Abstract
The regulation of replication is essential to preserve genome integrity. Mms1 is part of the E3 ubiquitin ligase complex that is linked to replication fork progression. By identifying Mms1 binding sites genome-wide in Saccharomyces cerevisiae we connected Mms1 function to genome integrity and replication fork progression at particular G-rich motifs. This motif can form G-quadruplex (G4) structures in vitro. G4 are stable DNA structures that are known to impede replication fork progression. In the absence of Mms1, genome stability is at risk at these G-rich/G4 regions as demonstrated by gross chromosomal rearrangement assays. Mms1 binds throughout the cell cycle to these G-rich/G4 regions and supports the binding of Pif1 DNA helicase. Based on these data we propose a mechanistic model in which Mms1 binds to specific G-rich/G4 motif located on the lagging strand template for DNA replication and supports Pif1 function, DNA replication and genome integrity.
INTRODUCTION
The preservation of genome stability is a major challenge for eukaryotic cells. The faithful duplication of DNA is necessary to maintain the integrity of the genome. During DNA replication, stalled replication forks are a major challenge to genome integrity, because they can cause DNA breakage. Mechanisms that repair or bypass stalled replication forks need to be activated in order to maintain genome stability (reviewed in (1)).
Recent studies indicate that Rtt101, Mms1 and Mms22 form the ubiquitin ligase Rtt101Mms1/Mms22 (2). All three components are required for replication fork progression in cells treated with the alkylating agent methyl methanesulfonate (MMS), which causes replicative stress (2,3). In line with this, it has been shown that Mms1 and Mms22 are required for homologous recombination (HR) at stalled forks, but not at HO endonuclease-induced double-strand-break (DSB) sites (4). Furthermore, it was shown that Mms1 binds near early origins of replication if replication forks are stalled by hydroxyurea (HU) and that this binding is dependent on the other two ligase components, Rtt101 and Mms22 (5).
In addition to the traditional regions (replication fork barriers) where DNA replication stalls (reviewed in (6,7)), DNA secondary structures such as G-quadruplex (G4) structures were shown to influence DNA replication fork progression (reviewed in (8,9)). G4 structures are guanine-rich, four-stranded structures that can form within nucleic acids if a defined nucleotide sequence, called G4 motif, is present (reviewed in (8,10)). Although the existence of G4 structures has long been controversial, increasing amounts of in vitro and in vivo data, such as immunostaining (11–13) and the detection of these structures by small molecules (14–16), now support a model in which G4 structures form in vivo. G4s can positively and/or negatively affect biological processes, such as telomere maintenance, gene expression, epigenetic regulation and DNA replication initiation (reviewed in (8,10,17)). In vitro and in vivo experiments have demonstrated that G4 structures hamper DNA replication and are prone for mutations and deletions (reviewed in (18)). Experiments in Saccharomyces cerevisiae, Caenorhabditis elegans and human cells have shown that formation of these structures leads to genomic instability (19–25). If a G4 structure is formed, helicases or translesion synthesis proteins are required to unwind or bypass these structures, respectively, to permit replication fork progression (reviewed in (26)). Yeast and human DNA helicases such as Pif1 and FANCJ bind to G4 motifs in vivo, prevent replication fork stalling, and support genome stability (19–25). The current model is that the replication machinery slows down when it encounters a G4 structure and that helicases (e.g. Pif1 in yeast) unwind them efficiently (21). In this case, the replication fork has to restart behind the obstacle leaving a gap behind (reviewed in (26)).
Currently, it is unclear how the Rtt101Mms1/Mms22 ligase recognizes stalled forks and if it acts at all stalled forks or only at specific loci. Here, we show that Mms1 binds to specific G-rich motifs located on the lagging strand template for DNA replication in the genome. Replication forks stall at these G-rich motifs in mms1 cells, and this G-rich motif has the potential to form G4 structures in vitro. Additional genetic assays show that Mms1 binding sites promote gross chromosomal rearrangements in the absence of Mms1. Chromatin immunoprecipitation (ChIP) experiments indicate that the observed replication and genome stability defects are due to lower Pif1 binding to these G-rich/G4 motifs in the absence of Mms1. Our data suggest a new mechanistic model of how Mms1 supports DNA replication at specific G-rich sequences that could form G4 structures.
MATERIALS AND METHODS
Strains, constructs and media
All yeast strains are listed in Supplementary data 1. All experimental strains are derivatives of the RAD5+ version of W303 (R. Rothstein) or YPH background (27). Deletions eliminated entire ORFs and were created according to (27). Epitope tagging to generate Mms1-Myc13, Pif1-Myc13 and DNA Pol2-Myc13 was carried out as described (28). Myc-tagged proteins were expressed from endogenous loci and promoters. The pif1-m2 mutant was created as described (29).
In vitro folding of G4
The folding was performed according to (30). To confirm G4 formation the samples were controlled by circular dichroism (CD) measurements.
Circular dichroism
15–20 μg DNA was subjected to CD measurements at 25°C using a Jasco J-810 spectropolarimeter (Jasco). The parameters were: continuous scanning mode (200–350 nm), accumulation 10, scanning speed 100 nm/min, response 0.25 s, band width 2 nm, and a data pitch of 0.2 nm.
Western analysis
Proteins for western analysis were isolated according to a protocol by Foiani (31). Western analysis was performed according to standard protocols. The primary antibodies for c-Myc (Clontech) and Hsp60 (Abcam) were used according to the manufacturer's protocol. Hsp60 served as a reference protein. As secondary antibody, we used a HRP-coupled antibody (Santa Cruz Biotechnology). Proteins were detected by chemiluminescence. Quantification was performed using Image Lab (BioRad).
Gross chromosomal rearrangement assay
The gross chromosomal rearrangement (GCR) assay was performed as published (32) with minor modifications. Briefly, seven colonies per strain were grown for 48 h. Cells were plated on two different plates: YEPD as a reference plate and on a FOA/CAN selective plate. After incubation colonies were counted on both plates and the GCR rate was determined via fluctuation analysis using FALCOR and the MSS maximum likelihood method (33).
Chromatin immunoprecipitation (ChIP)
ChIP of asynchronous and synchronous samples was essentially performed as described (21). For ChIP-seq the average length was 200 bp using a M220 Focused-ultrasonicator (Covaris) and for conventional ChIP the DNA was sheared to an average length of 250 bp using a Branson sonifier W250-D (50% amplitude, 50% duty cycle, 5 × 5 pulses) (Supplementary data 2A–C). The applied parameters for Covaris were 75 W, 25 duty and 200 cycles/burst for 20 min. C-Myc antibody was obtained from Clontech. Primers used for qPCR (Cycler and SYBR Green, Biorad) are listed in Supplementary data 3. For genome-wide sequencing, DNA was treated according to manufacturer's instructions (NEBNext ChIP-seq Library Prep Master Mix Set for Illumina, NEB) and submitted to deep sequencing (Illumina Nextseq500 sequencer). Obtained sequence reads were aligned to the yeast reference genome (sacCer3) with BOWTIE (34). After alignment, the number of reads was normalized to the sample with the lowest number of reads. Binding regions were identified by using the program ‘Model-based Analysis for ChIP-Seq (MACS 2.0)’ with default settings, -no model option, and -extsize 180, which correlates to the minimal fragment size (35). See Supplementary data 4 for all Mms1 peaks. The ChIP input sample was used as a control. MEME-based motif elicitation was used to identify a consensus motif (36) within the FASTA file from the binding regions identified by MACS 2.0. G4 motifs were identified using a script previously published (37). Overlap of binding sites and qPCR regions with G4 motifs and genes was determined using bedtools’ window command (38). A window size of 400 bp was used when evaluating overlap of binding regions and 500 bp when evaluating qPCR regions. To determine the overlap with genomic features we took the annotations from saccharomyces_cerevisiae_R64-2-1_20150113.gff. To examine if the qPCR regions contain G4 motifs on the leading and/or lagging strand, we identified the closest ARS to the G4 motifs using bedtools and determined strand specificity.
Arrest of yeast cells
Cells were arrested in G1 phase according to (39) and in S and G2 phase according to (40). FACS analysis to confirm cell cycle arrest was performed as described (39) using a FACSCanto II (BD).
Endogenous mutation of G4 using Cre-lox
G4 Chr VI (253592-255049) was mutated using Cre-Lox recombination. The mutated G4 motif was synthesized (Sigma) and cloned into pUG6 plasmid (41). Mutated G4 motif and the Marker were amplified and transformed into three different yeast strains (Pol2-Myc, Pol2-Myc mms1, Mms1-Myc). Via recombination, the mutated G4 with the Marker and the Cre-LoxP sites were integrated at the original sites of the G4 motif Chr VI. Next, plasmid pSH65 was transformed carrying a galactose-inducible Cre recombinase. After galactose induction, the Cre recombinase cuts at the loxP sites thereby removing the kanMX cassette leaving the mutated G4 motif and loxP site behind.
RESULTS
Mms1 binds genome-wide to G-rich motifs
To understand the interaction of Mms1 and stalled replication forks, we aimed to identify Mms1 binding sites genome-wide. We performed ChIP followed by genome-wide sequencing analysis (ChIP-seq) using endogenous Myc-tagged Mms1. As in a previous study, Mms1 was endogenously tagged at its C-terminus (5). To study the functionality of Myc-tagged Mms1, we used the published sensitivity of mms1 cells toward 0.01% MMS (42). A spot assay on plates containing 0.01% MMS showed that cells with Myc-tagged Mms1 show a mild growth defect compared to untagged cells (Supplementary data 5). However, this defect is not as severe as that of mms1 cells.
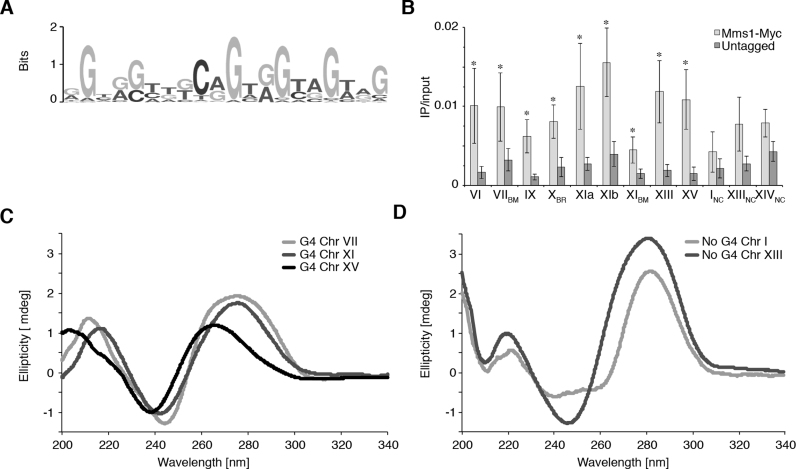
Asynchronous yeast cultures expressing Myc-tagged Mms1 were crosslinked with formaldehyde and processed for ChIP-seq. Using MACS 2.0 we identified 71 chromosomal sites with sequence enrichment, which we defined as Mms1 binding sites (see Supplementary data 4 for peaks). We compared the binding regions of Mms1 to annotated genomic features (e.g. centromeres and repeats). We did not detect significant association of Mms1 with any of the tested sites. To explore the binding specificity of Mms1 we searched for a consensus binding motif (BM) among Mms1 binding regions using MEME-based motif elicitation (36). The most significant binding motif is a 20 bp long G-rich motif (Figure 1A) (E-value: 2.5e–66). 23 of the 71 binding regions (BR) (32.4%) contain this BM at least once. Interestingly, the average GC-content of the Mms1 binding region was 50%, which is significantly higher than expected (P < 0.001) from the average GC content of the S. cerevisiae genome (∼38% GC).
Figure 1.
Mms1 associates with G-rich regions in vivo. (A) G-rich binding motif of Mms1 identified by MEME-ChIP search. (B) Validation and characterization of ChIPseq data. Using Myc-tagged Mms1 conventional ChIP and qPCR was performed using primers directed against endogenous regions. Regions are indicated below graph. Detail on regions and primers are listed in Supplementary data 3. Three regions identified as Mms1 binding sites by ChIPseq were tested (Chr VIIBM, XBR, XIBM) as well as nine additional genomic regions. Sites that contain the specific G-rich binding motif identified by MEME are marked with BM, additional G-rich binding regions identified by ChIP-seq with BR and negative controls with NC. Plotted are IP/input values as mean value ± standard deviation (SD). N≥3 biological replicates. Statistical significance compared to untagged cells was determined by Student's t-test. *P < 0.05. (C) CD analysis of the three Mms1 binding regions. (D) CD analysis of the two negative controls. The CD spectrum of a folded parallel G4 structure has a specific maximum and minimum (264 nm and 243 nm, respectively) which differs from the maximum and minimum of B-DNA (245 and 290 nm, respectively) (62,63). See Supplementary data 7 for details of analyzed regions and sequences. Shown are the ellipticity (in mdeg) values. This analysis demonstrates that G-rich Mms1 binding regions, harboring a G4tract2, can form G4 DNA structures.
The G-rich nature of the Mms1 BM suggested that some Mms1 binding regions might have the potential to form G4 structures. The MEME-based search was not developed to identify G4 motifs, so we applied a script that discovers G4 motifs to all 71 binding sites (37). We identified a G4 motif of the consensus sequence GGG(N)≤25GGG(N)≤25GGG(N)≤25GGG (G4tract3) in 15.5% (11/71) of Mms1 binding regions. Because the BM contained several GG di-nucleotides (Figure 1A) and G4 structures have the potential to form with fewer than three stacked tetrads (43–46), we also searched for a relaxed version of the G4 motif, GG(N)≤7GG(N)≤7GG(N)≤7GG (G4tract2). In this search, we found that 61 of the 71 Mms1 binding regions (86%) contain a G4tract2 within a window of ∼ 400 bp, which corresponds to the maximal DNA shearing size (Supplementary data 2A and B).
In order to get more insight into the binding properties of Mms1, and especially the potential contribution of G4 motifs, we performed ChIP-qPCR experiments using the same endogenous Myc-tagged Mms1 strain as for ChIPseq. For the qPCR, we chose 12 different regions in the yeast genome. Out of these 12 sites, three were Mms1 binding regions and seven exhibited the presence of a G4tract2. In addition, we selected two negative control regions based on the ChIP-seq data. For this and all subsequent ChIP experiments, IP values of each experiment were normalized to its input values. We considered it a positive binding if Mms1 levels were at least three times higher than those of the untagged control. In concordance with our earlier results, Mms1 bound three regions selected by ChIP-seq (Chr VIIBM, XBR, XIBM) with two regions harboring the MEME specific BM (Figure 1B). Strikingly, six out of seven regions with a G4tract2 (Chr VI, IX, XIa, XIb, XIII, XV) were also identified as Mms1 BR. Mms1 did not significantly bind to regions on Chr I, Chr XIV and an additional region from Chr XIII. Due to the lack of significant Mms1 binding, we treated these three regions as negative controls (NC) in the remaining analyses. To elucidate why Mms1 binds to specific regions, we further analyzed the ChIP-seq data. This analysis revealed that all regions which are bound by Mms1, using ChIP and qPCR, harbor one or multiple G4tract2 or even G4tract3 motifs (see Supplementary data 6 for details). However, the NC regions Chr INC and Chr XIIINC also harbor a G4tract2 and a G4tract3 motif, respectively. Considering the distance to the next ARS and the strand location of G4tract2 motifs, we identified that all G-rich/G4 motifs bound by Mms1 are located on the DNA strand replicated by the lagging strand machinery and have a mean loop length smaller than eight nucleotides. The NC regions have either no such G-rich/G4 motifs or those are not located on the lagging strand template (Supplementary data 6). In summary, ChIP-seq and ChIP-qPCR revealed that Mms1 binds only to G-rich regions, if they harbor at least one G4tract2 located on the lagging strand template for DNA replication. For simplicity, all regions that show significant binding of Mms1 either by ChIP-seq and/or ChIP-qPCR will be named in the text and figures as a BR.
To determine whether our identified G-rich binding regions can form G4 structures in vitro, we performed CD measurements on three of the 71 binding regions of Mms1. By these measurements, we confirmed that the regions from Chr VIIBR, XIaBR, and XVBR (all harboring a G4tract2 with a mean loop length < 8 nt, lagging strand template) folded into G4 structures in vitro (Figure 1C), whereas the regions from Chr INC and XIIINC (mean loop length > 8 nt, lagging strand template) did not (Figure 1D, Supplementary data 7). These results show that many regions bound by Mms1, and also those harboring G4tract2, have the ability to form G4 structures.
Mms1 binds in all cell cycle phases and independently of Rtt101 and Mms22
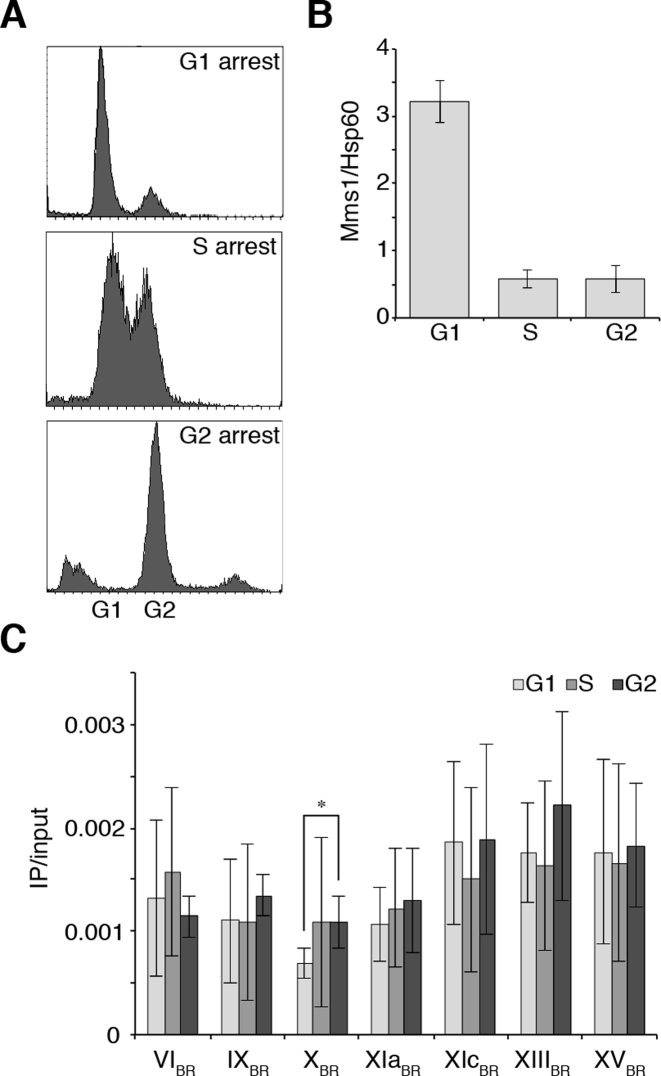
In order to determine the spatiotemporal function of Mms1, we determined in which cell cycle phase Mms1 binds to its target regions. First, we measured Mms1 levels at different cell cycle phases. Yeast cells expressing Myc-tagged Mms1 were arrested in G1, S, and G2 phase using different reagents (α factor, HU, and nocadozole, respectively). FACS analysis was performed to confirm cell cycle arrest (Figure 2A). Western blot analysis was performed to monitor Mms1 protein levels (Figure 2B, Supplementary data 8A). Although Mms1 protein levels peak in G1 phase (>5-fold more Mms1) Mms1 can be detected in all cell cycle phases (Figure 2B).
Figure 2.
Mms1 protein levels are highest in G1 phase and Mms1 binds throughout cell cycle. (A) FACS analysis of cells arrested in G1, S or G2 phase. Cells were arrested in G1 by treatment with α-factor, in S phase by HU and in G2 by nocadozole. (B) Western blot analysis of Myc-tagged Mms1 protein levels in G1, S and G2 phase. Level of Mms1 was quantified using Hsp60 as a reference protein. Shown are mean Myc-tagged Mms1 levels normalized to Hsp60 ± SD. N = 3 biological replicates. See Supplementary data 8A for the gel. (C) ChIP and qPCR analysis of Mms1-Myc to seven BR in G1-, S- and G2-phase. Plotted are IP/input values as means ± SD. N≥ 3 biological replicates. In most cases Mms1 binds similar in G1, S and G2 phase. Statistical significance compared to cells arrested in G1 phase was determined by Student's t-test. *P < 0.05.
To address the question when Mms1 binds to sites containing G4tract2 motifs, we performed ChIP and qPCR of Myc-tagged Mms1 cells arrested in G1, S, and G2 phase. The arrests were confirmed by FACS analysis (Supplementary data 8B). qPCR at Mms1 target regions Chr VIBR, IXBR, XBR, XIaBR, XIcBR, XIIIBR and XVBR revealed that Mms1 binds equally well throughout the cell cycle to all tested BR (Figure 2C).
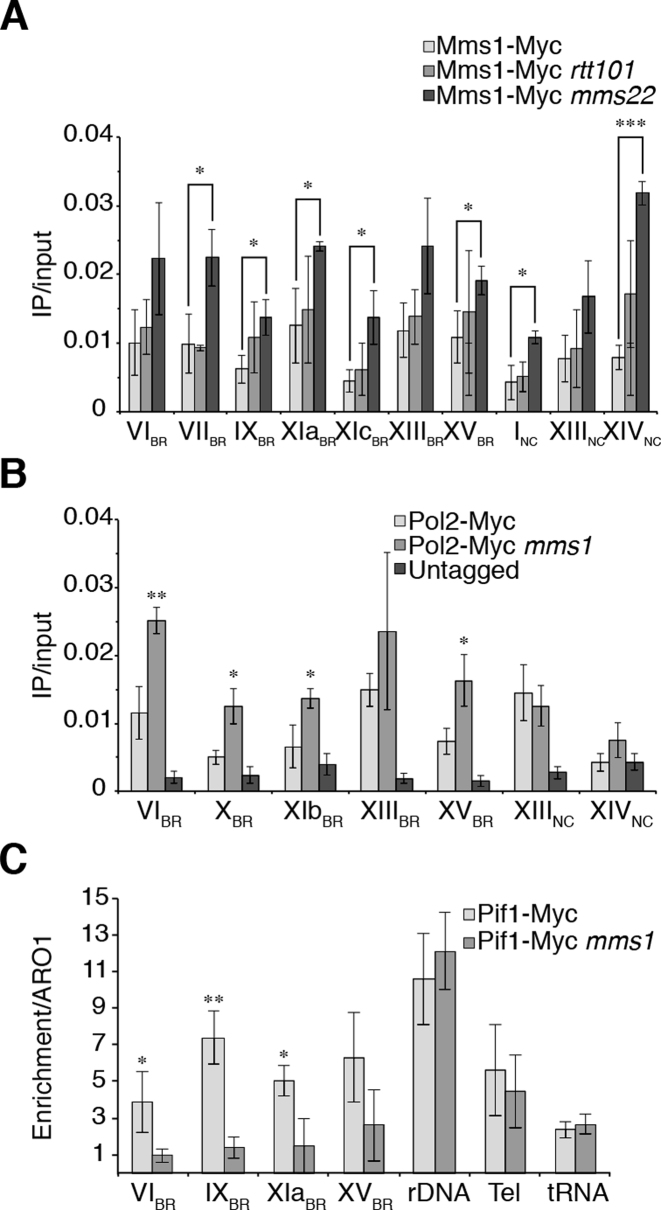
Previous work has shown that Mms1 interacts with Rtt101 and either Crt10 or Mms22 (2). So far the only process Crt10 was shown to be involved in is ribonucleotide reductase (RNR) gene expression (47), while Mms22 and Mms1 together with Rtt101 (Rtt101Mms1/Mms22) promote replication fork progression and HR at stalled replication forks (2–5). Due to the distinct functions of these two complexes, we hypothesized that the Rtt101Mms1/Mms22 complex could be recruited to our identified DNA target regions. In previous work, it was postulated that, in the Rtt101Mms1/Mms22 complex, Mms22 is the DNA interacting protein (2). Therefore, we first asked if Mms1 binding is dependent on Rtt101 or Mms22. We analyzed Mms1 binding in the absence of Mms22, Rtt101, and both proteins (MMS22/RTT101 double deletion) by ChIP-qPCR experiments. ChIP and qPCR analysis revealed that binding of Mms1 did not decrease in the rtt101 cells, but interestingly binding was significantly enriched in mms22 deficient cells (Figure 3A). This enhanced binding was observed at all tested regions including negative control regions. At Chr VIBR, VIIBR, IXBR, XIcBR, XIIIBR, INC, XIIINC and XIVNC we observed at least a 2-fold enriched binding of Mms1. In the mms22 rtt101 double mutant, Mms1 binding was similar to wild type or rtt101 cells (Supplementary data 8C).
Figure 3.
Mms1 binds independently of Rtt101 and Mms22, supports Pif1 binding at G4 motifs and by this promotes DNA replication. (A) ChIP and qPCR analysis of Mms1-Myc at seven BR and two NC. Binding of Mms1 was monitored in wild type (light), rtt101 (grey) and mms22 (dark) cells. Statistical significance compared to Myc-tagged Mms1 wild type cells. For details on regions see Supplementary data 3. (B) Replication fork progression was analyzed by detected DNA Pol2 binding levels at Mms1 binding sites. ChIP and qPCR of DNA Pol2 binding in wild type and mms1 cells was performed at five BR and two NC. Statistical significance compared to Myc-tagged DNA Pol2 wild type cells. (C) Binding of Pif1 DNA helicase was analyzed at four Mms1 binding regions (BR) in wild type and in mms1 cells. As control for Pif1 binding we used two known Pif1 binding sites, the replication fork barrier at the rDNA (rDNA) (54) and telomere VI-R (tel) (53) as well as one Pif1 independent site (tRNA). As done previously (53) IP/input values are compared to IP/input values of ARO1 where no Pif1 binds. Here, fold enrichment over ARO1 was plotted as mean value ± SD. For all ChIP; N ≥ 3 biological replicates. Statistical significance was determined by Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
To evaluate whether the observed difference in binding was due to changes in the total protein level of Mms1, we performed western blot analysis of Myc-tagged Mms1 and quantified the amount of Mms1 in wild type, rtt101, and mms22 cells using Hsp60 as a reference protein. The absence of Mms22 did not influence the abundance of Mms1, but Mms1 levels significantly increased in rtt101 cells (∼4-fold). (Supplementary data 8D,E). These data (ChIP and Western analysis) suggested that Rtt101 impacts Mms1 protein levels, but that it does not impact Mms1 binding to G-rich target regions. In Summary, these results suggest that Mms1 does not require Rtt101 or Mms22 for binding to G-rich/G4 sites located at the lagging strand template of DNA replication. Furthermore, it raises the possibility that Mms1 binds easier/better to G-rich/G4 regions without Mms22. If Mms1 acts as part of the E3 ligase, Mms22 is the preferred DNA binding protein as previously suggested (2).
Mms1 promotes progression of the stalled DNA replication fork
Previous work has shown that DNA replication is slowed in mms1 cells after MMS treatment, which stalls replication forks (2). Due to this strong connection between Mms1 function and DNA replication we further analyzed the contribution of Mms1 to replication fork progression. We aimed to investigate if replication fork progression is affected at the G-rich motifs identified as Mms1 binding sites in mms1 cells. DNA Polymerase 2 (DNA Pol2) is the catalytic subunit of DNA polymerase ε (48,49). It is assumed that regions with high DNA Pol2 levels are sites where DNA replication is slowed or stalled (21,50). Therefore, we performed ChIP-qPCR on asynchronous wild type and mms1 cultures that express endogenous Myc-tagged DNA Pol2. Binding of DNA Pol2 was monitored at five Mms1 binding regions (Chr VIBR, XBR, XIbBR, XIIIBR and XVBR) as well as two non-Mms1 binding regions (Chr XIIINC, XVNC) (Figure 3B). Binding of DNA Pol2 was significantly (P-value < 0.05, 1.8- to 2.5-fold) enriched at Chr VIBR, XBR, XIbBR and XVBR in mms1 cells. DNA Pol2 protein levels did not change in mms1 cells (Supplementary data 9A and B). Furthermore, we tested if DNA Pol2 binding was also elevated in rtt101, mms22 and rtt101 mms22 cells at these G-rich sites. ChIP experiments showed no elevated DNA Pol2 binding in these single or double mutants (Supplementary data 9C). In summary, elevated DNA Pol2 level are observed in the absence of Mms1 at G-rich motifs indicating that replication fork movement is slowed at these regions if Mms1 is absent. At the control regions (Chr XIIINC and XIVNC) DNA Pol2 occupancy was independent of Mms1, correlating with no Mms1 binding at these sites (see Figure 1B). Furthermore, these results further strengthen the point that Mms1 acts independently of Rtt101 and Mms22 at such sites.
Mms1 does not recruit Mre11 to its binding sites
Previous studies revealed that Mms1 is required for HR at stalled replication forks (4). Recently, it has been shown that HR proteins, such as Rad51 and BRCA1, regulate HR at G4 structures during DNA replication (51). Additionally, work in yeast showed that breakage near a stalled fork induces recombination (19,21). In the next set of experiments, we wanted to determine if Mms1 binding at its target regions also causes recruitment of HR factors due to stalled forks. We examined if Mms1 recruits Mre11, a component of the MRX complex involved in HR (reviewed in (52)). We performed ChIP of asynchronous yeast cells expressing endogenous Myc-tagged Mre11 in wild type and mms1 cells. Occupancy of Mre11 at Mms1 binding sites (from Figure 1B) was determined by qPCR, and the IP/input values were calculated (Supplementary data 10). We found that the association of Mre11 to all tested Mms1 binding regions was not dependent on Mms1, suggesting that Mms1 does not recruit HR factors to its target sites.
Mms1 and the Pif1 helicase act together at G-rich/G4 motifs
The Pif1 DNA helicase binds at the end of S phase to G4 motifs and supports DNA replication and promotes genome stability (19,21). The association of DNA Pol2 to G4 motifs is greatly increased in pif1-m2 mutants (21). In the next set of experiments, we tested if Mms1 and Pif1 act together at G4 structures. We re-analyzed genome-wide Pif1 binding sites (21) and checked for overlap with Mms1 peaks (Supplementary data 4). We identified 38 Mms1 binding regions that overlap Pif1 binding sites (Supplementary data 11). This is significantly more overlap than expected if these sites were randomly distributed across the genome (P = 0.001).
Replication fork progression is impeded at G4 sites in the absence of Pif1. Therefore, we tested whether Mms1 associates more strongly to G4 sites in pif1-m2 cells. In pif1-m2 mutants, expression of the nuclear isoform of Pif1 is disrupted, but the mitochondrial isoform of Pif1 is still expressed (29). We found that Mms1 binding did not change in pif1-m2 compared to wild type cells at all tested regions (Supplementary data 12A).
Previous work revealed that Pif1 binds at the end of S phase to G4 motifs (21) and that Mms1 binds throughout the cell cycle to specific G4tract2 motifs (Figure 2). We speculate that Mms1 binding supports Pif1 function at those sites. In this model, we expect reduced Pif1 binding to target regions in the absence of Mms1. In three independent ChIP experiments, we observed that Pif1 binding was more than two-fold reduced in the absence of Mms1 at G4tract2 motifs (these sites were previously also identified as Pif1 binding sites (21)) (Figure 3C). In contrast, Pif1 binding to three different control regions, telomere VI-R (Tel-VI-R (53)), the replication fork barrier at the rDNA repeat (rDNA (54)), and a tRNA gene (no Pif1 binding site (21)), was not altered. Pif1 levels in the cell did not change upon MMS1 deletion (Supplementary data 12B and C).
Mms1 binds to G4 structures
Our data led us to the hypothesis that Mms1 binds to G4 structures rather than to G-rich motifs. To determine whether Mms1 binds G-rich regions or G4 structures itself, we performed an affinity purification approach using biotinylated G4 structures or biotinylated control DNA sequences as bait. We used four G4 structures (Chr IG4-tract2, Chr IXG4tract3, Chr XIIIG4tract2, Oxy2G4tract4) as well as three controls (G-rich, non-G-rich, and a mutated G4 motif (95% identical to the G4 motif), none of which can form a G4 structure). The biotinylated oligodeoxynucleotides containing G4 motifs were folded into G4 structures. Control oligodeoxynucleotides, which cannot form a G4 structure, were treated in parallel. Formation of G4 structures as well as non-folding of the control sequences was confirmed by CD. Folded G4s and controls were incubated with total yeast protein lysate (from pif1-m2 cells: to reduce G4 unwinding potential in the lysate) in which Mms1 was endogenously tagged with a Myc tag. After wash steps and incubation with streptavidin-coupled beads, we isolated bound proteins. Western blot analysis revealed that Mms1 bound to all four G4 oligodeoxynucleotides but not to the three control oligodeoxynucleotides. This experiment showed that Mms1 did not bind to G-rich sequences in general or non-G-rich linear DNA (Supplementary data 13 lane 1–3), but that it specifically binds to G4 structures (Supplementary data 13, lanes 4–7).
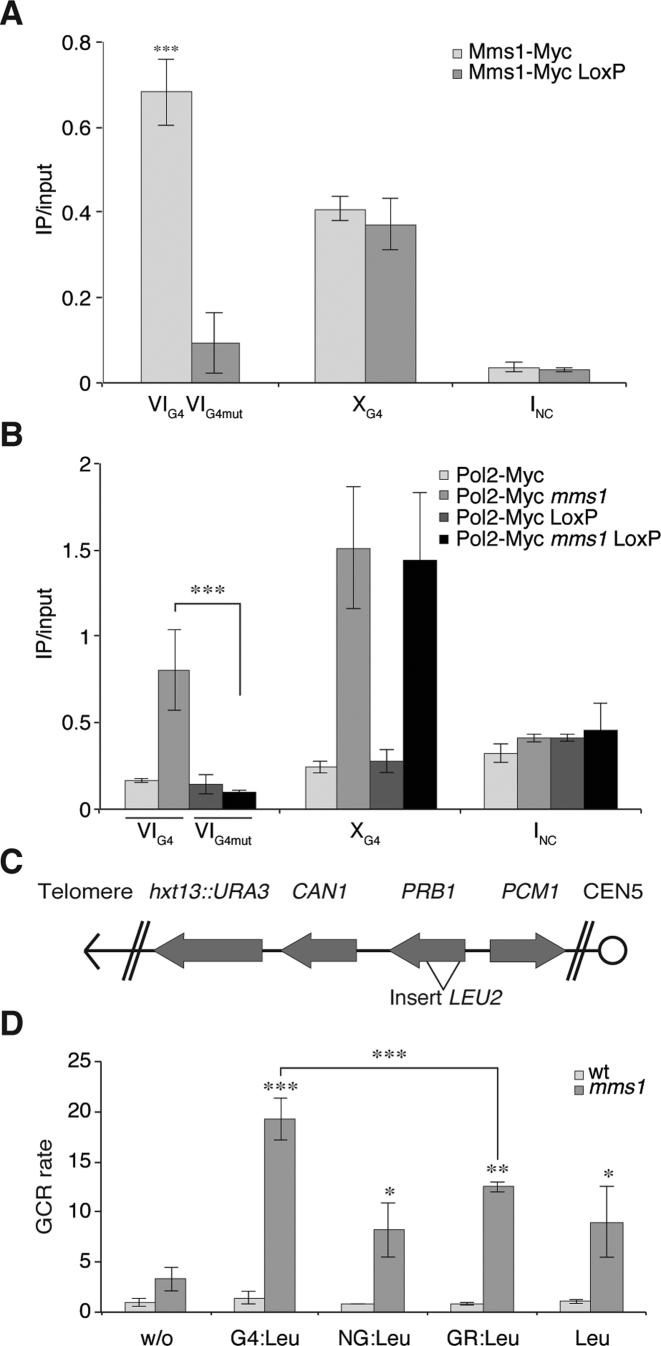
To further support the finding that Mms1 acts on G4 structures rather than just G-rich sequences, we mutated the G4 motif from Chr VI (IVG4) (GGGGCACACGTGCGGGAGTTTCAAAGGGGCAGAATAGTGGGGTTCAGGGG) by Cre-Lox recombination to abolish its G4-forming potential (Chr VIG4mut GGCGCACACGTGCGCGAGTTTCAAAGGCGCAGAATAGTGCGCTTCAGCCG). We endogenously tagged Mms1 in this strain. ChIP and qPCR analyses revealed that Mms1 binding is lost once the G4 motif is mutated (Figure 4A see VIG4). The binding of Mms1 to non-mutated G4 motif (XG4), in the same strain background was not altered (Figure 4A). This indicates that the G-rich nature of the motif is not responsible for Mms1 binding, but the G4 structure itself. To reveal if DNA replication pausing also depends on G4 motif at these sites, we endogenously tagged DNA Pol2 in the strain harboring the mutated G4 on Chr VI (VIG4mut). ChIP and qPCR analyses were performed in wild type and mms1 cells. As observed in Figure 3B, DNA Pol2 was enriched in mms1 at VIG4 and XG4. Interestingly, DNA Pol2 binding was no longer enriched at VIG4mut in mms1 cells upon mutation of the G4 motif (Figure 4B, black). Binding of DNA Pol2 in MMS1 cells did not change upon G4 mutation. These results further strengthen the hypothesis that Mms1 binds to G4 structures, including sequences with the G4tract2 motif as well as the conventional G4tract3 or G4tract4.
Figure 4.
Mms1 is necessary for genome stability, especially at G4 motifs. (A and B) Mms1 and DNA Pol2 binding to G4 and mutated G4 was monitored by ChIP and qPCR. VIG4 was mutated using Cre-LoxP (VIG4mut). Binding of Mms1 and DNA Pol2 was monitored at VIG4, XG4 and INC in the cre-loxP background as well as unaltered background. (A) As before (Figure 1) Mms1 binds to G4 motifs VIG4, XG4 (light). Binding of Mms1 to VIG is abolished upon mutation VIG4 (gray) using cre-LoxP. In the same background (cre-LoxP) Mms1 binding to XG4 is unaffected. (B) DNA Pol2 binding was analyzed in wild type and mms1 cells. In wild type cells, DNA Pol2 binds similarly to VIG4 and VIG4mut. As before (Figure 3B) in mms1 cells DNA Pol2 binding is enriched at VIG4 and XG4 (light grey), but upon mutation of VIG4mut binding of DNA Pol2 is reduced (black). DNA Pol2 binding in wild type and mms1 cells was not affected at XG4 in the cre-LoxP background. Plotted are the IP/input values as mean value ± SD. N ≥ 3 biological replicates. (C) Schematic of the genome region used in the GCR assay. The GCR rate was calculated by fluctuation analysis. (D) The GCR rate was determined in wild type (wt) (white) and mms1 (gray) cells. As inserts a G4 motif (G4-LEU2), a G-rich region (GR-LEU2), a non-G-rich region (NG-LEU2) as well as LEU2 marker were used (see Supplementary data 14 for additional information). Shown are mean values ± SD as fold enrichment over wild type without insert. n = 7 biological replicates, N≥3. Statistical significance compared to no insert strain. Significance was calculated by Student's t-test. *P < 0.05, **P < 0.05, ***P < 0.001.
Mms1 prevents genome instability
Our findings revealed that Mms1 binds to G-rich/G4 regions (Figure 1A and B) and supports Pif1 function (Figure 3C) at such sites. Previously, it was shown that, in the absence of Pif1, DNA replication is not only impeded, but genome stability is also challenged (19–23). We tested if genome instability at G4 motifs is increased in mms1 cells, in which Pif1 binding is reduced, using a previously published GCR assay (20,32). In this assay, we monitor G-rich versus G4 induced genome instability quantitatively. It allows us to measure complex genome rearrangement by simultaneous selection against two counter-selectable markers (URA3, CAN1) (20,55). We created four experimental strains in which a G4tract3 motif from Chr I (G4-LEU2), a G-rich region from Chr I (GR-LEU2), or a non-G-rich region from Chr VII (NG-LEU2) was inserted into the yeast genome on the left arm of Chr V, replacing the non-essential PRB1 gene (Figure 4C, see Supplementary data 14A for detailed information on regions and sequences). Because the insertions are done using a LEU2 gene, which harbors many G4tract2 on the template for lagging strand replication, that are Mms1 binding regions based on our ChIP and ChIP-seq data, we also inserted only the LEU2 gene into this region. The two counter-selectable markers (URA3 and CAN1) are located downstream of the PRB1 locus (Figure 4C). If the inserted sequence induces genomic instability, the markers are lost, and cells can grow on selective media. By counting the colonies on selective media compared to those on rich media plates, the GCR rate can be determined via fluctuation analysis (33). The GCR rate of the wild type cells without an insert was ∼0.1 × 10−9 events per generation, as published (56). The observed GCR rates, normalized to the rate of wild type without insert, are depicted in Figure 4D. None of the inserts induced a significant increase of the GCR rate in wild type cells. Interestingly, even without any insertion, the deletion of MMS1 caused an increased (3.3-fold) GCR rate over wild type without insert (Figure 4D). Normalizing these GCR rates by the rate obtained from mms1 without an insert, we measured 2.5- to 3.8-fold higher GCR rates using GR-LEU2, NG-LEU2, and the LEU2 gene as an insert. However, if a G4-LEU2 insert is present in mms1 cells, the GCR rate increased ∼19-fold compared to wild type cells (Figure 4D), which is significantly (P = 0.003) higher than in the presence of the other inserts. As mentioned above, the LEU2 gene contains a G4tract2 on the lagging strand template for replication and therefore increased GCR rates were expected for all inserts. Strikingly, insertion of an additional G4 motif resulted in an even higher GCR rate in mms1 cells (see Supplementary data 14B for data normalized against LEU2). Similarly, increased G4-dependent genome instability (GCR rates in pif1-m2 are without insert 76-fold and with a G4 insert 200-fold) was also detected using the same GCR assay in pif1-m2 cells (see (20)). These data suggest that loss of Mms1 causes genome instability and that this effect is even more severe if G4tract3 motifs are present.
DISCUSSION
In this study, we investigated the functional role of the ubiquitin ligase component Mms1 at G-rich/G4 regions during DNA replication. We demonstrated that Mms1 binds to G-rich regions, and more specifically to G4tract2 motifs on the template for lagging strand replication (Figure 1A,B, Supplementary data 6). CD experiments showed that a subset of the identified G4tract2 motifs form G4 structures in vitro (Figure 1C). Additionally, we observed that replication slows in mms1 cells at such sites, contributing to increased genome instability (Figures 3 and 4). Using ChIP experiments, we further revealed that Mms1 binding to G-rich/G4 sites supports Pif1 DNA helicase function at G4s (Figure 3C).
Based on our experiments and published data, we propose a new mechanistic model for G4 function during replication (Figure 5). Mms1 binds throughout the cell cycle to specific G-rich regions with a potential to form G4 structures. Interestingly, Mms1 does not bind to all G4 motifs it is specific for G4 motifs located on the lagging strand template. It is important to note that we cannot exclude the possibility that this binding is indirect. Regardless, once Mms1 binds to these G-rich/G4 regions it supports Pif1 binding at the end of S phase, allowing Pif1 to unwind structured DNA. Hence, replication fork progression and genome integrity are maintained. In the absence of Mms1, Pif1 binding is diminished, which results in replication fork pausing and genome instability. We present six results that support this model.
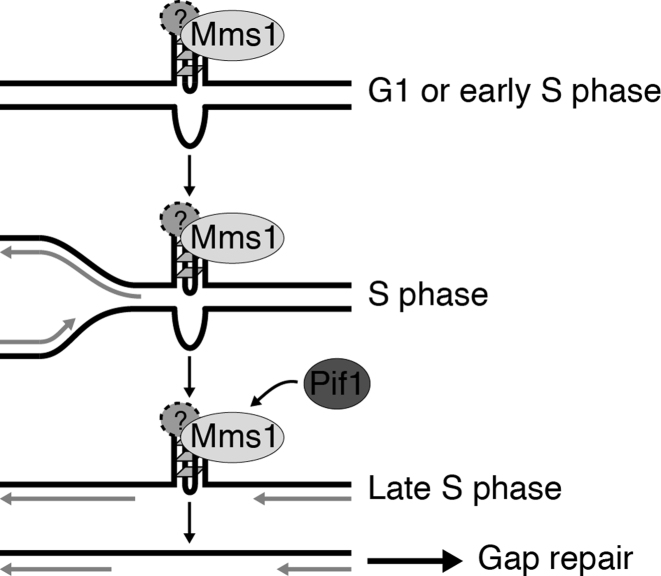
Figure 5.
Mechanistic model of Mms1 function at G4 structures. Mms1, perhaps in complex with an interaction partner, binds to G4 structures formed in G1 phase or in early S phase. It binds only to G4 structures form on the lagging strand template for DNA replication. During S phase, the replication fork passes the unsolved G4 structures leaving a gap behind. Mms1 enables (directly or indirectly) the binding of Pif1, which unwinds the G4 structure in late S phase. The resulting gap at the resolved G4 structure is then repaired via a so far unknown mechanism.
First, the consensus Mms1 binding motif is more G-rich than expected (P < 0.001) and most, 61 of 71 binding sites, harbor a G4tract2 motif located on the lagging strand template. Using CD analysis, we showed that a subset of Mms1 binding sites that harbor a G4tract2 can fold into stable G4s in vitro (Figure 1C). Second, Mms1 binding to G4tract2 as well as G4tract3 motifs with a potential to form G4s was shown by ChIP and qPCR (Figure 1B). The low abundance of Mms1 (57) and the unpredictable nature of G4 structure formation in vivo (8) supports the notion that Mms1 likely binds to more G4 sites in vivo than identified by ChIP-seq. This hypothesis is supported by ChIP-qPCR experiments that showed robust Mms1 binding to G4tract2 and G4tract3 motifs that were not discovered by ChIP-seq (Figure 1B, Chr VIBR, IXBR, XIaBR, XIbBR, XIIIBR, XVBR).
Third, we showed that Mms1 binding sites overlap significantly with Pif1 binding regions (P = 0.001, Supplementary data 11). Furthermore, in the absence of Mms1, less Pif1 binds to G-rich/G4 motifs (Figure 3C). This result indicates that either Mms1 binding supports Pif1 function at G4 structures or that Mms1 itself stabilizes G4 structures that are in need of unwinding by Pif1 helicase. From the current state of knowledge, we exclude the second scenario because without Mms1 more genome instability is observed (GCR assay, Figure 4D). Interestingly, Mms1 has a preference for G-rich/G4tract2 on the lagging strand template whereas Pif1 binds to G4tract3 motifs with no preference for leading and the lagging strand templates (21). This suggests that G4 structure regulation is not as straight forward as initially thought and that other factors or proteins will be found in the future that support Pif1 function at other G4 structures.
Fourth, in the absence of Mms1, DNA replication slows at G4/G-rich motifs (Figure 3B) and genome instability increases (Figure 4D). This is in line with the fact that without Mms1 less Pif1 binds to G4 structures (Figure 3C), consequently G4 structures are not unwound, thus DNA replication is impeded and genome instability is observed using a yeast genetic assay (GCR). Interestingly, if a G4tract3 motif was inserted in addition to the G4tract2 the GCR rate was further increased (Figure 4D). Previously, it was shown that in the absence of Pif1, GCR rate increases 2.5-fold upon addition of G4 motifs (increase from 76-fold in without insert to 200-fold with G4 insert) (20). The higher GCR rates in pif-m2 compared to the GCR rate in mms1 can be explained by the observation that without Mms1 Pif1 binding is reduced at G4 structures but not abolished and some Pif1 still binds to G4s. Additionally, without Pif1 the GCR rate (without insert) is already over 35-fold higher than in mms1 cells. This might be due to the fact that Pif1 is a multifunctional helicase involved in multiple steps important for genome stability (58). Nevertheless, the GCR results agree with the finding that more stable G4 structures, harboring a G4tract3, are more challenging for genome stability (Piazza et al., 2015) than G4tract2.
Fifth, we show that at G4 motifs located on the lagging strand template Mms1 is the core component which recognizes the G4 motif and supports Pif1 function. This function is independent of the E3 ligase complex (Figures 1 and 3A and Supplementary data 8). ChIP experiments showed that Mms1 binding is dependent on neither Mms22 nor Rtt101, which suggests that Mms1 is either binding the region itself or via a so far unknown interaction partner. However, we did not identify other Mms1 interaction partners by co-IP followed by mass spectrometry. In two independent approaches, we could only identify Rtt101 as a significant and relevant binding partner (Supplementary data 15). We think it is unlikely that Mms22 or Rtt101 support Mms1 binding at such sites, because deletion of one or both proteins did not result in reduced ChIP signals of Mms1-Myc (Figure 3A, Supplementary data 8) nor in replication fork pausing (Supplementary data 9). This conclusion is in contrast to previous publications where it was suggested that Mms1 binds via Mms22 to DNA (2,5,59). This difference could be explained by different experimental set ups and the fact that we look at a very specific target (G4 motifs on the lagging strand) of Mms1.
Sixth, our results show for the first time that Mms1 binds to G4 structures rather than to G-rich DNA regions (Supplementary data 13). Mms1 binding is even lost upon disruption of the G4 forming potential (Figure 4A). Consequently, DNA replication is independent of Mms1, and thus Pif1, if the G4 motif is mutated (Figure 4B). These results and the observed finding that Mms1 binds throughout the cell cycle to these G4 motifs indicate that G4 structures form already before the onset of S phase. This argument is supported by the fact that G4 structures can form in duplex DNA in vivo (14), but also raises the question how and why G4 structures form prior to S phase.
Previous work in yeast using a human minisatellite sequence had shown that only G4s with very short loops challenge genome stability. In this manuscript, we show that G4s with either longer loops or shorter G-tracts located on the laggings strand template are bound by Mms1 and that Mms1 assist DNA replication and genome stability by supporting Pif1 function here. This is in agreement with other publications showing that in the absence of helicases (e.g. Pif1, FANCJ, Dog-1) genome stability is at risk at G4 motifs (20,21,23–25,60,61). Our data strengthen the hypothesis that G4 regulation is done by specilized helicases and that a specific helicase regulates G4 unwinding at a specific loci or during a specific mechanism (e.g. Pif1 during DNA replication). In summary, the data presented here highlights the still mysterious nature of G4 structure, function and regulation in vivo, and also calls for further future experiments. Our study provides new insights into G4 structure regulation during replication. In yeast it was shown that Pif1 DNA helicase is responsible for genome stability at G4 structures. We provide data that, depending on the location of the G4 and maybe other unknown factors, G4 structures are unwound/unfolded with the support of other proteins. We revealed, that Mms1 supports Pif1 binding to G4 structures located on the lagging strand template for replication, leading to the question, which other proteins support Pif1 function at other G4 loci.
ACCESSION NUMBER
The accession numbers for the ChIPseq data reported in this paper are NCBI GEO: GSE98524.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elmar Wolf and Stefan Juranek for help with ChIP-seq experiments; the group of Boris Pfander and Martin Eilers for help with FACS analysis; the group of Hermann Schindelin for help with CD analysis; the group of Andreas Schlosser for mass spectrometry analysis; Matt Bochman, Silvia Götz and Sabrina Bartsch for providing some strains; Hinke Kazemier for experimental support and Michael Chang, Stefan Juranek and Satya Pandey for careful reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council Starting Grant [638988-G4DSB] and Emmy Noether Program of the Deutsche Forschungsgemeinschaft [DFG PA1649/3-1] to KP. Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barbour L., Xiao W.. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 2003; 532:137–155. [DOI] [PubMed] [Google Scholar]
- 2. Zaidi I.W., Rabut G., Poveda A., Scheel H., Malmstrom J., Ulrich H., Hofmann K., Pasero P., Peter M., Luke B.. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008; 9:1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luke B., Versini G., Jaquenoud M., Zaidi I.W., Kurz T., Pintard L., Pasero P., Peter M.. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr. Biol.: CB. 2006; 16:786–792. [DOI] [PubMed] [Google Scholar]
- 4. Duro E., Vaisica J.A., Brown G.W., Rouse J.. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair. 2008; 7:811–818. [DOI] [PubMed] [Google Scholar]
- 5. Vaisica J.A., Baryshnikova A., Costanzo M., Boone C., Brown G.W.. Mms1 and Mms22 stabilize the replisome during replication stress. Mol. Biol. Cell. 2011; 22:2396–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rothstein R., Michel B., Gangloff S.. Replication fork pausing and recombination or ‘gimme a break’. Genes Dev. 2000; 14:1–10. [PubMed] [Google Scholar]
- 7. Edenberg E.R., Downey M., Toczyski D.. Polymerase stalling during replication, transcription and translation. Curr. Biol.: CB. 2014; 24:R445–R452. [DOI] [PubMed] [Google Scholar]
- 8. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mendoza O., Bourdoncle A., Boule J.B., Brosh R.M. Jr., Mergny J.L.. G-quadruplexes and helicases. Nucleic Acids Res. 2016; 44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A.. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr., Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lam E.Y., Beraldi D., Tannahill D., Balasubramanian S.. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013; 4:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P.. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012; 8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muller S., Kumari S., Rodriguez R., Balasubramanian S.. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010; 2:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarsounas M., Tijsterman M.. Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol. 2013; 425:4782–4789. [DOI] [PubMed] [Google Scholar]
- 19. Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A.. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011; 30:4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paeschke K., Capra J.A., Zakian V.A.. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.P., Phan A.T., Nicolas A.. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015; 34:1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ribeyre C., Lopes J., Boulé J., Piazza P., Guédin A., Zakian V., Mergny J.-L., Nicolas A.. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009; 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K.. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008; 283:36132–36139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kruisselbrink E., Guryev V., Brouwer K., Pontier D.B., Cuppen E., Tijsterman M.. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol.: CB. 2008; 18:900–905. [DOI] [PubMed] [Google Scholar]
- 26. Cea V., Cipolla L., Sabbioneda S.. Replication of Structured DNA and its implication in epigenetic stability. Front. Genet. 2015; 6:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sikorski R.S., Hieter P.. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989; 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Driessche B., Tafforeau L., Hentges P., Carr A.M., Vandenhaute J.. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast. 2005; 22:1061–1068. [DOI] [PubMed] [Google Scholar]
- 29. Schulz V.P., Zakian V.A.. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994; 76:145–155. [DOI] [PubMed] [Google Scholar]
- 30. Bachrati C.Z., Hickson I.D.. Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol. 2006; 409:86–100. [DOI] [PubMed] [Google Scholar]
- 31. Cotterill S. Practical Approach Series 199. 1999; Oxford; NY: Oxford University Press; 281. [Google Scholar]
- 32. Putnam C.D., Kolodner R.D.. Determination of gross chromosomal rearrangement rates. Cold Spring Harbor Protoc. 2010; 2010, doi:10.1101/pdb.prot5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hall B.M., Ma C.X., Liang P., Singh K.K.. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009; 25:1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Capra J.A., Paeschke K., Singh M., Zakian V.A.. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010; 6:e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Azvolinsky A., Dunaway S., Torres J., Bessler J., Zakian V.A.. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006; 20:3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amberg D.C., Burke D., Strathern J.N., Burke D. Cold Spring Harbor Laboratory . Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2005. 2005; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 41. Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H.. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996; 24:2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hryciw T., Tang M., Fontanie T., Xiao W.. MMS1 protects against replication-dependent DNA damage in Saccharomyces cerevisiae. Mol. Genet. Genomics: MGG. 2002; 266:848–857. [DOI] [PubMed] [Google Scholar]
- 43. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wieland M., Hartig J.S.. Investigation of mRNA quadruplex formation in Escherichia coli. Nat. Protoc. 2009; 4:1632–1640. [DOI] [PubMed] [Google Scholar]
- 45. Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Ame. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 46. Qin M., Chen Z., Luo Q., Wen Y., Zhang N., Jiang H., Yang H.. Two-quartet G-quadruplexes formed by DNA sequences containing four contiguous GG runs. J. Phys. Chem. B. 2015; 119:3706–3713. [DOI] [PubMed] [Google Scholar]
- 47. Fu Y., Xiao W.. Identification and characterization of CRT10 as a novel regulator of Saccharomyces cerevisiae ribonucleotide reductase genes. Nucleic Acids Res. 2006; 34:1876–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hamatake R.K., Hasegawa H., Clark A.B., Bebenek K., Kunkel T.A., Sugino A.. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem. 1990; 265:4072–4083. [PubMed] [Google Scholar]
- 49. Morrison A., Araki H., Clark A.B., Hamatake R.K., Sugino A.. A third essential DNA polymerase in S. cerevisiae. Cell. 1990; 62:1143–1151. [DOI] [PubMed] [Google Scholar]
- 50. Azvolinsky A., Giresi P., Lieb J., Zakian V.. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 2009; 34:722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zimmer J., Tacconi E.M., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A. et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol. Cell. 2016; 61:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skoneczna A., Kaniak A., Skoneczny M.. Genetic instability in budding and fission yeast-sources and mechanisms. FEMS Microbiol. Rev. 2015; 39:917–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Phillips J.A., Chan A., Paeschke K., Zakian V.A.. The pif1 helicase, a negative regulator of telomerase, acts preferentially at long telomeres. PLoS Genet. 2015; 11:e1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ivessa A.S., Zhou J.-Q., Zakian V.A.. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000; 100:479–489. [DOI] [PubMed] [Google Scholar]
- 55. Schmidt K.H., Pennaneach V., Putnam C.D., Kolodner R.D.. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 2006; 409:462–476. [DOI] [PubMed] [Google Scholar]
- 56. Chen C., Kolodner R.D.. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 1999; 23:81–85. [DOI] [PubMed] [Google Scholar]
- 57. Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M.. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014; 11:319–324. [DOI] [PubMed] [Google Scholar]
- 58. Bochman M.L., Sabouri N., Zakian V.A.. Unwinding the functions of the Pif1 family helicases. DNA Repair. 2010; 9:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vejrup-Hansen R., Mizuno K., Miyabe I., Fleck O., Holmberg C., Murray J.M., Carr A.M., Nielsen O.. Schizosaccharomyces pombe Mms1 channels repair of perturbed replication into Rhp51 independent homologous recombination. DNA Repair. 2011; 10:283–295. [DOI] [PubMed] [Google Scholar]
- 60. Lemmens B., van Schendel R., Tijsterman M.. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015; 6:8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheung I., Schertzer M., Rose A., Lansdorp P.M.. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002; 31:405–409. [DOI] [PubMed] [Google Scholar]
- 62. Paramasivan S., Rujan I., Bolton P.H.. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007; 43:324–331. [DOI] [PubMed] [Google Scholar]
- 63. Del Villar-Guerra R., Gray R.D., Chaires J.B.. Characterization of quadruplex DNA structure by circular dichroism. Curr. Protoc. Nucleic Acid Chem. 2017; 68, doi:10.1002/cpnc.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.