Abstract
High-throughput sequencing methods are generating enormous amounts of genomic data, giving unprecedented insights into human genetic variation and its relation to disease. An individual human genome contains millions of Single Nucleotide Variants: to discriminate the deleterious from the benign ones, a variety of methods have been developed that predict whether a protein-coding variant likely affects the carrier individual's health. We present such a method, DEOGEN2, which incorporates heterogeneous information about the molecular effects of the variants, the domains involved, the relevance of the gene and the interactions in which it participates. This extensive contextual information is non-linearly mapped into one single deleteriousness score for each variant. Since for the non-expert user it is sometimes still difficult to assess what this score means, how it relates to the encoded protein, and where it originates from, we developed an interactive online framework (http://deogen2.mutaframe.com/) to better present the DEOGEN2 deleteriousness predictions of all possible variants in all human proteins. The prediction is visualized so both expert and non-expert users can gain insights into the meaning, protein context and origins of each prediction.
INTRODUCTION
High-throughput sequencing methods are generating a huge amount of genomic data (1), and full genome and exome sequencing techniques have greatly enhanced our knowledge about human genetic variation (2–4)). Since information is available for healthy individuals (5) as well as for patients suffering from various diseases (6), it is possible to analyse the differences between neutral variants, which are present in a control population and are thus considered benign, and deleterious variants, which lead to a disease phenotype. This copious data stream requires efficient processing and annotation to be useful, and in the case of human genetic diseases, efficient computational methods are necessary to reduce the number of observed variants to a few that are likely causative for the phenotype under scrutiny (2,3,7). Many computational tools for identifying these deleterious variants have been developed so far (8). Although these tools tackle the prediction of the pathogenicity of variants from different angles, they generally use Machine Learning (ML) or statistical methods to learn a discrimination between deleterious and neutral Single Nucleotide Variants (SNVs) resulting in amino-acid substitutions at the protein level (9–13). These predictors generally consider different molecular or protein-level aspects that are closely related to possible mechanisms of pathogenicity, such as the evolutionary conservation of the mutated position (9,10), the stability change upon mutation (14), possible structural alterations (9,14) and functional annotations using GO terms (13). With DEOGEN (12) we introduced contextualization of the target variants by combining sources of information related to different biological scales, with the aim to better represent the complexity of the relationship between variants and their pathogenicity. We integrated information such as the molecular effect of the variant, the relevance of the mutated gene, its known interaction and the pathways in which it is involved. We showed that this approach improves the quality of the final predictions, but our model did not explain how the final deleteriousness score relates to the other variants on the same protein and where it programmatically originates from.
We here present DEOGEN2, a predictor of missense SVNs for human proteins that is freely accessible at http://deogen2.mutaframe.com/. DEOGEN2 positively compares with other state-of-the-art methods (9,12,15–17) through the further incorporation of various sources of contextual information, such as early folding predictions and protein domain-oriented features. This performance improvement is confirmed on an independent dataset, evidencing the robustness of DEOGEN2. In addition, we provide interactive visualisation approaches to (i) explain how the deleteriousness score relates to all other variants within that protein and (ii) break down the origin of the score, so enabling the non-expert user to situate the prediction result in its wider biological context. This development enables geneticists and clinicians to move beyond the identification of new disease-causing variants towards their interpretation.
MATERIALS AND METHODS
Datasets
The main training and testing Humsavar16 dataset for DEOGEN2 is based on the February 2016 version of Humsavar (18). From the original 73 266 variants mapped on 12 335 proteins, 7375 unclassified variants were discarded and 27 606 deleterious SNVs and 38 285 neutral SNVs retained. In addition, an independent Blind dataset based on Testing Dataset I proposed in (8), which contains 120 deleterious variants extracted from 57 Nature Genetics publications and 124 neutral variants from (5,8), was created by filtering out 30 proteins also present in Humsavar16, so ensuring complete independence between these datasets.
Features
The DEOGEN2 features are listed in Table 1; we present only the new and improved features compared to DEOGEN (12).
Table 1. Summary of the features used in DEOGEN2.
| Feature | Status | Short name | Code |
|---|---|---|---|
| PROVEAN score (16) | From version 1 | PROV | PR |
| Conservation Index (12,13) | Improved | CI | CI |
| Mutant/wildtype log-odd ratio (12) | Improved | LOR | LO |
| Early Folding predictions | New | EF | EF |
| PFAM log-odd score (17) | New | PF | PF |
| Interaction patches annotation (19) | New | INT | IN |
| RVIS (20) | New | RVIS | RV |
| GDI (21) | New | GDI | GD |
| Recessiveness index (22) | From version 1 | REC | RE |
| Gene essentiality (23) | From version 1 | ESS | ES |
| Pathway log-odd score (12,13) | Extended data | PATH | PA |
Evolutionary-based features
The evolutionary-based CI (Conservation Index) and LOR (Log-Odd Ratio) scores (12) were improved by switching to hhBlits (24) from JackHmmer (25) to generate Multiple Sequence Aligments (MSAs), resulting in (i) MSAs that are faster to compute, (ii) the retrieval of more distant homologs and (iii) generally smaller alignments. The hhBlits MSAs were computed with one iteration and E-value =10−4, and we added a pre-filtering step to the MSAs before computing CI and LOR scores by removing the sequences with <0.3 coverage and 0.3 Sequence Identity (SI) from the LOR calculation and with <0.1 coverage and 0.1 SI from the CI computation (see Supplementary Section S1 and Figures S1 and S2).
Early folding predictions
Disruptions to the protein folding process are likely to deactivate the function of proteins that should fold. We added a prediction of protein Early Folding (EF) residues as a feature in DEOGEN2 to explore this type of molecular information in variant-effect prediction. This method uses predictions from DynaMine (26) in combination with a Support Vector Machine (SVM) model trained on the data from the Start2Fold dataset (27) to make its predictions (under review). For each variant the EF is calculated and the difference with the wild type prediction taken as a measure for the impact on the initial steps of the protein's folding process (see Supplementary Figure S3).
Domain-oriented feature
PfamScan (28) was used to obtain domain boundaries and other PFAM (29) annotations such as coiled-coil regions, repeats, motifs or disordered regions. Similar to (12,13,17), the training set was used in cross-validation settings to learn the log-odd ratio of the probability of observing a deleterious or a neutral variant on each PFAM entry (see Supplementary Section S2 for details). This PF scores provide an indication of the tendency of these entries to be involved in deleterious phenotypes (see Supplementary Figure S4).
Interaction patches
Annotations on 11 471 known interaction patches for 3627 human proteins were obtained from the INstruct database (19). For every pair of interacting proteins, INstruct contains an indication of where the interaction patch in each protein, identified from the structure of the complex, is situated in the sequence. Information about whether the variant occurs on a known interaction patch (or not) was included as the INT feature (see Supplementary Figure S5).
Gene-oriented features
The Residual Variation Intolerance Score (RVIS) (20) and Gene Damage Index (GDI) (21) scores improve the contextualization of whether a variant affects a gene/protein important for human health. GDI is a gene-level metric of the cumulative mutational damage that each gene carries in the general population (21). Since natural selection acts on genes in function of their relevance for human health, the most frequently mutated genes are more likely to harbor nearly-neutral variants, while variants on least damaged genes are more likely to be disease-related (21). RVIS (20) ranks the likelihood of the genes to be disease-causing by comparing the amount of functional variation carried by each gene with respect to the genome-wide average. Genes with low common functional variation are more subject to purifying selection than genes with a higher than average mutational burden, highlighting their functional relevance. (see Supplementary Figures S6 and S7)
Pathway-oriented feature
We updated the snp&Go (13) inspired pathway log-odd score with data from version 31 of ConsensusPathDB (30), which now contains 4012 human pathways and 131 216 proteins. As previously (12), we learned the log-odd scores for each pathway in cross-validation settings to obtain new PATH scores that are a proxy of the pathways’ sensitivity to deleterious variants.
Machine Learning and Validation
We contextualized each variant with the features described above, so obtaining 11-dimensional feature vectors for each variant. We used the scikit-learn (31) implementation of a Random Forest (32) classifier with 200 trees. The performances on the Humsavar16 dataset were computed in strict 10-fold cross-validation settings. To reduce possible over-fitting due to homologies between proteins in different cross-validation sets, we ensured that proteins in each set share <25% sequence similarity with the proteins in the other nine sets. The RF model was analysed using the treeinterpreter library (https://github.com/andosa/treeinterpreter), which decomposes each prediction into its feature contributions (see Supplementary Section S3). The final model used in the webserver has been trained on the entire Humsavar16 dataset.
Visualization
The server framework uses the lexicon visualization library, a collection of micro-libraries written using javascript (es5) and the D3 library (v3.5.17) (33). Each micro library operates on JSON formatted input. Programmatic access to the methods within each visualization object allows user-control and automated synchronization between different objects.
RESULTS
Extended contextualization improves the predictions
The relationship between a variant and the phenotypic outcome at the human individual level involves an extensive network of interactions and feedback that spans different biological scales, from the molecular to organs. An accurate prediction of the deleteriousness of a variant should encompass such information: we therefore improved and extended the multi-level biological contextualization of the target variants and the affected proteins within our model (12). In particular, we reduced the requirements in terms of homologous sequences in the MSAs to compute the CI and LOR features and improved the REC, ESS and PATH knowledge based features due to dbNFSP (34) and ConsensusPathDB (30) updates. We also introduced new strategic pieces of information as features that (i) provide information about the relevance of the mutated residues for the initial steps of the protein's folding process (EF), (ii) quantify the sensitivity of the domain affected by the mutation to deleterious variants (PF) and (iii) improve the gene-level assessment of the relevance of the mutated protein for human health. The incremental contributions of these features are shown in Supplementary Table S1 and the distributions of their contributions in the final prediction are shown in Supplementary Figures S8–S18.
Comparison with other methods
We compared the DEOGEN2 performance with the current state-of-the-art on the Humsavar16 dataset for 10 predictors, and on the Blind dataset for 14 predictors. The DEOGEN2 performances on Humsavar16 (Table 2) show that it has the highest Balanced Accuracy (BAC) and the highest MCC, alongside with metaSVM. Note that the performances of the other methods were extracted from dbNSFP (34) and may be over-estimated since their training set may overlap with Humsavar16, whereas the DEOGEN2 performances were computed in strict cross-validation settings. DEOGEN2 also performs 7% better than our previous DEOGEN model in terms of MCC. On the independent Blind dataset (8), DEOGEN2 has the highest BAC and the highest MCC (Table 3), with the performances of 13 predictors as reported in (8), plus the recently published M-CAP (35) included.
Table 2. Comparison of DEOGEN2 cross-validated performances with state of the art predictors on the Humsavar16 dataset.
| Method | Sen | Spe | Bac | Pre | MCC |
|---|---|---|---|---|---|
| PolyPhen2 | 85 | 73 | 79 | 71 | 57 |
| LRT | 84 | 70 | 77 | 70 | 54 |
| MutationTaster | 94 | 70 | 82 | 70 | 63 |
| MutationAssessor | 81 | 71 | 76 | 69 | 52 |
| fatHMM | 78 | 85 | 82 | 80 | 63 |
| PROVEAN | 82 | 75 | 79 | 72 | 57 |
| metaSVM | 83 | 93 | 88 | 90 | 76 |
| fatHMM-MKL | 94 | 54 | 74 | 61 | 51 |
| SIFT | 85 | 68 | 77 | 67 | 53 |
| PON-P2 | 86 | 83 | 84 | 80 | 69 |
| VEST3 | 88 | 87 | 87 | 82 | 74 |
| DEOGEN | 77 | 92 | 84 | 85 | 71 |
| DEOGEN2 | 89 | 88 | 89 | 84 | 76 |
DEOGEN and DEOGEN2 scores have been computed in-house with a stratified 10-folds cross-validation. DEOGEN2 uses the MCC-optimal deleteriousness threshold >0.45. PON-P2 predictions have been obtained from its web server. All the other scores have been extracted from the 3.2 version of dbNSFP (34).
Table 3. Predictor performances on the Blind dataset.
| Method | Sen | Spe | Bac | Pre | MCC |
|---|---|---|---|---|---|
| SIFT | 68 | 75 | 72 | 79 | 43 |
| PolyPhen2 (HVAR) | 88 | 67 | 78 | 78 | 57 |
| LRT | 88 | 66 | 77 | 78 | 57 |
| Mutation Taster | 94 | 74 | 84 | 83 | 70 |
| Mutation Assessor | 70 | 80 | 75 | 83 | 49 |
| FatHMM | 55 | 91 | 73 | 90 | 48 |
| GERP++ | 77 | 72 | 75 | 80 | 49 |
| PhyloP | 76 | 73 | 75 | 80 | 49 |
| SNAP | 53 | 70 | 62 | 63 | 23 |
| SNP&GO | 55 | 94 | 75 | 89 | 53 |
| MutPred | 74 | 81 | 78 | 79 | 55 |
| CONDEL | 71 | 73 | 72 | 72 | 44 |
| CADD phred | 79 | 74 | 77 | 81 | 53 |
| M-CAP | 93 | 68 | 81 | 74 | 63 |
| DEOGEN | 46 | 96 | 71 | 92 | 48 |
| DEOGEN2 | 87 | 86 | 87 | 87 | 73 |
Visualization of the results
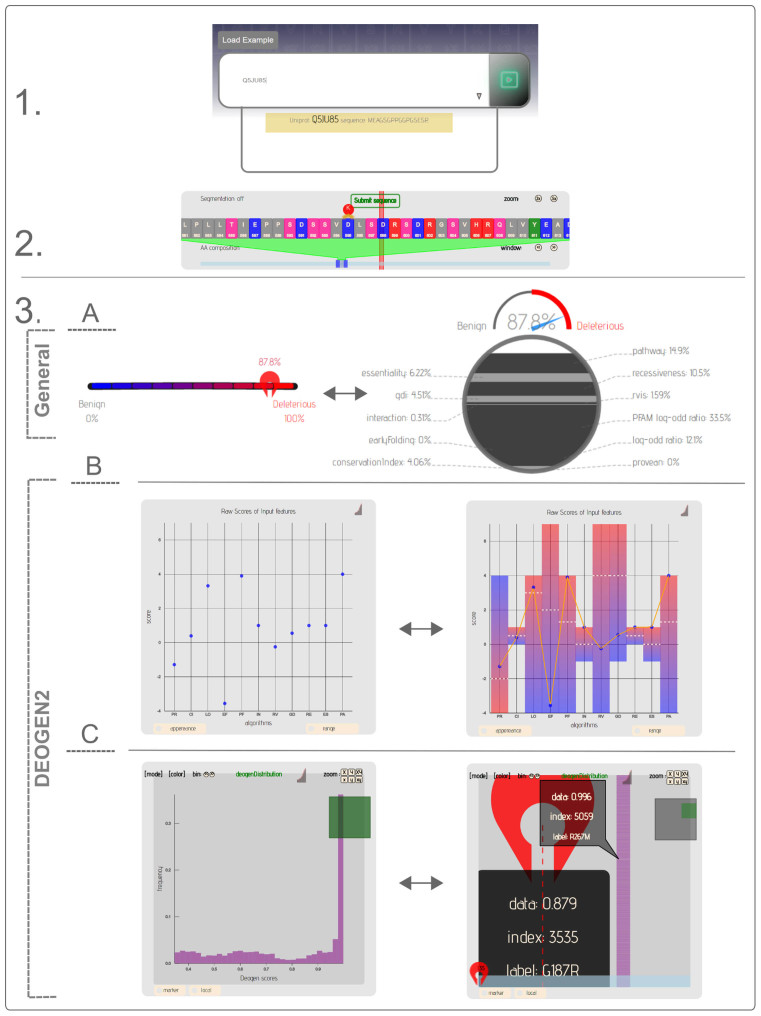
The DEOGEN2 visualisation process and graphs are described in Figure 1. A sample variant can be loaded by clicking on the ‘Load Example’ button: this displays the N45S variant in Transforming Growth Factor-Beta Receptor 1 (TGFBR1), with Uniprot ID P36897 and RefSeq ID NP_004603.1. Variants of this protein have been linked to Loeys-Dietz Syndrome and susceptibility to multiple self-healing squamous epithelioma (36,37), and we will discuss the information that the web server provides around this example of a disease-causing variant.
Figure 1.
Overview of the DEOGEN2 web server visualization. (1) The user starts to enter a Uniprot ID or sequence, which activates a dropdown list from which a human protein is selected. After pressing the play button, the user can navigate the sequence (2) to create and submit a variant for this sequence. After pressing ‘Submit sequence’, the variant is visualized in the page report (3) which contains two sections. The General section displays the change between the wild-type and variant amino acid, with the chemical structures of both shown, and the difference between the amino acids expressed on (A) the dashboard as a percentage; clicking on the percentage bar will show the breakdown of these components. The DEOGEN2 section shows the DEOGEN2 score with (B) a breakdown of the contribution of each machine learning feature, so informing the user about which contextual information was most important to reach the final score, and an overview of the raw features scores used as input for the machine learning. Section (C) shows the distribution of all the variant scores in this protein, including in a heat map format (not shown). Information on data points is obtained by hovering over them, the visualization can be changed by clicking on the buttons or the graph icon in the top right corner.
The ‘General’ section of the page report (Figure 1.3) shows that the amino acids are relatively similar; both are polar and hydrophilic amino acids with only a small size difference. The calculation of the percentages here is based on a normalised score of the differences in the BLOSUM62 matrix change, the amino acid size, hydrophobicity and charge. Despite the similarity of the amino acids, the ‘DEOGEN2’ section shows that this variant is likely deleterious with an overall score of 0.927 (0 is benign, 1 is deleterious, with the prediction cutoff point at 0.5). The dashboard highlights the key features that contributed to this score; clicking on the percentage bar will display a breakdown of these components.
The first graph underneath (Figure 1.Breakdown) shows the machine learning contributions in detail: on the x-axis each feature code is listed (Table 1), while the y-axis indicates the contribution of the corresponding feature toward the final decision: positive values vote toward deleteriousness, negative values toward a benign variant (see Supplementary Section S3). Clicking on the top-right graph button will link the points to better illustrate the decision profile for the variant of interest. In the case of variant N45S, the plot indicates that, among the evolutionary features, PROVEAN (PR) pushed the decision towards the deleterious class while CI (CI) and LOR (LO) slightly pushed towards neutrality. This behavior highlights the complementarity of the different evolutionary features used in our model: in this case the wider sequence context (PR) points to deleteriousness of the variant, not its individual position in the sequence. The contribution of the PFAM log-odd score (PF) strongly pushed the decision towards the deleterious class, due to the fact that N45S variant falls into the Activin receptor domain (PF01064.20), whose sensitivity to deleterious variants is reasonably high (log-odd score of 1.79). Among the protein-oriented features, only REC (RE) provided a noticeable contribution, indicating this protein is encoded by a gene that can cause recessive disorders when homozygously lost.
The next graph shows the raw feature scores that are the input for the prediction algorithm. Next, the distribution of the scores of all the variants in the same protein is shown (Figure 1.Distribution), with the y-axis showing the frequency and the x-axis the overall DEOGEN2 score. Clicking on the marker button enables the user to evaluate where their variant is situated in this distribution; in the case of TGFBR1, the protein contains variants predicted as deleterious but a larger proportion has scores below the 0.5 threshold and are therefore likely benign. By clicking on the ‘Local’ button the user can scroll through the sequence to see where the most deleterious or benign regions are located. The sequence position can be identified by hovering over the yellow highlighted sections. For example, the region around residue 193 is highly deleterious, whereas variants in the region around residue 360 are likely benign. Finally, this information is represented in a heat map that visualizes all variants for each sequence position. This enables the user to identify ‘hot spots’ in the sequence, and pinpoint which amino acid variants are predicted as the most deleterious.
DISCUSSION AND CONCLUSION
With the continuing performance improvement of variant effect predictors, partially due to the integration of increasing amounts of data at different biological levels, we think the time is right to shift their focus to the what, how and where, so generating understanding about what the variant means. With the DEOGEN2 webserver, we provide visualization of the meaning, protein context and origins of each prediction to enable the user to make a more informed decision or hypothesis about a particular variant instead of having to rely on a single all-encompassing score.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agency for Innovation by Science and Technology in Flanders (IWT) (to D.R.); M.R. is Research Director at the FNRS Fund for Scientific Research; the European Regional Development Fund (ERDF) and Brussels-Capital Region-Innoviris within the framework of the Operational Programme 2014–2020 [ERDF-2020 project ICITY-RDI.BRU]. Funding for open access charge: The ERDF-2020 project ICITY-RDI.BRU grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C.. Ten years of next-generation sequencing technology. Trends Genet. 2014; 30:418–426. [DOI] [PubMed] [Google Scholar]
- 2. Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J.. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011; 12:745–755. [DOI] [PubMed] [Google Scholar]
- 3. Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E.. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 2013; 14:681–691. [DOI] [PubMed] [Google Scholar]
- 4. Johnston J.J., Biesecker L.G.. Databases of genomic variation and phenotypes: existing resources and future needs. Hum. Mol. Genet. 2013; 22:R27–R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. 1000 Genomes Project Consortium Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A.. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N.. Human gene mutation database (HGMD®): 2003 update. Hum. Mutat. 2003; 21:577–581. [DOI] [PubMed] [Google Scholar]
- 7. Pabinger S., Dander A., Fischer M., Snajder R., Sperk M., Efremova M., Krabichler B., Speicher M.R., Zschocke J., Trajanoski Z.. A survey of tools for variant analysis of next-generation genome sequencing data. Brief. Bioinform. 2014; 15:256–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X.. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015; 24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adzhubei I., Daniel M.J., Sunyaev S.R.. Predicting functional effect of human missense mutations using PolyPhen2. Curr. Protoc. Hum. Genet. 2013; doi:10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng P.C., Henikoff S.. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B., Krishnan V.G., Mort M.E., Xin F., Kamati K.K., Cooper D.N., Mooney S.D., Radivojac P.. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009; 25:2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raimondi D., Gazzo A. M., Rooman M., Lenaerts T., Vranken W.F.. Multi-level biological characterization of exomic variants at the protein level significantly improves the identification of their deleterious effects. Bioinformatics. 2016; 32:1797–1804. [DOI] [PubMed] [Google Scholar]
- 13. Calabrese R., Capriotti E., Fariselli P., Martelli P.L., Casadio R.. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009; 30:1237–1244. [DOI] [PubMed] [Google Scholar]
- 14. De Baets G., Van Durme J., Reumers J., Maurer-Stroh S., Vanhee P., Dopazo J., Schymkowitz J., Rousseau F.. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012; 40:D935–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D.. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010; 7:575–576. [DOI] [PubMed] [Google Scholar]
- 16. Choi Y., Sims G.E., Murphy S., Miller J. R., Chan A.P.. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012; 7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J., Gaunt T. R.. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mut. 2013; 34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. UniProt-Consortium The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010; 38:D142–D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meyer M.J., Das J., Wang X., Yu H.. INstruct: a database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics. 2013; 29:1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B.. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013; 9:e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G. et al. . The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13615–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B. et al. . A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012; 335:823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Georgi B., Voight B.F., Buan M.. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013; 9:e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Remmert M., Biegert A., Hauser A., Söding J.. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012; 9:173–175. [DOI] [PubMed] [Google Scholar]
- 25. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cilia E., Pancsa R., Tompa P., Lenaerts T., Vranken W.F.. From protein sequence to dynamics and disorder with DynaMine. Nat. Commun. 2013; 4:2741. [DOI] [PubMed] [Google Scholar]
- 27. Pancsa R., Varadi M., Tompa P., Vranken W.F.. Start2Fold: a database of hydrogen/deuterium exchange data on protein folding and stability. Nucleic Acids Res. 2016; 44:D429–D434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mistry J., Bateman A., Finn R.D.. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007; 8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamburov A., Stelzl U., Lehrach H., Herwig R.. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013; 41:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. et al. . Scikit-learn: machine learning in Python. JMLR. 2011; 12:2825–2830. [Google Scholar]
- 32. Breiman L. Random forests. Mach. Learn. 2001; 45:532. [Google Scholar]
- 33. Bostock M., Ogievetsky V., Heer J.. D3 data-driven documents. IEEE Trans. Visual. Comp. Graph. 2011; 17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 34. Liu X., Wu C., Li C., Boerwinkle E.. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human non-synonymous and splice site SNVs. Hum. Mutat. 2016; 37:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jagadeesh K., Wenger A., Berger M., Guturu H., Stenson P., Cooper D., Bernstein J., Bejerano G.. M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016; 48:1581–1586. [DOI] [PubMed] [Google Scholar]
- 36. Goudie D.R., D’Alessandro M., Merriman B., Lee H., Szeverenyi I., Avery S., O’Connor B.D., Nelson S.F., Coats S.E., Stewart A. et al. . Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat. Genet. 2011; 43:365–369. [DOI] [PubMed] [Google Scholar]
- 37. Loeys B.L., Schwarze U., Holm T., Callewaert B.L., Thomas G.H., Pannu H., De Backer J.F., Oswald G.L., Symoens S., Manouvrier S. et al. . Aneurysm syndromes caused by mutations in the TGF-beta receptor. N. Engl. J. Med. 2006; 355:788–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.