Abstract
Diverse forms of unwanted signal variations in mass spectrometry-based metabolomics data adversely affect the accuracies of metabolic profiling. A variety of normalization methods have been developed for addressing this problem. However, their performances vary greatly and depend heavily on the nature of the studied data. Moreover, given the complexity of the actual data, it is not feasible to assess the performance of methods by single criterion. We therefore developed NOREVA to enable performance evaluation of various normalization methods from multiple perspectives. NOREVA integrated five well-established criteria (each with a distinct underlying theory) to ensure more comprehensive evaluation than any single criterion. It provided the most complete set of the available normalization methods, with unique features of removing overall unwanted variations based on quality control metabolites and allowing quality control samples based correction sequentially followed by data normalization. The originality of NOREVA and the reliability of its algorithms were extensively validated by case studies on five benchmark datasets. In sum, NOREVA is distinguished for its capability of identifying the well performed normalization method by taking multiple criteria into consideration and can be an indispensable complement to other available tools. NOREVA can be freely accessed at http://server.idrb.cqu.edu.cn/noreva/.
INTRODUCTION
Metabolomics aims at understanding biological and disease processes by systematic profiling of all metabolites in the studied organisms or biological samples (1–3). At present, mass spectrometry (MS) has become one of the most widely applied platforms for sensitively and reproducibly detecting thousands of metabolites from cells, tissues and bio-fluids (4–6), which substantially facilitates biomarker identification (7–9), pathological study (10–12) and drug discovery (13–15). In MS-based metabolomic analysis, various forms of unwanted experimental and biological variations including technical errors in the raw omics data may significantly hamper the identification of differential metabolic profiles, and in turn affect the effectiveness of metabolomics analysis (16–18). To remove these unwanted variations to the maximum extent, several processes such as signal drift correction (5,18), batch effect removal (19,20) and normalization (21,22) have been extensively employed.
Signal drift and batch effect are frequently encountered in metabolic profiling, especially the long-term and large-scale one whose time span is usually several months or even years (5,23). To correct signal drifts and remove batch effects, quality control sample (QCS) over the entire time course of large-scale study has been applied to concatenate data of multiple analytical blocks into a single dataset (24,25) and been recognized as an essential measurement in pre-processing large-scale metabolomics data (26). Moreover, normalization is recommended to be further employed (27), the aim of which is to improve the differential profile by detecting and decreasing unwanted variations arising from errors in sample preparation (28) and other biological fluctuations (21,27). Normalization is now widely considered as an integral part of data processing (29) and ≥19 methods (Supplementary Table S1) are utilized for MS-based metabolomics data (18,28,30). These methods (Supplementary Methods) can be grouped into two classes (31). Methods in the first class, such as Pareto (32), tend to reduce heteroscedasticity among metabolites, while the rest, like MSTUS (30), aim at removing the sample-to-sample variations. Besides the QCS-based correction followed by data normalization as mentioned above, several popular normalization methods based on the internal standard (IS) and/or quality control metabolite (QCM) are also widely used in current metabolomics studies (Supplementary Table S1). These methods include CCMN (33), NOMIS (34), RUV-2 (28), RUV-random (18) and SIS (35). Particularly, CCMN, NOMIS and SIS are methods based on single or multiple ISs and capable of removing unwanted experimental variations (18). Meanwhile, the RUV methods using QCM are constructed to remove overall unwanted variations (including both experimental and biological variations) in one-go (18,28).
Because of the significantly varied theories underlying each normalization method, different methods can produce conflicting results for the same dataset (21,36) and the suitability of a method is reported to be greatly dependent on the various nature of the analyzed datasets (37). Therefore, it is necessary to distinguish the best performed one from other methods for a given dataset. However, single criterion is not sufficient to assess the suitability of those methods and collective consideration of multiple criteria is recommended to ‘thoroughly’ evaluate each method from different perspectives (36). In particular, five well-established criteria are currently available by assessing (a) method's capability of reducing intragroup variation among samples (37), (b) method's effect on differential metabolic analysis (36), (c) method's consistency of the identified metabolic markers among different datasets (38), (d) method's influence on classification accuracy (28,39,40) and (e) level of correspondence between normalized and reference data (36). Taken together, a comprehensive performance evaluation by multiple criteria is essential for assessing the suitability of normalization methods.
Several powerful pipelines for analyzing the metabolomics data are currently available online, where various normalization methods are provided as one step in their analysis chain. These online pipelines include XCMS (41), MetaboAnalyst (42), Normalyzer (37), MetaDB (43), MetDAT (44), MSPrep (45), Metabolomics Workbench (46) and Workflow4Metabolomics (47). Most of these online pipelines focus on normalization service without performance evaluation. Normalyzer (37) and MetaPre (22) offer the function of outcome assessment using single criterion a and d, respectively (22,37), but none of them employ multiple criteria for the performance evaluation. Because 7 out of those 24 methods popular in MS-based metabolomics (listed in Supplementary Table S1) are not covered by either Normalyzer or MetaPre (Supplementary Table S2), it is not feasible to evaluate their performances. Moreover, some of the important tools and approaches are not provided by available online pipelines. These include the QCM-based removal of overall unwanted variation (18) and sequential strategy integrating QCS-based correction and normalization (27). Therefore, it is necessary to provide a publicly available service for comprehensively and comparatively evaluating the normalization performance of those methods used in MS-based metabolomics study.
In this work, an online tool NOREVA, designed for not only normalizing the MS-based metabolomics data but also comparatively evaluating the suitability of different normalization methods from various perspectives, was constructed and maintained at http://server.idrb.cqu.edu.cn/noreva/. The NOREVA could conduct normalization using all 24 methods mentioned above, and provided evaluation report by collectively considering 5 different criteria for assessing the normalization performance. Moreover, the originality and usefulness of this novel service were extensively exhibited by 4 case studies in the last section of this work. In summary, NOREVA aimed at normalizing MS-based metabolomics data, and distinguishing the best performed method from the others based on multiple evaluation criteria, which provided valuable guidance to the selection of suitable algorithm in metabolomics data analysis.
MATERIALS AND METHODS
Methods used for signal correction and data normalization
A univariate approach termed QC-based robust LOESS signal correction (QC-RLSC) to correct signal drift and remove batch effect from given large-scale metabolomics data (5), was provided in NOREVA by integrating the statTarget package (48) in the R software. Moreover, 24 methods in total popular for MS-based metabolomics data normalization were provided including Auto Scaling, CCMN, Contrast, Cubic Splines, Cyclic Loess, EigenMS, Level Scaling, Linear Baseline Scaling, Log-transform, Mean Normalization, Median Normalization, MSTUS, NOMIS, Pareto Scaling, Power Scaling, PQN, Quantile, Range Scaling, RUV-2, RUV-random, SIS, Total Sum, Vast Scaling and VSN. Detailed descriptions of all these 24 methods could be found in Supplementary Methods.
Criteria and measures used for evaluating the normalization performance
Five well-established criteria available for assessing the normalization performance were provided in NOREVA. (a) Method's capability of reducing intragroup variation among samples (37). Common measures of intragroup variability including pooled coefficient of variation (PCV), pooled estimate of variance (PEV) and pooled median absolute deviation (PMAD) were adopted under this criterion to evaluate variation between samples (36). A lower value (illustrated by boxplots) of these three measures denotes more thorough removal of experimentally induced noise and indicates a better performance. Moreover, the relative log abundance (RLA) plots (28) used to measure possible variations, clustering tendencies, trends and outliers across groups or within group were also provided. Boxplots of RLA were used to visualize the tightness of samples across or within group(s). The median in boxplots would be close to zero and the variation around the median would be low (29). In addition, the principal component analysis (PCA) was also used to visualize differences across groups. The more distinct group variations indicate better performance of the applied normalization method.
(b) Method's effect on differential metabolic analysis (36). The differential significance of metabolites between two groups measured by P-values was calculated by limma package (49). The distribution of P-values and clustering dendrogram and heatmap plots based on differential metabolites were provided (39). Methods would be considered as well-performed when a uniform distribution of P-values and an obvious differentiation between two groups in dendrogram and heatmap were both achieved.
(c) Method's consistency of the identified metabolic markers among different datasets (38). Under this criterion, a consistency score was defined to quantitatively measure the overlap of identified metabolic markers among different partitions of a given dataset (38). The higher consistency score represents the more robust results in metabolic marker identification for that given dataset.
(d) Method's influence on classification accuracy (28,39,40). Under this situation, receiver operating characteristic (ROC) curve together with area under the curve (AUC) values based on support vector machine (SVM) were provided. First, differential metabolic features were identified by partial least squares discriminant analysis (PLS-DA). Second, the SVM models were constructed based on these differential features identified. After k-folds cross validation, a method with larger area under the ROC curve and higher AUC value was recognized as well performed.
(e) Level of correspondence between normalized and reference data (36). Additional experimental data were frequently generated as references to validate or adjust prior result of metabolomics analysis (50). These references could be spike-in compounds and various molecules detected by quantitative analysis (50). Here, log fold changes (logFCs) of concentration between two groups were calculated, and the level of correspondence between normalized data and references was then estimated. The performance of each method could be reflected by how well the logFCs of normalized data corresponded to what were expected based on references (36). Moreover, a boxplot illustrating these variations was provided. The preferred median in boxplot would be zero with minimized variations.
In summary, criterion (a) aimed at estimating the methods’ capacity on reducing intragroup variations among samples by various measures; criterion (b) emphasized the influence on differential metabolic analysis; criterion (d) relied on the classification strategies; and identification of metabolic biomarkers was required by criteria (c and d). Different from these four criteria (a–d) general and useful for exploratory metabolomics study, criterion (e) required the prior knowledge of metabolites’ concentrations used as standard test sets. It was necessary to emphasize that selection of the appropriate methods could result in bias and overfitting if the audiences chose the method that gave results closest to their wanted ones.
All those criteria and the corresponding measures mentioned above were fully provided and functional in NOREVA. Each criterion made the performance assessment possible from its own perspective, and the combination of multiple criteria could therefore provide a comprehensive evaluation on the studied method. Evaluation results of all these criteria and measures were directly displayed on the web page, and fully downloadable from the website as separate reports. Detailed descriptions on these criteria together with their corresponding measures could be found in Supplementary Methods.
Implementation details
The NOREVA website is deployed on server with 128GB RAM, and CPU E7-4820 × 32 cores running the CentOS Linux v6.5 operating system, the Apache Tomcat servlet container and the Apache HTTP web server v2.2.15 (http://httpd.apache.org). The web interface was constructed using R v3.2.2 and R package Shiny v0.13.1 running on the Shiny-server v1.4.1.759 (http://www.rstudio.com/shiny). Several R packages were utilized in the background processes including affy, AUC, DiffCorr, DT, e1071, fastlo, ggfortify, ggsci, impute, limma, metabolomics, MetNorm, png, RcmdrMisc, rmarkdown, ropls, shiny, shinyBS, shinydashboard, shinyRGL, statTarget and vsn. NOREVA website can be readily accessed by all users with no login requirement, and by a variety of popular web browsers including Google Chrome, Mozilla Firefox, Safari and Internet Explorer (10 or later).
Comparing with standalone applications, the web-based servers were expected to be slower due to the cost of web connection and the shared nature of computational resources (51). To test the time cost of NOREVA, a large-scale metabolomics dataset MTBLS28 (52) with >1000 samples (469 patients and 536 controls) and 1807 metabolic features was collected. On the one hand, the cost of web connection was evaluated by uploading MTBLS28 data to NOREVA from different universities around the world (Supplementary Table S3). As shown, the time costs were acceptable (within 5 min). On the other hand, the calculation time of different normalization methods was further assessed by processing MTBLS28 and the time costs of the majority of these methods were <5 min with just one (EigenMS) exceeding 10 min (Supplementary Table S3). In summary, the network and hardware architectures of current NOREVA made it suitable for processing large-scale metabolomics dataset.
Required data formats in the input files
The file required at the beginning of NOREVA analysis should provide a sample-by-feature matrix in a csv format. For the analysis of metabolomics data with QC samples, the sample ID, batch ID, class of samples and injection order are sequentially provided in the first four columns of input file. Names of these columns must be kept as ‘sample’, ‘batch’, ‘class’ and ‘order’ without any changes during the entire analysis. The sample ID is uniquely assigned according to users’ preference; the batch ID refers to different analytical blocks or batches, and is labeled with ordinal number, e.g. 1, 2, 3, …; the class of samples indicates two sample groups and QC samples (the name of sample groups is different, and QC samples are all labeled as ‘NA’); the injection order is strictly follow the sequence of experiment. For experiments ignoring QC preparation and metabolomics dataset without QC samples, only sample ID and class of samples are required in the first two columns of the input file and are kept as ‘SampleName’ and ‘Label’. In the column of class of samples, ‘NA’ is not labeled to any sample due to the absence of QC samples. In the following columns of both types of input file, the raw peak intensities across all samples without logarithmic scaling are further provided. Unique IDs of each metabolite are listed in the first row of the csv file. For metabolomics studies based on IS and QCM, the required format of the input file is the same as that of input dataset without QC samples. Moreover, input file in correct format could be readily generated based on results of several popular tools such as XCMS online (41). Example file strictly following the above requirements can be directly downloaded from NOREVA's ‘Analysis’ panel. The uploaded csv file could be separated by comma, tab or semicolon.
To evaluate methods based on the last criterion, additional file providing information of the reference metabolites (e.g. spike-in compounds) is needed. In this file, the sample ID and the class of samples are required in the first two columns. Their names are provided as ‘sample’ and ‘class’. The sample ID is also uniquely assigned according to users’ preference and the class of samples indicates two sample groups of different name. Example file can also be downloaded from NOREVA.
Benchmark datasets collected for the validation case studies of this work
In order to test the utility of NOREVA, three benchmark datasets MTBLS59 (50), MTBLS79 (53) and MTBLS146 (54) collected from the MetaboLights (55) and two GC-MS-based metabolomics datasets (28,33) were collected and used in four case studies respectively in this work. These studies included: (α) collective evaluation of methods’ normalization performance on dataset MTBLS79, (β) assessment of methods’ normalization performance based on the spike-in metabolites of MTBLS59, (γ) evaluation of QC-RLSC's effect on correcting the signal drifts in MTBLS146 and (δ) assessment of IS- and QCM-based methods on removing unwanted variations in two GC-MS-based metabolomics datasets.
For the case studyα: MTBLS79 comprised of 20 cardiac tissue extracts analyzed repeatedly by direct infusion MS (DIMS) in eight batches across 7 days, together with a concurrent set of QC samples. In total, 48 metabolites were measured for each extract. This dataset was originally designed to test the efficacy of a batch-correction algorithm, and could serve as a benchmark for DIMS metabolomics. In the case studyβ: MTBLS59 presented metabolic spectra (each containing 1632 metabolites) of apple extracts detected by the ultra-performance liquid chromatography MS (UPLC-MS). This set of data consisted of 10 control samples and 3 spiked datasets of the same size, where the spiked compounds were added in different concentrations. MTBLS59 could therefore be used as benchmark dataset for performance assessment by comparing spiked ‘true’ markers with normalized results. For case studyγ: MTBLS146 provided women's profiles of 1312 metabolites based on LC-MS analysis. This set of data contained a total of 180 pregnant women divided into 6 subgroups of 30 individuals according to their variation in gestational weeks. Multiple batches and 39 QC samples in this dataset made it a good benchmark data for evaluating the performance of QC-RLSC correction used in NOREVA. In the last case studyδ: two sets of GC/TOF-MS based metabolomics data were collected. The first set of data (33) consisted of 42 samples mixed with 35 metabolites and 11 isotope-labeled internal standards (ISs), and the second set (28) provided 185 profiles of 33 metabolites including 9 QCMs identified by De Livera et al. These datasets could thus be used as benchmarks for assessing the performances of IS-based and QCM-based normalization methods. Detailed information of these five datasets were also provided on the website of the MetaboLights (55) and in the related publications (28,33).
RESULTS AND DISCUSSION
Web-service and operating procedure of NOREVA
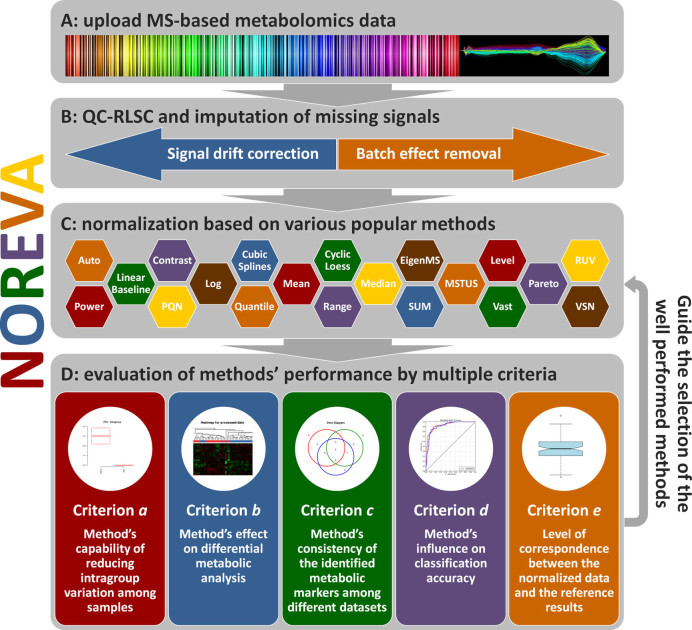
From the users’ perspective, the analysis implemented in NOREVA could be summarized into four steps: (i) upload of metabolomics data, (ii) data pre-processing, (iii) data normalization and (iv) performance evaluation. The general workflow of NOREVA integrating all these steps was illustrated in Figure 1. Detailed user manual and website demo were systematically provided in NOREVA's ‘Manual’ panel.
Figure 1.
The general workflow of NOREVA. (A) Uploading of mass spectrometry (MS)-based metabolomics data with or without IS, QCM and quality control sample (QCS); (B) Data pre-processing by QC-RLSC and imputation of missing signals; (C) Data normalization based on the studied methods; (D) Performance evaluation by multiple criteria.
In the step of metabolomics data upload, dataset with or without QCS, IS and QCM could be accepted. NOREVA stood out among available tools by providing sequential strategy integrating the QCS-based signal correction with normalization methods (19,47) and removing unwanted variations based on IS and QCM (18,28). Secondly, data pre-processing corrected signals by QC-RLSC (5) and missing value imputation (56). QC-RLSC provided various choices of filtering criteria, smoothing parameters and regression models for dataset with QC samples. Meanwhile, popular imputing algorithms (e.g. KNN, median values and minimum values) were further provided to fill missing signals. In the third step, data normalization integrated 24 methods popular in MS-based metabolomics. The resulting data matrix normalized by given methods was displayed and downloadable from the corresponding web page. In addition, the boxplot illustrating data distribution before and after normalization was provided. Finally, during performance evaluation, five distinct criteria were applied to evaluate methods from different perspectives. Dozens of measures representing the normalization performance were assessed by numerical values or illustrated by statistical graphics. After all those four steps shown in Figure 1, a report containing evaluation results was generated and downloadable in the format of PDF, HTML and DOC. In the case of normalizing large dataset, time cost on data processing would be expensive; the function of delivering evaluation reports via e-mail was thus required. In NOREVA, this function was made possible by simply typing an e-mail address in the panel of ‘Generate Evaluation Report’.
Case studies illustrating the new biological insights provided by NOREVA
(α) Collective evaluation of methods’ normalization performance
Dataset (53) used in this case study was a well-defined benchmark for the DIMS metabolomics. This dataset was analyzed by following the same procedure as demonstrated in the previous section, and the first four criteria (a–d) were chosen for performance evaluation. Table 1 showed their evaluation results. For each criterion, only one of the most representative measures was selected (a full list of results for all measures in each criterion was also demonstrated in Supplementary Table S4). On one hand, the performance of different methods evaluated by the same criterion varied significantly. To take PMAD in criterion a as an example, its values for 19 methods varied from 0.006 (for MSTUS) to 2.72 (for Vast Scaling), indicating substantial variations in performance among those 19 methods. On the other hand, the performance ranks of the same method assessed by different criteria also varied greatly. The worst method mentioned above (Vast Scaling), for example, was ranked even higher than the best (MSTUS), when considering criterion c. Thus, it is essential to first understand the nature of studied biological problem, which could then facilitate the selection of the most appropriate criterion before performance evaluation. In other words, only when researchers selected the proper criterion, could the identification of the well performed methods be meaningful for answering that biological question. Moreover, if the nature of a biological problem asked for a collective assessment based on multiple criteria, the services provided by NOREVA further made it distinguished from other available tools.
Table 1. Evaluation results of four criteria on benchmark dataset MTBLS79 (selected measure under each criterion was shown in bracket).
| Criterion (a) | Criterion (b) | Criterion (c) | Criterion (d) | |
|---|---|---|---|---|
| (PMAD) | (distribution of P-value) | (consistency) | (AUC) | |
| Auto scaling | 0.8360 | Good | 14.6500 | 0.8344 |
| Contrast | 0.7797 | Fair | 9.7500 | 0.6250 |
| Cubic splines | 0.1393 | Excellent | 13.7500 | 0.8322 |
| Cyclic loess | 0.3188 | Good | 15.6500 | 0.8356 |
| EigenMS | 0.1799 | Good | 16.4000 | 0.8010 |
| Level scaling | 0.2890 | Good | 15.1000 | 0.8345 |
| Linear baseline | 0.6035 | Fair | 6.3000 | 0.7072 |
| Log-transform | 0.1349 | Good | 14.7500 | 0.8168 |
| Mean | 0.3100 | Good | 14.7500 | 0.8213 |
| Median | 0.3100 | Good | 14.5500 | 0.8177 |
| MSTUS | 0.0064 | Good | 14.3500 | 0.8405 |
| Pareto scaling | 0.5320 | Good | 14.9500 | 0.8344 |
| Power scaling | 0.1660 | Good | 14.9500 | 0.8314 |
| PQN | 0.3260 | Good | 13.7000 | 0.8309 |
| Quantile | 0.2989 | Excellent | 13.8000 | 0.8119 |
| Range scaling | 0.1573 | Good | 15.3500 | 0.8344 |
| Total sum | 2.4336 | Fair | 14.7000 | 0.7538 |
| Vast scaling | 2.7200 | Good | 15.0000 | 0.8344 |
| VSN | 0.5626 | Excellent | 13.7500 | 0.8373 |
The way calculating those measures was described in ‘Materials and Methods’ section and ‘Supplementary Methods’ section. Besides of quantitative measures, qualitative ones such as distribution of P-value were also evaluated and three performance levels were provided (Excellent, Good and Fair). Qualitative measures were evaluated by visual inspection and examples illustrating how those three performance levels were assigned were shown in Supplementary Figure S1.
(β) Assessment of methods’ normalization performance based on the spike-in metabolites
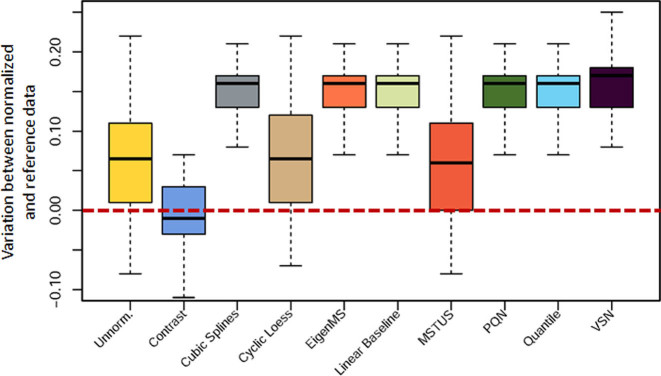
Dataset (50) used in the second case study was also a benchmark dataset for performance assessment by comparing the spiked ‘true’ markers with the normalized results. The analysis procedure was also specified in the previous section, but criterion e was selected this time for performance evaluation. The variations in logFCs between the normalized results and the spike-in metabolites were represented by boxplots in Figure 2. This type of boxplots was previously used by Risso et al. (39). As illustrated in Figure 2, only one method (Contrast) stood out from the rest by making the normalized data closer to the reference ones, which therefore effectively preserving the ‘true’ biological variations. Besides of the spike-in compounds, various ‘true’ markers detected by quantitative analysis and other analytical technics could be also uploaded to NOREVA as the golden standards for performance evaluation.
Figure 2.
Difference between logFCs of the normalized results across various methods and those of the spike-in metabolites (used here as the gold standard). Only one method (Contrast) led to unbiased logFC estimates and thus effectively preserved the true biological variations.
(γ) Evaluation of QC-RLSC's effect on signal drifts correction
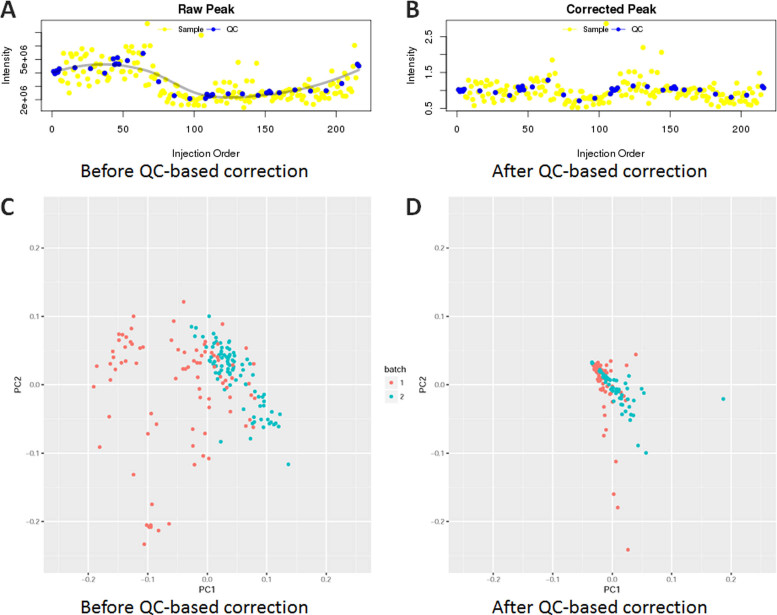
Dataset (54) used in the third case study was a widely tested sample dataset for signal drifts correction and batch effects removal. Effects of QC samples on signal correction were frequently reflected by the corrected peak area plots (5) and PCA (28). In this case, QC-RLSC (5) was applied to correct signals between two analytical batches. In contrast to Figure 3A (before QCS-based correction), intensities of an example metabolite M72T126 in Figure 3B (after correction) were greatly corrected. In particular, the intensities of QC samples in Figure 3B lay in a more straight line comparing to that before correction. Moreover, Figure 3C and D shown the first two principal components of the data before and after correction. Signal variations between two analytical batches were clearly evident in Figure 3C and were effectively suppressed by signal correction (shown in Figure 3D). These results demonstrated the extensive power of NOREVA on signal drift correction, which made it a functional tool for analyzing metabolomics data.
Figure 3.
Evaluation of QC-RLSC's effect on signal drifts correction. (A and B) performance evaluation based on the intensity of an example metabolite M72T126. In contrast to M72T126΄s intensity before QCS-based correction (A), the results after correction were greatly corrected (B) by lining QC samples (blue dots) in a more straight line. (C and D) The first two principal components of the dataset MTBLS146 before and after QCS-based correction. Signal variations between two analytical batches were clearly evident (C) and were effectively suppressed by signal correction (D).
(δ) Assessment of IS- and QCM-based methods on removing unwanted variations
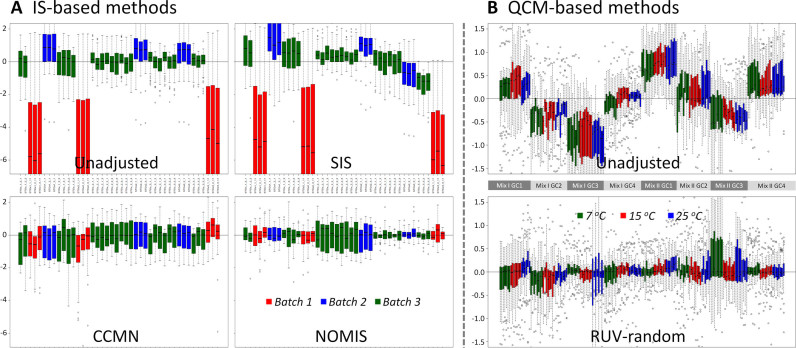
Benchmark datasets (28,33) used in this case study were applied to assess the performance of IS- and QCM-based methods by the commonly accepted measure–RLA plots (18,28). In this case study, the performance of three IS-based methods (CCMN, NOMIS and SIS) was evaluated by the corresponding RLA plots before and after their normalization. As shown in Figure 4A, compared with the RLA plots of unadjusted data, the plots after the normalization by CCMN and NOMIS resulted in a median closer to zero and lower variations around the median. Because the normalization by single IS was reported to be sensitive to its own obscuring variation (34), the performance of methods based on multiple ISs (CCMN and NOMIS) was found (Figure 4A) to be much better than that of single IS (SIS). Moreover, as a QCM-based method, the performance of RUV-random was analyzed by RLA plot before and after its normalization (Figure 4B). As illustrated, this method performed very well on removing unwanted experimental variations (28).
Figure 4.
Performance assessment of (A) three IS-based normalization methods and (B) one QCM-based normalization method on removing unwanted variations by the RLA plots before and after normalization. Parameters used in this case study for the RUV-random method were set as k = 3 and λ = 0.03.
CONCLUSION
NOREVA was developed to enable performance evaluation of various normalization methods from multiple perspectives. It complemented other available tools by providing (1) an integrated analysis based on five well-established criteria for more comprehensive evaluation and (2) the most complete set of normalization methods with unique features of removing overall unwanted variations based on QCMs and allowing QCS-based correction sequentially followed by normalization. Because of the substantial similarities among different types of OMIC data (such as sparsity, high dimension, systematic bias and so on), it was also feasible to extend the scope of NOREVA from MS-based metabolomics to other OMICs studies such as proteomics and nuclear magnetic resonance-based metabolomics. With the advent of precision medicine and big data era, NOREVA and other available tools could collectively contribute to various aspects of life science research, such as pathological study, drug discovery, biomarker identification and so on.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Precision Medicine Project of the National Key Research and Development Plan of China [2016YFC0902200]; Innovation Project on Industrial Generic Key Technologies of Chongqing [cstc2015zdcy-ztzx120003]; Fundamental Research Funds for Central Universities [10611CDJXZ238826, CDJZR14468801, CDJKXB14011, 2015CDJXY]. Funding for open access charge: Precision Medicine Project of the National Key Research and Development Plan of China [2016YFC0902200].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fessenden M. Metabolomics: small molecules, single cells. Nature. 2016; 540:153–155. [DOI] [PubMed] [Google Scholar]
- 2. Idle J.R., Gonzalez F.J.. Metabolomics. Cell Metab. 2007; 6:348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu F., Qin C., Tao L., Liu X., Shi Z., Ma X., Jia J., Tan Y., Cui C., Lin J. et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12943–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Southam A.D., Weber R.J., Engel J., Jones M.R., Viant M.R.. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat. Protoc. 2016; 12:310–328. [DOI] [PubMed] [Google Scholar]
- 5. Dunn W.B., Broadhurst D., Begley P., Zelena E., Francis-McIntyre S., Anderson N., Brown M., Knowles J.D., Halsall A., Haselden J.N. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011; 6:1060–1083. [DOI] [PubMed] [Google Scholar]
- 6. Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z.. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016; 44:D1069–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roberts L.D., Gerszten R.E.. Toward new biomarkers of cardiometabolic diseases. Cell Metab. 2013; 18:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buas M.F., Gu H., Djukovic D., Zhu J., Onstad L., Reid B.J., Raftery D., Vaughan T.L.. Candidate serum metabolite biomarkers for differentiating gastroesophageal reflux disease, Barrett's esophagus, and high-grade dysplasia/esophageal adenocarcinoma. Metabolomics. 2017; 13:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J. et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012; 40:D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newgard C.B. Metabolomics and metabolic diseases: where do we stand. Cell Metab. 2017; 25:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weiss R.H., Kim K.. Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol. 2012; 8:22–33. [DOI] [PubMed] [Google Scholar]
- 12. Kaddurah-Daouk R., Krishnan K.R.. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology. 2009; 34:173–186. [DOI] [PubMed] [Google Scholar]
- 13. Zhang A., Sun H., Wang X.. Mass spectrometry-driven drug discovery for development of herbal medicine. Mass Spec. Rev. 2016; 9999:1–14. [DOI] [PubMed] [Google Scholar]
- 14. Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016; 15:473–484. [DOI] [PubMed] [Google Scholar]
- 15. Zhu F., Han B., Kumar P., Liu X., Ma X., Wei X., Huang L., Guo Y., Han L., Zheng C. et al. Update of TTD: therapeutic target database. Nucleic Acids Res. 2010; 38:D787–D791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weckwerth W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003; 54:669–689. [DOI] [PubMed] [Google Scholar]
- 17. van den Berg R.A., Hoefsloot H.C.J., Westerhuis J.A., Smilde A.K., van der Werf M.J.. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006; 7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Livera A.M., Sysi-Aho M., Jacob L., Gagnon-Bartsch J.A., Castillo S., Simpson J.A., Speed T.P.. Statistical methods for handling unwanted variation in metabolomics data. Anal. Chem. 2015; 87:3606–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia J., Sinelnikov I.V., Han B., Wishart D.S.. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015; 43:W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S.Y., Kuo C.H., Tseng Y.J.. Batch Normalizer: a fast total abundance regression calibration method to simultaneously adjust batch and injection order effects in liquid chromatography/time-of-flight mass spectrometry-based metabolomics data and comparison with current calibration methods. Anal. Chem. 2013; 85:1037–1046. [DOI] [PubMed] [Google Scholar]
- 21. Craig A., Cloarec O., Holmes E., Nicholson J.K., Lindon J.C.. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal. Chem. 2006; 78:2262–2267. [DOI] [PubMed] [Google Scholar]
- 22. Li B., Tang J., Yang Q., Cui X., Li S., Chen S., Cao Q., Xue W., Chen N., Zhu F.. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci. Rep. 2016; 6:38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zelena E., Dunn W.B., Broadhurst D., Francis-McIntyre S., Carroll K.M., Begley P., O’Hagan S., Knowles J.D., Halsall A., Consortium H. et al. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Anal. Chem. 2009; 81:1357–1364. [DOI] [PubMed] [Google Scholar]
- 24. van der Kloet F.M., Bobeldijk I., Verheij E.R., Jellema R.H.. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009; 8:5132–5141. [DOI] [PubMed] [Google Scholar]
- 25. Brunius C., Shi L., Landberg R.. Large-scale untargeted LC-MS metabolomics data correction using between-batch feature alignment and cluster-based within-batch signal intensity drift correction. Metabolomics. 2016; 12:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hendriks M.M.W.B., van Eeuwijk F.A., Jellema R.H., Westerhuis J.A., Reijmers T.H., Hoefsloot H.C.J., Smilde A.K.. Data-processing strategies for metabolomics studies. Trends Analyt. Chem. 2011; 30:1685–1698. [Google Scholar]
- 27. Gagnebin Y., Tonoli D., Lescuyer P., Ponte B., de Seigneux S., Martin P.Y., Schappler J., Boccard J., Rudaz S.. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal. Chim. Acta. 2017; 955:27–35. [DOI] [PubMed] [Google Scholar]
- 28. De Livera A.M., Dias D.A., De Souza D., Rupasinghe T., Pyke J., Tull D., Roessner U., McConville M., Speed T.P.. Normalizing and integrating metabolomics data. Anal. Chem. 2012; 84:10768–10776. [DOI] [PubMed] [Google Scholar]
- 29. De Livera A.M., Olshansky M., Speed T.P.. Roessner U, Dias DA. Statistical analysis of metabolomics data. Metabolomics Tools for Natural Product Discovery: Methods and Protocols. 2013; 1055:Springer Science+Business Media, LLC; 291–307. [DOI] [PubMed] [Google Scholar]
- 30. Warrack B.M., Hnatyshyn S., Ott K.H., Reily M.D., Sanders M., Zhang H., Drexler D.M.. Normalization strategies for metabonomic analysis of urine samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009; 877:547–552. [DOI] [PubMed] [Google Scholar]
- 31. Kohl S.M., Klein M.S., Hochrein J., Oefner P.J., Spang R., Gronwald W.. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics. 2012; 8:146–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eriksson L., Antti H., Gottfries J., Holmes E., Johansson E., Lindgren F., Long I., Lundstedt T., Trygg J., Wold S.. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal. Bioanal. Chem. 2004; 380:419–429. [DOI] [PubMed] [Google Scholar]
- 33. Redestig H., Fukushima A., Stenlund H., Moritz T., Arita M., Saito K., Kusano M.. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal. Chem. 2009; 81:7974–7980. [DOI] [PubMed] [Google Scholar]
- 34. Sysi-Aho M., Katajamaa M., Yetukuri L., Oresic M.. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007; 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gullberg J., Jonsson P., Nordstrom A., Sjostrom M., Moritz T.. Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 2004; 331:283–295. [DOI] [PubMed] [Google Scholar]
- 36. Valikangas T., Suomi T., Elo L.L.. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2016; 2016, doi:10.1093/bib/bbw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chawade A., Alexandersson E., Levander F.. Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 2014; 13:3114–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X., Gardiner E.J., Cairns M.J.. Optimal consistency in microRNA expression analysis using reference-gene-based normalization. Mol. Biosyst. 2015; 11:1235–1240. [DOI] [PubMed] [Google Scholar]
- 39. Risso D., Ngai J., Speed T.P., Dudoit S.. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014; 32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gromski P.S., Xu Y., Hollywood K.A., Turner M.L., Goodacre R.. The influence of scaling metabolomics data on model classification accuracy. Metabolomics. 2015; 11:684–695. [Google Scholar]
- 41. Gowda H., Ivanisevic J., Johnson C.H., Kurczy M.E., Benton H.P., Rinehart D., Nguyen T., Ray J., Kuehl J., Arevalo B. et al. Interactive XCMS online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014; 86:6931–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia J.G., Wishart D.S.. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011; 6:743–760. [DOI] [PubMed] [Google Scholar]
- 43. Franceschi P., Mylonas R., Shahaf N., Scholz M., Arapitsas P., Masuero D., Weingart G., Carlin S., Vrhovsek U., Mattivi F. et al. Metadb a data processing workflow in untargeted MS-based metabolomics experiments. Front. Bioeng. Biotechnol. 2014; 2:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biswas A., Mynampati K.C., Umashankar S., Reuben S., Parab G., Rao R., Kannan V.S., Swarup S.. MetDAT: a modular and workflow-based free online pipeline for mass spectrometry data processing, analysis and interpretation. Bioinformatics. 2010; 26:2639–2640. [DOI] [PubMed] [Google Scholar]
- 45. Hughes G., Cruickshank-Quinn C., Reisdorph R., Lutz S., Petrache I., Reisdorph N., Bowler R., Kechris K.. MSPrep–summarization, normalization and diagnostics for processing of mass spectrometry-based metabolomic data. Bioinformatics. 2014; 30:133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., Edison A., Fiehn O., Higashi R., Nair K.S. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016; 44:D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giacomoni F., Le Corguille G., Monsoor M., Landi M., Pericard P., Petera M., Duperier C., Tremblay-Franco M., Martin J.F., Jacob D. et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015; 31:1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luan H., Liu L.F., Meng N., Tang Z., Chua K.K., Chen L.L., Song J.X., Mok V.C., Xie L.X., Li M. et al. LC-MS-based urinary metabolite signatures in idiopathic Parkinson's disease. J. Proteome Res. 2015; 14:467–478. [DOI] [PubMed] [Google Scholar]
- 49. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franceschi P., Masuero D., Vrhovsek U., Mattivi F., Wehrens R.. A benchmark spike-in data set for biomarker identification in metabolomics. J. Chemom. 2012; 26:16–24. [Google Scholar]
- 51. Lee D.Y., Saha R., Yusufi F.N., Park W., Karimi I.A.. Web-based applications for building, managing and analysing kinetic models of biological systems. Brief. Bioinform. 2009; 10:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mathe E.A., Patterson A.D., Haznadar M., Manna S.K., Krausz K.W., Bowman E.D., Shields P.G., Idle J.R., Smith P.B., Anami K. et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014; 74:3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kirwan J.A., Weber R.J., Broadhurst D.I., Viant M.R.. Direct infusion mass spectrometry metabolomics dataset: a benchmark for data processing and quality control. Sci. Data. 2014; 1:140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luan H., Meng N., Liu P., Fu J., Chen X., Rao W., Jiang H., Xu X., Cai Z., Wang J.. Non-targeted metabolomics and lipidomics LC-MS data from maternal plasma of 180 healthy pregnant women. Gigascience. 2015; 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haug K., Salek R.M., Conesa P., Hastings J., de Matos P., Rijnbeek M., Mahendraker T., Williams M., Neumann S., Rocca-Serra P. et al. MetaboLights–an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013; 41:D781–D786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stegle O., Parts L., Piipari M., Winn J., Durbin R.. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012; 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.