Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) constitute a rapidly growing class of natural products with diverse structures and bioactivities. We have developed RiPPMiner, a novel bioinformatics resource for deciphering chemical structures of RiPPs by genome mining. RiPPMiner derives its predictive power from machine learning based classifiers, trained using a well curated database of more than 500 experimentally characterized RiPPs. RiPPMiner uses Support Vector Machine to distinguish RiPP precursors from other small proteins and classify the precursors into 12 sub-classes of RiPPs. For classes like lanthipeptide, cyanobactin, lasso peptide and thiopeptide, RiPPMiner can predict leader cleavage site and complex cross-links between post-translationally modified residues starting from genome sequences. RiPPMiner can identify correct cross-link pattern in a core peptide from among a very large number of combinatorial possibilities. Benchmarking of prediction accuracy of RiPPMiner on a large lanthipeptide dataset indicated high sensitivity, specificity, accuracy and precision. RiPPMiner also provides interfaces for visualization of the chemical structure, downloading of simplified molecular-input line-entry system and searching for RiPPs having similar sequences or chemical structures. The backend database of RiPPMiner provides information about modification system, precursor sequence, leader and core sequence, modified residues, cross-links and gene cluster for more than 500 experimentally characterized RiPPs. RiPPMiner is available at http://www.nii.ac.in/rippminer.html.
INTRODUCTION
Ribosomally synthesized and post-translationally modified peptides (RiPPs) constitute a large class of natural products with diverse structures and bioactivities (1). The logic that binds these structurally diverse peptides into one class is their biosynthesis. RiPPs are typically biosynthesized from a ribosomal peptide consisting of a leader and core segment. A variety of modifying enzymes encoded by neighboring genes carry out extensive post-translational modifications (PTMs) in the core peptide, followed by cleavage of the leader peptide (2). Some of the well known RiPP classes are lanthipeptides, lasso peptides, cyanobactins and thiopeptides (Supplementary Figure S1). Lanthipeptides consist of core region rich in Ser/Thr and Cys residues which are post-translationally modified to form intramolecular lanthionine/methyl-lanthionine (Lan/MeLan) linkages. This intramolecular cross-linking is a two step process, where Ser/Thr are dehydrated to form 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb) and subsequently linked to Cys thiols in a Michael-type addition to form Lan/MeLan. In case of cyanobactins, a single precursor peptide can give rise to more than one core peptide. Core peptides are N-to-C macrocyclized and often contain azol(in)e rings derived from heterocyclization of Ser/Thr/Cys residues with the carbonyl group of the preceding amino acid residue. The core peptide in lasso peptides consists of 16–21 residues where the N-terminal amino group is condensed to the carboxyl side chain of glutamate or aspartate, present at eighth or ninth position, to form a macrolactam ring (3). The C-terminal tail is trapped within the N-terminal macrolactam ring. Thiopeptides are peptide macrocycles containing multiple azole rings derived from Cys, Thr and Ser residues; and a six-membered nitrogenous ring derived from intramolecular cross-linking between two Dha and a carbonyl group. Recent spurt in genome sequencing has revealed that a huge number of genetically encoded RiPPs remain uncharacterized (4). During the last decade computational methods have played a crucial role in genome guided discovery of secondary metabolites. Powerful bioinformatics methods (5) have been developed by several groups for analysis of polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS) which are involved in biosynthesis of polyketides and nonribosomal peptides–the two major classes of secondary metabolites. Bioinformatics resources like antiSMASH (6), PRISM (7), SBSPKS (8), SMURF (9), ClusterMine360 (10), DoBISCUIT (11), NP.Searcher (12), NRPSPredictor (13), NaPDoS (14) have played major role in the analysis of secondary metabolite gene clusters, identification of catalytic domains in PKS/NRPS megasynthases and prediction of their substrate specificities. Such computational methods have facilitated discovery of new metabolites by genome mining (15).
In contrast to the large number of bioinformatics resources available for analysis of biosynthetic clusters of polyketides and non-ribosomal peptides, there are relatively fewer computational tools available for analysis of RiPP gene clusters. The diversity and complexity of PTMs and cross-links in RiPPs has been a major impediment in development of bioinformatics tools for deciphering chemical structure of RiPPs by genome mining. A recent update of antiSMASH contained improved lanthipeptide detection and prediction of leader peptide cleavage site (16). The lanthipeptide structure prediction tool antiSMASH still has limitations in distinguishing unmodified Ser/Thr from modified residues (Dha/Dhb) and in identifying Dha/Dhb residues which are cross-linked to Cys residues to form Lan/MeLan linkages. Very recently Skinnider et al. (17) have developed a novel computational tool RiPP-PRISM for charting ribosomally synthesized natural product chemical space. They have demonstrated its utility in genome guided discovery of a new molecule from a rare family of RiPP. RiPP-PRISM uses a hypothetical structure enumeration approach, similar to RiPP-Quest (18) and algorithms used by Zhang et al. (19), for identification of RiPP natural products in combination with mass spectrometry data. RiPP-PRISM has limited utility in deciphering chemical structures of RiPPs from genomic information or in absence of mass spectrometry data. This prompted us to develop alternate computational methods which can predict chemical structures of RiPPs using genomic sequences of RiPP, even in absence of mass spectrometric data.
Rapid increase in the numbers of experimentally characterized RiPPs and availability of sequence and chemical structure information of large number of RiPPs in well curated databases like BAGEL (20), Bactibase (21), MIBiG (22) and Thiobase (23) present the opportunity for developing knowledge based approaches for deciphering RiPP chemical structure using sequence information. However, the complexity of RiPP chemical space arising from PTMs and cross-links demand powerful computational algorithms which can decode sequence to chemical structure relationships. The success of machine learning-based methods like NRPSPredictor (13) in exploring chemical space of non-ribosomal peptides prompted us to explore machine learning methods for predicting chemical structures of RiPPs. We describe here the development, benchmarking and usage of RiPPMiner web server, which derives its predictive power from support vector machine (SVM) and random-forest (RF) classifiers trained on a well curated database of more than 500 experimentally characterized RiPPs. Given a query sequence, RiPPMiner uses SVM to distinguish RiPP precursors from other small proteins and classify the precursors into 12 sub-classes of RiPPs. For classes like lanthipeptide, cyanobactin, thiopeptide and lasso peptide models based on SVM and RF are used to predict the leader cleavage site, final complex cross-linking and post-translationally modified residues in the core peptide. To the best of our knowledge RiPPMiner is the only software which is currently available for predicting complex chemical structures of lanthipeptides, lasso peptides, cyanobactins and thiopeptides starting from sequences of the corresponding RiPP genes.
METHODS AND IMPLEMENTATION
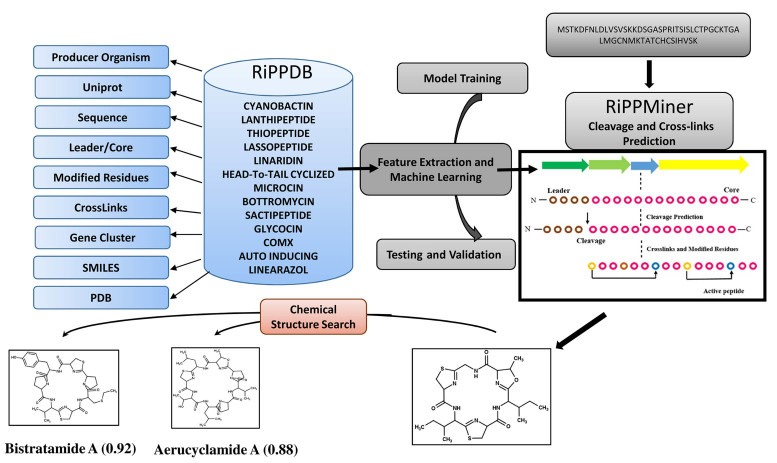
RiPPMiner web server consists of two major components, the backend database RiPPDB and query interface RiPPMiner (Figure 1). The backend database catalogs information on experimentally characterized RiPPs, while the query interface has been developed based on analysis of the sequence and chemical structure data of RiPPs using a machine learning approach.
Figure 1.
Schematic diagram depicting overall organization of RiPPMiner web server The RiPPMiner web server consists of two major components, the backend database RiPPDB and query interface RiPPMiner. The backend database catalogs information on experimentally characterized ribosomally synthesized and post-translationally modified peptides (RiPPs), while the query interface has been developed based on machine learning based analysis of the sequence and chemical structure data of known RiPPs in RiPPDB.
Compilation of data on RiPPs with known chemical structure
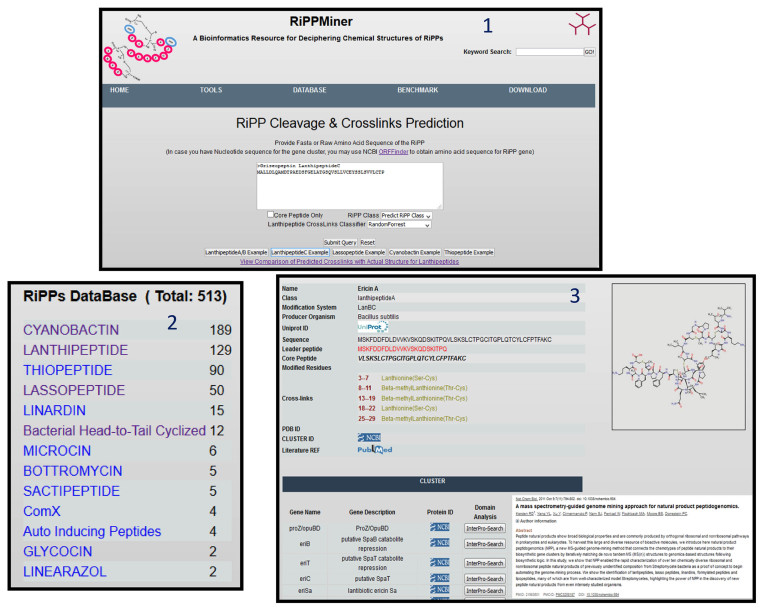
Since RiPPMiner uses a knowledge based approach for prediction of cleavage and cross-links, it requires a well curated database of experimentally characterized RIPPs with known chemical structures. Even though in silico analysis of microbial genomes reveal the presence of very large number of RiPP biosynthetic gene clusters (BGCs), only for a small fraction of them the subclass, leader cleavage site and complete chemical structure with cross-links and modified residues have been experimentally characterized. Information about RiPPs with known chemical structure were retrieved from databases like Bactibase (21), BAGEL (20), Thiobase (23), MIBiG (22) repository and entries in UniProt (24). In addition information about several entries was also compiled based on extensive literature search and Supplementary Data provided in a publication on RiPP-PRISM (17). Currently RiPPDB has information about 513 RiPPs from 13 RiPP classes (Figure 2). The major RiPP families like lanthipeptides, cyanobactins, thiopeptides and lasso peptides have more than 50 entries, while other seven RiPP families have 15 or less number of entries. For each entry, RiPPDB has cataloged name and chemical structure of the secondary metabolite, RiPP subclass, amino acid sequence of the precursor polypeptide, cleavage site, sequences of leader and core peptides, modification system, modified residues, list of cross-linked residues/type of cross-link, information about the corresponding gene cluster in NCBI and PUBMED ID of the publication which reports the experimental characterization (Figure 2). Links are also provided to the corresponding entries in UniProt, NCBI and PDB (if crystal or nuclear magnetic resonance structures are available for the secondary metabolite). In comparison to related databases, RiPPDB not only has information about maximum number of RiPPs, but also has information about maximum number of features about each of the characterized RiPPs.
Figure 2.
Screenshots depicting query interface of RiPPMiner and various of known RiPPs present in RiPPDB The textbox for providing sequence of the RiPP for prediction of cleavage site and cross-links (Panel 1). Statistics on number of RiPPs in each of the 11 RiPP classes (Panel 2). Screenshot depicting various features of experimentally characterized RiPPs cataloged in RiPPDB using lathipeptide Ericin A as example (Panel 3).
Machine learning for identification of RiPP and prediction of class, cleavage and cross-links
Machine learning approach was used to develop a computational method for distinguishing RiPP precursor sequences from other small proteins, classifying the precursors into 12 sub-classes of RiPPs, predicting cleavage site for leader peptide and also for predicting the final cross-linked chemical structure of the RiPP. We briefly describe here the method for feature extraction and training of SVM and RF classifiers using the well curated RiPPDB, while details of the method are provided in Supplementary Methods. SVMLight (Version 6.02) package (http://svmlight.joachims.org/) was used for developing SVM models, while RF classifiers were trained using WEKA Version 3.6.14 (http://www.cs.waikato.ac.nz/ml/weka/citing.html).
Identification of RiPPs and prediction of RiPP Class
RiPP analysis tools like antiSMASH and RiPP-PRISM use Hidden Markov Model (HMM) profiles of modifying enzymes present in the RiPP BGCs to predict the RiPP class. Unlike these tools, RiPPMiner uses a machine learning model trained using the amino acid sequence of the RiPP gene alone to identify RiPPs and then predict RiPP class. RiPPMiner first distinguishes RiPPs from other proteins and peptides using a SVM model. This SVM model for RiPP identification has been trained using 293 known RiPPs as positive dataset and 8140 genome encoded non-RiPP polypeptides (size lower than 100 amino acids) as negative data. The negative dataset included entries from SWISS-Prot having length similar to RiPPs, e.g. 30s ribosomal proteins, matrix proteins and cytochrome b proteins etc. The feature vectors for the SVM model consisted of amino acid composition and dipeptide frequencies. Benchmarking of this RiPP identification methods on an independent dataset (not included in training) using two-fold cross-validation approach indicated Sensitivity, Specificity, Precision and MCC values of 0.93, 0.90, 0.90 and 0.85 respectively (details in Supplementary Methods). This indicates good predictive power of the SVM model for distinguishing between RiPPs and non-RiPPs.
For prediction of RiPP class or sub-class a Multi Class SVM was trained using the amino acid composition and dipeptide frequencies as feature vectors. During the training of the Multi Class SVM for prediction of RiPP class, available RiPP precursor sequences belonging to a given class (e.g. lasso peptide) were used as positive set, while RiPPs belonging to all other classes were used as negative set. As described in Supplementary Methods apart from major RiPP classes, classifiers were also trained for RiPP subfamilies like class A, B and C lanthipeptides. It may be noted that RiPPDB and RiPPMiner follow the lanthipeptide classification as per BAGEL and Bactibase, but the corresponding modifying enzymes have been classified as class I, II, III and IV in recent literature on biosynthesis of lanthipeptides. Since several RiPP classes had very few members, the prediction accuracy was benchmarked using Leave One Out (LOO) method. Based on analysis of the receiver operator characteristic (ROC) curves suitable score cutoff was chosen for RiPP identification and classification. The prediction interface for RiPP identification and prediction of class accepts amino acid sequence of RiPP precursor in fasta format as input.
Prediction of cleavage site
Out of the four major RiPP classes which had more than 50 experimentally characterized RiPPs in RiPPDB, SVM models for prediction of cleavage sites could be developed for lanthipeptides, cyanobactins and lasso peptides. As information on complete precursor sequences of thiopeptide were insufficient, predictive model for cleavage prediction could not be developed. In order to develop SVM for prediction of cleavage site for lanthipeptides, 12 mer peptide sequences centered on the cleavage sites were extracted from a set on 115 lanthipeptide precursor sequences with known cleavage pattern. This resulted in a positive dataset of 103 unique 12 mer peptides harboring the cleavage site at the center, while all other unique 12 mer peptides in these 115 lanthiepeptides constituted the negative dataset as they lacked the cleavage site. Feature vectors for each of these 12 mers consisted of concatenation of 20 dimensional vectors corresponding to each of the 20 amino acids. SVM model for prediction of cleavage site was developed and benchmarked using 2-fold cross validation approach, where half of the data were used in training and the other half was used in testing as described in details in Supplementary Methods. SVM models were also developed for prediction of cleavage site in cyanobactin and lasso peptides, as discussed in Supplementary Methods. Based on analysis of the ROC curves suitable score cutoff was chosen for prediction of cleavage sites in lanthipeptides and lasso peptides.
Prediction of cross-links
The algorithm for prediction of cross-links and deciphering complete chemical structure of RiPP has been implemented for lanthipeptides, lasso peptide, cyanobactins and thiopeptides. The prediction of lanthionine linkages in lanthipeptides have been carried out using a machine learning approach. In order to develop machine learning based classifiers for prediction of lanthionine linkages, a dataset of 93 lanthipeptides having known chemical structures were taken from RiPPDB. For each lanthipeptide in this set, sequence of the core peptide was scanned for strings or sub-sequences of the type Ser/Thr-(X)n-Cys or Cys-(X)n-Ser/Thr to enumerate all theoretically possible cyclization patterns. Out of these sequence strings, the strings corresponding to Ser/Thr-Cys or Cys-Ser/Thr pairs which were linked by lanthionine bridges in the lanthipeptides were included in the positive set, while all other strings were included in the negative set. Supplementary Figure S2 gives a schematic depiction of the steps involved in obtaining positive and negative set sub-sequences for the lanthipeptide nisin. After following similar procedure for all the peptides in the dataset and removal of duplicate strings, a positive dataset of 218 unique strings and a negative dataset of 1358 unique strings were extracted. Feature vectors were obtained for these datasets using amino acid composition and dipeptide frequency. Using these feature vectors SVM and RF classifiers were trained and benchmarked using LOO as well as 2-fold cross validation approach (Supplementary Methods). Based on analysis of ROC curves obtained from benchmarking studies, suitable score cutoffs were chosen for cross-link prediction using SVM or RF classifiers. Even though these classifiers can distinguish positive and negative set sub-sequences with reasonably high accuracy, in some cases overlapping cross-links are predicted. For example, same Ser/Thr being cross-linked to more than one Cys or vice versa. Since such overlapping lanthionine cross-links are not chemically feasible, the program only retains the cross-link with highest score and removes other cross-links. Apart from lanthionine linkages, labionine linkages are also known to occur in class III (class C in RiPPDB) lathipeptides. Analysis of known labioinine containing lanthipeptides revealed that, labionine cross-links can be predicted based on occurrence of S-X-X-[S/T]-(X) 3-5-C motif. Hence, motif based method has been implemented for labionine cross-link prediction in addition to machine learning approach. For thiopeptides, the cross-links have been predicted based on occurrence of SC-(X)n-CSC or SC-(X)n-[C/S]SSSS motif, where Ser residues marked in bold are post-translationally modified to Dha and are then cross-linked via formation of nitrogen containing six membered rings (Supplementary Methods). In case of lasso peptides and cyanobactins, the cross-link prediction algorithm involves a combination of machine learning and motif based prediction as described in Supplementary Methods. For the various different types of cross-link predictions carried out using RiPPMiner, the software can also generate chemical structures of the cross-linked RiPP in SMILES (simplified molecular-input line-entry system) format using an in-house developed code. Apart from SMILES the software also provides image of the predicted chemical structure using MarvinSketch tool of JChem software version 17.2.13.0 from Chemaxon (https://www.chemaxon.com/).
Search interfaces for similar sequences and similar chemical structures
RiPPMiner provides two user friendly interfaces for sequence similarity search and for search of similarities in chemical structures. Sequence similarity search has been implemented using local version of NCBI BLAST 2.2.30+ (25). Given a query sequence, pairwise BLAST searches are carried out against the sequences of precursor polypeptides or core polypeptide sequences of various RiPPs stored in RiPPDB. It may be noted that out of the 513 RiPP entries cataloged in RiPPDB, precursor polypeptide sequences consisting of leader and core regions are available for only 296 RiPPs. For large number of cyanobactins and thiopeptides, only the sequence for the core peptide or final cross-linked RiPP structure is available. The chemical structure similarity search has been implemented in RiPPMiner using local version of the Openbabel tool kit (v2.3.1) (26) and chemical structure similarity is measured as Tanimoto score. Structure similarity search can also be carried out for chemical structures of RiPPs predicted by RiPPMiner.
Implementation of web server
Web interface of RiPPMiner is implemented on a LINUX server using Perl CGI, PHP, HTML CSS, Java script, JQuery and apache web server. MySQL has been used for the backend database RiPPDB.
RESULTS
Usage of RiPPMiner for prediction and analysis
We describe here, a typical use of RiPPMiner web server for prediction of RiPP class, cleavage site and cross-links using amino acid sequence of a RiPP precursor polypeptide as input and also for comparison of the predicted RiPP chemical structure with other experimentally characterized RiPPs cataloged in RiPPDB. The homepage of RiPPMiner gives a brief overview of various types of analysis which can be carried out in text as well as pictorial form. The TOOLS link on the RiPPMiner homepage provides access to various prediction and search interfaces of RiPPMiner, while DATABASE link leads to the characterized RiPPs cataloged in RiPPDB (Figure 2, Panel 1). As can be seen in Figure 2, RiPPDB provides statistics on number of RiPPs in each of the 13 RiPP classes (panel 2) and upon selecting a given RiPP (e.g. lathipeptide Ericin A) the user can visualize chemical structure of the RiPP and also various other features like leader and core peptide sequences, modified residues, cross-links and neighboring genes in the RiPP gene cluster (panel 3). The RiPP entry page in RiPPDB also provides links to external databases like UniProt, NCBI nucleotide and PUBMED. Since we have integrated information from other databases, currently RiPPDB provides comprehensive information on maximum number of features on a given RiPP compared to other databases like MIBiG, BAGEL and Bactibase.
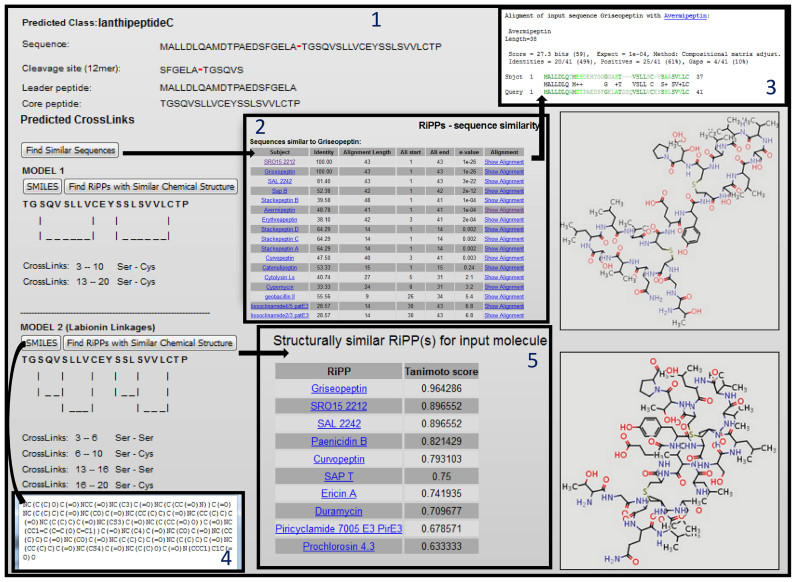
The panel 1 in Figure 2, shows the screenshot for the interface for prediction of cleavage and cross-links. The user can paste the amino acid sequence of the RiPP gene in the textbox or provide sequence of the core peptide only. If complete precursor sequence is provided, the software will first identify whether the input sequence is a RiPP precursor or not, then predict the RiPP class and subsequently apply the appropriate cross-link prediction rule. On the other hand, if core peptide is provided as input, the user should click the checkbox and select the RiPP class. Since cross-link predictions for lanthipeptides have been implemented using both SVM and RF classifiers, in case of lanthipeptides the user has the choice to select SVM or RF, though RF is default (Figure 2, panel 1). Panel 1 in Figure 3 shows typical output screen of RiPPMiner for cleavage and cross-link prediction for a lanthipeptide. The precursor sequence of Griseopeptin was used as input so that the user can easily validate the predictions by comparing the results to Griseopeptin entry in RiPPDB. As can be seen, RiPPMiner correctly identifies the RiPP class as lanthipeptide C, predicts the correct cleavage site and predicts two models for the cross-linked lanthipeptide. Model 1 contains lanthionine linkage, while the model 2 has the labionine linkage. The class C lanthipeptides like Griseopeptin, Erythreapeptin and Avermipeptin etc are known to have alternate cross-linked forms; hence multiple models are predicted by RiPPMiner. However, for other lanthipeptides single cross-linked structures are predicted. For easy visualization, RiPPMiner shows the lanthionine/labionine cross-links between modified Ser and Cys as dashed lines in the core peptide sequence, but the SMILES link and the images depict the chemical structures with all the atomic details and relevant bonds connecting the atoms. It may be noted that the current version of RiPPMiner does not predict other modified residues (e.g. Dha, Dhb) which are not cross-linked as the prediction accuracy has not been correctly benchmarked. Apart from chemical structure, the output page also provides links to SMILES code (panel 4, Figure 3) and other links for identifying similar sequences and similar chemical structures in RiPPDB. Upon clicking the ‘RiPPs with similar chemical structure’ link, the program provides the list of RiPPs with corresponding Tanimoto scores and links are also provided to the corresponding entries in RiPPDB (panel 5). As expected Griseopeotin is listed as the top hit but Tanimoto score is 0.96 because the predicted peptide does not contain all of the modified residues. Upon clicking the ‘Similar Sequences’ link (panel 1) the program provides list of other RiPPs having similar precursor sequences, their percentage identity, e-value (panel 2, Figure 3) and links for visualizing the alignments (panel 3). It is interesting to note that, apart from Griseopeptin, SRO15 2212 is also listed as lanthipeptide having 100% sequence identity with the query sequence (panel 2, Figure 3), while it's Tanimoto score with the predicted structure is 0.89 (panel 5). This demonstrates utility of RiPPMiner in identifying examples of identical RiPP precursor sequences giving rise to RiPPs with different cross-links.
Figure 3.
Typical output screen of RiPPMiner for cleavage and cross-link prediction for a lanthipeptide and subsequent analysis of the results screenshot depicting predicted RiPP class, cleavage site, cross-links and chemical structures for a lanthipeptide (Panel 1). Search results for RiPPs in RiPPDB having similar precursor sequence as the query RiPP (Panel 2). BLAST alignment of the query RiPP sequence with matching RiPPs in RiPPDB (Panel 3). SMILES code for the predicted cross-linked structure (Panel 4). Results of search for known RiPPs having chemical structure similarity to the predicted cross-linked structure (Panel 5).
Apart from lanthipeptides, RiPPMiner can predict cross-links for thiopeptides, lasso peptides and cyanobactins. Supplementary Figure S3 shows screenshots depicting results for cross-link prediction for two thiopeptides, when core peptide sequence is given as input. The dashed line connecting the Ser residues in the core peptide represents cross-links involving nitrogen containing six-membered rings, while complete chemical structures with cross-links and modified Cys residues as thiazoles are shown as images. It is interesting to note that motif based approach is able to correctly predict the cross-links in thiopeptides. The program currently modifies all Cys residues to thiazoles, while oxazole or Dha modifications of Ser residues are not predicted. Supplementary Figures S4–5 show typical examples for cleavage and cross-link prediction by RiPPMiner for lasso peptides and cyanobactins.
Benchmarking of prediction accuracy
The various machine learning based methods for identification of RiPP, prediction of RiPP class, cleavage site and cross-links implemented in RiPPMiner web server have also been extensively benchmarked. Prediction accuracy of each machine learning based method has been tested by computing area under the curve (AUC) values for ROC curves using LOO as well as 2-fold cross validation approach. The detailed benchmarking results and ROC curves are available to the users under the BENCHMARK link of RiPPMiner server, while Table 1 summarizes the benchmarking results for all the different types of predictions and details are provided in Supplementary Tables S1–4. As can be seen most predictions have used LOO as well as 2-fold cross validation, but only LOO method has been used for RiPP class prediction, cleavage and cross-link prediction of lasso peptides and cleavage site prediction of cyanobactins as number of test cases were less. It is encouraging to note that, for all predictions high AUC values have been obtained both for LOO as well as 2-fold cross validation. Even though classifiers have been validated using LOO as well as 2-fold method, RiPPMiner server uses LOO classifiers to provide maximum benefit of training for predictions on experimentally uncharacterized RiPPs. Table 1 also shows sensitivity, specificity, MCC and precision values for the score cut off at which predictions are carried out by RiPPMiner server.
Table 1. Summary of benchmarking results.
| Prediction type | Classifier type | Cross validation | AUC-ROC | Sensitivity | Specificity | MCC | Precision |
|---|---|---|---|---|---|---|---|
| RiPP identification | SVM | 2-FOLD | 0.96 | 0.93 | 0.90 | 0.85 | 0.90 |
| RiPP class | Multi Class SVM | LOO | 0.79 | 0.98 | 0.78 | ||
| Lanthipeptide cleavage site | SVM | LOO | 0.97 | 0.71 | 0.99 | 0.69 | 0.69 |
| SVM | 2-FOLD | 0.97 | |||||
| Lanthipeptide cross-links | RF | LOO | 0.90 | 0.72 | 0.95 | 0.73 | 0.68 |
| RF | 2-FOLD# | 0.81 | |||||
| RF | 2-FOLD* | 0.92 | |||||
| SVM | LOO | 0.81 | 0.57 | 0.94 | 0.63 | 0.54 | |
| SVM | 2-FOLD# | 0.76 | |||||
| SVM | 2-FOLD* | 0.87 | |||||
| Lasso peptide cleavage and cross-link | SVM | LOO | 0.99 | In 83% (50 out of 60) of the test cases correct prediction was in top rank, while in 92% of the test cases correct prediction was in top two ranks. | |||
| Cyanobactin core peptide | SVM RSII@ | LOO | 0.96 | Correct prediction could be done in all cases in a dataset consisting of 21 fragments with heterocycle rings and 7 fragments without heterocycle rings. | |||
| SVM RSIII@ | LOO | 0.95 | |||||
| Thiopeptide | Motif Based | Correct cross-links could be predicted in 28 out of 35 thiopeptides | |||||
In case of RiPP class prediction sensitivity, specificity and MCC values indicated by underline are average over all 12 RiPP classes.
#For validation of lanthipeptide prediction the dataset has been divided into two halves at cyclizable fragment level (i.e. sub-sequences of the type Ser/Thr-(X)n-Cys or Cys-(X)n-Ser/Thr).
*For validation of lanthipeptide prediction the dataset has been divided into two halves at lanthipeptide level.
@ Each core sequence of cyanobactin is flanked by an N-terminal recongnition sequence (RSII) and a C-terminal recognition sequence (RSIII).
The performance of the RiPPMiner server for cleavage and cross-link prediction of lanthipeptides is note worthy as it involves distinguishing a small number of positive dataset (correct cross-link or cleavage site) from a very large number of negative dataset. Supplementary Figure S6 shows comparison of results from RiPPMiner, antiSMASH and RiPP-PRISM for prediction of cleavage and cross-links in lanthipeptides using nisin as an example. As can be seen, antiSMASH only predicts cleavage site correctly, but no cross-links are predicted and all Ser/Thr residues are predicted to be modified. RiPP-PRISM predicts a set of 50 different cross-linked structures as it uses exhaustive enumeration approach. On the other hand, RiPPMiner provides a single prediction and correctly predicts the RiPP class, cleavage site and cross-link. We have also benchmarked the performance of RiPPMiner for prediction of cross-links in 93 lanthipeptides using a 2-fold cross validation approach. Table 1 shows ROC-AUC values for this 2-fold cross validation. Since only half of the 93 lanthipeptide structures are used for training, the results for 45 lanthipeptides in the test set are essentially blind predictions. Benchmarking page of RiPPMiner server provides a link to compare the predicted cross-links for these 45 lanthipeptides with the cross-links in the actual structures of these lanthipeptides. Supplementary Table S4 shows a summary of these results. As can be seen RiPPMiner can predict 109 and 886 true positive (TP) and true negative cross-links respectively out of 154 and 917 positive and negative cross-links in these 45 lanthipeptides. This corresponds to a true positive rate (TPR) of 71% at a false positive rate (FPR) of 3.4%.
RiPPMiner could also predict correct cross-links in 28 out of the 35 thiopeptides using the motif based approach. Similarly in case of cyanobactins correct prediction could be done in all cases in a dataset consisting of 21 fragments with heterocycle rings and 7 fragments without heterocycle rings. In our benchmarking for prediction of cross-links in lasso peptides, 83% of the test cases correct prediction was in top rank, while in 92% of the test cases correct prediction was within top two ranks. While our work was under review, Tietz et al. reported a machine learning based tool RODEO (27) for identifying precursors of lasso peptides and predicting leader-core cleavage sites. They have verified their findings by experimentally characterizing five novel lasso peptide structures, namely AnaA, LagA, CitA, MooA and LpeA. Interestingly RiPPMiner can correctly predict leader cleavage, disulfide bridge formation and cyclization for all these five newly characterized lasso peptides. It may be noted that RODEO currently does not carry out prediction of cross-links in lasso peptides.
These results convincingly demonstrate the superior performance of RiPPMiner and power of the machine learning approach. However, real predictive ability of a tool like RiPPMiner can only be evaluated by experimental characterization of several peptides from different classes.
DISCUSSION
RiPPs constitute a major class of natural products known for their bioactivities. Though a number of tools have been developed for genome mining of RiPPs, its diversity and complex cross-linking has challenged the development of complete structure prediction tools. Prediction of cross-linked structure can help in synthetic biology efforts to characterize new bioactive members of RiPP family. To this end we have developed RiPPMiner, a bioinformatics tool to classify RiPPs and predict their cross-linked structures. RiPPMiner uses the power of machine learning models trained on a manually curated database of 500+ known RiPPs, cataloged in backend database RiPPDB, to identify RiPPs, predict the RiPP class, leader cleavage sites of RiPPs, their cross-links and modified residues. RiPPMiner has been widely tested on a variety of test cases and extensively benchmarked. RiPPMiner is a unique resource which can predict complex chemical structures of several classes of RiPPs starting from genome sequences.
AVAILABILITY
http://www.nii.ac.in/rippminer.html. This website is free and open to all users and there is no login requirement. Command line version of RiPPMiner is also available as a standalone tool from ‘Download’ link in the RiPPMiner web server.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Departmentof Biotechnology, Government of India (to National Institute of Immunology, New Delhi); Department of Biotechnology, India under BTIS [BT/BI/03/009/2002 to D.M.]; COE [BT/COE/34/SP15138/2015 to D.M.]; Council of Scientific & Industrial Research, India Fellowship (to N.S.). Funding for open access charge: National Institute of Immunology, New Delhi (to D.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013; 30:108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ortega M.A., van der Donk W.A.. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 2016; 23:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maksimov M.O., Pan S.J., James Link A.. Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 2012; 29:996–1006. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Q., Doroghazi J.R., Zhao X., Walker M.C., van der Donk W.A.. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in actinobacteria. Appl. Environ. Microbiol. 2015; 81:4339–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Medema M.H., Fischbach M.A.. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015; 11:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Muller R., Wohlleben W. et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015; 43:W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L., Wyatt M.A., Magarvey N.A.. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015; 43:9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anand S., Prasad M.V., Yadav G., Kumar N., Shehara J., Ansari M.Z., Mohanty D.. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010; 38:W487–W496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khaldi N., Seifuddin F.T., Turner G., Haft D., Nierman W.C., Wolfe K.H., Fedorova N.D.. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010; 47:736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conway K.R., Boddy C.N.. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013; 41:D402–D407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ichikawa N., Sasagawa M., Yamamoto M., Komaki H., Yoshida Y., Yamazaki S., Fujita N.. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013; 41:D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M.H., Ung P.M., Zajkowski J., Garneau-Tsodikova S., Sherman D.H.. Automated genome mining for natural products. BMC Bioinformatics. 2009; 10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rottig M., Medema M.H., Blin K., Weber T., Rausch C., Kohlbacher O.. NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011; 39:W362–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ziemert N., Podell S., Penn K., Badger J.H., Allen E., Jensen P.R.. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012; 7:e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Challis G.L. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology. 2008; 154:1555–1569. [DOI] [PubMed] [Google Scholar]
- 16. Blin K., Kazempour D., Wohlleben W., Weber T.. Improved lanthipeptide detection and prediction for antiSMASH. PLoS One. 2014; 9:e89420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skinnider M.A., Johnston C.W., Edgar R.E., Dejong C.A., Merwin N.J., Rees P.N., Magarvey N.A.. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6343–E6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohimani H., Kersten R.D., Liu W.T., Wang M., Purvine S.O., Wu S., Brewer H.M., Pasa-Tolic L., Bandeira N., Moore B.S. et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014; 9:1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Q., Ortega M., Shi Y., Wang H., Melby J.O., Tang W., Mitchell D.A., van der Donk W.A.. Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Jong A., van Hijum S.A., Bijlsma J.J., Kok J., Kuipers O.P.. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006; 34:W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hammami R., Zouhir A., Le Lay C., Ben Hamida J., Fliss I.. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 2010; 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015; 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J., Qu X., He X., Duan L., Wu G., Bi D., Deng Z., Liu W., Ou H.Y.. ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS One. 2012; 7:e45878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open babel: an open chemical toolbox. J. Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tietz J.I., Schwalen C.J., Patel P.S., Maxson T., Blair P.M., Tai H.C., Zakai U.I., Mitchell D.A.. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nature Chem. Biol. 2017; 13:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.