Abstract
Sophisticated and interactive visualizations are essential for making sense of the intricate 3D structures of macromolecules. For proteins, secondary structural components are routinely featured in molecular graphics visualizations. However, the field of RNA structural bioinformatics is still lagging behind; for example, current molecular graphics tools lack built-in support even for base pairs, double helices, or hairpin loops. DSSR (Dissecting the Spatial Structure of RNA) is an integrated and automated command-line tool for the analysis and annotation of RNA tertiary structures. It calculates a comprehensive and unique set of features for characterizing RNA, as well as DNA structures. Jmol is a widely used, open-source Java viewer for 3D structures, with a powerful scripting language. JSmol, its reincarnation based on native JavaScript, has a predominant position in the post Java-applet era for web-based visualization of molecular structures. The DSSR-Jmol integration presented here makes salient features of DSSR readily accessible, either via the Java-based Jmol application itself, or its HTML5-based equivalent, JSmol. The DSSR web service accepts 3D coordinate files (in mmCIF or PDB format) initiated from a Jmol or JSmol session and returns DSSR-derived structural features in JSON format. This seamless combination of DSSR and Jmol/JSmol brings the molecular graphics of 3D RNA structures to a similar level as that for proteins, and enables a much deeper analysis of structural characteristics. It fills a gap in RNA structural bioinformatics, and is freely accessible (via the Jmol application or the JSmol-based website http://jmol.x3dna.org).
INTRODUCTION
Sophisticated analysis and visualizations are essential for making sense of the intricate 3D structures of macromolecules, providing insights into their diverse functions in essential biological processes. For proteins, secondary structural components (e.g. α-helices and β-strands) are readily accessible via the de facto standard DSSP algorithm (1) and routinely featured in general-purpose molecular graphics programs, such as Jmol/JSmol (2,3), the PyMOL molecular graphics system (version 1.8, Schrödinger, LLC), and the NGL viewer (4). However, the field of RNA structural bioinformatics is still lagging behind; for example, most current molecular graphics tools lack built-in support even for base pairs, double helices or hairpin loops.
In addition to general-purpose 3D molecular viewers, quite a few tools are dedicated to RNA structures. Jalview (5) is a program for editing, visualizing, and analyzing multiple sequence alignments. VARNA (6) allows for interactive drawing and editing of RNA secondary structures. Assemble (7) combines RNA secondary structure design with 3D modeling. Moreover, RNApdbee (8) extracts RNA secondary structure from 3D coordinates, and presents it in planar diagrams. None of these tools, however, has the capability of selecting common RNA structural features and highlighting them in a 3D context interactively. The work presented here fills a gap in RNA structural bioinformatics, serving a huge user base both in research and education.
DSSR, Dissecting the Spatial Structure of RNA, is a stand-alone, command-line program written in ANSI C (9). The single-file DSSR binary executable is tiny (∼1MB) and self-contained (with zero dependencies), with an efficient and robust performance. DSSR is a component of the 3DNA suite of programs (10,11) for the analysis, rebuilding, and visualization of 3D nucleic acid structures. It has been designed to streamline the analysis and annotation of RNA tertiary structures using only an mmCIF or PDB file as input. DSSR has a comprehensive and unique set of functionalities for the analysis of RNA 3D structures (12,13). The program automatically identifies and annotates base pairs (14,15), detects higher-order coplanar base associations (triplet, quadruplet, etc.), finds coaxially stacked helices, characterizes loops of various types (hairpin, bulge, internal, and junction), and categorizes pseudoknots of arbitrary complexity, among numerous other functionalities (9). Moreover, DSSR-derived RNA secondary structures are available in three commonly used file formats: ViennaRNA package dot-bracket notation (.dbn) (16), Mfold connect table (.ct) (17), and CRW base-pair sequence (.bpseq) (18).
Jmol (2) is a widely used, open-source Java viewer for 3D structures, featuring a powerful and flexible scripting language. JSmol (3), its HTML5/JavaScript equivalent, has a predominant position in the post Java-applet era for web-based visualization of molecular structures. The DSSR-Jmol integration presented here makes salient features of DSSR readily accessible using either Jmol or JSmol, equivalently. In this paper, we use the term ‘Jmol’ to refer to the general features of both Jmol and JSmol, and ‘JSmol’ to refer to specific features of the web-based version of Jmol.
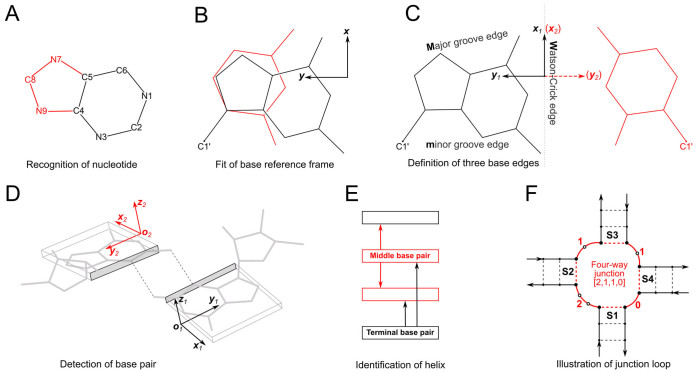
Figure 1 provides definitions of key nucleic acid structural components in DSSR that are currently accessible through Jmol. The DSSR web server accepts 3D coordinate files (in mmCIF or PDB format) initiated from a Jmol session. It returns DSSR-derived structural features in JSON (JavaScript Object Notation) for parsing and visualization via a dedicated structured query language (Jmol SQL for DSSR, see the Supplementary Data). This seamless combination of DSSR and Jmol brings the molecular graphics of 3D RNA structures to a similar level as that for proteins, and enables a much deeper analysis of structural characteristics.
Figure 1.
Definitions of key nucleic acid structural components in DSSR [reproduced from Figure 1 of reference (9)]. (A) Nucleotides are recognized using standard atom names and base planarity. This method works for both the standard bases (A, C, G, T and U), and those of modified nucleotides, regardless of their tautomeric or protonation states. (B) Bases are assigned a standard reference frame (25) that is independent of sequence identity: purines and pyrimidines are symmetrically placed with respect to the sugar. (C) The standard base frame is derived from an idealized Watson-Crick base pair, and defines three base edges (Watson-Crick, minor groove, and Major groove) that are used to classify pairing interactions. (D) Base pairs are identified from the co-planarity of base rings and the occurrence of hydrogen bonds. This geometric algorithm can find canonical (Watson-Crick and G–U wobble) as well as non-canonical pairs. Higher-order (three or more) co-planar base associations, termed multiplets, are also detected. (E) Helices are defined by stacking interactions of base pairs, regardless of pairing type (canonical or otherwise) or backbone connectivity (covalently connected or broken). A helix consists of at least two base pairs. The same algorithm is applied to identify continuous base stacks that are outside of helical regions, by using bases instead of pairs as the assembly unit. Nucleotides not involved in base-stacking interactions are collected into one separate group. A stem is defined as a special type of helix, made up of canonical pairs and with a continuous backbone along each strand. Coaxial stacking is defined by the presence of two or more stems within one helix. An isolated canonical pair is one that is not contained within a stem. (F) ‘Closed’ loops of various types (hairpin, bulge, internal, and junction loops) are delineated by stems or isolated pairs, and specified by the lengths of the intervening, consecutive nucleotide segments. A kissing-loop motif entails formation of one or more canonical pairs between the bases in different hairpin loops. Single-stranded segments that lie outside loops are separately listed.
In order to highlight the capabilities of DSSR in the context of Jmol, we have developed the DSSR-Jmol website http://jmol.x3dna.org, which has been in active service since 2014 and has been extensively tested by the DSSR and Jmol user communities. By adhering to web standards, it works in all modern browsers on various computer/operating systems (including handheld devices, such as tablets and smart phones). The interface is simple and intuitive, and new users can get started easily. It also allows power users to take full advantage of Jmol scripting via a command-line and console window.
IMPLEMENTATIONS
The DSSR-Jmol integration is facilitated via the DSSR - -json=ebi option, which takes 3D coordinates initiated from a Jmol session and renders output in the lightweight and structured JSON data-interchange format (http://www.json.org). Residues are specified in the easy-to-parse unit identifier format (http://www.bgsu.edu/research/rna/help/rna-3d-hub-help/unit-ids.html) proposed by the Leontis-Zirbel RNA structural bioinformatics group. DSSR-derived structural features are then queried via a flexible and powerful structured query language, Jmol SQL for DSSR (see below and the Supplementary Data). The work presented here corresponds to DSSR version 1.6.8 (released on 28 March 2017), and Jmol version 14.15.1 (released on 27 April 2017).
Accessible features
In the current DSSR-Jmol integration, 16 essential RNA structural features derived from DSSR are directly accessible to Jmol (Figure 1). Table 1 lists the names of these features, using the classic yeast phenylalanine tRNA structure (19) as an example (PDB id: 1ehz). Importantly, these 16 names can serve as keywords in the Jmol scripting language for selecting residues. For example, the following command script highlights the three hairpin loops in 1ehz: select hairpins; color red. Table 1 also showcases the way that annotations are structured for a base pair and a nucleotide.
Table 1. DSSR-derived features and DSSR-specific selections in Jmol, using yeast phenylalanine tRNA (1ehz) as an example.
| Accessible features (16 keys) | ["pairs", "multiplets", "helices", "stems", "isoCanonPairs", "coaxStacks", "hairpins", "bulges", "iloops", "junctions", "kissingLoops", "ssSegments", "stacks", "nonStack", "hbonds", "nts"] |
| Actual counts (1ehz) | {"pairs":34, "multiplets":4, "helices":2, "stems":4, "isoCanonPairs":1, "coaxStacks":2, "hairpins":3, "junctions":2, "kissingLoops":1, "ssSegments":1, "stacks":11, "nonStack":4, "hbonds":118, "nts":76} |
| Base pair (G1–C72) | {"index":1, "nt1":"|1|A|G|1||||", "nt2":"|1|A|C|72||||", "bp":"G-C", "name":"WC", "Saenger":"19-XIX", "LW":"cWW", "DSSR":"cW-W"} |
| Nucleotide (2MG10) | {"nt_name":"2MG", "nt_id":"|1|A|2MG|10||||","is_modified":true, "chi":169.599, "puckering":"C3΄-endo"} |
| Jmol (SQL) selections | SELECT hairpins |
| SELECT within(dssr, "nts WHERE is_modified") | |
| SELECT within(dssr, "pairs WHERE name != 'WC'") |
DSSR web service
The web service is hosted at Columbia University, via a single-file PHP script that calls the DSSR command-line tool for back-end analysis. The DSSR web service runs independently of Jmol, and accepts atomic coordinates in mmCIF or PDB format. The service also takes a four-letter PDB id to automatically fetch the corresponding mmCIF formatted coordinate file from the RCSB PDB (20) (see the Supplementary Data). For an NMR ensemble, only the first model is analyzed by default. For X-ray crystal structures, the asymmetric unit is analyzed. The DSSR algorithm works for both DNA and RNA, either in isolation or in their complexes with proteins. The server has an 18MB-size limit for uploaded coordinate files. For DNA/RNA structures with <300 nucleotides, it takes DSSR less than one second to run (see Table 1 of reference (9)).
Jmol SQL for DSSR
The structured query language (SQL) implemented in Jmol incorporates JSON as a subset. As a result, DSSR-derived annotations can be searched directly in Jmol using an SQL-like syntax. After loading a structure with DSSR annotations, the _M.dssr associative array contains all annotations in the JSON output from DSSR (see the Supplementary Data). Specifically, this array holds the 16 keys from DSSR (see Table 1) for residue selections. The syntax is: select @{_M.dssr.key}, where key is nts, pairs, or hairpins, etc., as listed in Table 1. For example, select @{_M.dssr.hairpins} has the same functionality as the two shorthand forms: select hairpins noted above or select within(dssr, 'hairpins'). Moreover, an individual hairpin can be selected by appending an array index: e.g. select @{_M.dssr.hairpins[1]} or select within(dssr, 'hairpins..1') picks the first hairpin loop (i.e. the D-loop in 1ehz). The power of Jmol SQL shines when fine-grained characteristics from DSSR (such as base-pair types) are queried, as illustrated in Table 1 and detailed in the Supplementary Data.
Java-based Jmol application
The DSSR web service can be directly accessed via the Jmol script console in two ways. The Jmol command load =1ehz/dssr specifies that the 1ehz model should be loaded from RCSB PDB, with DSSR-derived annotations retrieved from Columbia. Alternatively, the syntax load 1ehz.cif; calculate structure dssr allows one to do the same with local mmCIF or PDB data.
The JSmol web interface to DSSR
The http://jmol.x3dna.org (Figure 2) site provides a simple web interface that has the same scripting capability. In this case, the scripting is carried out by JavaScript calls to JSmol, initiated by users clicking buttons on the page. The website is written in JavaScript and HTML5, complying with web standards. The advantage of the web interface is that typical users do not need to know anything about Jmol scripting to benefit from the DSSR-Jmol integration. In addition, this page can be cloned and adapted easily by interested web developers. This paper focuses on the web interface to illustrate some of the functionalities that the DSSR-Jmol integration brings to the table, using a few examples (see below).
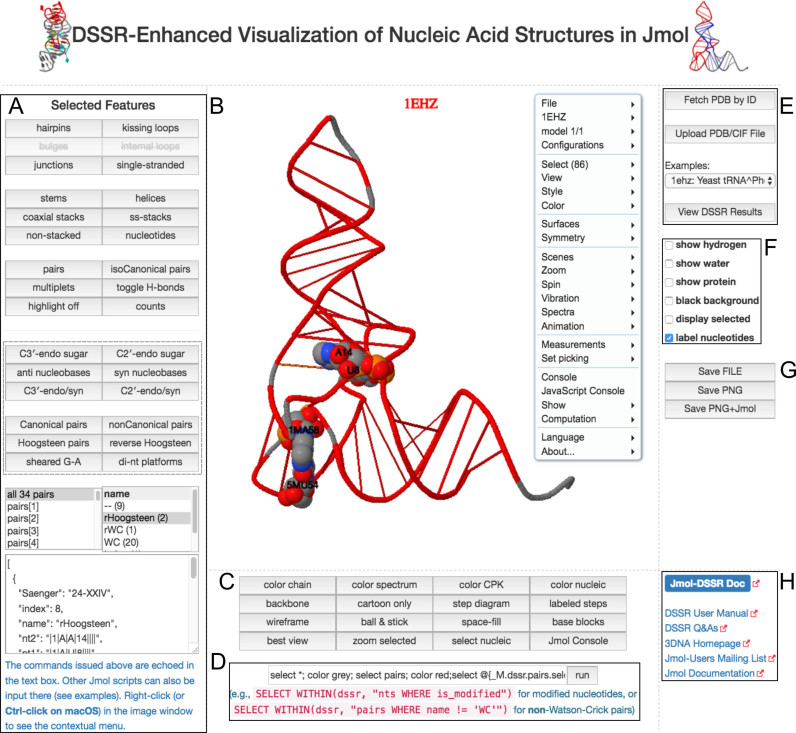
Figure 2.
A screenshot of the DSSR-JSmol web interface, highlighting the two reverse Hoogsteen pairs (U8–A14 and 5MU54–1MA58) of yeast phenylalanine tRNA (PDB id: 1ehz). (A) DSSR-derived structural features integrated into Jmol. (B) The main JSmol viewer canvas for visualization and interactive manipulations. (C) Common representation styles for selected structural features. (D) A simple text input field for advanced users to enter (short) Jmol script commands. (E) Structure input by PDB id, file upload (drag-and-drop), or selecting one from the twelve sample RNA structures. (F) Utilities to toggle between two states for six common cases. (G) Export of coordinate file or PNG image. (H) Links to online resources for DSSR and Jmol.
Figure 2 presents a screenshot of the DSSR-JSmol web interface divided into eight functional blocks (A–H), using yeast phenylalanine tRNA (PDB id: 1ehz) as an example. Since 1ehz does not have bulges and internal loops, the two corresponding buttons are grayed out (A). Clicking the ‘counts’ button provides further details of the analysis. The dashed-box provides buttons for querying some common characteristics of nucleotides or base pairs. The commands issued in (A) are echoed in the text input field in (D), providing users with illustrations of how to script Jmol themselves. The main JSmol viewer canvas (B) highlights the two reverse Hoogsteen pairs rendered as space-filling spheres. The context menu in (B) provides many options as in the Jmol application. Further details and scripting functionality are available by opening a Jmol console (C). For power users, it provides the most flexibility. The full DSSR-JSON output as received by JSmol can be viewed (E). The PNG+Jmol option (G) is worth mentioning: it creates an image file with enough attached data to allow it to be loaded back into Jmol, reproducing the 3D model exactly as it appears in the image. The ‘Jmol-DSSR Doc’ hyperlink (H) points to the Supplementary Data (in PDF), which is the manual of the DSSR-Jmol integration.
SAMPLE APPLICATIONS
The web interface is simple and intuitive so new users can get started quickly. Sophisticated results can be achieved by a combination of clicking the buttons, using the context menu, and issuing Jmol scripting commands. Figure 3 showcases different structural features in distinct representation styles that are enabled by the DSSR-Jmol integration and facilitated by the web interface, using five representative RNA structures as examples.
Figure 3.
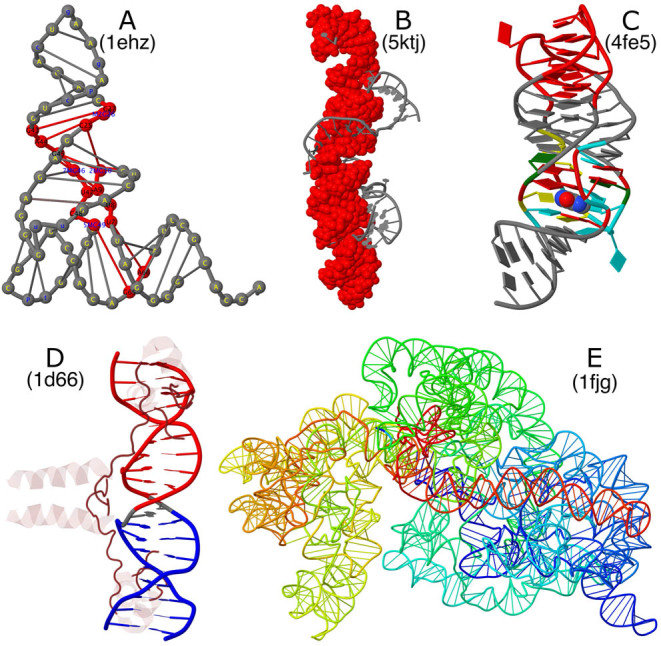
Sample molecular images illustrating selected RNA structural features enabled by the DSSR-Jmol integration. (A) Structure of yeast phenylalanine tRNA (PDB id: 1ehz) in step diagram with bases labeled, highlighting (in red) the [2,1,5,0] four-way junction loop in 3D. (B) Structure of the Pistol self-cleaving ribozyme (PDB id: 5ktj), showcasing (in red) the vertical helix in space-filling representation. The helix is composed of six short stems stabilized via coaxial stacking interactions. (C) Structure of the xpt-pbuX guanine riboswitch in complex with hypoxanthine (PDB id: 4fe5) in ‘base blocks’ representation. The three-way junction loop encompassing the metabolite (in space-filling representation) is color-coded by base identity: A, red; C, yellow; G, green; U, cyan. The loop-loop interaction (a kissing-loop motif) at the top is highlighted in red. (D) Structure of the yeast GAL4 protein-DNA complex (PDB id: 1d66) in Jmol cartoon representation. DNA is color-coded by DSSR stems, and protein is in translucent brown. The DNA helix is broken into two stems due to a non-Watson-Crick pair (G11 with C28 in gray) in the middle. (E) Structure of the Thermus thermophilus 30S ribosomal subunit in complex with the antibiotics (PDB id: 1fjg) using a step diagram. The 16S ribosomal RNA is color-coded in spectrum with the 5΄-end in blue and the 3΄-end in red.
Figure 3A emphasizes the four-way junction loop that links the two helical arms (‘L-shape’) together in the 3D structure of yeast phenylalanine tRNA (19). The 16-nucleotide ‘closed’ loop (colored red, and labeled with residue name and sequence number) corresponds to the central roundabout in the classic cloverleaf secondary structure diagram of tRNA. The 14 modified nucleotides in the structure are automatically detected by DSSR. They are colored blue, and labeled in lower case one-letter abbreviations outside the junction (e.g. the blue ‘g’ for OMG34 in the anticodon hairpin loop at the very top). The ‘step diagram’ draws rods to connect nucleotides of base pairs identified by DSSR.
Figure 3B highlights (in space-filling representation in red) the coaxial-stacked helix made up of six short stems in the X-ray crystal structure of Pistol, a self-cleaving ribozyme (21). The helix is 41 base pairs long, and curved, but with uninterrupted stacking interactions. The asymmetric unit of a crystal structure is analyzed, in this case consisting of two molecules.
Figure 3C shows the structure of the xpt-pbuX guanine riboswitch in complex with the metabolite hypoxanthine (22) in ‘base blocks’ representation. The three-way junction loop encompasses the metabolite via a network of base stacking and pairing interactions. DSSR further identifies a quadruplet that involves the ligand (HPA101) with U47, U51 and C74. The two hairpin loops highlighted in red (at the top) form a kissing-loop motif via two Watson-Crick pairs (G37–C61 and G38–C60). The kissing-loop motif creates a pseudoknot. In DSSR, it is also characterized as a four-way junction loop, with the pseudoknotted stem counted twice.
Figure 3D showcases the structure of the yeast GAL4 protein-DNA complex (23) in cartoon presentation, with DNA color-coded by DSSR stems and protein in translucent brown. Due to the non-Watson-Crick pair between G11 and C28 in the middle, the DNA helix is broken into two stems. Based on the atomic coordinates of PDB id 1d66, C28 is in the unusual syn instead of the common anti conformation. In DSSR, the pair between G11 and C28 is designated G+C, and classified as cWH according to the Leontis-Westhof notation (14) and cW+M following the DSSR scheme (see Figure 1C). The syn nucleobase of C28 turns out to be a modeling mistake (Stephen C. Harrison, personal communication). Flipping the base around the glycosidic bond of C28 by 180° will result in a proper G11–C28 Watson-Crick pair, and a single continuous stem. The list of examples on the website (Figure 2E) contains two entries for 1d66: one as directly downloaded from the RCSB PDB, and the corrected one with the C28 syn nucleobase flipped on the fly by JSmol.
Finally, Figure 3E illustrates the structure of the Thermus thermophilus 30S ribosomal subunit in complex with the antibiotics (24) using a step diagram. The structure (PDB id: 1fjg) contains two RNA chains: the 16S ribosomal RNA (chain A, 1507 nucleotides), and a fragment of messenger RNA (chain X, 6 nucleotides). On the website, it takes ∼1 minute to process this ribosomal structure, with two RNA molecules (1513 nucleotides) and 20 proteins.
CONCLUSIONS
The DSSR-Jmol integration bridges the DSSR command-line analyzing tool and the Jmol molecular viewer together via a simple JSON interface and a powerful query language. Users can now select DSSR-derived RNA structural features (such as base pairs, double helices, and various loops) as easily as they can select protein α-helices and β-strands. Moreover, fine-grained characteristics of these features can be queried via Jmol SQL for DSSR, allowing for selections such as Hoogsteen pairs, or nucleotides with a C2΄-endo sugar pucker and a syn nucleobase conformation. Notably, the novel representation styles (step diagram and base blocks) and coloring schemes, as demonstrated in Figure 3, bring RNA visualization to an entirely new level.
This work fills a gap in RNA structural bioinformatics, and it also provides an example for integrating DSSR-derived features into other molecular graphics programs. The website described here is fully functional, useful to researchers, educators, and students alike. Furthermore, it can serve as a starting point for anyone who wishes to develop additional interactive web-based resources involving nucleic acid structures.
We aim to continuously support, refine, and expand the DSSR-Jmol integration and the website (http://jmol.x3dna.org). Potential new features include: handling multi-model PDB files, integrating a dynamic viewer of RNA secondary structures (6), and simplifying visualization of large RNA 3D structures.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Wilma K. Olson and Jessalyn Lu for proofreading the manuscript, and the DSSR and Jmol user communities for testing the web interface and providing us with feedback. We appreciate the comments and suggestions of the three anonymous reviewers that helped improve and clarify this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM096889 to X.J.L.]. Funding for open access charge: NIH [R01GM096889].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kabsch W., Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983; 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 2. Hanson R.M. Jmol—-a paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 2010; 43:1250–1260. [Google Scholar]
- 3. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next-generation web-based representation of 3D molecular structure as applied to Proteopedia. Israel J. Chem. 2013; 53:207–216. [Google Scholar]
- 4. Rose A.S., Hildebrand P.W.. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015; 43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview Version 2—-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jossinet F., Ludwig T.E., Westhof E.. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010; 26:2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antczak M., Zok T., Popenda M., Lukasiak P., Adamiak R.W., Blazewicz J., Szachniuk M.. RNApdbee—-a webserver to derive secondary structures from PDB files of knotted and unknotted RNAs. Nucleic Acids Res. 2014; 42:W368–W372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu X.J., Bussemaker H.J., Olson W.K.. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu X.J., Olson W.K.. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008; 3:1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butcher S.E., Pyle A.M.. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011; 44:1302–1311. [DOI] [PubMed] [Google Scholar]
- 13. Moore P.B. Structural motifs in RNA. Annu. Rev. Biochem. 1999; 68:287–300. [DOI] [PubMed] [Google Scholar]
- 14. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saenger W. Principles of Nucleic acid Structure. 1984; NY: Springer-Verlag. [Google Scholar]
- 16. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorith. Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zuker M., Mathews D.H., Turner D.H.. Barciszewski J, Clark BFC. RNA Biochemistry and Biotechnology. 1999; 90:Kluwer Academic Publishers; 11–43. [Google Scholar]
- 18. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi H., Moore P.B.. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000; 6:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen L.A., Wang J., Steitz T.A.. Crystal structure of Pistol, a class of self-cleaving ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Batey R.T., Gilbert S.D., Montange R.K.. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004; 432:411–415. [DOI] [PubMed] [Google Scholar]
- 23. Marmorstein R., Carey M., Ptashne M., Harrison S.C.. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992; 356:408–414. [DOI] [PubMed] [Google Scholar]
- 24. Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V.. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000; 407:340–348. [DOI] [PubMed] [Google Scholar]
- 25. Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey S.C., Heinemann U., Lu X.J., Neidle S., Shakked Z. et al. . A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001; 313:229–237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.