SUMMARY
Almost all known forms of fast chemical synaptic transmission require the synaptic hub protein Munc13. This essential protein has also been implicated in mediating several forms of use-dependent plasticity, but the mechanisms by which it controls vesicle fusion and plasticity are not well understood. Using the C. elegans Munc13 ortholog UNC-13, we show that deletion of the C2B domain, the most highly conserved domain of Munc13, enhances calcium-dependent exocytosis downstream of vesicle priming, revealing a novel autoinhibitory role for the C2B. Furthermore, C2B inhibition is relieved by calcium binding to C2B, while the neighboring C1 domain acts together with C2B to stabilize the autoinhibited state. Selective disruption of Munc13 autoinhibition profoundly impacts nervous system function in vivo. Thus, C1–C2B exerts a basal inhibition on Munc13 in the primed state, permitting calcium- and lipid-dependent control of C1–C2B to modulate synaptic strength.
INTRODUCTION
Synaptic transmission exemplifies a highly regulated mode of vesicle fusion, and its proper modulation ensures optimal functioning of the nervous system. The fusion of neurotransmitter-containing vesicles with the presynaptic plasma membrane is powered by the assembly of the neuronal SNAREs: Synaptobrevin/VAMP on the synaptic vesicle (SV), and SNAP-25 and Syntaxin on the plasma membrane (Jahn and Fasshauer, 2012; Südhof and Rothman, 2009). Several proteins coordinate this process, including Munc13, Munc18, NSF, αSNAP, Synaptotagmin, and Complexin (Chapman, 2008; Rizo and Südhof, 2012; Rizo and Xu, 2015; Südhof, 2012, 2013). Two factors control the amount of neurotransmitter released by an action potential: the number of available or “primed” SVs (denoted as the readily releasable pool, or RRP) and the release probability of a given SV (Pvr) (Neher, 2015). Both of these factors are influenced by the concentration of presynaptic calcium, and both are substrates for modulation and use-dependent plasticity of the synapse (Fioravante and Regehr, 2011). What are the likely candidate proteins that control these aspects of synaptic transmission? One promising candidate is Munc13, which regulates RRP size by catalyzing SV priming (Richmond et al., 1999; Rizo and Xu, 2015). In addition, Munc13 affects post-priming aspects of SV fusion such as Pvr, and has been proposed to drive superpriming (Lee et al., 2013; Lou et al., 2008; Madison et al., 2005; McEwen et al., 2006). Finally, Munc13 is also required for certain calcium-dependent forms of synaptic plasticity and neuromodulatory enhancement of synaptic strength through lipid signaling pathways (de Jong and Fioravante, 2014; Lipstein et al., 2013; Rhee et al., 2002). Revealing the mechanistic foundation of Munc13 function will therefore shed light on many key aspects of synaptic transmission and its plasticity.
Munc13 is required for almost all known forms of chemical synaptic communication in both vertebrates and invertebrates (Augustin et al., 1999; Brose et al., 1995; Richmond et al., 1999; Rizo and Südhof, 2012), and its Syntaxin-interacting MUN domain is essential for Munc13 function (Basu et al., 2005; Liu et al., 2016; Madison et al., 2005; Stevens et al., 2005). Several conserved protein domains flank the MUN domain and are thought to mediate interactions with lipids, calcium, and other proteins as a means of controlling Munc13 function and differentially regulating distinct SV pools (Hu et al., 2013; Rizo and Xu, 2015; Zhou et al., 2013). The most conserved of these modules is a tandem C1–C2 domain N-terminally adjacent to the MUN domain and present in all known neuronal Munc13 homologs, sharing about 70% sequence identity between rat and C. elegans. Much like the C1–C2 tandem domains of conventional PKCs, the C1 domain of Munc13 binds diacylglycerol (DAG) and beta phorbol esters (β-PEs) (Betz et al., 1998; Igumenova, 2015; Rhee et al., 2002; Shen et al., 2005). The neighboring C2 domain (C2B) binds calcium and anionic phospholipids with a preference for both PI(4)P and PI(4,5)P2 (Shin et al., 2010).
Despite the work done on the C1 and C2B domains of Munc13, the mechanisms by which these modulatory domains alter Munc13 function remain enigmatic. The structure of the C1–C2B tandem domain together with the MUN domain was recently determined, and mutations designed to disrupt several domain interfaces revealed a variety of effects on synaptic transmission and short-term plasticity (Xu et al., 2017), highlighting the functional importance of these regulatory domains. Using a combination of genetics, biochemistry, and electrophysiology, we found that the C1–C2B module mediates a previously unknown inhibitory function in the C. elegans Munc13 ortholog UNC-13. In particular, removal of either the C1 or C2B domain enhanced calcium-triggered synaptic transmission whereas deletion of both domains almost eliminated synaptic transmission. Moreover, calcium liganding to C2B released autoinhibition, while a highly conserved linker between the C2B and MUN domains played a crucial role in maintaining the C1–C2B autoinhibition. The specific loss of UNC-13 autoinhibition significantly disrupted nervous system function, highlighting its physiological relevance. We propose that a combination of elevated calcium, PIP/PIP2 levels, and DAG production boosts Munc13 activity downstream of vesicle priming and enhances secretion by releasing C1–C2B from its inhibitory conformation. This calcium- and lipid-dependent relief of inhibition conveys a direct coupling of activity and modulatory signaling to SV fusion, and provides mechanistic insight into the post-priming roles of Munc13 in controlling synaptic strength.
RESULTS
The UNC-13 C2B Domain Inhibits Calcium-Triggered Neurotransmitter Release
Of the six well-defined protein domains of Munc13-1/2, C2B shows the highest sequence conservation, with over 80% identity and 95% similarity to the C. elegans ortholog, UNC-13 (Figures 1A and S1A). In particular, all five aspartates required for calcium binding are also conserved across phylogeny (Figures 1B and S1B). To investigate the role of C2B in regulation of Munc13 function in vivo, a unc-13 null mutant C. elegans strain was rescued with a single copy of either full-length UNC-13 (scUNC-13(+)) or a version of the protein lacking its C2B domain (ΔC2B) expressed throughout the nervous system. The long isoform (UNC-13L) was chosen since this isoform suffices to restore synaptic function and behavior (Hu et al., 2013). Nervous system function was assessed by monitoring the locomotion of individual animals moving on an agar plate. unc-13(s69) null mutants were completely immobile whereas single-copy neuronal expression of UNC-13 in the null mutant restored wild-type (WT) locomotion (Figures 1C and 1D). Rescue of the null mutant with ΔC2B partially restored locomotion, suggesting that UNC-13 function is impaired in the absence of C2B. Note that locomotor speed is not directly related to NMJ synaptic strength as both increases and decreases in acetylcholine (ACh) secretion have been correlated with impaired locomotion (Martin et al., 2011; McEwen et al., 2006; Richmond et al., 2001; Wang et al., 2001).
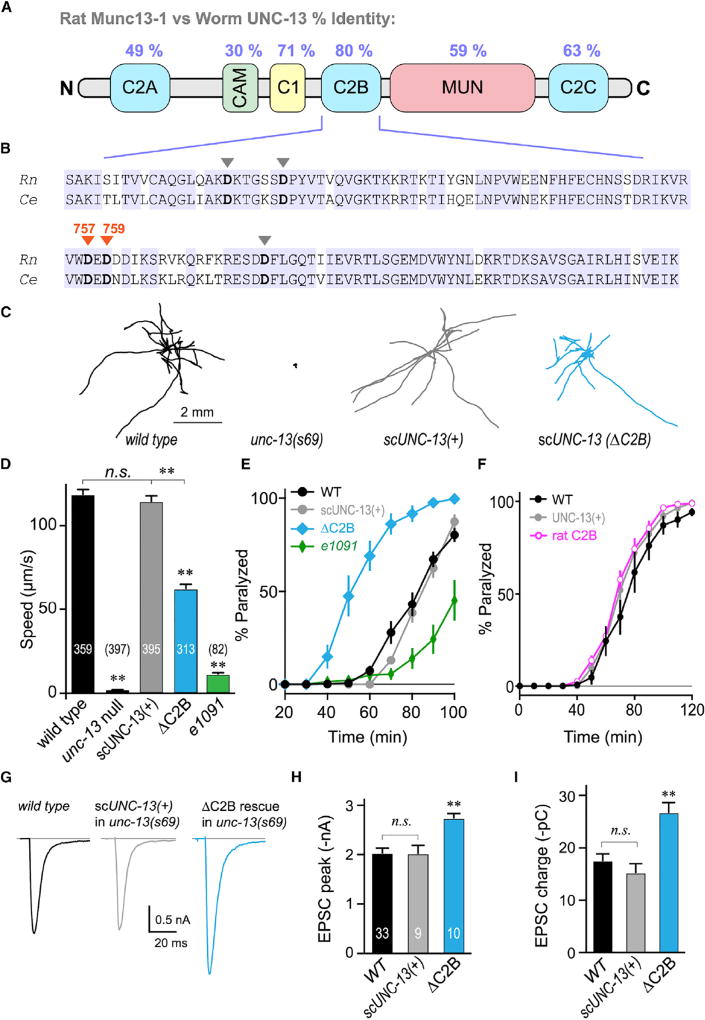
Figure 1. The UNC-13 C2B Domain Is Required for Normal Locomotion and Inhibits ACh Secretion.
(A) Cartoon depicting the six major protein domains of the canonical Munc13 protein along with protein sequence identity between rat and worm.
(B) C2B sequence alignment for rat Munc13-1 and worm UNC-13L. Identical residues are highlighted (blue) and the five calcium-binding aspartates are indicated with arrowheads. The two loop 3 aspartates mutated in this study are specified (orange).
(C) Ten representative locomotion trajectories collected over a 1 min interval are plotted for WT (black), unc-13(s69) null mutant, unc-13(s69) rescued with a single-copy integrant of full-length UNC-13 (scUNC-13(+); gray), and unc-13(s69) rescued with a single-copy UNC-13 integrant lacking its C2B domain (ΔC2B; blue).
(D) Average locomotion rates are compared for the same four strains as well as the hypomorph unc-13(e1091). Note that scUNC-13(+) and ΔC2B are expressed in the unc-13(s69) null background. The number of animals analyzed is indicated in the graph.
(E) Average paralysis time courses for wild-type (WT; black), unc-13(s69) expressing a full-length rescue (scUNC-13(+); gray), unc-13(e1091) hypomorph (green), and unc-13(s69) expressing UNC-13 lacking its C2B domain (ΔC2B; blue).
(F) Average paralysis time courses for wild-type (WT; black) and unc-13(69) rescued with an extrachromosomal array of either full-length UNC-13 (UNC-13(+); gray) or UNC-13 containing the rat Munc13-1 C2B domain (rat C2B; pink).
(G) Average traces of stimulus-evoked EPSCs for WT (black), single-copy rescue in unc-13(s69) (scUNC-13(+); gray), and ΔC2B rescue in unc-13(s69) (ΔC2B; blue).
(H and I) Average EPSC peak amplitude (H) and cumulative charge transfer (I) for the same three genotypes.
Errors bars are mean ± SEM (**p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer test for multiple comparisons). Strains: N2, BC168, CB1091, JSD805, JSD925, JSD1038, and JSD1039.
The impact of the C2B domain on ACh secretion in vivo was assessed using acute sensitivity to the cholinesterase inhibitor aldicarb by quantifying the rate of paralysis upon exposure to 1 mM aldicarb. Numerous studies have established that impairment of ACh release decreases sensitivity to aldicarb and slows the rate of paralysis, whereas hypersecretory mutations accelerate paralysis (Mahoney et al., 2006; Martin et al., 2011; Miller et al., 1996; Nurrish et al., 1999; Rand and Russell, 1985). Because unc-13 null mutants do not move, they cannot be evaluated by this assay. However, rescue of the unc-13 null allele with scUNC-13(+) fully restored WT aldicarb sensitivity, while the partial loss-of-function unc-13(e1091) mutant paralyzed more slowly (Figure 1E). Synaptic ACh release is reduced by more than 90% in the unc-13(e1091) mutant, consistent with the delayed paralysis observed here (Hu et al., 2013; Richmond et al., 1999). Remarkably, ΔC2B transgenic animals paralyzed significantly faster than WT, suggestive of an enhancement of ACh secretion. Replacement of worm C2B with rat Munc13-1 C2B completely restored WT secretory function in vivo, further highlighting the profound conservation of function between these two distantly related orthologs (Figure 1F). Rescue of unc-13 with a single-copy integrated transgene or overexpression using an extrachromosomal array produced the same degree of rescue, so the mode of transgene expression does not alter the impact of C2B on synaptic function (Figures S2D–S2F). Thus, locomotion is considerably impaired in ΔC2B animals, and this disruption of proper nervous system function originates from excessive rather than insufficient neurotransmitter secretion.
We examined the specific impact of C2B on calcium-triggered fusion by recording stimulus-evoked excitatory postsynaptic currents (EPSCs) at the neuromuscular junction in dissected WT versus UNC-13(ΔC2B) animals in 1 mM external calcium (Figures 1G and S2A–S2C). Both the peak current amplitude and the total charge elicited by a single stimulus were significantly increased in the absence of the C2B domain, supporting the finding that synaptic transmission is enhanced in the absence of this critical protein domain (Figures 1H and 1I). Furthermore, the decay kinetics of the average EPSC were slower in the absence of C2B as a result of increased dispersion of the individual release events since there was no difference in the rise and decay kinetics of WT and ΔC2B unitary EPSCs (Figures S2D–S2F, 3E, and 3F) (Diamond and Jahr, 1995). Notably, the increased secretion observed in ΔC2B animals did not appear to arise from a significant increase in the pool of primed SVs since there was no relative difference in sucrose-triggered secretion between synapses rescued with scUNC-13(+) versus ΔC2B (Figures S2I–S2K). In contrast, loss of unc-13 almost entirely eliminated the primed vesicle pool as previously observed (Madison et al., 2005; Richmond et al., 1999; Zhou et al., 2013). Thus, the increased fusion observed in ΔC2B animals appeared to originate from a post-priming function of UNC-13. Behavioral assays performed while electrically silencing the motor neurons further indicated that calcium-triggered secretion was enhanced in ΔC2B animals (Figure S3). Taken together, these in vivo and in vitro synaptic assays indicate an unexpected enhancement of neurotransmitter secretion when the C2B domain of UNC-13 was removed, and the dysregulation of UNC-13 significantly impacted nervous system function.
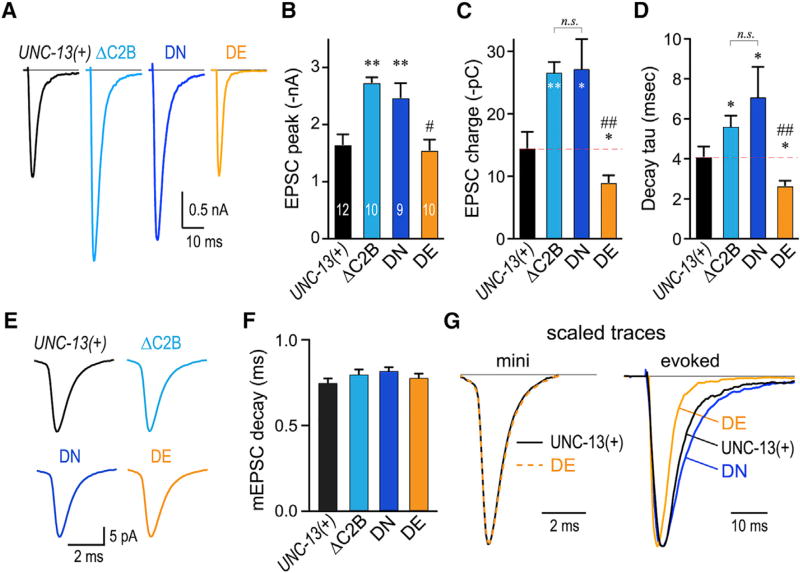
Figure 3. Calcium-Triggered Release Is Inhibited by the Calcium-Free Mimetic C2B and Stimulated by Calcium-Bound C2B Mimetic.
(A) Averaged stimulus-evoked EPSC traces for unc-13(s69) animals rescued with transgenes encoding full-length UNC-13 (UNC-13(+); black), ΔC2B (light blue), worm DN (navy), or worm DE (orange).
(B–D) Summary of peak currents (B), cumulative charge (C), and EPSC decay time constants (D) for all four genotypes.
(E) Average miniature EPSC (mEPSC) traces.
(F) Summary of average mEPSC decay time constant (single exponential fit) for the four genotypes.
(G) Average UNC-13(+) (black) and DE (orange) rescue mEPSC traces (left) and average evoked EPSC (right) for same transgenic animals scaled and superimposed. DN (navy) is also superimposed for comparison. Note the difference in scale bar time.
Errors bars are mean ± SEM with experiment numbers included in bars. *p < 0.05, **p < 0.01; n.s., not significant; #, significant difference from ΔC2B and DN, but not UNC-13(+) with p < 0.01; ##; significant difference from ΔC2B and DN with p < 0.01. Multiple comparisons using ANOVA and Tukey-Kramer statistics. Strains: JSD805, JSD835, JSD849, and JSD1038.
Mutations in C2B Calcium-Binding Loop 3 Mimic the Calcium-Bound and Calcium-Free States
The Munc13-1/2 C2B domain binds anionic phospholipid membranes containing PI(4)P and PI(4,5)P2 in a calcium-dependent manner (Shin et al., 2010), so calcium may modify the inhibitory function of C2B by translocating this region of Munc13 to the plasma membrane or altering its mode of membrane binding. Prior studies have identified specific structural changes within mammalian C2B upon calcium binding via five crucial aspartate residues distributed along two regions termed loop 1 and loop 3 (Figure 2A) (Shin et al., 2010). These five aspartates are perfectly conserved in C2B across all known Munc13 orthologs (Figure S1B), and they coordinate two calcium ions in the bound state. Two of the aspartates in loop 1 (D705 and D711) have been mutated in previous studies, while less is known about the three aspartates in loop 3 (Shin et al., 2010). Of particular interest, the major structural change in the bound state of C2B is the formation of a short alpha helix in loop 3 (Figure 2A). To explore the impact of calcium liganding in vivo, we first sought to create novel C2B variants that mimicked either the calcium-liganded or calcium-free conformational state of loop 3 using a combination of in silico modeling and in vitro biochemistry. Two neighboring aspartates on loop 3 (D757 and D759) interact with both bound calcium ions, and protein structure prediction software (Kelley et al., 2015) indicated that mutations in these residues would impact loop 3 helix formation. In particular, we chose to replace these aspartates with glutamates to preserve the net electrostatic charge in this region of C2B while simultaneously eliminating the calcium-binding pocket. A similar strategy was reported for synaptotagmin 1 C2A (Striegel et al., 2012). Computational predictions of secondary structure suggested that the glutamate substitutions favor the calcium-unbound state. In contrast, replacing the same loop 3 aspartates with neutralizing asparagines was predicted to stabilize the bound conformation, potentially mimicking the calcium-liganded state both structurally and electrostatically (Figure S4). To assess these predictions biochemically, recombinant WT rat Munc13-1 C2B domain (WT) was compared with glutamate and asparagine substitutions in loop 3 (respectively referred to as the DE or DN mutants hereafter). Circular dichroism (CD) spectroscopy of all three variants indicated that they were well folded with a characteristic β sheet spectrum (Figure 2B) (Shin et al., 2009). Moreover, thermal melting curves monitored by CD exhibited a steep transition typical of well-folded proteins, and the transition temperature range (50°C–60°C) was similar to that previously reported for C2 domains (Figure 2C) (Shin et al., 2009). Interestingly, the DN variant displayed a sharper melting transition between 55°C and 60°C compared to the WT and DE variants in the absence of calcium (Figure 2C), suggesting that this variant may adopt a more cooperatively folded state comparable to the calcium-bound state, as predicted by the structural model (Figure S4).
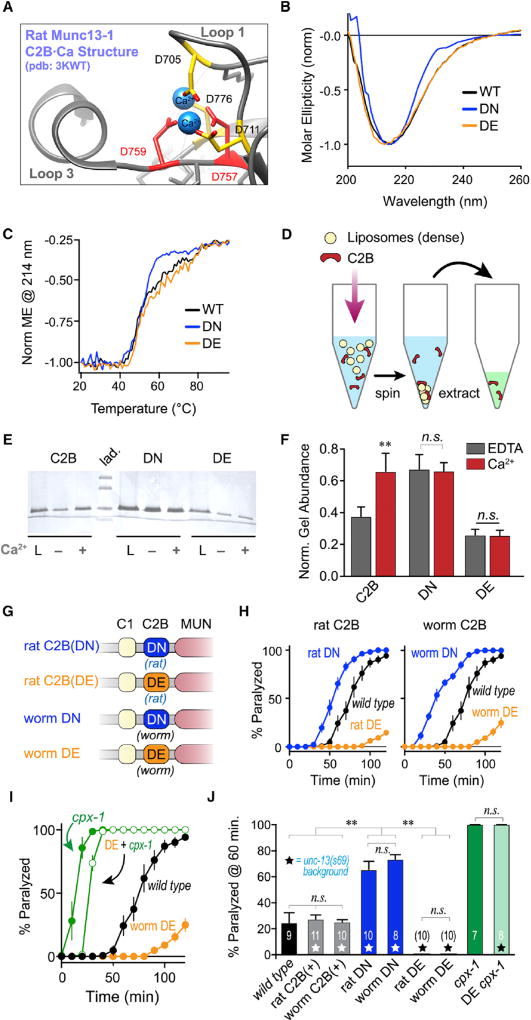
Figure 2. Calcium Binding Releases C2B Inhibition for Both Rat and Worm C2B.
(A) The calcium-binding pocket of rat Munc13-1 C2B (PDB: 3KWT) from Shin et al. (2010) with loop 3 aspartates indicated in red.
(B) CD spectra of recombinant rat Munc13-1 C2B: WT (black), D757N + D759N (DN; navy), and D757E + D759E (DE; orange). Data are normalized to the ellipticity value at 214 nm.
(C) Thermal denaturation over a 70°C temperature range was quantified using the CD value at 214 nm for each C2B variant as indicated.
(D) Schematic of the heavy liposome co-sedimentation assay as described in the STAR Methods.
(E) Representative Coomassie-stained SDS-PAGE gel for the total protein loaded (L) and the protein pellets recovered in EDTA (−) or 100 µM calcium (+) for WT (C2B), DN, and DE variants.
(F) Quantification of protein bands for n = 4 independent co-sedimentation assays across the three C2B variants in the presence (red) or absence (gray) of calcium.
(G) Cartoon depicting four UNC-13 variants used to rescue the unc-13 null mutant: rat Munc13-1 C2B harboring the loop 3 DN substitution (rat DN) or DE substitution (rat DE), or the worm UNC-13 C2B with the DN (worm DN) or DE (worm DE) substitution.
(H) Left: average aldicarb paralysis time course for WT (black) versus rat DN (navy), and rat DE (orange) transgenic animals expressed in unc-13(s69). Right: similar average time courses for transgenic animals expressing worm DN (navy) and worm DE (orange) in unc-13(s69).
(I) Average aldicarb time course for WT (black), worm DE variant in unc-13 null mutant (orange), cpx-1 null mutant (green filled), and the DE rescue variant in a cpx-1 null background (green open).
(J) Summary of aldicarb paralysis assays quantified at 60 min for the strains described in (H) and (I). Transgenic strains in the unc-13 null background are indicated with stars and the number of independent assays for each strain is indicated on the graph.
Errors bars are mean ± SEM (**p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer). Strains: N2, JSD14, JSD835, JSD849, JSD850, JSD925, JSD974, JSD1022, and JSD1038.
To examine the impact of these mutations on calcium-dependent versus calcium-independent membrane binding, each of the three rat C2B variants was incubated with heavy liposomes containing sucrose along with a mixture of neutral and anionic phospholipids including PIP2 (see STAR Methods) in the presence and absence of 100 µM calcium (Figure 2D). After centrifugation and extraction of the phospholipids, the co-sedimenting protein fraction was recovered and quantified using SDS-PAGE. WT C2B exhibited a degree of membrane binding even in the absence of calcium, and 100 µM calcium significantly enhanced this binding as previously observed (Figures 2E and 2F) (Shin et al., 2010). By comparison, DN exhibited maximal membrane binding irrespective of calcium. In contrast, DE displayed minimal membrane binding in both low and high calcium. Based on these results, we conclude that the loop 3 DN variant mimics a constitutively calcium-liganded C2B while the DE variant stabilizes the C2B-unliganded state even in the presence of calcium.
Calcium Binding to C2B Releases Inhibition of Calcium-Triggered Fusion
The three rat C2B structural variants were introduced into the worm UNC-13 protein in place of its native C2B sequence, and rescuing transgenic animals were generated in the unc-13 null mutant background (Figure 2G). While the WT rat C2B variant fully restored WT ACh secretion based on aldicarb sensitivity as noted earlier (Figures 1F and 2J), the constitutively liganded mimic rat C2B(DN) (rat DN) enhanced secretion to a similar degree as the C2B deletion variant (Figures 1E and 2H). In stark contrast, the calcium-free mimetic rat C2B(DE) transgenic animals (rat DE) exhibited significantly lower levels of ACh secretion. Similar transgenic animals were generated expressing the worm version of the bound- and apo-state mimetics (hereafter referred to as worm DN and worm DE), and these animals closely paralleled the rat C2B transgenic strains, confirming the functional conservation of the loop 3 ligand mimetic mutations (Figures 2H and 2J). A previous study examined the impact of a lysine-to-tryptophan substitution in loop 1 of the rat Munc13-2 C2B domain and found that this mutation enhanced calcium-triggered fusion in hippocampal autaptic cultures (Shin et al., 2010). This loop 1 lysine is deeply conserved across phylogeny, so we examined the worm K835W (KW) substitution in unc-13(s69) (Figures S5A and S5B). Notably, the KW substitution phenocopied deletion of the C2B domain based on aldicarb sensitivity (Figures S5C and S5D). As KW enhances the membrane-binding affinity of rat C2B, this observation is consistent with the loop 3 DN substitution described here (Shin et al., 2010). Mimicking the constitutively bound C2B domain produced a similar phenotype to the C2B domain deletion, so calcium binding appears to relieve a basal inhibition, perhaps by driving translocation of the C2B domain to the plasma membrane. A calcium-binding-deficient C2B domain, in contrast, appears to be locked into its autoinhibitory mode.
Although we showed that both the rat DN and rat DE variants preserved the folding of the C2B domain in vitro, an alternative interpretation of the lower ACh secretion observed in worm DE transgenic animals is that this mutation disrupted global UNC-13 folding, localization, or exocytic function in vivo, mimicking other severe loss-of-function alleles. To control for this possibility, we examined the worm DE variant in a complexin null cpx-1 mutant. Spontaneous (calcium influx independent) release is highly elevated in worm synapses lacking complexin (Wragg et al., 2013). This hypersecretion is entirely dependent on UNC-13 function as almost no secretion is observed in unc-13 cpx-1 double mutants (Zhou et al., 2013). Also, loss of complexin does not increase secretion in the unc-13(e1091) partial loss-of-function mutant (Figure S6A), further indicating that the additional fusion events generated in the absence of complexin specifically depend on UNC-13 exocytic function. When complexin was removed in worm DE animals, ACh secretion strongly increased to similar levels observed in cpx-1 mutants, as expected for a fully functional UNC-13 protein (Figures 2I and 2J). Thus, the worm DE mutant does not impair UNC-13 but rather stabilizes C2B inhibition of calcium-triggered release. Moreover, the calcium-independent fusion observed in the absence of complexin appears to be only slightly affected by the DE mutation. Perhaps C2B regulation is more strongly coupled to calcium-triggered fusion. Alternatively, loss of complexin may destabilize the inhibited state of UNC-13, contributing to the enhanced release observed even in the absence of calcium (Wragg et al., 2013).
To examine the impact of DN and DE mutations on calcium-triggered secretion, stimulus-evoked EPSCs were recorded in worm DN and worm DE transgenic animals. Evoked secretion was enhanced in the DN variant to a similar degree as deleting the entire C2B domain (Figures 3A–3C). Interestingly, the peak current was similar between WT and DE animals, while the total synaptic charge was significantly decreased in DE animals as a result of accelerated EPSC decay kinetics (Figure 3D). In contrast, EPSC decay was slowed either in the absence of C2B or in the calcium-binding mimetic DN variant. Faster kinetics could arise from either faster decay of unitary (or miniature) EPSCs (mEPSCs) or by a decrease in the temporal dispersion of release across the population of fusion events comprising the evoked EPSC (Chen and Regehr, 1999; Diamond and Jahr, 1995). As the mEPSC kinetics were indistinguishable between WT and DE animals, C2B appears to inhibit a slow component of calcium-triggered release (Figures 3E–3G). Of note, different isoforms of UNC-13 alter the kinetics of calcium-triggered release at the worm NMJ (Hu et al., 2013), supporting the notion that UNC-13 plays an important role in the coupling of calcium influx to vesicle fusion. The biochemical data, worm behavioral assays, and electrophysiological recordings taken together indicate that the C2B domain inhibits calcium-triggered fusion and this inhibition is relieved upon calcium binding, independent of SV priming by Munc13. This regulatory effect is observed even in single evoked stimuli, suggesting that Munc13 C2B exerts an inhibitory tone on the release machinery in the resting, primed state.
The C2B-MUN Linker Region Is Required for Inhibition by C2B
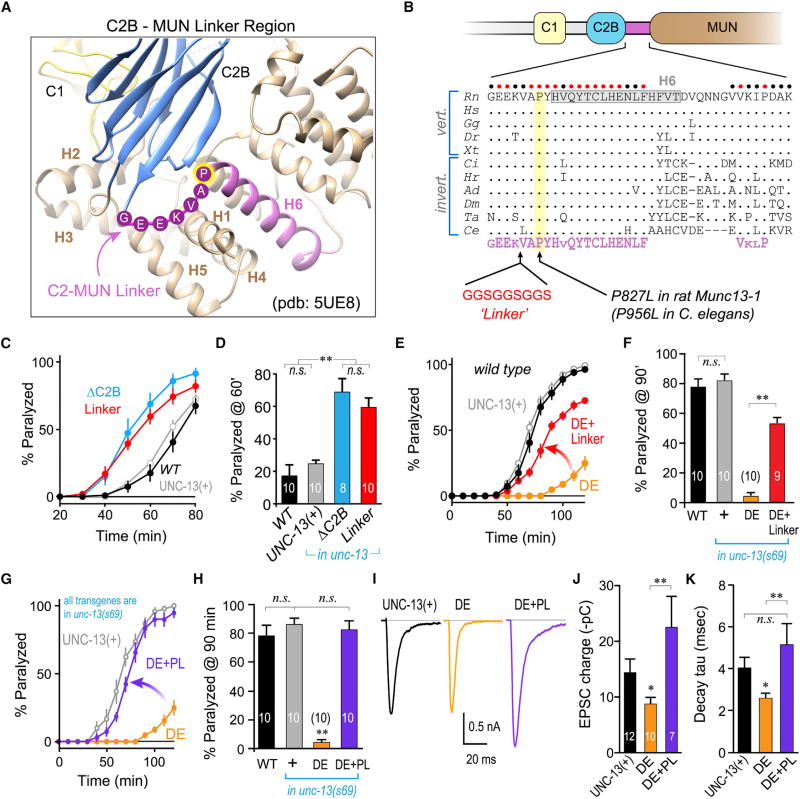
The C2B domain is directly coupled to the MUN domain through a short linker region, and this stretch of amino acids is markedly conserved across phylogeny, suggestive of a functional role beyond simply providing a flexible linkage (Figures 4A and 4B). A recent structural study of this region revealed that the C2B-MUN linker (CML) region comprises an alpha helical stretch (H6) bounded by a conserved GEEKVAPYH motif emerging from the β sheet structure of the C2B domain (Figure 4A) (Xu et al., 2017). We reasoned that a structured linker may play a role in maintaining autoinhibitory interactions between the C2B and MUN domains. We inserted a small flexible stretch of residues in the middle of the CML upstream of the conserved VAPYH motif to disrupt potential CML-mediated coupling between the C2B, CML, and MUN domains (“Linker”; Figure 4B). This disruption of the CML indeed enhanced ACh secretion to a similar degree as deleting the entire C2B domain (Figures 4C and 4D). Furthermore, CML disruption bypassed much of the constitutive inhibition imparted by DE and restored UNC-13 function to nearly WT levels (Figures 4E and 4F). Of particular interest, a point mutation in one of the conserved residues in the CML region of the human Munc13-1 ortholog UNC13A was recently linked to severe autism and neurological dysfunction (Lipstein et al., 2017). The proline residing just N-terminal to H6 (P827 in rat Munc13-1 or P956 in worm UNC-13) was mutated to a leucine (PL), and this substitution strongly enhanced neurotransmitter secretion in rodent hippocampal cultures and in worm neurons (Lipstein et al., 2017). We introduced P956L into the UNC-13(DE) variant (DE+PL) and found that this point mutant occluded the constitutive DE inhibition both by behavioral and electrophysiological assays (Figures 4G–4K). These data suggest that the human autism PL mutation destabilizes the inhibitory function of the unliganded C2B domain, thereby driving a hypersecretory state with profound effects on the CNS. Moreover, the impact of this C2B negative regulatory function is deeply conserved at least between nematodes and mammals.
Figure 4. The C2B-MUN Linker Domain Is Conserved and Required for C2B Inhibition.
(A) Ribbon diagram of the C2B-MUN interface crystal structure (Xu et al., 2017) using PDB: 5UE8, illustrating the C1 domain (yellow), C2B (blue), and the C2B-MUN linker (CML) region (purple). The six helices surrounding the C1 and C2B domains are labeled H1–H6 and the conserved residues in the flexible region at the start of the rat Munc13-1 CML are indicated.
(B) Schematic of Munc13 C1–C2B-MUN and protein sequence alignment (periods indicate sequence identity) for a broad collection of vertebrate and invertebrate species: Ce (Caenorhabditis elegans), Ta (Trichoplax adherens), Ci (Ciona intestinalis), Hr (Helobdella robusta), Ad (Anopheles darlingi), Dm (Drosophila melanogaster), Gg (Gallus gallus), Rn (Rattus norvegicus), Hs (Homo sapiens), Dr (Danio rerio), and Xt (Xenopus tropicalis). Circles above the sequence indicate perfectly conserved (red) or strongly conserved (black) residues across species. The consensus sequence motif is indicated below (purple).
(C) Aldicarb paralysis time course for WT (black), and unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); gray), ΔC2B (blue), or UNC-13 harboring an insertion within the C2B-MUN linker (Linker; red).
(D) Summary of paralysis at 60 min for the same strains.
(E) Aldicarb paralysis time course for WT (black), and unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); gray), worm DE (DE; orange), or the worm DE variant harboring the CML linker insertion (DE+Linker; red).
(F) Summary of paralysis at 90 min for the same strains.
(G) Aldicarb paralysis time course for unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); gray), worm DE (DE; orange), or the worm DE variant also containing a P956L point mutation (DE+PL; purple).
(H) Summary of paralysis at 90 min for the same strains.
(I) Average evoked EPSCs recorded in unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); black), worm DE (DE; orange), or worm DE with P956L (DE+PL; purple).
(J and K) Average cumulative EPSC charge transfer (J) and EPSC decay time constant (K) for the same three strains. Experiment number is indicated on the graph.
Errors bars are mean ± SEM (*p < 0.05, **p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer). Strains: N2, JSD805, JSD849, JSD885, JSD892, JSD1030, and JSD1038.
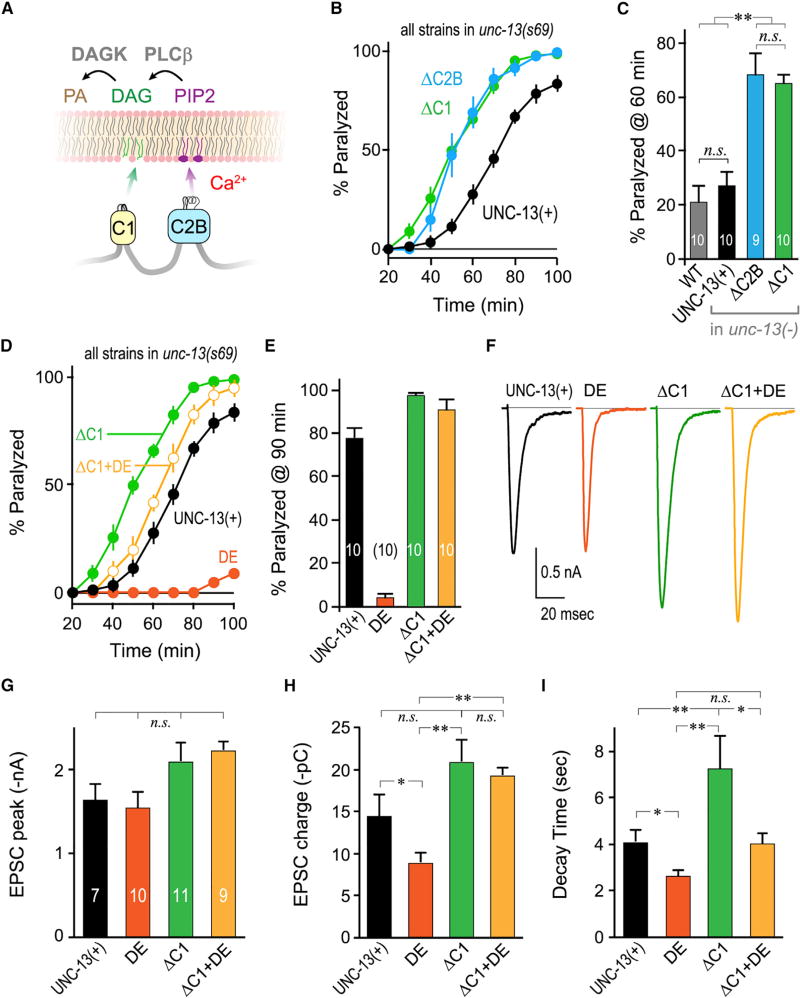
C1 Acts in Concert with C2B to Inhibit Neurotransmitter Secretion
Another well-established regulatory domain of Munc13 is its phorbol-ester-binding C1 domain adjacent to the C2B domain (Figure 5A) (Ahmed et al., 1992; Basu et al., 2007; Betz et al., 1998; Rhee et al., 2002). Functionally, the Munc13 C1 domain is a significant β-PE target at the synapse (Basu et al., 2007; Lou et al., 2008; Nurrish et al., 1999; Rhee et al., 2002) and, along with PKCαβγ isoforms, is required for the enhanced secretion observed at many synapses upon treatment with β-PEs. This enhancement is thought to arise from an increase in vesicular release probability rather than an expansion of the RRP (Lou et al., 2005). Notably, the C1 H567K mutation rendered Munc13 insensitive to β-PE treatment while also increasing release probability (Basu et al., 2007; Rhee et al., 2002). As this mutation is likely to unfold the C1 domain, several studies have speculated that disrupting C1 may have eliminated an inhibitory function rather than a stimulatory function of this domain (Basu et al., 2007; Lipstein et al., 2013; Liu et al., 2016; Rhee et al., 2002; Xu et al., 2017). However, it is difficult to differentiate between a point mutation mimicking the DAG-bound state of C1 versus simply disrupting C1 domain function, so the distinction between H567K and lipid activation of C1 remains unclear. Does C1 act in concert with C2B to inhibit Munc13? At least two possible mechanisms could couple C1 membrane binding to C2B-mediated inhibition. First, C1 recruitment to the plasma membrane could act as an indirect signal to release C2B inhibition, perhaps by bringing Munc13 to a particular location or recruiting another binding partner to Munc13 that interacts with the C2B (Nurrish et al., 1999). Alternatively, C1 could directly participate in stabilizing an autoinhibited conformation akin to conventional PKCs. In the first scenario, impairing or removing C1 should stabilize C2B inhibition, akin to the C2B DE mutation. In contrast, the second scenario predicts that loss of C1 would destabilize C2B inhibition. To distinguish between these possibilities, we examined the impact of complete C1 domain deletion on neurotransmitter secretion. Remarkably, removal of C1 produced a similar increase in secretion as the C2B deletion, supporting direct participation of C1 in autoinhibition (Figures 5B and 5C). Moreover, deletion of C1 almost completely reversed the C2B inhibition observed in DE transgenic animals by behavioral assays (Figures 5D and 5E), and by electrophysiological recordings (Figures 5F–5I), suggesting that C1 is required for stabilizing C2B inhibition in the calcium-free state. We examined the ability of the DAG mimetic phorbol-12-myristate-13 acetate (PMA) to reverse the constitutive inhibition in DE transgenic animals. Upon treatment with PMA, enhanced secretion was almost completely restored in DE transgenic animals (Figures S5C and S5D). The impact of endogenously generated DAG rather than exogenous phorbol esters was examined by analyzing mutant animals lacking the DAG kinase θ ortholog DGK-1 (Nurrish et al., 1999). DAG kinase normally limits the extent of DAG signaling by converting DAG to phosphatidic acid (Ishisaka and Hara, 2014; Tu-Sekine and Raben, 2011), and dgk-1 DAG kinase mutants exhibit hypersecretion consistent with chronic high levels of DAG in the absence of this elimination pathway (Nurrish et al., 1999). Consistent with the PMA results, elevated DAG reversed the effects of UNC-13 inhibition in DE transgenic animals (Figures S6E and S6F). Interestingly, although removal of the C1 domain reversed the C2B(DE)-mediated inhibition of neurotransmitter release as measured by total EPSC charge transfer, it did not have much impact on the kinetics of release (Figure 5H versus Figure 5I), suggesting that these functions of Munc13 can be separated within the C1–C2B module. Thus, manipulations that recruit C1 to the plasma membrane appear to release the inhibitory effect of C2B on UNC-13 and therefore couple lipid signaling to the inhibitory regulation of Munc13.
Figure 5. Removal of the C1 Domain Relieves Inhibition by the C2B Domain.
(A) Cartoon of C1 and C2B membrane interactions depicting C1 binding to DAG (green) and C2B binding to PIP/PIP2 (purple) in the presence of calcium. Phospholipase C beta (PLCβ) converts PIP2 to DAG while DAG kinase (DAGK) converts DAG to phosphatidic acid (PA).
(B) Aldicarb time course for unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); black), ΔC2B (blue), or UNC-13 lacking its C1 domain (ΔC1; green).
(C) Summary of aldicarb paralysis at 60 min for the same strains as well as WT (gray).
(D) Average aldicarb paralysis time courses for unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); black), worm DE (orange), ΔC1 (green), or UNC-13 lacking its C1 domain and also harboring the C2B DE mutation (ΔC1+DE; light orange).
(E) Summary of paralysis at 90 min following aldicarb treatment for the same strains.
(F) Average evoked EPSC traces for the same four strains.
(H–I) Average peak EPSC amplitude (G), cumulative EPSC charge transfer (H), and EPSC decay time constant (I) for the same strains.
Errors bars are mean ± SEM, and experiment number is given within the bars. *p < 0.05, **p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer. Strains: N2, JSD805, JSD849, JSD1038, JSD1067, and JSD1069.
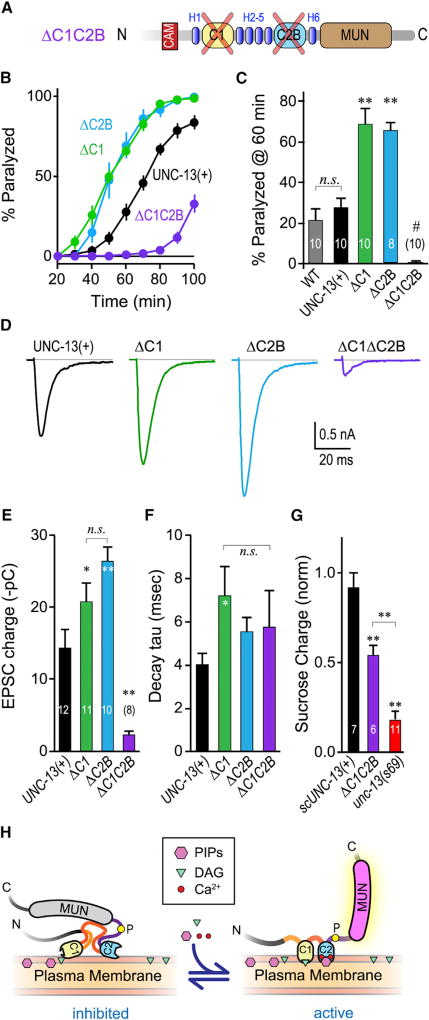
The C1–C2B Module Is Required to Support Calcium-Triggered Vesicle Fusion
To explore the relationship between C1 and C2B, we rescued unc-13 null mutants with a UNC-13 variant lacking both its C1 and C2B domains while preserving the interleafed alpha helical domains (H1–H8) in this region of the protein (Figures 4A and 6A). Strikingly, secretion was strongly decreased both by behavioral assays and synaptic recordings (Figures 6B–6F). The deletion of both domains could in principle disruptUNC-13 protein stability or synaptic localization. However, significant sucrose-evoked SV fusion was observed in the ΔC1ΔC2B transgenic animals (Figure 6G), indicating that this variant ofUNC-13 retained some priming function despite the loss of both domains. In addition, ΔC1ΔC2B animals were somewhat mobile (unlike unc-13(s69) null mutants), thereby permitting analysis by acute aldicarb sensitivity (Figure 6B). This observation indicates that a critical fusogenic role of the C1–C2B module is maintained in the absence of either individual domain but impaired when both domains are removed. As both C1 and C2B provide some membrane binding even in the absence of their respective ligands, perhaps some of this impairment arises from a loss of plasma membrane tethering (Liu et al., 2016). The relative decrease in sucrose-evoked charge transfer is consistent with a significant decrease in the readily releasable pool (Figure 6G), but may not entirely explain the ~90% decrease in EPSC charge transfer. The data presented here support a model in which C1 and C2B act together to inhibit Munc13 function, and this inhibition is reversed when C1–C2B is recruited to and/or properly positioned on the plasma membrane by calcium, PIP, and DAG (Figure 6H). However, loss of both lipid-binding domains from the module severely disrupts calcium-triggered fusion, revealing multiple functionalities for this highly conserved Munc13 regulatory domain.
Figure 6. Loss of Both C1 and C2B Nearly Eliminates Calcium-Triggered Fusion.
(A) Schematic of the ΔC1 ΔC2B double domain deletion. The two domains were removed while leaving the surrounding helical regions (H1–H8) intact to minimize global disruption of UNC-13 structure.
(B) Average aldicarb time course for unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); black), ΔC2B (blue), ΔC1 (green), or ΔC1ΔC2B (purple).
(C) Summary of paralysis at 60 min for these four strains as well as WT animals (gray).
(D–F) Average evoked EPSCs (D), average EPSC charge transfer (E), and EPSC decay time constant (F) for the same four strains.
(G) Sucrose-evoked charge (3 s integration; see STAR Methods) for unc-13(s69) (red) alone or rescued with a single-copy full-length UNC-13 (scUNC-13(+); black) or ΔC1ΔC2B (purple). Total charge is normalized to WT.
(H) Model of the C1–C2B protein module in the unliganded (inhibited) state versus the liganded (active) state. The conserved linker region proline is indicated with a yellow circle. Lipid and calcium binding to C1–C2B induces a conformational change that activates a fusogenic function of UNC-13.
Errors bars are mean ± SEM, and experiment number is given within the bars. *p < 0.05, **p < 0.01; #, significantly different from all other strains with p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer. Strains: N2, BC168, JSD805, JSD1038, JSD1039, JSD1064, and JSD1067.
DISCUSSION
The experiments described above provide several lines of evidence that the C1–C2B tandem domain of Munc13 functions in part to inhibit calcium-triggered neurotransmitter release. First, in the absence of either C1 or C2B, neurotransmitter secretion is enhanced rather than impaired. Second, mutations that mimic the calcium-bound state of C2B and enhance its calcium-independent membrane binding phenocopy the complete domain deletion in vivo. Third, mutations that stabilize the calcium-unbound state of C2B inhibit exocytosis, and this constitutive inhibition can be suppressed by phorbol esters, removal of the C1 domain, or disrupting the linker between C2B and the MUN domain. And finally, simultaneous removal of both membrane-binding domains severely reduces calcium-triggered fusion. Notably, selective disruption of C2B modulation of Munc13 function markedly impairs nervous system function in the worm, highlighting the physiological impact of proper Munc13 regulation beyond its permissive role for vesicle fusion. And from an evolutionary point of view, the only protein families generally found to have tandem C1–C2 domains (in this order) are Munc13 and conventional PKCs (see STAR Methods). Our current study suggests a form of functional conservation of a C1–C2 tandem domain in stabilizing a refractory state of either conventional PKC or Munc13 that is activated in the presence of several distinct signaling molecules.
Impact of C2B Loop 1 versus Loop 3 Mutations on Membrane Binding
The Munc13 C2B domain has been implicated in the enhancement of neurotransmitter secretion in cultured hippocampal neurons from mouse (Shin et al., 2010) as well as in proteoliposome fusion assays conducted in vitro (Liu et al., 2016). Compared to previous investigations of the C2B domain of Munc13, our approach in the present study is unique in two ways. First, the complete deletion of either the C1 or C2B domain has not been studied in synapses to our knowledge. Second, the impact of loop 3 aspartates has not been studied previously in vitro or in vivo, and manipulating the loop 3 aspartates permitted the creation of bound-state and unbound-state mimetics. In the Shin et al. study, by comparison, two calcium-binding aspartates in Munc13-2 C2B loop 1 were replaced with asparagines, thereby disrupting calcium-dependent phospholipid binding. This disruption had little effect on basal synaptic transmission but largely eliminated synaptic potentiation following a high-frequency stimulus train. We speculate that this loop 1 DN substitution is similar to our loop 3 DE substitution in that C2B membrane binding is weakened and no longer calcium dependent. Consistent with this notion, phorbol esters could enhance secretion in the loop 1 DN mutants similar to the effects of phorbol esters and the DAG kinase mutant observed here with the loop 3 DE mutation. One notable difference is that the loop 3 DE mutations described here produce a stronger inhibitory effect on single stimuli compared to the loop 1 DN mutations utilized by Shin et al. Perhaps the loop 3 DE substitution enhances inhibition because the charge density near the calcium-binding site remains negative (mimicking the calcium-free state), whereas the loop 1 DN mutations neutralized some of the negative charge. The loop 1 KW mutation in Shin et al. increased the membrane-binding affinity of C2B and also boosted calcium-triggered fusion at the synapse. Consistent with this observation, we found that the same mutation in worm UNC-13 also leads to an enhancement of secretion similar to our loop 3 DN mutation. Overall, the Shin et al. study is consistent with our model in which C1 and C2B inhibit basal neurotransmitter release and calcium boosts release by relieving this inhibition. Our own experiments strongly support this model and provide a new framework for interpreting the results of previous studies on C1 and C2B of Munc13, but further exploration of the loop 1 and loop 3 mutations in mammalian and worm synapses will be needed for a rigorous comparison of Munc13 and UNC-13 functional conservation.
Role of Munc13 C1 Domain in Synaptic Vesicle Fusion
Engagement of Munc13 C1 by phorbol esters is thought to increase secretion by elevating release probability rather than increasing the number of primed vesicles (Basu et al., 2007; Lou et al., 2008). Moreover, in the H567K C1 mutant, the RRP actually decreased in size (Basu et al., 2007; Rhee et al., 2002). Because manipulations of the C1 domain increased both spontaneous and calcium-triggered vesicle fusion, this domain was proposed to lower the energy barrier of fusion, irrespective of the mechanism by which fusion was triggered (Basu et al., 2007). A refined interpretation consistent with past observations is that the C1 domain normally adds to the fusion energy barrier by inhibiting a post-priming function of Munc13 function under resting conditions, and when DAG drives C1 translocation to the PM, the energy barrier is lowered by removing an inhibitory interaction. Indeed, the H567K mutant displayed enhanced spontaneous fusion and elevated release probability under resting conditions, analogous to the observations reported here with a deletion of the entire C1 domain (Basu et al., 2007). The experiments presented here indicate that either DAG engagement or deletion of C1 suppresses the strong inhibition imparted by the C2B domain and support the notion that C1 engagement is coupled to release of C2B inhibition. Together with the results on Munc13 C1 from previous studies, these findings can be explained by postulating that the C1 and C2B domains act in concert to stabilize an inhibited state of Munc13 in the absence of membrane recruitment signals such as calcium, PIPs, and DAG (Figure 6H).
Does Munc13 Further Modify a Primed Synaptic Vesicle?
At a mechanistic level, two possibly overlapping functions have been proposed for Munc13 at the synapse. One well-studied aspect of Munc13 function is its role in catalyzing the assembly of the SNARE complex via Syntaxin-MUN interactions during SV docking/priming (Ma et al., 2013; McEwen et al., 2006; Richmond et al., 2001; Rizo and Südhof, 2012; Rizo and Xu, 2015; Südhof, 2013; Yang et al., 2015). Recent structural and functional evidence that describes a tripartite interface of C1, C2B, and the MUN domain further supports the concept that rearrangements in C1–C2B could exert forces on the MUN domain and affect its function (Xu et al., 2017). However, the Syntaxin-MUN interaction occurs relatively far from this tripartite interface and thus would not likely be directly affected by C1–C2B conformational changes (Xu et al., 2017; Yang et al., 2015). In addition, the MUN domain and flanking C2 domains may act to tether SVs to the plasma membrane (Liu et al., 2016). Both of these functions may contribute to the observation that, in the absence of Munc13, the RRP is eliminated, correlating with the disappearance of SVs that are directly adjacent to the plasma membrane (<5 nm) (Augustin et al., 1999; Imig et al., 2014; Madison et al., 2005; Richmond et al., 1999; Rizo and Xu, 2015). Besides tethering SVs and opening Syntaxin, does Munc13 directly affect the probability that a primed vesicle will fuse? Each SV in the RRP has an intrinsic fusogenicity, and this property is apparently heterogeneous within the synapse (Ariel et al., 2013; Herman and Rosenmund, 2015; Lee et al., 2013; Nakamura et al., 2015; Rosenmund et al., 1993; Schlüter et al., 2006; Taschenberger et al., 2016). In particular, a subset of primed vesicles has been proposed to be in a “superprimed” state with a significantly higher release probability than the remainder of the SVs (Lee et al., 2013; Schlüter et al., 2006; Taschenberger et al., 2016). A recent study revealed that the superprimed pool of SVs is enhanced by phorbol esters, and this pool was transiently expanded following high-frequency stimulation, thereby contributing to post-tetanic potentiation (PTP) (Taschenberger et al., 2016). Our data along with previous studies support an important post-priming function of Munc13 (Basu et al., 2007; Lou et al., 2008; Madison et al., 2005; Rhee et al., 2002). Given the impact of phorbol esters on both super-priming and Munc13 disinhibition, perhaps the disinhibited Munc13 corresponds to the molecular state of superprimed SVs. Interestingly, PTP is thought to reflect a transient increase in release probability rather than RRP size, and the conditions that trigger PTP can activate conventional PKCs by relieving C1–C2 autoinhibition (de Jong and Fioravante, 2014; Fioravante et al., 2014; Korogod et al., 2007; Lipstein et al., 2012; Rosenmund et al., 2002). Thus, the disinhibition of Munc13 that we describe here may underlie the connection between PTP and superpriming. We further speculate that brief high-frequency bursts of activity drive corresponding increases in calcium and DAG, which will transiently promote disinhibition of Munc13 in newly primed vesicles and enhance their release probability. Another possible mechanism for superpriming may be “positional priming” of a subset of SVs within close proximity of voltage-gated calcium channels (Lee et al., 2013). However, if direct C2B interactions with the synprint domains of presynaptic calcium channels drive positional priming, one might expect C2B deletions to impair calcium-channel binding and subsequently to increase rather than decrease the distance between SVs and their neighboring calcium channels (Calloway et al., 2015). Thus, the enhanced vesicular release probability observed in the absence of C1–C2B is probably not due to the elimination of a calcium-channel-binding interaction. A more extensive exploration of the post-priming effects of Munc13 will be required to better understand its multifaceted roles in controlling release probability and particular forms of short-term plasticity.
Conclusion
Our work reveals a novel inhibition of Munc13 by C1–C2B that profoundly impacts nervous system function. In addition to these intramolecular interactions, the C1–C2B module may interact with other synaptic proteins to regulate SV fusion, and the net increase in SV fusion in the absence of C2B could emerge from the aggregate impact of disrupting several control systems. This scenario may account for the prominent decrease in release in the double C1–C2B domain deletion, for instance. Regardless of these potential interactions, the results presented here demonstrate the role of an Munc13 protein module in determining synaptic strength and provide a physical link to key regulatory molecules such as calcium and DAG. Future studies on the structure of Munc13 and on its numerous protein and lipid interactions at the active zone will further uncover the mechanistic basis for the control of synaptic transmission.
STAR⋆METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○C. elegans
-
○Rescue Constructs
-
○
- METHOD DETAILS
-
○Aldicarb Sensitivity Assays
-
○Assessing calcium-triggered secretion in vivo
-
○Worm Tracking and Analysis
-
○Electrophysiology
-
○Impact of C2B removal on tonic neurotransmitter release
-
○Protein Purification
-
○CD Spectroscopy
-
○Liposome Preparation
-
○Co-sedimentation Assay
-
○Protein Structure Modeling
-
○C1–C2 Domain Conservation Analysis
-
○
QUANTIFICATION AND STATISTICAL ANALYSIS
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeremy Dittman (jed2019@med.cornell.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans
C. elegans strains were maintained and genetically manipulated as previously described (Brenner, 1974). Animals were raised at 20°C on nematode growth media seeded with OP50. See Key Resources Table for full list of transgenic animals used. Single-copy transgenes were introduced into the EG6699 strain genome using transposon-based mos1-mediated single copy insertion (mosSCI) method, as previously described (Frøkjaer-Jensen et al., 2008). All UNC-13 expression constructs coded the long-form isoform of UNC-13 (UNC-13L) (Hu et al., 2013).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Aldicarb | Ultra Sci. | ULPST-940 |
| Phorbol 12-myristate 13-acetate | Enzo Life Sci. | 89161-402 |
| Histamine Chloride | Alfa Aesar | AAAL09198-14 |
| Phosphatidylcholine (egg PC) | Avanti | 840051C |
| Phosphatidylserine (PS) | Avanti | 840032C |
| Phosphatidylethanolamine (PE) | Avanti | 840022 |
| Brain PI(4,5)P2 | Avanti | 840046X |
| Cholesterol | VWR | 97061-660 |
| Experimental Models: Organisms/Strains | ||
| wild type | Caenorhabditis Genetics Center | N2 |
| unc-13(e1091) I | Caenorhabditis Genetics Center | CB1091; RRID: WB-STRAIN:CB1091 |
| cpx-1(ok1552) I | Caenorhabditis Genetics Center | JSD14 |
| unc-13(e1091) cpx-1(ok1552) I | this paper | JSD746 |
| unc-13(s69) I | Caenorhabditis Genetics Center | BC168; RRID: WB-STRAIN:BC168 |
| slo-1(js379) V | Caenorhabditis Genetics Center | NM1968; WB-STRAIN:NM1968 |
| dgk-1(nu62) X | Caenorhabditis Genetics Center | KP1097: WB-STRAIN:KP1097 |
| tauEx260[Punc-17::HisCl1] | this paper | JSD733 |
| ttTi5605 II; unc-119(ed3) III; oxEx1578[Peft-3::gfp+Cbr-unc-119] | Caenorhabditis Genetics Center | EG6699; RRID: WB-STRAIN:EG6699 |
| cpx-1; tauEx277[Punc-17::HisCl1] | this paper | JSD758 |
| slo-1; tauEx347[Punc-17::HisCl1] | this paper | JSD895 |
| ΔC2B= Psnb-1::UNC-13(ΔC2B) single copy integrant in unc-13(s69) | this paper | JSD0805 |
| KW = Psnb-1::UNC-13(K835W) in unc-13(s69) | this paper | JSD0830 |
| DN = Psnb-1::UNC-13(D886N, D888N) in unc-13(s69) | this paper | JSD0835 |
| DE = Psnb-1::UNC-13(D886E, D888E) in unc-13(s69) | this paper | JSD0849 |
| cpx-1;DE = Psnb-1::UNC-13(D886E, D888E) in unc-13(s69) | this paper | JSD0850 |
| Linker = Psnb-1::UNC-13(CML GGS Linker insert) in unc-13(s69) | this paper | JSD0885 |
| ΔC2B+His = Psnb-1::UNC-13(ΔC2B) + Punc-17::HisCl1 in unc-13(s69) | this paper | JSD0891 |
| DE+Linker = Psnb-1::UNC-13(D886E, D888E, CML GGS Linker) in unc-13(s69) | this paper | JSD0892 |
| rat C2B = Psnb-1::UNC-13(rat Munc13-1 C2B) in unc-13(s69) | this paper | JSD0925 |
| rat DE = Psnb-1::UNC-13(rat Munc13-1 C2B D757E D759E) in unc-13(s69) | this paper | JSD0974 |
| rat DN = Psnb-1::UNC-13(rat Munc13-1 C2B D757N D759N) in unc-13(s69) | this paper | JSD1022 |
| DE+PL = Psnb-1::UNC-13(D886E, D888E, P956L) in unc-13(s69) | this paper | JSD1030 |
| UNC-13(+) = Psnb-1::UNC-13L in unc-13(s69) | this paper | JSD1038 |
| scUNC-13(+) = Psnb-1::UNC-13L single copy integrant in unc-13(s69) | this paper | JSD1039 |
| dgk-1;DE = Psnb-1::UNC-13(D886E, D888E) in unc-13(s69) | this paper | JSD1060 |
| ΔC1ΔC2B = Psnb-1::UNC-13(ΔC1ΔC2B) in unc-13(s69) | this paper | JSD1064 |
| ΔC1 = Psnb-1::UNC-13(ΔC1) in unc-13(s69) | this paper | JSD1067 |
| ΔC1+DE = Psnb-1::UNC-13(ΔC1, D886E, D888E) in unc-13(s69) | this paper | JSD1069 |
| Recombinant DNA | ||
| Psnb-1::UNC-13L | this paper | JP942 |
| Psnb-1::UNC-13L MosSCI | this paper | JP647 |
| Psnb-1::UNC-13L (DC2B) MosSCI | this paper | JP679 |
| Psnb-1::UNC-13L(K835W) | this paper | JP768 |
| Psnb-1::UNC-13L(D886N,D888N) | this paper | JP772 |
| Psnb-1::UNC-13L(D886E, D888E) | this paper | JP786 |
| Psnb-1::UNC-13L(CML GGS Linker) | this paper | JP812 |
| Psnb-1::UNC-13L(D886E, D888E, CML GGS Linker) | this paper | JP815 |
| Psnb-1::UNC-13L(rat Munc13-1 C2B) | this paper | JP845 |
| Psnb-1::UNC-13L(rat Munc13-1 C2B D757E D759E) | this paper | JP881 |
| Psnb-1::UNC-13L(rat Munc13-1 C2B D757N D759N) | this paper | JP925 |
| Psnb-1::UNC-13L(D886E, D888E, P956L) | this paper | JP941 |
| Psnb-1::UNC-13L(ΔC1) | this paper | JP975 |
| Psnb-1::UNC-13L(DC1 DC2B) | this paper | JP971 |
| Psnb-1::UNC-13L(DC1, D886E, D888E) | this paper | JP976 |
| pET_SUMO Munc13-1 C2B | Calloway et al., 2015 | JP718 |
| pET_SUMO Munc13-1 C2B D757E D759E | this paper | JP918 |
| pET_SUMO Munc13-1 C2B D757N D759N | this paper | JP919 |
| Punc-17::HisCl1 | Pokala et al., 2014 | JP673 |
| Software and algorithms | ||
| Igor Pro | Wavemetrics | Version 6 |
Rescue Constructs
For all rescue experiments in this study, we expressed variants of the UNC-13L isoform (specifically isoform ZK524.2f or UniProt: P27715-7) under the snb-1 promoter. NheI and KpnI sites were introduced at the 5′ and 3′ ends respectively, and a NotI site was included just before the stop codon for potential C-terminal fusion constructs. The 5′ sequence begins with GCTAGCAAAAATGGATGACGTTGG… The 3′ sequence ends with …GTCAAAACATCAATCGAACAGCGGCCGCCTAAGGTACC. The NheI-KpnI fragment encoding UNC-13L was 5476 bases in length. The C1 domain deletion removed 51 residues beginning with PHNF. The C2B deletion removed 151 residues beginning with KITL and ending in the C2B-MUN Linker region …FLAA. In the double deletion construct UNC-13L(ΔC1ΔC2B), these same two deletion boundaries were used, thereby preserving seven of the eight helical stretches surrounding C1 and C2B as described in the recent crystal structure (Xu et al., 2017).
METHOD DETAILS
Aldicarb Sensitivity Assays
To measure aldicarb sensitivity, 20–25 young adult animals were placed on agar plates containing 1 mM aldicarb (Watson International, China). Worms were scored for paralysis at ten minute intervals for 2 hr. Each genotype was tested (blind) approximately ten times and paralysis curves were generated by averaging paralysis time courses for each plate as described previously (Dittman and Kaplan, 2008). To measure aldicarb sensitivity in the presence of the DAG mimic phorbol 12-myristate 13-acetate (PMA), worms were incubated for two hours on plates containing 0.25 µg/µL PMA (Enzo Life Sciences), and then transferred to plates containing 0.25 µg/µL PMA and 1mM aldicarb (Ultra Scientific) and worms were scored for paralysis at ten minute intervals for 2 hr. As PMA was dissolved in ethanol, these assays were compared to ethanol controls. To measure aldicarb sensitivity in the presence of histamine, worms were incubated for thirty minutes on plates containing 10 mM histamine, and then transferred to plates containing 10mM histamine and 1mM aldicarb and worms were scored for paralysis at ten minute intervals for 2 hr.
Assessing calcium-triggered secretion in vivo
The electrophysiological data described in Figures 1 and S2 indicate that calcium-triggered fusion was enhanced when the C2B domain of UNC-13 was removed. To investigate the contribution of calcium-triggered fusion to the observed hypersecretion phenotype in vivo, motor neurons were electrically silenced in intact behaving animals. The Drosophila HisCl histamine-gated chloride channel was expressed in cholinergic neurons to reversibly silence motor neurons in intact animals as described previously (Pokala et al., 2014; Wragg et al., 2017). Sensitivity to aldicarb was scored in the presence and absence of sub-maximal histamine in wild-type animals, and a significant delay in paralysis was driven by partial electrical silencing of cholinergic motor neurons (Figure S3). Two control strains were examined to calibrate the impact of electrical silencing in this assay. First, complexin null mutants (cpx-1) rapidly paralyze due to a high rate of spontaneous SV fusion even in the absence of calcium (Hobson et al., 2011; Martin et al., 2011; Wragg et al., 2013), and therefore electrical silencing should have little effect on the rate of paralysis in cpx-1 animals. In contrast, slo-1 BK potassium channel mutants paralyze rapidly in the presence of aldicarb due to prolonged depolarization and elevated calcium-triggered fusion in the absence of this critical repolarizing current (Wang et al., 2001). Thus, electrical silencing should counteract the enhanced secretion observed in the slo-1 mutant. Both predictions were confirmed by monitoring paralysis kinetics following treatment with histamine (Figures S3B and S3C). Notably, histamine also significantly slowed paralysis in the ΔC2B animals, demonstrating that the increased secretion was predominantly driven by a calcium-dependent secretory process in intact behaving animals (Figures S3D and S3E).
Worm Tracking and Analysis
To measure the locomotion of strains, 70–80 young adult worms were placed onto an agar plate devoid of OP50, washed with M9, and transferred to a second plate without OP50. The worms were allowed to rest for one hour before imaging. One minute digital videos of individual animals were captured (5 Hz frame rate) on an ORCA-05G CCD camera (Hamamatsu) mounted on a Motic stereomicroscope (SMZ-143 FBLED) using 10× magnification. The center of mass was recorded for each animal on each video frame using custom object tracking software in MATLAB (Ramot et al., 2008). Average speed was determined for each animal and then pooled to form a population average and standard error for each genotype.
Electrophysiology
Whole-cell voltage-clamp recordings on body-wall muscles were performed on dissected adult C. elegans as previously described (Hu et al., 2012; Richmond et al., 1999). Worms were superfused in an extracellular solution containing 127 mM NaCl, 5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 20 mM glucose, 1 mM CaCl2, and 4 mM MgCl2, bubbled with 5% CO2, 95% O2 at 20°C. Whole-cell recordings were carried out at −60 mV using an internal solution containing 105 mM CsCH3SO3, 10 mM CsCl, 15 mM CsF, 4 mM MgCl2, 5 mM EGTA, 0.25 mM CaCl2, 10 mM HEPES, and 4 mM Na2ATP, adjusted to pH 7.2 using CsOH. Under these conditions, we only observed endogenous acetylcholine EPSCs. Stimulus-evoked EPSCs were stimulated by placing a borosilicate pipette (5–10 µm) near the ventral nerve cord (one muscle distance from the recording pipette) and applying a 0.4 ms, 85 µA square pulse using a stimulus current generator (WPI). All unstimulated events recorded in voltage clamp were categorized as individual fusion events since spontaneous action potentials (large synchronous release events) do not typically occur at the worm NMJ (Madison et al., 2005; Richmond et al., 1999). For estimating the readily releasable pool size, a pipette containing 0.5M sucrose solution was placed at the end of the patched muscle cell, and a 20 psi, 5 s pressure pulse was applied by Picospritzer to create a rapid jump in osmolarity at the neuromuscular junction (< 200 msec latency on average). The integrated charge transfer was computed as a function of time throughout the sucrose delivery, and the charge accumulation was corrected for the baseline holding current and spontaneous fusion events prior to sucrose application. In unc-13(s69) mutants, there was no detectable charge accumulation over the first several seconds following the osmolarity jump, consistent with previous studies (Madison et al., 2005; Richmond et al., 1999; Zhou et al., 2013).
Impact of C2B removal on tonic neurotransmitter release
Neuromuscular cholinergic synapses in wild-type and transgenic animals were directly recorded by voltage-clamping muscles in dissected animals. Under these conditions, endogenous neuronal activity normally generates a moderate degree of neurotransmitter release referred to as tonic excitatory postsynaptic currents (tonic EPSCs), and the majority of these fusion events are calcium-triggered (Hobson et al., 2011; Liu et al., 2009; Martin et al., 2011; Richmond et al., 1999). All cholinergic fusion events require UNC-13 because in the absence of UNC-13, tonic activity is completely eliminated (Hu et al., 2013; Madison et al., 2005; Richmond et al., 1999; Zhou et al., 2013). In transgenic animals expressing UNC-13(ΔC2B), tonic activity was increased by ~30% relative to wild-type animals whereas no changes in unitary EPSC amplitude were detected (Figures S2A–S2C). Since the tonic release rate is composed of both calcium-triggered and true spontaneous fusion events, this increase in the tonic rate may have arisen from an excess of either calcium-triggered or calcium-independent fusion. However, increased stimulus-evoked release (Figures 1G–1I) and enhanced sensitivity to electrical silencing (Figure S3) indicate that calcium-triggered release is boosted in the absence of C2B.
Protein Purification
Munc13-1 C2B domain (residues 675–820) was cloned into the pET SUMO vector with no additional sequence tags. The D757N/D759N and D757E/D759E loop 3 mutants were generated via site-directed mutagenesis of the WT construct. Proteins were expressed in bacterial cultures and purified as previously described (Wragg et al., 2013). Briefly, BL21(DE3) E. coli cells were transformed with the relevant plasmid and grown in LB media to an optical density of ~0.6. Cells were induced with isopropyl-β-D-thiogalactoside (IPTG) and grown for four hours before collection. Cells were lysed by sonication on ice, supernatants were clarified by centrifugation at 40,000 r.p.m. for 45 minutes, and SUMO-tagged fusion protein was purified from supernatant on a Ni-NTA column and dialyzed into 20 mM Tris pH 8, 150 mM NaCl, 1 mM dithiothreitol. The His6-SUMO affinity tag was cleaved by SUMO protease and purified C2B domain was isolated through a second run over a Ni-NTA column. Protein was stored for in 20 mM Tris pH 8, 350 mM NaCl, and 1cmM dithiothreitol at 4°C. CD experiments were performed in the storage buffer, with the addition of 100 µM EDTA. Protein concentration was estimated by comparing Coomassie-stained gel band intensities for C2B dilutions with a set of BSA standards.
CD Spectroscopy
Far-ultraviolet CD spectroscopy experiments were performed on an AVIV Biomedical Model 410 CD Spectrometer. CD spectra were obtained from 200 to 260 nanometers (nm) at 25°C with a wavelength step of one nm, an averaging time of two seconds, and with nine averaged scans per sample. For the melting curves, the wavelength was held at 214 nm and the temperature increased from 20°C to 96°C at an increment of 2°C, and averaging time of one second, and with three averaged scans per sample. A cell path length of 0.02 cm was used, with protein concentrations at 100 µM. Data for the C2B constructs were converted to mean residue molar ellipticity.
Liposome Preparation
Lipids were obtained from Avanti Polar Lipids and stored at −20°C. Lipids were mixed in the concentration of 44% PC, 32% PE, 13% PS, 10% cholesterol, and 1% PI(4,5)P (mole percentage), and dried under N2 gas and residual solvent was removed under vacuum for 1chour. After resuspension in buffer containing 0.5M sucrose, LUVs were prepared by ten cycles of freezing in liquid nitrogen and thawing in warm water followed by repetitive extrusion through 400 nm pore-size polycarbonate films (21 times each) using a 1 mL Avanti Mini-Extruder from Avanti Polar Lipids. The liposomes underwent overnight dialysis into the co-sedimentation buffer containing no sucrose (see below). Vesicles were stored at 4°C and used within one week. Lipid concentration was estimated based on the amounts of starting lipid.
Co-sedimentation Assay
Protein stocks were spun at top speed in a table-top centrifuge for ten minutes before being used in the co-sedimentation assay, to remove proteins that had become insoluble. The proteins were mixed with liposomes to a final concentration of 4 µM protein, 7.5 µM liposomes, 100 mM NaCl and 20 mM Tris pH 8, with either 100 µM EDTA or 100 µM CaCl2 in a final volume of 70 µL. The protein-liposome mix was incubated for 30 min at room temperature, then spun at 50,000 g for 30 min. The supernatant was removed and the liposome pellet was dissolved in chloroform:methanol (1:2 (v/v)). The precipitated proteins were recovered by centrifugation (50,000 g for 30 min), resuspended in 30 µL of 2 × SDS sample buffer, and analyzed by SDS-PAGE and Coomassie Blue staining. Coomassie-stained gels were quantified using ImageJ.
Protein Structure Modeling
The Phyre2 web portal for protein modeling, prediction and analysis was used to predict the effects of residue changes in the C2B domain. As previously described (Kelley et al., 2015), for any candidate mutations, the sequence of the entire rat C2B domain with the residue changes were entered into the Phyre2 web portal, and the “Intensive” mode was used to predict structure. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
C1–C2 Domain Conservation Analysis
The C1–C2B sequence of C. elegans UNC-13L (ZK524.2a) was queried using both NCBI Conserved Domain Architecture Retrieval Tool (CDART) and Simple Modular Architecture Research Tool (SMART). Of all protein sequences containing both a C1 and C2 domain (and ignoring small protein fragments), a large majority possessed either MHD/DUF1041 domains (unc13-related) or a PKC-related S_TKc domain. Of the remaining sequences not associated with either family, most did not have adjacent C1 and C2 domains in this order. For example, 5906 sequences were retrieved in CDART containing both C1 and C2, but only 8 sequences (corresponding to three predicted proteins) harbored adjacent C1 and C2 domains, while also failing to possess a DUF1041 or kinase domain. These exceptional sequences are found in two parasitic fungi (Phytophthora infestans [NCBI: XP_002895965.1] and Albugo laibachii [NCBI: CCA25035.1]) and a single-celled eukaryote (Capsaspora owczarzaki [NCBI: XP_004365979.2]). Thus, at least within Metazoa, the C1–C2 module appears to be unique to proteins belonging to either the conventional PKCs or Munc13-related proteins.
QUANTIFICATION AND STATISTICAL ANALYSIS
For C. elegans locomotion and electrophysiology data presented here, the number of independent measurements (n) refers to the number of animals. For the aldicarb sensitivity assays, n refers to independent measurements on plates containing approximately 20 animals each. Student’s t test was used to compute significance for pairwise comparisons and the Tukey–Kramer method for multiple comparisons as a post hoc test following a one-way ANOVA. Error bars represent ± SEM. The aldicarb data for wild-type worms in Figure 1F were also used in Figure 2J. The aldicarb data for rat C2B in Figure 1F were also used in Figures 2H and 2J. The aldicarb data for ΔC2B in Figure 1E were also used in Figures 4C and S4C.
Supplementary Material
Highlights.
The C1–C2B module of Munc13 inhibits calcium-triggered fusion of primed vesicles
Calcium and removal of C1 release the inhibition to boost synaptic transmission
A conserved linker region between C2B and MUN is crucial for stabilizing C2B inhibition
Regulation of Munc13 through C1–C2B is required for proper nervous system function
Acknowledgments
We thank Tim Ryan, David Eliezer, Cori Bargmann, Josh Kaplan, Daniel Parisotto, Steven Nurrish, David Snead, Géraldine Gouzer, Nat Calloway, and Trudy Ramlall for help, advice, and critically reading the manuscript. We also thank Cori Bargmann and Navin Pokala for the HisCl plasmid, and Lucy Skrabanek for help with the protein domain conservation analysis. This work was supported by the NIH grant R01-GM095674 (J.S.D.), MSTP T32GM007739 (F.M.), Australian Research Council DP160100849 (Z.H.), and NHMRC APP1122351 (Z.H.).
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2017.07.015.
AUTHOR CONTRIBUTIONS
Conceptualization, F.M. and J.S.D.; Methodology, F.M., J.S.D., and Z.H.; Investigation, F.M., H.L., and Z.H.; Writing, F.M. and J.S.D.; Funding Acquisition, F.M., J.S.D., and Z.H.
References
- Ahmed S, Maruyama IN, Kozma R, Lee J, Brenner S, Lim L. The Caenorhabditis elegans unc-13 gene product is a phospholipid-dependent high-affinity phorbol ester receptor. Biochem. J. 1992;287:995–999. doi: 10.1042/bj2870995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel P, Hoppa MB, Ryan TA. Intrinsic variability in Pv., RRP size, Ca(2+) channel repertoire, and presynaptic potentiation in individual synaptic boutons. Front. Synaptic Neurosci. 2013;4:9. doi: 10.3389/fnsyn.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin I, Rosenmund C, Süudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, Grishin NV, Rosenmund C, Rizo J. A minimal domain responsible for Munc13 activity. Nat. Struct. Mol. Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- Basu J, Betz A, Brose N, Rosenmund C. Munc13-1 C1 domain activation lowers the energy barrier for synaptic vesicle fusion. J. Neurosci. 2007;27:1200–1210. doi: 10.1523/JNEUROSCI.4908-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Ashery U, Rickmann M, Augustin I, Neher E, Süudhof TC, Rettig J, Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Süudhof TC. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J. Biol. Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- Calloway N, Gouzer G, Xue M, Ryan TA. The active-zone protein Munc13 controls the use-dependence of presynaptic voltage-gated calcium channels. eLife. 2015;4 doi: 10.7554/eLife.07728. http://dx.doi.org/10.7554/eLife.07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- Chen C, Regehr WG. Contributions of residual calcium to fast synaptic transmission. J. Neurosci. 1999;19:6257–6266. doi: 10.1523/JNEUROSCI.19-15-06257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AP, Fioravante D. Translating neuronal activity at the synapse: presynaptic calcium sensors in short-term plasticity. Front. Cell. Neurosci. 2014;8:356. doi: 10.3389/fncel.2014.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JS, Jahr CE. Asynchronous release of synaptic vesicles determines the time course of the AMPA receptor-mediated EPSC. Neuron. 1995;15:1097–1107. doi: 10.1016/0896-6273(95)90098-5. [DOI] [PubMed] [Google Scholar]
- Dittman JS, Kaplan JM. Behavioral impact of neurotransmitter-activated G-protein-coupled receptors: muscarinic and GABAB receptors regulate Caenorhabditis elegans locomotion. J. Neurosci. 2008;28:7104–7112. doi: 10.1523/JNEUROSCI.0378-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravante D, Regehr WG. Short-term forms of presynaptic plasticity. Curr. Opin. Neurobiol. 2011;21:269–274. doi: 10.1016/j.conb.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravante D, Chu Y, de Jong AP, Leitges M, Kaeser PS, Regehr WG. Protein kinase C is a calcium sensor for presynaptic short-term plasticity. eLife. 2014;3:e03011. doi: 10.7554/eLife.03011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Rosenmund C. On the brink: a new synaptic vesicle release model at the calyx of held. Neuron. 2015;85:6–8. doi: 10.1016/j.neuron.2014.12.038. [DOI] [PubMed] [Google Scholar]
- Hobson RJ, Liu Q, Watanabe S, Jorgensen EM. Complexin maintains vesicles in the primed state in C. elegans. Curr. Biol. 2011;21:106–113. doi: 10.1016/j.cub.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Hom S, Kudze T, Tong XJ, Choi S, Aramuni G, Zhang W, Kaplan JM. Neurexin and neuroligin mediate retrograde synaptic inhibition in C. elegans. Science. 2012;337:980–984. doi: 10.1126/science.1224896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Tong XJ, Kaplan JM. UNC-13L, UNC-13S, and Tomosyn form a protein code for fast and slow neurotransmitter release in Caenorhabditis elegans. eLife. 2013;2:e00967. doi: 10.7554/eLife.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igumenova TI. Dynamics and membrane interactions of protein kinase C. Biochemistry. 2015;54:4953–4968. doi: 10.1021/acs.biochem.5b00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig C, Min SW, Krinner S, Arancillo M, Rosenmund C, Südhof TC, Rhee J, Brose N, Cooper BH. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 2014;84:416–431. doi: 10.1016/j.neuron.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Ishisaka M, Hara H. The roles of diacylglycerol kinases in the central nervous system: review of genetic studies in mice. J. Pharmacol. Sci. 2014;124:336–343. doi: 10.1254/jphs.13r07cr. [DOI] [PubMed] [Google Scholar]
- Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korogod N, Lou X, Schneggenburger R. Posttetanic potentiation critically depends on an enhanced Ca(2+) sensitivity of vesicle fusion mediated by presynaptic PKC. Proc. Natl. Acad. Sci. USA. 2007;104:15923–15928. doi: 10.1073/pnas.0704603104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Ho WK, Neher E, Lee SH. Superpriming of synaptic vesicles after their recruitment to the readily releasable pool. Proc. Natl. Acad. Sci. USA. 2013;110:15079–15084. doi: 10.1073/pnas.1314427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipstein N, Schaks S, Dimova K, Kalkhof S, Ihling C, Kölbel K, Ashery U, Rhee J, Brose N, Sinz A, Jahn O. Nonconserved Ca(2+)/calmodulin binding sites in Munc13s differentially control synaptic short-term plasticity. Mol. Cell. Biol. 2012;32:4628–4641. doi: 10.1128/MCB.00933-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipstein N, Sakaba T, Cooper BH, Lin KH, Strenzke N, Ashery U, Rhee JS, Taschenberger H, Neher E, Brose N. Dynamic control of synaptic vesicle replenishment and short-term plasticity by Ca(2+)-calmodulin-Munc13-1 signaling. Neuron. 2013;79:82–96. doi: 10.1016/j.neuron.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Lipstein N, Verhoeven-Duif NM, Michelassi FE, Calloway N, van Hasselt PM, Pienkowska K, van Haaften G, van Haelst MM, van Empelen R, Cuppen I, et al. Synaptic UNC13A protein variant causes increased neurotransmission and dyskinetic movement disorder. J. Clin. Invest. 2017;127:1005–1018. doi: 10.1172/JCI90259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hollopeter G, Jorgensen EM. Graded synaptic transmission at the Caenorhabditis elegans neuromuscular junction. Proc. Natl. Acad. Sci. USA. 2009;106:10823–10828. doi: 10.1073/pnas.0903570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Seven AB, Camacho M, Esser V, Xu J, Trimbuch T, Quade B, Su L, Ma C, Rosenmund C, Rizo J. Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife. 2016;5 doi: 10.7554/eLife.13696. http://dx.doi.org/10.7554/eLife.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Scheuss V, Schneggenburger R. Allosteric modulation of the presynaptic Ca2+ sensor for vesicle fusion. Nature. 2005;435:497–501. doi: 10.1038/nature03568. [DOI] [PubMed] [Google Scholar]
- Lou X, Korogod N, Brose N, Schneggenburger R. Phorbol esters modulate spontaneous and Ca2+-evoked transmitter release via acting on both Munc13 and protein kinase C. J. Neurosci. 2008;28:8257–8267. doi: 10.1523/JNEUROSCI.0550-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JM, Nurrish S, Kaplan JM. UNC-13 interaction with syntaxin is required for synaptic transmission. Curr. Biol. 2005;15:2236–2242. doi: 10.1016/j.cub.2005.10.049. [DOI] [PubMed] [Google Scholar]
- Mahoney TR, Luo S, Nonet ML. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 2006;1:1772–1777. doi: 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- Martin JA, Hu Z, Fenz KM, Fernandez J, Dittman JS. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr. Biol. 2011;21:97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen JM, Madison JM, Dybbs M, Kaplan JM. Antagonistic regulation of synaptic vesicle priming by Tomosyn and UNC-13. Neuron. 2006;51:303–315. doi: 10.1016/j.neuron.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Harada H, Kamasawa N, Matsui K, Rothman JS, Shigemoto R, Silver RA, DiGregorio DA, Takahashi T. Nanoscale distribution of presynaptic Ca(2+) channels and its impact on vesicular release during development. Neuron. 2015;85:145–158. doi: 10.1016/j.neuron.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. Merits and limitations of vesicle pool models in view of heterogeneous populations of synaptic vesicles. Neuron. 2015;87:1131–1142. doi: 10.1016/j.neuron.2015.08.038. [DOI] [PubMed] [Google Scholar]
- Nurrish S, Ségalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron. 1999;24:231–242. doi: 10.1016/s0896-6273(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc. Natl. Acad. Sci. USA. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Russell RL. Molecular basis of drug-resistance mutations in C. elegans. Psychopharmacol. Bull. 1985;21:623–630. [PubMed] [Google Scholar]
- Rhee JS, Betz A, Pyott S, Reim K, Varoqueaux F, Augustin I, Hesse D, Südhof TC, Takahashi M, Rosenmund C, Brose N. Beta phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs. Cell. 2002;108:121–133. doi: 10.1016/s0092-8674(01)00635-3. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Süudhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu. Rev. Cell Dev. Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- Rizo J, Xu J. The synaptic vesicle release machinery. Annu. Rev. Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Clements JD, Westbrook GL. Nonuniform probability of glutamate release at a hippocampal synapse. Science. 1993;262:754–757. doi: 10.1126/science.7901909. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Sigler A, Augustin I, Reim K, Brose N, Rhee JS. Differential control of vesicle priming and short-term plasticity by Munc13 isoforms. Neuron. 2002;33:411–424. doi: 10.1016/s0896-6273(02)00568-8. [DOI] [PubMed] [Google Scholar]
- Schlüuter OM, Basu J, Süudhof TC, Rosenmund C. Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J. Neurosci. 2006;26:1239–1246. doi: 10.1523/JNEUROSCI.3553-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Guryev O, Rizo J. Intramolecular occlusion of the diacylglycerol-binding site in the C1 domain of munc13-1. Biochemistry. 2005;44:1089–1096. doi: 10.1021/bi0476127. [DOI] [PubMed] [Google Scholar]
- Shin OH, Xu J, Rizo J, Süudhof TC. Differential but convergent functions of Ca2+ binding to synaptotagmin-1 C2 domains mediate neurotransmitter release. Proc. Natl. Acad. Sci. USA. 2009;106:16469–16474. doi: 10.1073/pnas.0908798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Lu J, Rhee JS, Tomchick DR, Pang ZP, Wojcik SM, Camacho-Perez M, Brose N, Machius M, Rizo J, et al. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat. Struct. Mol. Biol. 2010;17:280–288. doi: 10.1038/nsmb.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DR, Wu ZX, Matti U, Junge HJ, Schirra C, Becherer U, Wojcik SM, Brose N, Rettig J. Identification of the minimal protein domain required for priming activity of Munc13-1. Curr. Biol. 2005;15:2243–2248. doi: 10.1016/j.cub.2005.10.055. [DOI] [PubMed] [Google Scholar]
- Striegel AR, Biela LM, Evans CS, Wang Z, Delehoy JB, Sutton RB, Chapman ER, Reist NE. Calcium binding by synaptotagmin’s C2A domain is an essential element of the electrostatic switch that triggers synchronous synaptic transmission. J. Neurosci. 2012;32:1253–1260. doi: 10.1523/JNEUROSCI.4652-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süudhof TC. The presynaptic active zone. Neuron. 2012;75:11–25. doi: 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süudhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschenberger H, Woehler A, Neher E. Superpriming of synaptic vesicles as a common basis for intersynapse variability and modulation of synaptic strength. Proc. Natl. Acad. Sci. USA. 2016;113:E4548–E4557. doi: 10.1073/pnas.1606383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu-Sekine B, Raben DM. Regulation and roles of neuronal diacylglycerol kinases: a lipid perspective. Crit. Rev. Biochem. Mol. Biol. 2011;46:353–364. doi: 10.3109/10409238.2011.577761. [DOI] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Wragg RT, Snead D, Dong Y, Ramlall TF, Menon I, Bai J, Eliezer D, Dittman JS. Synaptic vesicles position complexin to block spontaneous fusion. Neuron. 2013;77:323–334. doi: 10.1016/j.neuron.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wragg RT, Parisotto DA, Li Z, Terakawa MS, Snead D, Basu I, Weinstein H, Eliezer D, Dittman JS. Evolutionary divergence of the C-terminal domain of complexin accounts for functional disparities between vertebrate and invertebrate complexins. Front. Mol. Neurosci. 2017;10:146. doi: 10.3389/fnmol.2017.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Camacho M, Xu Y, Esser V, Liu X, Trimbuch T, Pan YZ, Ma C, Tomchick DR, Rosenmund C, Rizo J. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife. 2017;6 doi: 10.7554/eLife.22567. http://dx.doi.org/10.7554/eLife.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang S, Sheng Y, Zhang M, Zou W, Wu L, Kang L, Rizo J, Zhang R, Xu T, Ma C. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat. Struct. Mol. Biol. 2015;22:547–554. doi: 10.1038/nsmb.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Stawicki TM, Goncharov A, Jin Y. Position of UNC-13 in the active zone regulates synaptic vesicle release probability and release kinetics. eLife. 2013;2:e01180. doi: 10.7554/eLife.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.