Abstract
Genome-wide transcriptome profiling has enabled non-supervised classification of tumours, revealing different sub-groups characterized by specific gene expression features. However, the biological significance of these subtypes remains for the most part unclear. We describe herein an interactive platform, Minimum Spanning Trees Inferred Clustering (MiSTIC), that integrates the direct visualization and comparison of the gene correlation structure between datasets, the analysis of the molecular causes underlying co-variations in gene expression in cancer samples, and the clinical annotation of tumour sets defined by the combined expression of selected biomarkers. We have used MiSTIC to highlight the roles of specific transcription factors in breast cancer subtype specification, to compare the aspects of tumour heterogeneity targeted by different prognostic signatures, and to highlight biomarker interactions in AML. A version of MiSTIC preloaded with datasets described herein can be accessed through a public web server (http://mistic.iric.ca); in addition, the MiSTIC software package can be obtained (github.com/iric-soft/MiSTIC) for local use with personalized datasets.
INTRODUCTION
Correlated gene expression has been studied to infer biological function, as genes whose expression is highly correlated across a large number of samples often participate in a common biological process or signalling pathway (1,2). In particular, this approach can yield valuable information on the mechanisms of tumourigenesis. Genes that display coordinated changes in expression levels between samples may be targeted by genetic events resulting in altered expression or activity of common epigenetic regulators, transcription factors or miRNAs; alternatively, when correlated genes occupy a discrete chromosomal locus, their altered expression may result from localized chromosomal aberrations (deletion or amplification). The precision of gene correlation analyses has been further boosted by the recent advent of transcriptome sequencing (RNA-Seq), which offers greater fidelity and dynamic range compared to gene expression microarrays (3,4). The availability of large public datasets for collections of normal or tumour tissues (e.g. datasets produced by The Cancer Genome Atlas and the International Cancer Genome Consortium, (5–7)) also offers unique opportunities to compare gene correlation patterns between different cancers to illuminate the specificity or generality of the mechanisms of tumourigenesis at play.
Analytic tools such as GOMiner (8), Gene Set Enrichment Analysis (GSEA) (9) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) (10) have been developed to enable biological interpretation of gene lists derived from transcriptome profiles. However, most currently available methods or platforms for gene expression correlation analysis do not readily enable annotation of gene clusters with information relevant to the potential mechanisms underlying clustering (e.g. enrichment in gene sets associated with specific cytobands or containing sites or chromatin regions bound by specific transcription factors), or clinical annotation of subsets of samples determined by expression of genes identified as potential markers through correlation analysis. Furthermore, there is a lack of tools for the systematic comparison of gene correlation structures between different datasets.
The complexity of gene coexpression networks constructed from genome-wide expression profiles is an obstacle to a useful visualization of correlation structures enabling interactive analyses at different levels of granularity. The large number of nodes and edges invariably produces a ‘hairball’ in which dense local features are obscured. Using more stringent thresholds for retaining edges on the other hand removes gene interactions, and the information that they impart, from the network. Existing tools designed to address these issues tend to focus on providing methods for automated identification of sub-graphs or clusters presenting pre-defined criteria. This approach is typified by Cytoscape, which includes a number of functionalities to explore networks based on expression datasets (11). The combination of a dendrogram and a heat map (12,13), the dominant visual representation for gene expression data, is also unwieldy in the case of human expression profiling, where the number of elements to cluster is >20 000. Even when preselecting a few hundred genes, it is difficult to reconcile the dendrogram with coloured patterns seen in the heat map. When lists of co-expressed genes are needed, the dendrogram is ‘cut’ at a given similarity threshold and gene clusters are recovered from the resulting disjoint trees. The selection of a similarity threshold is typically done globally for a dendrogram, making it difficult to present clusters forming at different thresholds. The dendrogram and heat map approach also suffers from the inability to graphically represent correlation-based comparisons between datasets.
Here, we report the development of a new software tool, Minimum Spanning Trees Inferred Clustering (MiSTIC), which addresses a critical unmet need, namely the availability of intuitive software for tapping the power of gene expression correlations and integrative analysis in large quantitative datasets to reveal biological insights in the mechanisms of cell differentiation and tumourigenesis. MiSTIC offers a simultaneous view of all gene correlations in a set of transcriptomes through the use of minimum spanning trees (14) and a radial projection of the associated hierarchical clustering using an icicle representation (15). Further, MiSTIC was designed to easily navigate back and forth between representations of gene correlation at the level of a global dataset (icicle view), of a gene cluster (by zooming in on individual clusters in the icicle) or of individual genes or transcripts (single gene correlations and pair-wise scatterplots), enabling both global and targeted analyses. MiSTIC maximizes the value of high-resolution RNA-Seq gene expression correlations by providing an interactive interface for visualizing, analyzing and integrating these data with other types of information (including gene ontology, chromosomal location, microarrays, ChIP-Seq, TFBS predictions). Importantly, this interface also enables representation of pair-wise comparisons of dataset correlative structures. Finally, MiSTIC also enables sorting of sample cohorts into sub-populations based on expression levels of genes identified as potential markers through correlation analysis, and performs enrichment analysis in clinical annotations to better characterize groups of tumours thus identified.
Several examples are shown where MiSTIC revealed dataset-specific clusters of gene expression and helped formulate testable hypotheses on the mechanisms responsible for cluster formation in RNA-Seq datasets. Our examples are chosen to illustrate the capacity to refine existing biological knowledge, reveal novel connections and extend molecular classification in both acute myeloid leukaemia (AML) and breast cancer.
MATERIALS AND METHODS
Datasets
A total of 27 different RNA-Seq datasets were used in this study (Supplementary Table S1). Dataset 1 consists of RNA-Seq results from 152 human AML samples from the Leucegene project (Gene Expression Omnibus accession numbers GSE49642, GSE52656, GSE62190, GSE67040, GSE67039). Dataset 2, also called ‘Leucegene AML NK’, represents a subset of dataset 1 and consists of normal karyotype specimens. Taking into consideration that AML specimens are often heavily contaminated with non-leukemic cells (16), we took significant care to select samples with a high proportion of blasts such that the average blast count (before Ficoll gradient separation) was 85% with a minimal value of 65% (Supplementary Table S2). This value is higher than that observed with the TCGA NK-AML dataset (mean 69%, minimum 30%). Not surprisingly, this selection process resulted in the inclusion of a large proportion of AML without maturation (WHO classification) or AML-M1 (FAB classification) representing over 50% of the cases (Supplementary Table S2). RNA-Seq data of 17 normal CD34+ cord blood specimens (dataset 5: purity 70–86% CD34+) are also presented as the normal counterpart of NK-AML, which contained on average 26 ± 15% (range 0–99%, Supplementary Table S2) CD34-positive cells. Datasets 3, 4 and 6–24 are from The Cancer Genome Atlas (TCGA). Dataset 25 is normalized microarray expression data (Affymetrix U133 plus 2.0) from the Cancer Cell Line Encyclopedia (CCLE) (17). Datasets 26 and 27 both correspond to the GNE cell line RNA-Seq dataset from Genentech (rpkm or variance-stabilized data) (18).
Leucegene transcriptome dataset
Sample preparation and RNA-Seq data processing were performed as previously described (19). Briefly, total RNA from AML patient bone marrow or blood samples (∼5 million cells) was isolated using TRIzol (Invitrogen) as recommended by the manufacturer, and then further purified using RNeasy columns (Qiagen). Sample quality was assessed using Bioanalyzer RNA Nano chips (Agilent). Transcriptome libraries were generated from 4 μg total RNA using the TruSeq RNA Sample Prep Kit (v2) (Illumina) following the manufacturer's protocols. Paired-end (2 × 100 bp) sequencing was performed using the Illumina HiSeq2000 machine running TruSeq v3 chemistry. Cluster density was targeted at ∼600–800k clusters/mm2. Two transcriptomes were sequenced per lane. Sequence data were mapped to the reference genome (hg19) using the Illumina Casava 1.8 package and Elandv2e mapping software. An average of 144 million ± 51 million mapped reads were obtained in this dataset (Supplementary Table S2). All the patient samples used in this study were collected by the Leukemia Cell Bank of Quebec (BCLQ) with informed consent and approval of the project by the Research Ethics Board of the Maisonneuve-Rosemont Hospital and Université de Montréal.
Access to public datasets
Publicly available TCGA datasets (LAML tumour; BRCA tumour, normal; COAD tumour; KIRC tumour, normal, LUAD tumour, normal) were downloaded via the Cancer Genome Atlas data portal (https://tcga-data.nci.nih.gov/tcga/). RPKM values (as computed by TCGA and referred to as IlluminaHiSeq_RNASeq) were used in these analyses. The AML clinical data matrix (also downloaded via the TCGA data portal) was used to subset the TCGA AML dataset to normal karyotype samples. The BRCA clinical data matrix was used to subset the TCGA BRCA dataset to ER+, HER2+ and triple negative. The Luminal A and B subsets were identified with the PAM50 signature and the genefu R/Bioconductor package (20). The BRCA miRNA dataset was downloaded from the Broad Institute's Genome Data Analysis Center (http://gdac.broadinstitute.org/runs/stddata__2013_07_15/data/BRCA/20130715/gdac.broadinstitute.org_BRCA.Merge_mirnaseq__illuminahiseq_mirnaseq__bcgsc_ca__Level_3__miR_gene_expression__data.Level_3.2013071500.0.0.tar.gz). The normalized expression data from CCLE dataset was downloaded from GEO (GSE36133) using the R Bioconductor package GEOquery. The RPKM and variance-stabilized data from the GNE dataset were obtained from Array Express (E-MTAB-2706) and from the Klijn et al.'s supplementary data website (http://research-pub.gene.com/KlijnEtAl2014/).
Correlation analysis in MiSTIC
We have implemented MiSTIC as a JavaScript front-end performing AJAX queries to a web server built using the Pyramid web framework in Python. Data rather than graphical representations are sent from the server in JSON format with graphical representation being built on the client browser. This allows for a high level of interactivity. Adding new datasets to MiSTIC is achieved by pre-computation on the server-side of correlation matrices, minimum spanning trees and graph layouts. These steps are implemented in Python and C++, launched from the command-line and typically require a few minutes (e.g. 22 min, Intel i7, 2.7 GHz for a dataset of 60 samples).
Expression data is presented to MiSTIC in matrix form following quantification analysis. Expression values can be subjected to log or rank transformation before computing the correlation matrix (Pearson's correlation coefficient, r). The log transformation is implemented as a started-log with log10(1000x + 1), where x is the untransformed expression values. The small weight of the constant was chosen to minimize compression in the low range of expression values. The minimum spanning tree is then constructed with Kruskal's algorithm (14)), using (1 – r) as a distance. Algorithmically, the limiting step is, in most cases, the computation of the minimum spanning tree, which is done in O(n2 log n) where n is the number of genes. With datasets containing a large number of samples, the limiting step can shift to computing the correlation matrix as it requires time in O(n2m) where m is the number of samples.
Enrichment analysis in gene sets and clinical features
In interfaces where subsets of genes (Icicle and MST representation, Figure 1) or patient samples (scatter plots, Figure 1) are selected, an interactive enrichment analysis is performed using Fisher's exact test and categories that are enriched in the selection with a P-value <0.05 are reported. A q value after correction for multiple testing (false discovery rate, Benjamini–Hochberg) is also presented as a more stringent evaluation of statistical significance; note however that this correction does not take into consideration the related character of different gene lists. Clinical features associated with patient samples are treated as categorical variables. By selecting appropriate thresholds, continuous clinical features are converted into categorical variables (ex. age > 60 versus age ≤ 60), as often performed for analysis of clinical data. This enables enrichment analysis and visualization of these categories in the scatterplots as for discrete categories.
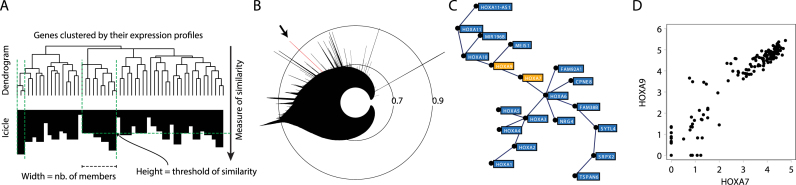
Figure 1.
Visualization of gene expression correlations at different levels of resolution in MiSTIC. (A) Conversion from a dendrogram representation (top) to a classical icicle (bottom). Only clusters of size 3 and above are shown in the icicle. The width and height of peaks indicate the cluster size and the similarity threshold at which it forms. A deep crevice between adjoining peaks indicates a lack of gene correlation between peaks. (B) The icicle is transformed using a power-scale for the similarity measure and circularizing the original plot, the angle corresponding to genes and the radius to similarity measures. The angle at which peaks emerge from the structure reflects the arbitrary ordering of the genes/clusters in the dendrogram. Radius values represent Pearson correlations. (C) Clicking on a peak (arrow in B) generates a graph representation of the corresponding cluster. (D) Selecting two nodes in the cluster (orange labels) and clicking on the scatterplot tab generates a scatterplot representation of samples according to levels of expression of selected genes.
Gene sets from published databases are listed in Supplementary Table S3 (adding new gene sets can be performed using command line tools on the server). ChIP-Seq gene sets contain lists of RefSeq and miRNA genes with at least one ChIP-Seq region associated with a specific TF within 25, 10 or 5 kb of their transcription start site (TSS). They were created with the IntersectBed program from BEDTools (21) using the refGene and wgRna annotation tracks of the hg19 UCSC Genome browser database (22). When necessary, the genomic coordinates of the ChIP-Seq regions were converted to hg19 coordinates with the liftOver program from the UCSC (22). Chromosome gene sets contain chromosome and cytoband-restricted lists of RefSeq and miRNA genes. They were created with the IntersectBed program from BEDTools (21) using the cytoBand, refGene and wgRna annotation tracks of the hg19 UCSC Genome browser database (22).
TFBS gene sets contain lists of genes with at least one predicted site within 10, 5 or 2.5 kb of the TSS of RefSeq and miRNA genes determined as previously described (23). Transcription factor binding sites were searched for in the reference human genome sequences (hg17, May 2004, UCSC Genome Browser database). Briefly, different windows (±10, ±5, ±2.5 kb) centered around the TSS of annotated genes in the refGene and wgRna annotation track were extracted from the genome. These sequences were screened with matrices from TRANSFAC Professional 2010.2 (24) for binding sites using a base matrix score of 65%. For each matrix, the average number of sites/gene was calculated in 5% increments between 65% and 100%. In order to avoid artefactual biases due to overly abundant or rare site frequencies, the scores with an average closest to 0.25 site/gene were used for each TF.
Scatterplot analyses and manipulation of highlight groups
In the multi-way scatterplot tool, samples can be selected (green dots) by direct clicking on each sample or by defining a selection area on the plot with the pointer. By using the ‘select a characteristic’ drop-down menu, the current selection will be updated to represent the set of samples sharing this characteristic. A selection can be stored in a ‘highlight group’ by using the ‘+’ icon of a given group. The ‘–’ icon will subtract the current selection from the group, the ‘trash can’ icon will remove all samples from the group and the ‘right arrow’ icon will replace the current selection by the samples labeled by the group. New highlight groups can be added using the ‘new group’ button.
The ‘Group settings’ panel appears when clicking on the group's symbol. A label can be applied to identify the nature of this group. Several features of the symbol can be adjusted: the shape, the fill colour (surface of the symbol), and the stroke colour and width (outline of the symbol). When a shape is selected or the fill colour or stroke colour/width are enabled, selected and saved, samples identified by this group will present these graphical features, defining its state. If ‘inherit’ is selected for any feature, then samples defined by this group will keep the same feature states as in previous highlight groups. This scheme enables four-way visual intersections by combining shape, fill and stroke colour and width linked to different highlight groups.
Since the final state of each graphical feature is dependent on the order of application of highlight groups, it is possible to modify this order by using the up and down arrow icons on the upper-right corner of each group. The ‘x’ icon shown beside the arrows is used to entirely remove a highlight group.
Using MiSTIC
A tutorial video is available from the Help section to guide users in the performance of the various analyses described above; in addition, help buttons provide operational instructions for each function. The MiSTIC source code is available at github.com/iric-soft/MiSTIC.
RESULTS AND DISCUSSION
Global visualization of genome-wide gene expression correlations via circularized icicles
For computational speed and clarity of the resulting representation, gene clustering in MiSTIC is performed using a minimum spanning tree approach (14), edges being created by detecting maximum correlations between pairs of nodes. The associated hierarchical clustering can be visualized using an icicle representation (15), wherein the width of each peak represents the number of its components at a given similarity threshold (Figure 1A). The circularization of the icicle and the use of a power scale for the similarity measure both magnify the useful region of the plot containing clusters of genes with highly similar expression profiles. Concentric lines in the circularized icicle histogram (Figure 1B) represent different levels of clustering, the similarity measure (indicated on the radius) increasing towards the outside of the icicle. Each peak at the periphery of the icicle represents a set of correlated genes (e.g. arrow, Figure 1B) organized in a single cluster or several aggregated clusters depending on the correlation thresholds at which genes are joined to the original cluster giving rise to each peak (see peak structure in Figure 1A). This representation removes the requirement for setting an arbitrary threshold to limit the dataset under investigation and provides a single-image view highlighting all gene expression clusters for any set of transcriptomes included in the MiSTIC server.
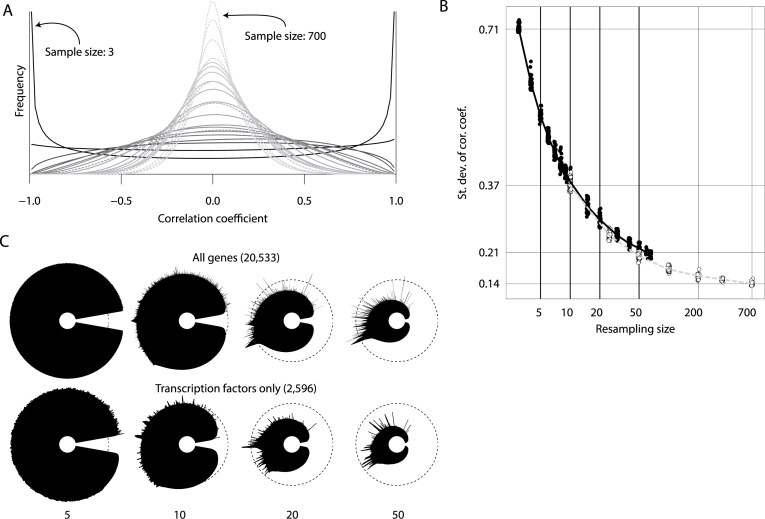
We used resampling of 69 Leucegene AML samples and of the TCGA breast tumour dataset (754 samples) to empirically explore the quantitative impact of sample size on the matrix of pairwise correlations and the qualitative impact on the resulting icicle plot. For each sample size explored, 50 randomly sampled datasets were prepared and correlation matrices built. As the sample size increased, the variance of the distribution of correlations for independent gene pairs decreased, converging upon matrices where most of the calculated correlations are close to zero (Figure 2A). This trend is clearly demonstrated by plotting the standard deviation of all pairwise correlations as a function of sample size (Figure 2B). The Leucegene AML and TCGA breast tumour datasets displayed similar behaviour when resampled at similar levels. Based on this analysis, our approach becomes adequate for datasets containing between 20 and 50 samples or when the standard deviation of the correlation reaches <0.25.
Figure 2.
Effect of sample size on icicle representation. Datasets with reduced sample size were obtained by resampling for both a subset of the Leucegene AML dataset (69 samples) and of the TCGA breast tumour dataset (754 samples). For each sample size explored, 50 datasets were prepared and correlation matrices built. (A) Distribution of correlation coefficients for one resampled dataset per sample size. Plain lines are used for resampled datasets derived from Leucegene AML and dashed lines for resampled datasets derived from TCGA breast tumours. The gray shade indicates the sample size, ranging from 3 (black) to 700 (light gray). (B) Standard deviation of correlation coefficients obtained from resampled datasets. The deviations shown on the vertical axis correspond to the average computed for the minimum and maximum sample size of both original datasets. Open circles: TCGA; dark circles: Leucegene (C) Icicle representations were built and displayed in MiSTIC for 5, 10, 20 and 50 samples with either all protein-coding genes or only genes coding for transcription factors. Note the increase in peak prominence as sample sizes increase to 50 specimens.
Figure 2C depicts icicles generated from sample sets of different sizes (5,10,20,50) using all protein-coding genes (n = 20 533) and illustrates that when too few samples are available, all edges of the spanning tree are correlations close to 1, making it impossible to identify true clusters among the noise. When the number of samples is increased to 50 samples, peaks emerge and become sharply defined. Similar results in terms of peak separation were obtained using a dataset with a smaller number of features, i.e. genes coding for transcription factors only (n = 2596), albeit with lower correlations.
Visualization of gene expression correlation and gene set enrichment: from whole genomes to individual genes
Icicles can be generated from minimum spanning trees for each dataset within MiSTIC (log representation is the default option) in the Dataset and icicle tab (Supplementary Figure S1A and B). Beyond the visualization of correlation peaks in the icicle view, it is possible to download the lists of genes associated with these peaks as an Excel table (Extract peaks tool). It is also possible to perform an enrichment analysis (Gene set enrichment tool) for pre-determined gene sets (categories ChIP-Seq, Chromosome, CNV, CPDB, GeneFamilies, GeneSigDB (25), Microarrays, MiRNA, MSigDB (9), Target genes, Transcription Factor Binding sites; see Supplementary Table S3). A gradient from red to grey is used to display the over-representation (determined by a Fisher's exact test) of genes from a selected set in each cluster (Supplementary Figure S1B). In addition, the localization of individual genes in the clusters/peaks forming the icicle can be visualized using the Locate tool (Supplementary Figure S1C).
Gene clusters in the icicle can be viewed individually by a simple click on the corresponding section of the icicle (e.g. peak highlighted by an arrow in Figure 1B, resulting in the graph representing the same cluster in Figure 1C); note that it is possible to zoom in on any region of the icicle to help select the desired feature. The minimum spanning tree edges show the highest correlations linking each member in the cluster, and allow a planar representation without creating a hairball (Figure 1C). Genes are displayed in an interactive layout that enables manual rearrangement (dynamic force-directed layout) for greater visibility. Labels display gene symbols as the default option but can be converted to full names with the Change label option or removed using the Toggle labels option to better display the topology of the network. A list of all genes in the cluster can be obtained by clicking on the Select all button. For each displayed cluster, a downloadable table automatically provides statistically significant enrichment results (Supplementary Figure S2A, right panel) using all gene sets listed in Supplementary Table S3. Clicking on individual gene/transcript labels in the cluster selects them (Supplementary Figure S2A, orange labels) and provides links to external gene databases (below the enrichment table) for the latest selected item. A list of selected genes (either from iterative manual selection in the cluster or from selection of a gene set in the enrichment table) can be downloaded using the Copy button (Supplementary Figure S2A, top left).
The pairwise correlation scatterplot representation can be accessed from the gene cluster representation by activating the Go to scatterplot button after manual selection of at least two genes, revealing specimen-specific expression values (log-transformed RPKM values) for all gene/transcript pairs (Figure 1D and Supplementary Figure S2B). It can also be directly accessed from MiSTIC's main menu by selecting the desired dataset and serially entering genes to analyze, resulting in a series of pair-wise scatterplot graphs. Specimens selected individually or as groups in one graph are automatically labelled also in all other pairwise graphs (see green dots in Supplementary Figure S2B) and are listed at the bottom of the page. This view also presents enrichment in patient/tumour annotations (tumour type, age of patient, survival, gene mutation status, etc.) for selected specimens in a sample term enrichment table (Supplementary Figure S2B, red arrow). This greatly facilitates the identification of novel prognostic genes or the characterization of clinically-relevant tumour subsets (see below). The simultaneous annotation of different subsets of specimens is enabled by storing selections iteratively using the + sign in the Highlight group toolbox (blue arrow in Supplementary Figure S2B). Samples are thus added to the group's sample list; group names and label colour/shape can be modified by clicking on the coloured dot for each group (red arrow in Supplementary Figure S2C). Views built with the scatterplot representation can be saved either as a pdf or as a link to the scatterplot html page.
The Single gene correlation tool, accessible from a separate tab in the main menu of MiSTIC (Supplementary Figure S2D, red arrow), displays for a single query gene a scatterplot of sorted correlations of this gene with respect to all others and identifies the most correlated and anti-correlated genes in the dataset. In this view, it is possible to visually assess enrichment of a given gene set as a barcode on the x-axis (Supplementary Figure S2D). Newly identified correlated or anti-correlated genes can be in turn visualized in the icicle using the Locate option (see above).
Thus, the visualization mode of gene correlations in MiSTIC is adaptable to functional annotation at different levels of analysis, thereby providing a flexible integration platform for dataset mining (please also refer to the video tutorial in the Help section).
Comparison of gene correlation clusters between different datasets
One powerful aspect of icicles is that pairs of datasets can be superimposed in one image for rapidly assessing conservation of expression clusters. A reference dataset is visualized and a second dataset can be chosen for comparative analysis in the Dataset comparison section (Supplementary Figure S1D). In this analysis, gene clusters corresponding to subsections of the peaks in the reference icicle are each compared to all gene clusters of the comparing dataset using Cohen's kappa coefficient (chosen for computational frugality due to the need to perform this comparison on demand). Cluster overlap is represented on the reference icicle view using a colour scale (from light to dark blue for low to high levels of overlap, see examples below; pointing the cursor on a cluster highlights the corresponding kappa coefficient). It is possible to locate genes within this cluster in the second dataset if desired using the Locate tool on the icicle corresponding to this dataset.
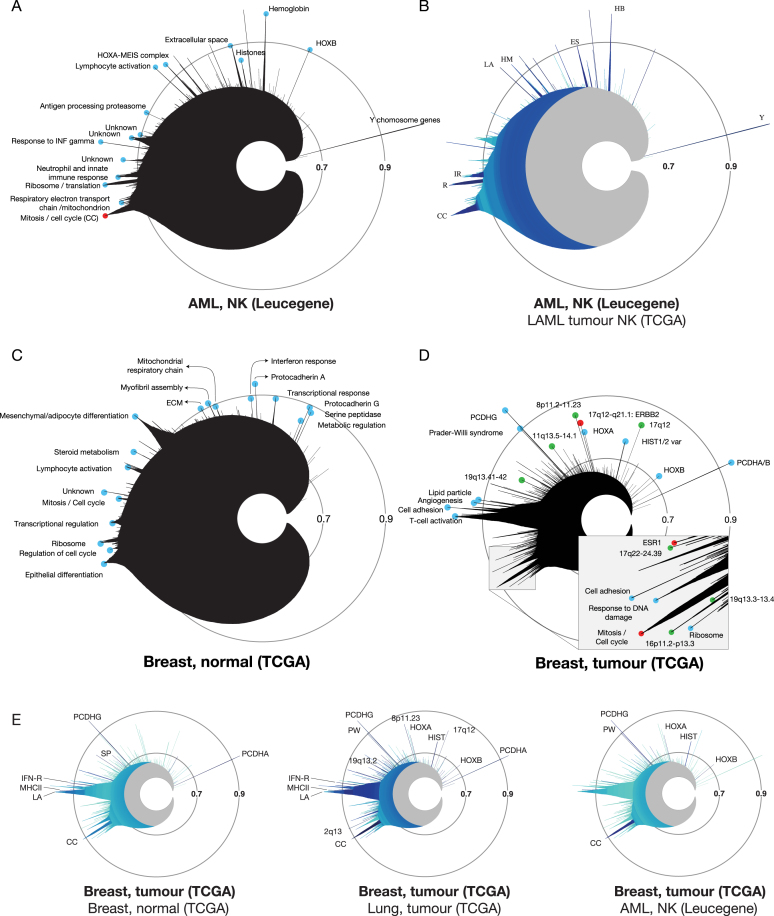
Superposition of the icicle plot derived from dataset 2 (Leucegene AML normal karyotype (NK), Figure 3A) with that of dataset 4 (TCGA AML NK) indicates that the vast majority of the expression clusters are validated in a second, unrelated experimental setup (Figure 3B). The strong correlation between both NK-AML datasets is illustrated by the deep blue colour of several peaks, which were enriched in Gene Ontology, MSigDB/GeneSigDB, or CPDB terms corresponding to cell cycle (CC), ribosome (R), innate immune response (IR), lymphocyte activation (LA), Hox-Meis (HM), extracellular space (ES) and haemoglobin (HB) (Figure 3A and B). On the other hand, superposition of normal CD34+ human cord blood cells (dataset 5, Supplementary Table S1) over Leucegene AML (several of which are also CD34+) indicates that the majority of peaks are light blue (low degree of conservation) with the exception of two correlation clusters, the ribosome (R) and the Y chromosome (Y) (Supplementary Figure S3A).
Figure 3.
Imaging and comparing gene expression clusters in cancer and normal tissues with MiSTIC. (A) Icicle of the Leucegene AML NK dataset (dataset 2, Supplementary Table S1). Blue circles identify named peaks. The mitosis/cell cycle peak is labeled in red. (B) Comparative icicle of Leucegene versus TCGA AML NK datasets (dataset 2 versus 4). Dark shades indicate that similar clusters are found in both datasets while light blue indicates lack of conservation. (C) Icicle of normal TCGA breast samples (dataset 6, Supplementary Table S1). (D) Icicle of the TCGA breast tumour dataset (dataset 7, Supplementary Table S1). Green circles correspond to indicated chromosomal loci. The mitosis/cell cycle peak and peaks corresponding to the ESR1 and ERBB2 gene clusters are labeled in red. (E) Representations of the TCGA breast cancer icicle highlighted for conservation with normal breast tissue (dataset 6), lung adenocarcinoma (dataset 23) or TCGA AML NK (dataset 4). Abbreviations: CC: cell cycle/mitosis, ES: extracellular space, IFN-R: interferon response, HB: haemoglobin, HM: Hox Meis, IR: immune response, LA: lymphocyte activation, MHCII: MHC class II antigen processing and presentation, HIST: histones, PW: Prader-Willi syndrome, R: ribosome, SP: serine-protease activity, TR: transcriptional regulation, Y: Y chromosome.
Similar to the comparison between leukaemia and normal CD34+ samples, comparison of the most correlated expression clusters in normal breast tissue (dataset 6, icicle in Figure 3C) with those of breast cancer (dataset 7, icicle in Figure 3D) also shows a lack of concordance between these 2 datasets (Figure 3E) with the exception of a few conserved clusters such as the protocadherin alpha/beta and gamma clusters on chromosome 5q31 (PCDHA/B, PCDHG), whose clustering is likely due to overlapping transcription, and clusters of genes associated with MHC class II (MHCII), cellular response to type I interferon (IFN-R) and serine-protease activity (SP) when analyzed for enrichment analysis with Gene Ontology, MSigDB/GeneSigDB and CPDB gene sets. Peaks conserved to a lesser extent include the cell cycle (CC) and lymphocyte activation (LA) correlation clusters. Similar observations were made when comparing kidney cancer and, to a lesser degree, lung cancer with normal tissues (datasets 20–23, not shown), supporting extensive gene expression reprogramming during tumourigenesis (26).
Interestingly, we found that similarity in expression clusters can be greater between different types of cancers than that observed between each cancer and its normal matched tissue (e.g. breast cancer versus normal breast or versus lung cancer, Figure 3E). This is especially true when comparing different types of solid tumours. However, comparison of breast cancer and leukaemia datasets revealed little conservation except for cell cycle (CC), HOXA and HOXB, Prader Willi (PW) syndrome and histones (HIST) clusters (Figure 3E).
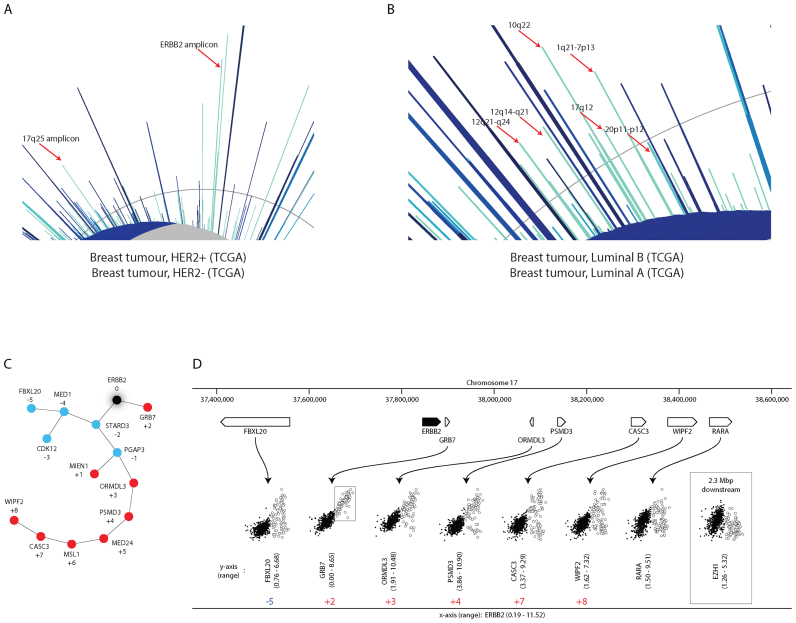
Because breast cancer is a heterogeneous disease comprising several subtypes, we examined whether some of the peaks differ between tumour subtypes by separately entering subsets of tumours identified as ER+, HER2+ and triple-negative by pathological analysis, or subtypes predicted by transcriptome analysis (e.g. luminal B versus luminal A tumours). Several peaks are indeed specific to certain subtypes (light blue peaks in Figure 4A and B). For instance, as expected, one of the peaks present in HER2+ but not conserved in HER2– tumours contains the ERBB2 gene, which encodes the HER2/Neu oncogenic protein (Figure 4A and C), suggesting this peak reflects the impact of chromosomal amplification on gene expression (see also below).
Figure 4.
Identification of clusters differentially represented in breast cancer subtypes. (A) The HER2+ breast cancer dataset (#10, Supplementary Table S1) derived from the TCGA breast cancer dataset (#7, Supplementary Table S1) is shown highlighted for conservation with the complementary HER2– breast cancer dataset (#11, Supplementary Table S1). (B) The luminal B breast cancer dataset (#15, Supplementary Table S1) derived from the TCGA breast cancer dataset is shown highlighted for conservation with the luminal A cancer dataset (#14, Supplementary Table S1). (C) Minimum spanning tree representation of the ERBB2 gene cluster. Numbers show the relative position of each gene in the cluster with respect to ERBB2 (taken as origin). Colours represent the location of the genes centromeric (blue) or telomeric (red) with respect to ERBB2. (D) Variations in correlation with ERBB2 gene expression across the 17q12-q21.1 locus. Scatter plots are shown for selected genes within the ERBB2 cluster, evidencing the progressive drop in correlation as the distance from ERBB2 increases. In addition, correlations with RARA, a gene found at the end of the large ERBB2 amplicon and EZH1, a gene situated well outside the amplicon, are also shown. Empty circles correspond to tumours with high expression of ERBB2. Between parentheses are minimum and maximum expression levels in log-transformed RPKM. Gene numbering is shown as in C.
Analyzing the basis for correlation clusters in cancer datasets
Cluster formation can be explained by at least three distinct mechanisms, which are illustrated below using examples selected from our analysis of AML and breast cancer datasets.
Cellular heterogeneity
Normal human breast is mainly composed of epithelial cells, adipocytes/fibroblasts and inflammatory cells (27). Genes known to be specifically expressed in these cell types form easily identifiable clusters in the icicle generated from the TCGA normal breast samples (Figure 3C), including an ‘epithelial differentiation’ peak, a ‘mesenchymal/adipocyte differentiation’ and a ‘lymphocyte activation’ peak (see Supplementary Table S4 for enrichment in relevant GO, MSigDB and CPDB terms).
Chromosome amplification or deletions and sex chromosome differences
Several clusters that are specific to breast cancer versus normal tissue could be readily attributed to recurrent amplification or deletion of specific chromosomal regions using enrichment analysis with chromosomal bands and with MSigDBgene lists and correspond to known chromosomal deletions or amplifications in breast cancer (28) (see green dots in Figure 3D for some examples). For instance, the ERBB2 cluster is enriched in genes located in chromosomal cytobands 17q12 (p = 2.33 × 10−17) and 17q21 (p = 5.32 × 10−8) and in MSigDB terms ‘NIKOLSKI breast cancer 17q11-q21 amplicon’ (p = 5.45 × 10−31) and ‘SMID breast cancer elevated in ERRB2+’ (p = 9.02 × 10−17). Interestingly, the ERBB2 cluster structure reflects the linear gene organization within the 17q12-21 locus (see numbers in Figure 4C and D). ERBB2 is best correlated with the closest genes compared to more distant ones; pair-wise gene correlations in the scatterplot format indicate that the percentage of tumours with high expression levels of ERBB2 also having high expression of another gene in the cluster decreases with the distance from ERBB2 (Figure 4D), compatible with heterogeneous limits of the amplification region (29). Thus, the correlation tree of amplification or deletion peaks readily provides information on the genes whose expression is affected by these defects.
Similarly, most of the peaks that differ between breast cancer subtypes contain genes in single or adjacent chromosomal bands, suggestive of amplifications or deletions in these regions being more frequent in one subtype versus the other. This is the case for instance for the ERBB2 amplicon in HER2+ versus HER2– tumours (Figure 4A). Comparison with copy number annotations of the TCGA breast cancer dataset (30) indicates that several other peaks are enriched in genes present in regions of recurrent amplification (e.g. 11q14.1, 12q15, 13q34, 19q13.42) or deletion (e.g. 2q37.3, 5q11,2, 6q15, 9q21.11, 10q23.31, 10q26.3, 12q24.31, 14q24.1, 16q24.3, 17q21.3). Note that enrichment in gene sets associated with GISTIC2 predicted regions of amplifications/deletions can be revealed directly in the icicle using the ‘CNV.BRCA1:1: genes in significant amplifications/deletions’ gene sets, or in the enrichment table for each selected cluster. In addition, several peaks associated with specific chromosome bands identified in luminal B, but not luminal A tumours (Figure 4B), were confirmed using the cBioportal for Cancer Genomics (6,7) as containing genes co-amplified with variable but often low frequencies, consistent with the greater genetic instability associated with the lumB versus lumA subtype (31). Of note, some amplification or deletion clusters in breast cancer are also conserved in other solid tumours, such as lung and colon cancer (Figure 3E, Supplementary Figure S3B). For instance, the 8q24.3 cluster is conserved between breast cancer and colon cancer (Supplementary Figure S3B), in keeping with the reported amplification of this region in both cancer types (32–34), also observed using cBioportal. However, gene expression clusters associated with discrete chromosomal bands occur infrequently in leukaemia (Figure 3A and E), in keeping with a different landscape of genetic defects compared to solid tumours (28).
Heterogeneity of sex chromosomes (rather than chromosome amplification or deletion) can also affect gene expression within tumours and result in cluster formation. The most striking example is the Y chromosome gene cluster in the Leucegene AML dataset (see for instance Figure 3A and B), which is not correlated with any other group of genes and results from sex variation in this set of patients. This correlation peak is observed in all datasets examined to date that contain patients of both sexes, although it is much smaller in the TCGA breast cancer dataset, likely due to the low number of male patients (7/754).
Transcriptional regulatory networks
Some peaks that correspond to genes found in discrete chromosomal bands cannot be attributed to chromosomal amplification/deletion or chromosomal heterogeneity in the tumour population. This is the case for the HOXA/B cluster(s) and their co-factors MEIS1/PBX3 in the AML dataset (Supplementary Figure S4A). Intriguingly, HOXA/B cluster genes do not correlate with their MEIS/PBX co-factors in lung tumours or other solid tumours (Supplementary Figure S4B and C), suggesting their association due to regulatory mechanisms specific to leukemic cells. Examination of the results of enrichment analysis with genes containing predicted TF binding sites (TFBS category) or regions associated with TFs in ChIP-Seq experiments (ChIP-Seq category) in the HOXA/B cluster in the TCGA AML NK dataset confirms enrichment of known retinoic acid target genes (35) (33.38-fold, p = 1.21 × 10−5), of genes with chromatin regions associated with RARγ in LoVo cells (3.07, p = 4.76 × 10−3) or with RARα in breast cancer cells (3.29-fold, p = 5.37 × 10−3) and of genes with predicted RAR binding sites conserved in three species (130.57, p = 1.75 × 10−9) (36), consistent with the role of retinoic acid signalling in regulating expression of HOXA/B cluster genes.
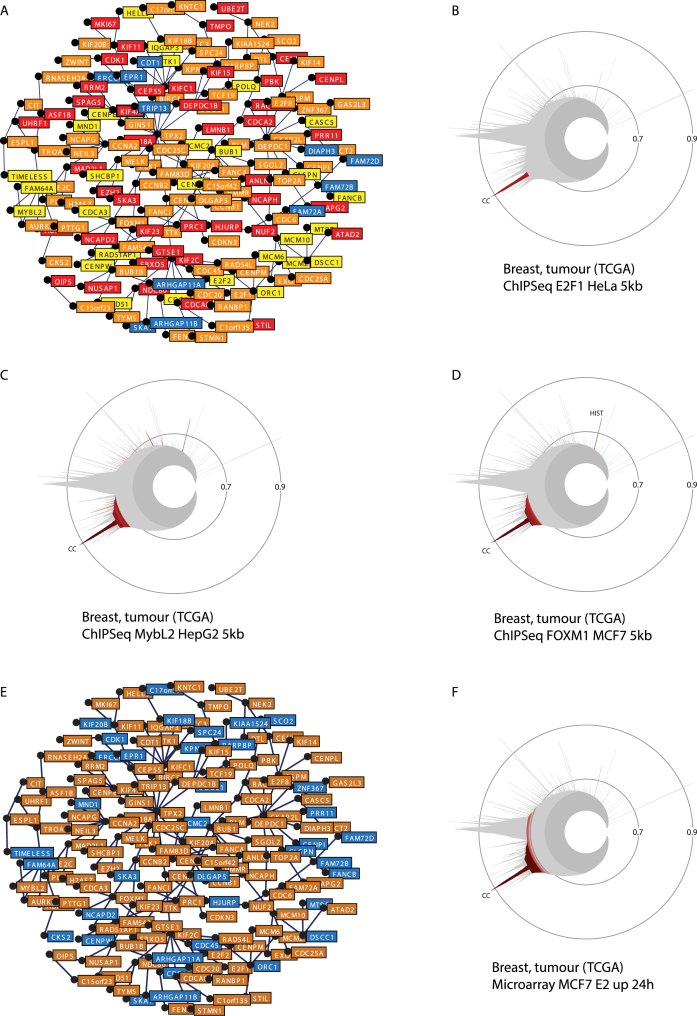
The conserved mitosis/cell cycle cluster (CC, Figures 3E and 5A and B, see also label-free version in Supplementary Figure S5) contains genes from a variety of chromosomal locations. It includes several TF genes, including E2F family members E2F1, E2F2 and E2F8 in breast cancer and E2F1 and E2F8 in leukaemia, as well as MYBL2 and FOXM1 in both datasets. This may indicate transcriptional regulation of cluster genes by these factors, in keeping with previous reports of their roles in cell cycle control (37,38). Entering the gene set type term ‘ChIP-Seq’ (Supplementary Table S3) in the search window of the enrichment table displayed in the breast cancer CC cluster view reveals enrichment in genes containing chromatin regions associated with E2F4, FOXM1 and MYBL2 within 5 kb of their TSS (range of 12- to 217-fold enrichment, p-values between 6 × 10−43 and 1 × 10−86, see representation in Figure 5A). The specificity of enrichment of these gene sets in the CC cluster versus other clusters in the leukaemia or breast cancer datasets can be analyzed by copying their names in the ‘Gene set enrichment box’ found in the icicle view (Supplementary Figure S1B). Enrichment in genes containing ChIP-Seq regions for E2F1, FOXM1 and MYBL2 within 5 kb of their TSS is mostly specific to the CC cluster (Figure 5B–D), with the exception of a few clusters, such as the ‘replication-dependent histones’ (HIST, also enriched with MYBL2 and FOXM1 ChIP-Seq regions, Figure 5B–D). MiSTIC can be exploited to further dissect the transcriptional regulatory signals that define this proliferation/cell cycle cluster. For example, when entering the gene set type term ‘microarray’ (Supplementary Table S3) in the search window of the CC cluster view, an enrichment in genes upregulated by 17β-estradiol in MCF7 cells at 24 h is revealed (126-fold, p = 7 × 10−152; Figure 5E and F). Indeed estrogens, acting via estrogen receptor alpha (ER, encoded by ESR1), are known to upregulate FOXM1 and E2F genes (23,39,40). In addition, both FOXM1 and MYBL2, which contain E2F1-associated regions in HeLa cells (Supplementary Table S5), are transcriptionally regulated by E2Fs (41,42), forming a transcriptional regulatory network that controls expression of CC cluster genes in response to extra-cellular signals such as estrogens in breast cancer cells.
Figure 5.
Transcriptional networks in the cell cycle/mitosis (CC) cluster. (A) Compilation of genes with ChIP regions associated with at least one of the three factors. Genes associated with one factor (yellow), with two factors (orange) or with three factors (red) are highlighted (see also Supplementary Table S5). (B–D) Specificity of the enrichment of gene sets associated with the presence of ChIP regions for E2F1, MYBL2 or FOXM1 in the CC cluster. (E) Enrichment analysis for the gene set ‘Microarray up MCF7 24 h E2’ from Bourdeau et al. (23) indicates that it is enriched in the CC cluster. Up-regulated estradiol target genes are highlighted in orange in the cluster representation. (F) Selective enrichment of the gene set ‘Microarray up MCF7 24 h E2’ in the CC cluster in the breast cancer icicle.
The examples described above derived from well-characterized correlation clusters illustrate that it is possible using gene set enrichment in MiSTIC to formulate testable hypotheses on the mechanisms responsible for cluster formation in RNA-Seq datasets. These hypotheses can be supported directly in MiSTIC by experimental annotations such as known amplification/deletions, genes regulated by signalling molecules in microarray/transcriptome sequencing experiments or genes with TF-associated flanking regions in ChIP-Seq experiments.
From correlation clusters to molecular classification of cancer
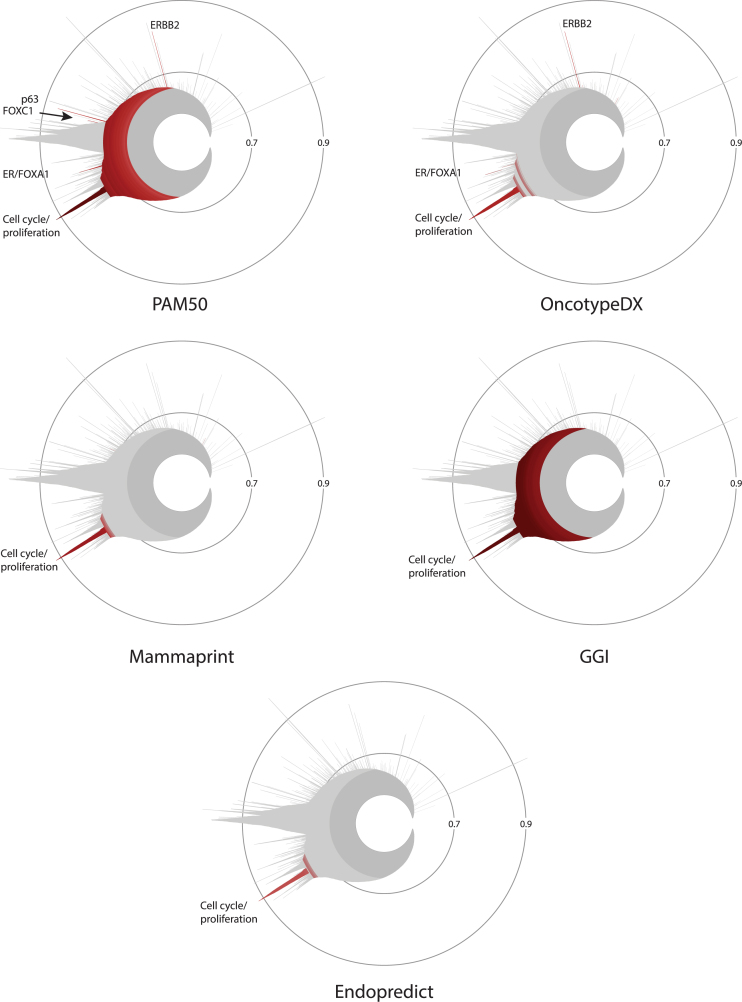
As correlation clusters are formed by sets of genes whose expression co-varies in the dataset, they represent potential biomarkers for the source of heterogeneity that created them (see above). Of particular interest are clusters reflecting genetic aberrations, biological subtypes and/or transcriptional networks. The transcriptome of breast tumours has been previously analyzed to uncover markers of biological subtypes and/or prognosis such as the PAM50, Oncotype DX, Mammaprint, Endopredict and GGI signatures (43–47). Enrichment analysis in the breast cancer icicle indicates that most prognostic gene signatures capture mainly the cell cycle proliferation peak (Figure 6). PAM50 and OncotypeDX are also enriched in the ER/FOXA1 (ER subcluster for OncotypeDx) and ERBB2 peaks. In addition, the PAM50 gene set, used for determination of molecular subtypes of breast tumours, is enriched in two other peaks, containing genes for transcription factors FOXC1 and p63, respectively. The latter two peaks are statistically enriched in gene sets associated with basal-like tumours (Supplementary Figure S6 and Tables S7 and S8). Note that these signatures may contain genes that are found in a high correlation cluster without yielding statistical enrichment, or have informative value despite low correlation with other genes at the transcriptional level; see for instance the localization of all genes in the PAM50 and Oncotype Dx signatures on the icicle using the Locate the genes of a set tool (Supplementary Figure S7). Nevertheless, this analysis reveals that a considerable portion of the correlation peaks is not represented in these signatures, and may contain information that is pertinent for either breast cancer classification or prognosis.
Figure 6.
Enrichment analysis of gene signatures used for breast cancer prognosis and subtype classification in the correlation clusters of the TCGA breast cancer icicle. Enrichment is visualized for the gene sets PAM50, Oncotype DX, Mammaprint, GGI and Endopredict.
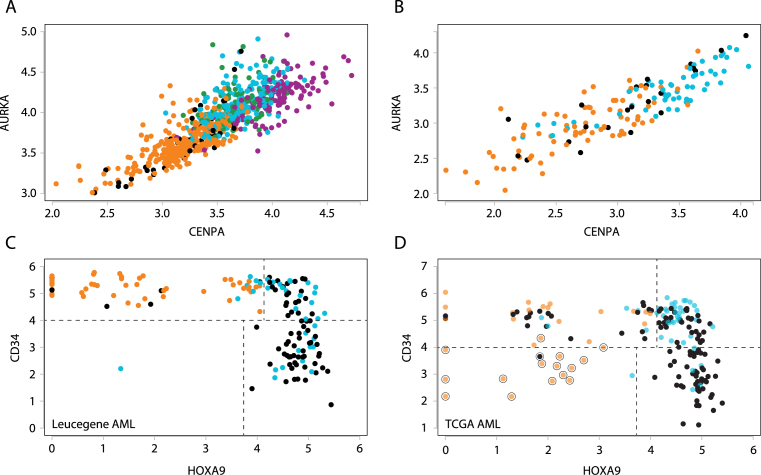
The multi-way scatterplot tool can readily reveal how tumours are distributed with respect to expression levels of selected biomarkers. Expression of genes in the proliferation cluster did not partition tumours in clearly distinct groups, yielding rather a continuum of tumours with increasing levels of markers such as AURKA and CENPA (Figure 7 and Supplementary Figure S8). However, tumours with low levels of both markers were enriched in luminal A tumours in the sample term enrichment analysis, while tumours with the highest expression levels of these markers were triple-negative and/or basal-like tumours (Supplementary Figure S8A and B). Highlighting tumours identified as luminal A, B, HER2+ and basal-like by PAM50 analysis using the Highlight groups tool (Supplementary Figure S8C) further indicates that HER2+ and luminal B tumours have intermediate expression levels of these markers (Figure 7A).
Figure 7.
Proliferative genes discriminate between intrinsic breast cancer subtypes and between blood and bone marrow leukaemia samples. (A) Samples in the breast cancer dataset (#7, Supplementary Table S1) were ordered according to expression levels of AURKA and CENPA. Tumours annotated as LumA, LumB, HER2+ and Basal-like were highlighted in different colours as shown in Supplementary Figure S8C. (B) Samples in the AML Leucegene dataset (#1, Supplementary Table S1) were ordered according to expression levels of AURKA and CENPA. Samples annotated as Blood or Bone Marrow were highlighted in different colours as shown in Supplementary Figure S10C. (C) Samples in the AML Leucegene dataset (#1, Supplementary Table S1) are presented according to expression levels of CD34 and HOXA9. Samples in favourable and adverse cytogenetics risk groups are respectively shown in orange and light blue. (D) Data from TCGA showing inclusion of promyelocytic AML (M3: large dots).
Contrary to the proliferation cluster genes, mRNA levels for ESR1 and ERBB2, two genes encoding breast cancer driver genes found in two separate correlation clusters (Figure 3D), define four distinct tumour populations, ESR1hi/ERBB2lo (∼70% of the TCGA breast cancer dataset), ESR1hi/ERBB2hi (∼7%), ESR1lo/ERBB2hi (∼5%) and ESR1lo/ERBB2lo (∼18%). Each of these populations can be highlighted in a different colour using the Highlight groups tool (Supplementary Figure S9A). Using the select characteristic tool, which is populated with tumour lists associated with specific clinical characteristics, it is possible to examine the coincidence of these groups with the ER and HER2 status of tumours as determined at pathological examination. There was a general very good overlap between the ESR1hi and ER+ tumours identified by immunohistology (Supplementary Figure S9B). Of interest, however, several ER+ tumours had in fact ESR1 mRNA levels as low as most ER– tumours (ER+ ESR1lo group, red, Supplementary Figure S9C). Levels of the ER target genes GREB1, CA12 and AGR3 are also mostly low in this selected tumour population (Supplementary Figure S9D and not shown), suggesting lack of ER signalling in the tumour fragment from which transcriptomes were generated. Conversely, several ER– tumours were found to have high levels of ER expression at the mRNA (Supplementary Figure S9E) and high levels of ER target gene expression (Supplementary Figure S9F), indicating that ER signalling is intact in the tumour fragment analyzed for transcriptome profiling. These discrepancies may be attributed either to variable standards of ER positivity by IHC, to a heterogeneity of ER status within the tumour sample, or to contaminating normal tissue leading to detection of ESR1 expression by RNA-Seq in spite of an ER– status by IHC. Similarly, there was a very good concordance between HER2– and ERBB2lo tumours, indicating a low level of false negatives. However, several tumours with a HER2+ clinical status had low levels of both ERBB2 and GRB7 mRNA (Supplementary Figure S9G–H, red tumours); most of these also did not have predicted amplification of the ERBB2 gene by CNV analysis (6,7,28), suggesting false positives or discrepancy between tumour fragments. Together, these results suggest that the mRNA levels of ESR1, ERBB2 and associated genes provide useful complementary diagnostic information.
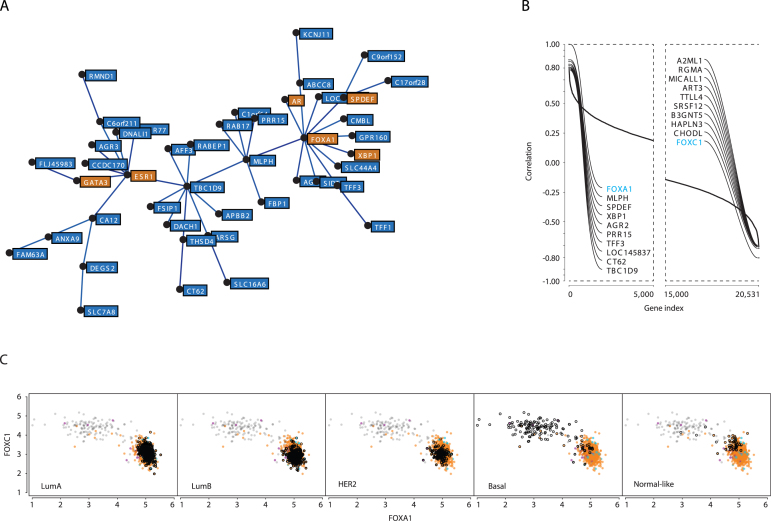
ESR1 is part of a gene cluster in breast cancer (Figure 8A) that is not conserved in any of the non-breast cancer datasets analyzed (list in Supplementary Table S1), indicative of a tissue-specific function. The breast cancer ESR1 cluster contains other genes encoding TFs that play roles in the control of luminal cell differentiation (48–51), such as GATA3, SPDEF and FOXA1 (Figure 8A). Genes encoding the XBP1 transcription factor (XBP1), an estrogen target gene (23), and the androgen receptor (AR) are also part of this cluster. A role of luminal TFs in regulating cluster genes is suggested by the fact that genes with ChIP-Seq binding sites for ER, FOXA1, GATA3 and/or SPDEF in MCF7 or for AR in LNCaP cells were enriched in the entire cluster (see Supplementary Table S6 for the presence of binding sites in the TSS flanking regions of cluster genes). Although a detailed analysis of the transcriptional network underlying formation of this cluster will require modulation of expression of each TFs and characterization of the impact on cluster genes (H.I. and S.M., in preparation), co-recruitment of these TFs at cluster genes is consistent with previous reports that FOXA1 and GATA3 enhance ER binding to chromatin (52,53). In addition, the waterfall plot tool revealed that FOXA1 gene expression is most anti-correlated with that of FOXC1 (blue labels, Figure 8B), whose expression is associated with triple-negative status (54). Strikingly, FOXA1 and FOXC1 mRNA expression levels defined two well-segregated breast tumour subsets (Figure 8C). The FOXA1loFOXC1hi group was enriched in basal-like tumours (Figure 8C, second last panel, open black dots), and the FOXA1hiFOXC1lo tumours included the lumA, lumB and HER2+ groups (Figure 8C, see samples highlighted by open back dots in each panel). This striking anti-correlation between two FOX transcription factor family members suggests negative cross-talk between their transcriptional networks. This hypothesis is in part supported by the recent observation that FOXC1 counteracts GATA3 activity and ER expression (55).
Figure 8.
FOXA1 and FOXC1 mRNA levels define two main sub-populations of tumours corresponding to basal-like versus other tumour types. (A) Minimal spanning tree for the ‘luminal’ cluster. Transcription factor-encoding genes, AR, ESR1, FOXA1, GATA3, SPDEF and XBP1 are highlighted in orange. (B) Waterfall analysis of FOXA1 most correlated and anti-correlated genes. FOXA1 and FOXC1 are highlighted in blue. (C). Breast tumours were sorted according to expression levels of FOXA1 and FOXC1, revealing two main groups of tumours. FOXA1hiFOXC1lo tumours include the lumA, lumB, HER2+ and normal-like groups, while the FOXA1loFOXC1hi group coincides with basal-like tumours.
These examples illustrate the usefulness of MiSTIC for exploring the significance of genes identified via correlation analyses as markers of heterogeneity within a tumour sample population, and for comparing different tumour subtypes via multidimensional tracking of biomarkers and enrichment analysis.
Correlation with clinical data
MiSTIC is also designed to interface clinical data with gene expression profiles. For example, in a scatterplot of AML samples (dataset 1, Leucegene AML) based on AURKA and CENPA expression levels, we observed in the sample term enrichment table that specimens expressing high levels of these two genes (or any other genes found in the proliferation peak) were collected from bone marrow aspiration whereas blood-derived specimens are low expressers (Supplementary Figure S10C). To better assess the impact of the tissue of origin (blood versus bone marrow) on proliferation gene expression profile, we selected these two clinical characteristics in MiSTIC and indeed observed a marked difference in expression levels of proliferative genes between these two groups (Figure 7B). These results suggest for the first time that leukaemia blasts in peripheral blood are much less proliferative than those in the bone marrow. Supporting these findings, it was previously reported that long-term culture-initiating cells (LTC-IC) are mostly quiescent in human blood while those derived from human bone marrow are cycling (56), suggesting that leukemic cell proliferation is under the same environmental control as that operating with normal cells. However, tumours associated with longer term survival (>3 years) did not cluster together in this representation (Supplementary Figure S10C, green dots), indicating that expression levels of proliferative genes have little prognostic value in leukaemia.
High levels of HOXA9 (57) or CD34 (58,59) have been proposed as risk predictors for AML. Unfortunately, each of these genes on its own fails to completely segregate favourable from intermediate to poor prognosis AML patients. Scatterplot analysis of these two genes however provides four distinctive populations (Figure 7C and D for the Leucegene and TCGA dataset). The first population consists of CD34hiHOXA9lo specimens (upper left quadrant in Figure 7C and D). This cohort of patients includes all (45 of 45; >100-fold enrichment, p = 8 × 10−28) favourable cytogenetic AML (t(8;21) and inv(16)) and all four specimens with CEBPA biallelic mutations which are also known for good prognosis AML (60). Not surprisingly, this group is enriched for long-term survivors (>3 years). The second population consists of CD34hiHOXA9hi specimens (upper right quadrant in Figure 7C and D) and is highly enriched for adverse cytogenetic risk (4.1-fold enrichment; p = 0.0007), tandem duplication in the MLL gene (20.8-fold enrichment; p = 0.001) and failure to achieve complete remission (3.8-fold enrichment; p = 0.0037). Importantly, this population is also enriched for TP53 mutated specimens (5.5-fold enrichment, p = 0.025). The subgroup characterized by CD34loHOXA9hi AML (lower right quadrant in Figure 7C and D) is highly enriched for NPM1 mutations (19.6-fold enrichment, p = 7 × 10−11) and for intermediate cytogenetic risk (9.9-fold enrichment; p = 8 × 10−10). The TCGA AML dataset reveals a fourth population of patients (lower left quadrant in Figure 7D) corresponding to cases of M3 AML (t(15;17)), a group yet represented in the current Leucegene dataset but of very good prognosis.
Analysis of other types of datasets with MiSTIC
Examples of analyses shown above were performed with RNA-Seq datasets. However, datasets generated using gene expression microarray platforms can also be used. For instance, we entered normalized expression data from the CCLE dataset, derived from Affymetrix U133 plus 2.0 microarray characterization of cell lines (17). Comparison with the GNE dataset, obtained by RNA-Seq of polyA+ transcriptomes, indicates a high degree of cluster conservation (Supplementary Figure S11A), in agreement with the reported high mean gene correlation between the two datasets despite differential representation of cell lines (18). This demonstrates that MiSTIC can be used for analysis of transcriptomes across different types of platforms. As expected from the heterogeneity of these tumour cell collections, conservation is weaker when comparing to individual cancer or normal tissues (e.g. Supplementary Figure S11B). However, it is possible to detect differentiation clusters such as the epithelial differentiation cluster observed in normal breast tissue (Supplementary Figure S11C). Genes within this cluster differentiate a group of cell lines enriched in the carcinoma annotation from one enriched in the hematopoietic-lymphoid origin annotation (Supplementary Figure S11D). However, variable proportions of carcinoma cell lines depending on the cancer type are associated with lack of epithelial cluster gene expression, suggesting loss of epithelial identity and acquisition of mesenchymal traits (Supplementary Figure S11E). Similar results were obtained with the GNE dataset (not shown).
It is also possible to construct icicles from miRNA RNA-Seq datasets. In this case, the number of features is lower, fewer peaks are formed and correlations reach lower values. Comparison across datasets is possible, identifying dataset-specific or common clusters (Supplementary Figure S12A). Enrichment analysis can be performed as above to assess whether miRNA clusters are linked to individual chromosomal loci (whether due to amplification/deletion or to coregulation of miRNA clusters) or coregulated by common TFs. Correlations between protein-coding genes and miRNAs can be explored by fusing these datasets. An example of this approach is shown in Supplementary Figure S12B–D, illustrating the anticorrelation between MIR200A/B/C, found in the epithelial cluster, and genes in the mesenchymal cluster and the enrichment in binding sites for mesenchymal genes ZEB1/2 in regulatory regions of epithelial cluster genes including MIR200C. In addition to miRNAs, other non-coding RNA whose expression can be quantified using RNA-Seq can also be used either alone or combined to protein coding genes to construct correlation networks. This will become all the more interesting in the future as these transcripts become more systematically detected, mapped and annotated.
Finally, quantitative profiles other than gene expression profiles can also be used in MiSTIC. For instance, we have verified that biological responses of AML specimens to small chemical compounds can be entered and analyzed in MiSTIC, revealing compound clustering based on their ability to selectively kill primary AML samples (61).
CONCLUSION
In conclusion, MiSTIC is a tool that visualizes and compares collections of gene expression profiles, instantly highlighting differences and similarities in gene clustering between cancer types or subtypes. Its integrative concept greatly improves the accessibility of complex datasets by end-users and enables the generation of hypotheses on mechanisms driving correlated gene expression. It should also facilitate identification of new prognostic markers and accelerate improvements in the molecular classification of cancers. Finally, MiSTIC is adaptable to the analysis of any collection of quantitative profiles.
AVAILABILITY
The MiSTIC software package can be obtained at github.com/iric-soft/MiSTIC.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Louis Gaboury and John H. White for critical reading of the manuscript. The authors wish to thank Patrick Gendron from the IRIC bioinformatics platform for data processing and Pierre Chagnon and Marianne Arteau at the IRIC genomics platform for RNA sequencing. The dedicated work of BCLQ staff, namely Claude Rondeau, Sylvie Lavallée and Giovanni D’Angelo, is also acknowledged. Hema Quebec graciously provided cord blood specimens. S.L., D.J.H., S.M. and G.S. designed research. S.L., T.S., D.L. and H.I. performed research. M.R., G.B., V.L., D.A.B., B.W., J.H. contributed new reagents/analytic tools, S.L., S.M. and G.S. analyzed data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Quebec [to G.S., J.H., S.L. and B.W.]; Terry Fox Foundation [2009-26 to G.S., J.H., S.L. and B.W.]; Canada Research Chair in the Molecular Genetics of Stem Cells [to G.S.]; Cancer Research Network of the Fonds de Recherche du Québec-Santé [to J.H.]; Australian Government National Health and Medical Research Council [Independent Research Institute Infrastructure Support Scheme to D.H.; program grant 1016647 to T.S.]; Canadian Cancer Society Research Institute [703961 to S.M.]; Canadian Imperial Bank of Commerce breast cancer research chair at Université de Montréal [to S.M.]. Funding for open access charge: The Canadian Cancer Society Research Institute and Genome Quebec.
Conflict of interest statement. None declared.
REFERENCES
- 1. Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B.. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998; 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bono H., Okazaki Y.. Functional transcriptomes: comparative analysis of biological pathways and processes in eukaryotes to infer genetic networks among transcripts. Curr. Opin. Struct. Biol. 2002; 12:355–361. [DOI] [PubMed] [Google Scholar]
- 3. Hong S., Chen X., Jin L., Xiong M.. Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res. 2013; 41:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iancu O.D., Kawane S., Bottomly D., Searles R., Hitzemann R., McWeeney S.. Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics. 2012; 28:1592–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. International Cancer Genome, C. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabe R.R., Bhan M.K., Calvo F., Eerola I. et al. International network of cancer genome projects. Nature. 2010; 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeeberg B.R., Feng W., Wang G., Wang M.D., Fojo A.T., Sunshine M., Narasimhan S., Kane D.W., Reinhold W.C., Lababidi S. et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003; 4:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 11. Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007; 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eisen M.B., Spellman P.T., Brown P.O., Botstein D.. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 14. Kruskal J.B., Jr On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956; 7:48–50. [Google Scholar]
- 15. Kruskal J.B., Landwehr J.M.. Icicle plots—better displays for hierarchical-clustering. Am. Stat. 1983; 37:162–168. [Google Scholar]
- 16. Vardiman J.W., Thiele J., Arber D.A., Brunning R.D., Borowitz M.J., Porwit A., Harris N.L., Le Beau M.M., Hellstrom-Lindberg E., Tefferi A. et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009; 114:937–951. [DOI] [PubMed] [Google Scholar]
- 17. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015; 33:306–312. [DOI] [PubMed] [Google Scholar]
- 19. Simon C., Chagraoui J., Krosl J., Gendron P., Wilhelm B., Lemieux S., Boucher G., Chagnon P., Drouin S., Lambert R. et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012; 26:651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haibe-Kains B., Schroeder M., Bontempi G., Sotiriou C., Quackenbush J.. genefu: relevant functions for gene expression analysis, especially in breast cancer. R package version 1.14.0. 2014; http://compbio.dfci.harvard.edu/. [Google Scholar]
- 21. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B. et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013; 41:D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bourdeau V., Deschenes J., Laperriere D., Aid M., White J.H., Mader S.. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008; 36:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006; 34:D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Culhane A.C., Schroder M.S., Sultana R., Picard S.C., Martinelli E.N., Kelly C., Haibe-Kains B., Kapushesky M., St Pierre A.A., Flahive W. et al. GeneSigDB: a manually curated database and resource for analysis of gene expression signatures. Nucleic Acids Res. 2012; 40:D1060–D1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suva M.L., Riggi N., Bernstein B.E.. Epigenetic reprogramming in cancer. Science. 2013; 339:1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watson C.J., Khaled W.T.. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008; 135:995–1003. [DOI] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacot W., Fiche M., Zaman K., Wolfer A., Lamy P.J.. The HER2 amplicon in breast cancer: Topoisomerase IIA and beyond. Biochim. Biophys. Acta. 2013; 1836:146–157. [DOI] [PubMed] [Google Scholar]
- 30. Center., B.I.T.G.D.A. SNP6 Copy number analysis (GISTIC2). Broad Institute of MIT and Harvard. Breast Invasive Carcinoma. 2016; doi:10.7908/C1NP23RQ. [Google Scholar]
- 31. Ades F., Zardavas D., Bozovic-Spasojevic I., Pugliano L., Fumagalli D., de Azambuja E., Viale G., Sotiriou C., Piccart M.. Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 2014; 32:2794–2803. [DOI] [PubMed] [Google Scholar]
- 32. Nakao M., Kawauchi S., Uchiyama T., Adachi J., Ito H., Chochi Y., Furuya T., Oga A., Sasaki K.. DNA copy number aberrations associated with the clinicopathological features of colorectal cancers: Identification of genomic biomarkers by array-based comparative genomic hybridization. Oncol. Rep. 2011; 25:1603–1611. [DOI] [PubMed] [Google Scholar]
- 33. Saha S., Bardelli A., Buckhaults P., Velculescu V.E., Rago C., St Croix B., Romans K.E., Choti M.A., Lengauer C., Kinzler K.W. et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001; 294:1343–1346. [DOI] [PubMed] [Google Scholar]
- 34. Bilal E., Vassallo K., Toppmeyer D., Barnard N., Rye I.H., Almendro V., Russnes H., Borresen-Dale A.L., Levine A.J., Bhanot G. et al. Amplified loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS One. 2012; 7:e38575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Balmer J.E., Blomhoff R.. Gene expression regulation by retinoic acid. J. Lipid Res. 2002; 43:1773–1808. [DOI] [PubMed] [Google Scholar]
- 36. Balmer J.E., Blomhoff R.. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J. Steroid Biochem. Mol. Biol. 2005; 96:347–354. [DOI] [PubMed] [Google Scholar]
- 37. Luporsi E., Andre F., Spyratos F., Martin P.M., Jacquemier J., Penault-Llorca F., Tubiana-Mathieu N., Sigal-Zafrani B., Arnould L., Gompel A. et al. Ki-67: level of evidence and methodological considerations for its role in the clinical management of breast cancer: analytical and critical review. Breast Cancer Res. Treat. 2012; 132:895–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koo C.Y., Muir K.W., Lam E.W.. FOXM1: From cancer initiation to progression and treatment. Biochim. Biophys. Acta. 2012; 1819:28–37. [DOI] [PubMed] [Google Scholar]
- 39. Stender J.D., Frasor J., Komm B., Chang K.C., Kraus W.L., Katzenellenbogen B.S.. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol. Endocrinol. 2007; 21:2112–2123. [DOI] [PubMed] [Google Scholar]
- 40. Millour J., Constantinidou D., Stavropoulou A.V., Wilson M.S., Myatt S.S., Kwok J.M., Sivanandan K., Coombes R.C., Medema R.H., Hartman J. et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010; 29:2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanada N., Lo H.W., Day C.P., Pan Y., Nakajima Y., Hung M.C.. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 2006; 45:10–17. [DOI] [PubMed] [Google Scholar]
- 42. Millour J., de Olano N., Horimoto Y., Monteiro L.J., Langer J.K., Aligue R., Hajji N., Lam E.W.. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol. Cancer Ther. 2011; 10:1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van 't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415:530–536. [DOI] [PubMed] [Google Scholar]
- 44. Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004; 351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 45. Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006; 98:262–272. [DOI] [PubMed] [Google Scholar]
- 46. Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009; 27:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Filipits M., Rudas M., Jakesz R., Dubsky P., Fitzal F., Singer C.F., Dietze O., Greil R., Jelen A., Sevelda P. et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011; 17:6012–6020. [DOI] [PubMed] [Google Scholar]
- 48. Kouros-Mehr H., Slorach E.M., Sternlicht M.D., Werb Z.. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006; 127:1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernardo G.M., Lozada K.L., Miedler J.D., Harburg G., Hewitt S.C., Mosley J.D., Godwin A.K., Korach K.S., Visvader J.E., Kaestner K.H. et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010; 137:2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kong S.L., Li G., Loh S.L., Sung W.K., Liu E.T.. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol. Syst. Biol. 2011; 7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buchwalter G., Hickey M.M., Cromer A., Selfors L.M., Gunawardane R.N., Frishman J., Jeselsohn R., Lim E., Chi D., Fu X. et al. PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells. Cancer Cell. 2013; 23:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S.. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011; 43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Theodorou V., Stark R., Menon S., Carroll J.S.. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013; 23:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ray P.S., Wang J., Qu Y., Sim M.S., Shamonki J., Bagaria S.P., Ye X., Liu B., Elashoff D., Hoon D.S. et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010; 70:3870–3876. [DOI] [PubMed] [Google Scholar]
- 55. Yu-Rice Y., Jin Y., Han B., Qu Y., Johnson J., Watanabe T., Cheng L., Deng N., Tanaka H., Gao B. et al. FOXC1 is involved in ERalpha silencing by counteracting GATA3 binding and is implicated in endocrine resistance. Oncogene. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iwama A., Wang M.H., Yamaguchi N., Ohno N., Okano K., Sudo T., Takeya M., Gervais F., Morissette C., Leonard E.J. et al. Terminal differentiation of murine resident peritoneal macrophages is characterized by expression of the STK protein tyrosine kinase, a receptor for macrophage-stimulating protein. Blood. 1995; 86:3394–3403. [PubMed] [Google Scholar]
- 57. Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999; 286:531–537. [DOI] [PubMed] [Google Scholar]
- 58. Rockova V., Abbas S., Wouters B.J., Erpelinck C.A., Beverloo H.B., Delwel R., van Putten W.L., Lowenberg B., Valk P.J.. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011; 118:1069–1076. [DOI] [PubMed] [Google Scholar]
- 59. Langer C., Radmacher M.D., Ruppert A.S., Whitman S.P., Paschka P., Mrozek K., Baldus C.D., Vukosavljevic T., Liu C.G., Ross M.E. et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008; 111:5371–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schlenk R.F., Dohner K., Krauter J., Frohling S., Corbacioglu A., Bullinger L., Habdank M., Spath D., Morgan M., Benner A. et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008; 358:1909–1918. [DOI] [PubMed] [Google Scholar]
- 61. Baccelli I., Krosl J., Boucher G., Boivin I., Lavallee V.P., Hebert J., Lemieux S., Marinier A., Sauvageau G.. A novel approach for the identification of efficient combination therapies in primary human acute myeloid leukemia specimens. Blood Cancer J. 2017; 7:e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.