Abstract
A considerable effort has been devoted to retrieve systematically information for genes and proteins as well as relationships between them. Despite the importance of chemical compounds and drugs as a central bio-entity in pharmacological and biological research, only a limited number of freely available chemical text-mining/search engine technologies are currently accessible. Here we present LimTox (Literature Mining for Toxicology), a web-based online biomedical search tool with special focus on adverse hepatobiliary reactions. It integrates a range of text mining, named entity recognition and information extraction components. LimTox relies on machine-learning, rule-based, pattern-based and term lookup strategies. This system processes scientific abstracts, a set of full text articles and medical agency assessment reports. Although the main focus of LimTox is on adverse liver events, it enables also basic searches for other organ level toxicity associations (nephrotoxicity, cardiotoxicity, thyrotoxicity and phospholipidosis). This tool supports specialized search queries for: chemical compounds/drugs, genes (with additional emphasis on key enzymes in drug metabolism, namely P450 cytochromes—CYPs) and biochemical liver markers. The LimTox website is free and open to all users and there is no login requirement. LimTox can be accessed at: http://limtox.bioinfo.cnio.es
INTRODUCTION
Considerable effort in the field of biomedical text mining has been devoted to molecular biology literature. The main focus has been on functionally relevant information for a particular entity type, mainly genes and proteins, their attributes and relationships between them, e.g. protein–protein interactions. In comparison, there are fewer text-mining systems available that focus on chemical compounds and drugs, despite the central importance of these bio-entities, not only for pharmacological and toxicological research but for biomedicine in general (1). A number of text mining efforts that tried to detect pharmacogenomics-related aspects, including the extraction of gene-drug associations, were published (2,3), and some attempts were made to extract drug–drug interactions from text (4,5). An important pioneering study, resulting from a collaboration between safety scientists from the Pharma company Pfizer and biocurators of the Comparative Toxicogenomics Database (CTD), revealed that text-mining results are useful to aid scientific literature curation of cardiovascular, neurological, renal and hepatic toxicities of drugs (6). Drug-induced liver injuries (DILIs) and associations of compounds to toxicological/toxicity endpoints have not been addressed extensively by literature mining since this attempt. However, drug-induced hepatotoxicity is of particular relevance for drug approval. It has motivated withdrawal of several drugs and is one of the major causes for drug attrition. Therefore, there is a pressing need for online text mining tools that may help to detect information related to toxic properties of chemical compounds and drugs. The mechanisms leading to drug-induced liver toxicity are particularly complicated, and hepatotoxicity poses a significant challenge for widely used predictive chemoinformatics approaches because of its implicit complexity. Some biochemical pathways and enzymes like cytochromes or aminotransferases play a central role in the characterisation of hepatotoxicity, and the importance of altered levels of certain serum enzymes (e.g. of alanine aminotransferase or aspartate aminotransferase) to detect hepatotoxicity was underlined by medical experts (7).
Attempts were made to construct structured knowledge bases that provide information on hepatotoxicity, like the CTD (8) and the LTKB (9) databases. Complementing high quality manual annotations, the systematic extraction of DILI relevant information can be obtained through text-mining methods. An early approach applied a commercial text mining pipeline (BioWisdom's Sofia platform) to recover statements associated to hepatotoxicity in the form of concept triplets from PubMed abstracts. These consisted of co-mentions of concept-relationship-concept triplets (10) of compounds with terms related to hepatobiliary anatomy/pathology.
To facilitate a more targeted retrieval of hepatotoxicity relevant information, we implemented an online text mining application called LimTox that extracts automatically toxicology relevant information from text, with special emphasis on drug-induced adverse hepatobiliary reactions.
SYSTEM OVERVIEW AND TEXT MINING PIPELINE
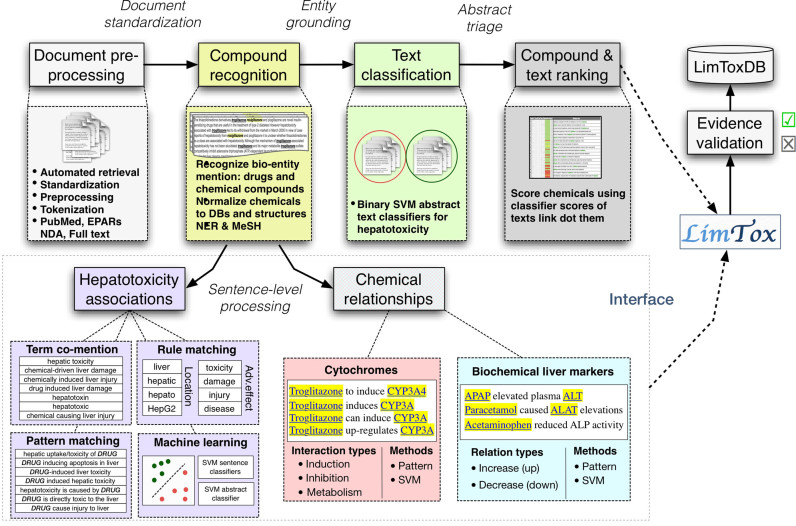
LimTox incorporates several text mining and information extraction components, which will be briefly introduced in this section. Further details regarding methodological aspects and evaluation settings can be found in the Additional Material sections. Figure 1 shows a schematic overview of the general LimTox flow chart.
Figure 1.
Simplified schematic flow chart of the LimTox system pipeline. This figure shows the various tasks that are part of the LimTox processing pipeline, from the initial document pre-processing to the detection of chemical entities to the hepatotoxicity text scoring approaches and relation extraction tasks.
Document selection and pre-processing
Four different document types were processed by LimTox, namely the entire set of PubMed abstracts, a collection of 13 234 full text articles (describing CYPs) as well as full text drug-related reports, i.e. 2145 European public assessment reports (EPARs) and 7738 New Drug Applications (NDAs). Full text PDF files were automatically converted into plain text, while for sentence boundary recognition and tokenisation the segtok (https://github.com/fnl/segtok) and sentence_splitter (https://github.com/fnl/sentence_splitter) python libraries, originally developed in our group, were used. The used sentence splitter supports processing both PubMed abstracts as well as full text articles. During the document standardisation step all input texts were converted into a common representation format.
Recognition of chemicals and drugs
The recognition of chemicals in running text is a key step required for subsequent semantic searches of chemicals and their association to chemically induced adverse reactions. The Additional Material 1 section describes in detail the used chemical entity tagging approaches. Linking documents to chemical entities by the LimTox system was done through two strategies. The main approach relied on the results obtained by running the ChemSpot tagger (11), a tool that recognizes both systematic and trivial chemical names and associates them to various chemical database identifiers. We applied a post-processing pipeline to remove frequent false positive chemical compound mentions. Chemical names were furthermore linked to structural representations by using name to structure conversion software (name-to-struct version 13.0). The ChEBI web service was exploited to retrieve structural information for mentions that were assigned to ChEBI identifiers by ChemSpot.
Scoring text for hepatotoxicity
Recent biomedical text mining evaluation efforts focusing on user interaction showed that one of the strongly desired end user requirements was the ability to filter/sort/rank the results according to different output criteria rather than obtaining a single static systems result (12). Support of various ranking strategies for associating texts, in particular sentences, to adverse reactions offers additional flexibility to online text mining tools. The importance of simple co-occurrence-based methods, as a sort of high recall baseline system complementing machine learning or natural language processing based approaches, was already previously highlighted by Jensen et al. (13).
Four of the most popular types of biomedical literature processing strategies are rule, pattern, dictionary/term and machine learning based text-processing methods (14), each showing particular strengths and weaknesses. The LimTox system, rather than returning a unique ensemble-based output, enables end users to directly exploit the underlying advantages of the each of these text-scoring methods. The Additional Material 2 section describes in more depth the used text scoring mechanisms.
Textual data contained in LimTox was processed at the level of abstracts (PubMed records) and individual sentences (PubMed records, full text articles and drug-related reports).
Term mention detection
A well-known functionality of biomedical search engines, sometimes regarded as a sort of baseline strategy, involves indexing of documents with keywords or terms that are representative for a particular topic of interest. Term co-occurrence results have the obvious advantage that humans can easily interpret them. Moreover they allow exploitation of quantitative occurrence statistics (13). Relevant lexical resources for hepatotoxicity were scattered/fragmented across different ontologies, gazetteers and thesauri. To support the hepatotoxicity-related term indexing process, we have built the LimTox lexicon. This lexicon covers adverse hepatobiliary vocabularies from multiple existing ontologies and terminologies, and therefore facilitates direct concept grounding. Moreover, the LimTox lexicon also incorporates lexical resources generated through a process of automatic term extraction. The used term extraction approach (see Additional Material 2 section) relied on the recognition of hepatotoxicity trigger words, hepatobiliary related trigger words and toxicity/adverse event related trigger words. The LimTox lexicon comprises 29 371 terms in total (4141 were manually validated).
Rule-based detection
Adverse hepatobiliary events can be expressed in running text in ways that go beyond a specific term or phrase of consecutive words. To handle such cases we extended the automatic hepatotoxicity term selection strategy (Additional Material 2 section) to process entire sentences instead of individual terms or phrases. Therefore, we have integrated a simple rule-based strategy that detects expressions referring to the target site, the so-called hepatobiliary trigger words (organ, tissue, cell type, molecular entities), together with the expressions referring to adverse, pathologic or toxicity assertions (toxicity/adverse event trigger words). Both types of trigger words had to be co-mentioned within sentences. This approach relied on 852 manually defined hepatotoxicity, 960 hepatobiliary and 552 toxicity trigger terms.
Pattern-based detection
The pattern-based approach required the initial co-occurrence of two semantic types, namely the agent (chemical compound or drug) and the target site (hepatobiliary system) of the adverse effect. LimTox used a set of 2926 manually constructed causal adverse effect relation text patterns (Additional Material 2 section). Those patterns essentially consisted of minimal text spans that expressed causal relations between chemicals and adverse hepatobiliary events. Some patterns were extracted from sentences of the SCAI corpus (15), while other were constructed semi-automatically using heuristic rules that took into account aspects such as frequency, length, part of speech information or presence of certain relation trigger words. LimTox patterns were used as templates to recognize positive matching phrases in the entire set of PubMed sentences.
Classifier strategy
A very popular approach to classify relevant text passages relies on using supervised machine learning methods. In particular Support Vector Machine (SVM) algorithms have generated competitive results for triage tasks posed at several BioCreative community challenges (biocreative.org). We trained machine learning-based abstract and sentence classifiers using SVM algorithms. The classifiers relied on word n-gram features and term frequency-inverse document frequency term weighting. We used classipy (https://github.com/fnl/classipy), a command-line tool originally developed in our lab to develop advanced text classifiers. The hepatotoxicity SVM abstract classifier was trained on a balanced set of 10 984 abstracts (16). As positive training data we used records describing drug-induced liver damage selected through either a keyword or a rule-based approach. The negative training set consisted of a random sample of PubMed abstracts of the same size. To determine the quality of the training data, we inspected manually a sample of 100 abstracts. About 83% of them corresponded to DILI relevant documents and another 12% to adverse liver events caused by alterations in genes and gene products. The resulting classifier model was applied to score the entire collection of abstracts contained in the PubMed database. For evaluation purposes we used several independent validation datasets to estimate whether the classifier was able to recover records labeled as hepatotoxicity relevant. The classifier was able to recover between 89.43 and 98.08% of the records annotated as DILI relevant from various datasets (see Additional Material 2 section).
Chemical relationships
The detection of relationships between chemicals and p450 cytochromes (CYPs) is of key relevance, as these enzymes play a fundamental role in the xenobiotic metabolism of drugs. LimTox incorporates a pipeline for automatic extraction of CYPs mentions and CYPs-chemical relations from sentences (see Additional Material 3 section). For recognition of CYPs mentions, a gazetteer lookup approach was applied, using a CYPs gene/protein gazetteer originally derived from the UniProt database. Additionally, CYP nomenclature guidelines were encoded into a rule-based recognition approach for the detection of non-continuous text strings referring to CYPs. The LimTox system detected three types of chemical-CYPs associations, namely induction, inhibition and metabolism relations. These relations were extracted by two complementary methods, a high precision pattern-based system and a machine learning-based sentence classifier approach.
LimTox also carried out the automatic extraction of relationships between chemicals and substances commonly measured in biochemical liver assays. A total of 17 types of liver marker mentions (13 proteins, 3 chemicals and 1 generic term) were automatically recognized in sentences co-mentioning chemical entities. A rule-based system was implemented to determine if there is an increase or decrease of liver markers following drug administration (details can be found in Additional Material 3 section).
LimTox incorporates negation handling by adapting a NegEx algorithm implementation in Python. Negation detection was applied to the results of the relation extraction pipelines, checking whether the chemical compound or its relation target (terms, CYPs, markers) corresponded to negated arguments recognized by NegEx.
FUNCTIONALITY AND USAGE
The LimTox system allows to recover information from different retrieval viewpoints, covering information related to cytochromes, markers associated to liver toxicity and genes for which alterations have been described in the presence of liver damage. The system is organized into six main search categories: keywords, chemical compounds, cytochromes, liver-damage markers, toxicological endpoints and genes. It processes a number of document sources, namely PubMed abstracts, full-text sentences and medical agency reports. Specific entity identifiers, for instance chemical identifiers from public available databases such as ChEBI or Drugbank, can be used to refine searches. LimTox also offers the possibility to use co-occurrence constraints by selecting sentences that co-mention at least one additional entity type. Each recognized entity mention is highlighted according to the entity type coloring criteria (chemical compounds are shown in yellow, cytochromes in light red, markers in turquoise, terms in violet and species in light green). LimTox offers support for manually validated information by end users in an open community curation-type annotation approach. Curated or manually validated evidence is indicated by a tick symbol while the initial query term is underlined. LimTox provides, where possible, outlinks to external databases to enrich the information retrieved for chemical compounds. Finally, results for chemical compounds, cytochromes and markers associated to liver damage can be curated at the sentence level from the results table. The curation process consists in confirming (checkbox) or denying (crossbox) the association between that entity and the hepatotoxic effect present in the sentence. In order to cope with synonyms and aliases for a given chemical entity, LimTox relies on a query expansion mechanism that makes use of an internal alias mapping table. For chemical entity hits, a result table provides alternative names, chemical structure visualisation and an entity interaction network (a graph of compound relationships), where the chemical compound query is displayed at the center of the graph as the primary node. The edges of the network represent relationships between the queried compound and associated entities.
Case studies
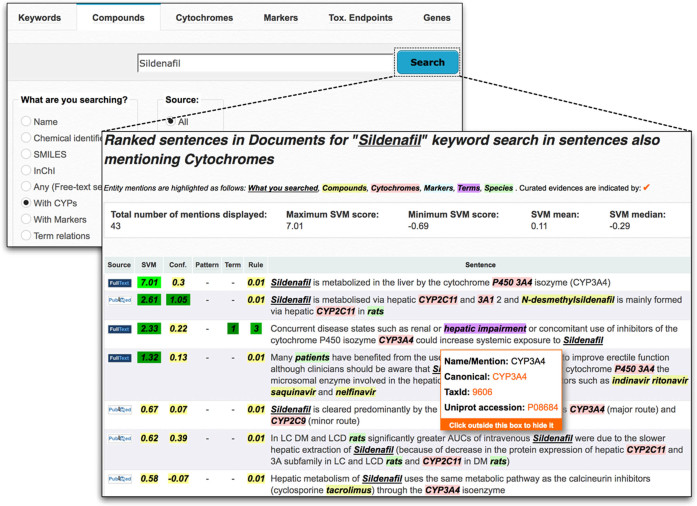
Case 1: Sildenafil search. As an example, to illustrate the kind of metabolism-related information supported by LimTox, we queried the system to retrieve documents with CYPs interacting with Sildenafil. LimTox returned a collection of descriptive sentences (Figure 2). Within the results, the user can quickly identify that CYP3A4 is the major CYP involved in Sildenafil metabolism, together with minor routes, including CYP2C9. This is in agreement with information reported in DrugBank (ID: DB00203) and literature (17). This search provided further details on other sildenafil-related compounds: primary metabolite (N-desmethylsildenafil), drug–drug interactions with CYP3A4 inhibitors (e.g. ketoconazole, itraconazole, ritonavir and indinavir) and other PDE5 inhibitors (vardenafil). Additionally, disease states (hepatic impairment) affecting Sildenafil pharmacokinetics were also anticipated. To gain further knowledge on the sildenafil-hepatic impairment connection, a LimTox search for specific hepatoxicity endpoints of sildenafil revealed pathologies (cirrhosis) and factors (age) leading to hepatic insufficiency that might impose recommendations on drug dosage administration. A broad vision of all compounds, CYPS, liver toxicity markers and toxicity terms associated with Sildenafil can be obtained by running a basic keyword search for Sildenafil and displaying its interaction network.
Figure 2.
Sildenafil search for CYPs relations using LimTox compound search interface. Example search output using as a query Sildenafil with the user restriction to return only those sentences that co-mention CYPs. The corresponding sentences returned by LimTox can be re-ranked by end users according to the various hepatotoxicity scoring criteria, that is using the SVM sentence classifier scores (SVM), the corresponding confidence scores of the SVM classifiers (conf.), the pattern-based approach (Pattern), the term-lookup method (Term) and the rule-based method (Rule). A simple mouse-over highlight allows the end user to get a short description of each method type, while clicking on the method generated a re-sorted output.
Case 2: Paracetamol search. Paracetamol (acetaminophen or APAP), when used at the recommended dose, is a safe analgesic/antipyretic drug. This is not the case for long-term use or acetaminophen overdose. In this context, LimTox can aid in finding the underlying toxicity mechanisms. By carrying out a LimTox search for chemical-marker relations using this drug, it became apparent that glutathione (GSH) depletion is one of the major causes of acetaminophen toxicity. This was highlighted by the large number of top-scoring hits and corroborated by the fifth-ranked sentence: ‘GSH depletion and oxidative stress are believed to be the main cause of APAP toxicity’.
IMPLEMENTATION AND USER TESTING
LimTox is mainly written in PHP using the web application framework Symfony2 (2.7.20, http://symfony.com). Results are provided using Apache web server, 2.4.7 version. Some of the scripts used to obtain compound images or to calculate Tanimoto distances between compounds were implemented using Python and the rdkit python module (www.rdkit.org). A PostgreSQL (9.3.15) relational database server stores all entities and the relation information. The RDKit PostgreSQL cartridge was installed to have built-in PostgreSQL chemical processing support. ElasticSearch 1.7.2 (www.elastic.co), a non-relational database, was used to index the documents. As a PHP client for ElasticSearch, we used Elastica (http://elastica.io). We used FosElasticaBundle to provide integration of ElasticSearch and Elastica with Symfony. LimTox runs on a server with 32 Intel(R) Xeon(R) CPU E5-2650 2.00GHz processors. The server has a total memory of 400GB, and 30GB as the maximum memory allocation pool for the Java Virtual Machine (JVM) that runs the ElasticSearch instance. LimTox was tested on the most commonly used web-browsers (i.e. Firefox, Chrome and Internet Explorer). Online help, including documentation and video tutorials, as well as an online survey and a contact link, are provided on the LimTox web. LimTox was tested both by academic and Pharma users in the context of the EU project eTOX. Test users included experts from medicinal chemistry, toxicology, chemoinformatics, bioinformatics and biology. Their feedback was captured in form of a structured user survey and used to improve the online functionalities of LimTox.
DISCUSSION AND CONCLUSION
Here we describe a text mining system called LimTox, which extracts relevant information for the characterisation of adverse hepatobiliary effects induced by chemical compounds. The system is available online and integrates various approaches, from co-occurrence of chemicals with hepatotoxicity relevant terms to rule-based and machine learning techniques for detecting compounds that cause induced liver toxicities. The system can be used as a topic-specific search engine. The detection of DILI-related articles and entities were compared to annotations from various databases (15). Beyond direct associations of chemicals to hepatotoxicity, two additional entity relation types important in toxicological and clinical settings were included into LimTox, that is chemicals-CYP and chemicals-liver marker relations. LimTox also enables more targeted searches for other adverse events, namely nephrotoxicity, cardiotoxicity, thyrotoxicity, phospholipidosis.
AVAILABILITY
LimTox is free and open to all users and there is no login requirement. LimTox can be accessed at: http://limtox.bioinfo.cnio.es.
Supplementary Material
ACKNOWLEDGEMENTS
Many thanks to Florian Leitner, Miguel Vazquez, Montse Cases, Nicole Dolker and Analia Lourenço for useful feedback and input.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
eTOX project [IMI-115002]; European Commission H2020 project OpenMinted [654021]; Plan de Impulso de las Tecnologías del Lenguaje de la Agenda Digital (PITL) of the Secretary of State of Telecommunications of the Spanish Ministry of Energy, Tourism and the Digital Agenda; ISCIII and ERDF [PT13/0001/00]. Funding for open access charge: European Commission H2020 project OpenMinted [654021].
Conflict of interest statement. None declared.
REFERENCES
- 1. Vazquez M., Krallinger M., Leitner F., Valencia A.. Text mining for drugs and chemical compounds: methods, tools and applications. Mol. Inform. 2011; 30:506–519. [DOI] [PubMed] [Google Scholar]
- 2. Clematide S., Rinaldi F.. Ranking relations between diseases, drugs and genes for a curation task. J. Biomed. Semantics. 2012; 3:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiegers T.C., Davis A.P., Cohen K.B., Hirschman L., Mattingly C.J.. Text mining and manual curation of chemical-gene-disease networks for the comparative toxicogenomics database (CTD). BMC Bioinformatics. 2009; 10:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Percha B., Garten Y., Altman R.B.. Discovery and explanation of drug-drug interactions via text mining. In Pacific symposium on biocomputing. Pac. Symp. Biocomput. 2012; 410. [PMC free article] [PubMed] [Google Scholar]
- 5. Tari L., Anwar S., Liang S., Cai J., Baral C.. Discovering drug–drug interactions: a text-mining and reasoning approach based on properties of drug metabolism. Bioinformatics. 2010; 26:i547–i553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis A.P., Wiegers T.C., Roberts P.M., King B.L., Lay J.M., Lennon-Hopkins K., Sciaky D., Johnson R., Keating H., Greene N. et al. A CTD–Pfizer collaboration: manual curation of 88 000 scientific articles text mined for drug–disease and drug–phenotype interactions. Database. 2013; bat080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Navarro V.J., Senior J.R.. Drug-related hepatotoxicity. N. Engl. J. Med. 2006; 354:731–739. [DOI] [PubMed] [Google Scholar]
- 8. Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J.. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2016; 45:D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen M.J., Vijay V., Shi Q., Liu Z.C., Fang H., Tong W.D.. ‘FDA-approved drug labeling for the study of drug-induced liver injury,’. Drug Discov. Today. 2011; 16:697–703. [DOI] [PubMed] [Google Scholar]
- 10. Fourches D., Barnes J.C., Day N.C., Bradley P., Reed J.Z., Tropsha A.. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem. Res. Toxicol. 2009; 23:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rocktäschel T., Weidlich M., Leser U.. ChemSpot: a hybrid system for chemical named entity recognition. Bioinformatics. 2012; 28:1633–1640. [DOI] [PubMed] [Google Scholar]
- 12. Wang Q., Abdul S.S., Almeida L., Ananiadou S., Balderas-Martínez Y.I., Batista-Navarro R., Campos D., Chilton L., Chou H.J., Contreras G. et al. Overview of the interactive task in BioCreative V. Database (Oxford). 2016; baw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen L.J., Saric J., Bork P.. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006; 7:119–129. [DOI] [PubMed] [Google Scholar]
- 14. Hunter L., Cohen K.B.. Biomedical language processing: what's beyond PubMed?. Mol. Cell. 2006; 21:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gurulingappa H., Rajput A.M., Roberts A., Fluck J., Hofmann-Apitius M., Toldo L.. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012; 45:885–892. [DOI] [PubMed] [Google Scholar]
- 16. Joachims T. Svmlight: Support vector machine. 1999; 19:University of Dortmund; http://svmlight.joachims.org/. [Google Scholar]
- 17. Huang S.A., Lie J.D.. Phosphodiesterase-5 (PDE5) inhibitors in the management of erectile dysfunction. Pharm. Ther. 2013; 38:407–419. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.