Scholar]




