Abstract
Ossifying fibromyxoid tumor (OFMT) is an uncommon mesenchymal neoplasm of uncertain differentiation and intermediate malignant potential. Recurrent gene fusions involving either PHF1 or BCOR have been found in 85% of OFMT, including typical and malignant examples. As a subset of OFMT still lack known genetic abnormalities, we identified two OFMTs negative for PHF1 and BCOR rearrangements, which were subjected to transcriptome analysis for fusion discovery. The RNA sequencing found a novel CREBBP-BCORL1 fusion candidate in an axillary mass of a 51 year-old male and a KDM2A-WWTR1 in a thigh mass of a 36 year-old male. The gene fusions were validated by RT-PCR and FISH in the index cases and then screened by FISH on 4 additional OFMTs lacking known fusions. An identical CREBBP-BCORL1 fusion was found in an elbow tumor from a 30 year-old male. Both OFMTs with CREBBP-BCORL1 fusions had areas of typical OFMT morphology, exhibiting uniform round to epithelioid cells arranged in cords or nesting pattern in a fibromyxoid stroma. The OFMT with KDM2A-WWTR1 fusion involved dermis and superficial subcutis, being composed of ovoid cells in a fibromyxoid background with hyalinized giant rosettes. The S100 immunoreactivity ranged from very focal to absent. Similar to other known fusion genes in OFMT, BCORL1, CREBBP and KDM2A are also involved in histone modification. In summary, we expand the spectrum of molecular abnormalities in OFMT with 2 novel fusions, CREBBP-BCORL1 and KDM2A-WWTR1, further implicating the epigenetic deregulation as the leading pathogenetic mechanism in OFMT.
Keywords: ossifying fibromyxoid tumor, CREBBP, BCORL1, KDM2A, WWTR1, endometrial stromal sarcoma
INTRODUCTION
Ossifying fibromyxoid tumor (OFMT) is a rare soft tissue neoplasm of uncertain histogenesis and intermediate risk of malignancy (rarely metastasizing).(Fletcher et al., 2013) It often affects middle-aged adults, with a median age of 49 years and a male-to-female ratio of 1.5:1.(Schneider et al., 2016) The most common presentation of OFMT is in the superficial or deep soft tissues of the extremities, and less commonly in the trunk or head and neck regions. First described in 1989 by Enzinger et al., OFMT is a well-circumscribed neoplasm often associated with an incomplete peripheral shell of bone, multilobular growth pattern, and uniform round to oval tumor cells arranged in cords or nests in a fibromyxoid stroma.(Enzinger et al., 1989) Following the recognition of this entity, tumors with atypical histology or more aggressive clinical behavior, including higher recurrent and metastatic rates, have been reported.(Kilpatrick et al., 1995; Zamecnik et al., 1997; Folpe and Weiss, 2003; Miettinen et al., 2008) However, the diagnostic criteria of atypical or malignant OFMT have been controversial, since they often lack the characteristic immunophenotype and/or peripheral ossification. Recent advances in our understanding of genetic abnormalities of OFMTs have shown that at least 85% of cases, including typical/benign, atypical, and malignant, harbor recurrent genetic translocations.(Gebre-Medhin et al., 2012; Graham et al., 2013; Antonescu et al., 2014) The most common molecular alterations are the PHF1-related fusions, involving various gene partners, such as EP400, MEAF6, and EPC1.(Gebre-Medhin et al., 2012; Graham et al., 2013; Antonescu et al., 2014) Rare cases harboring ZC3H7B-BCOR fusion have also been described.(Antonescu et al., 2014) A JAZF1-PHF1 fusion was also reported in a likely case of OFMT, described as a peculiar cardiac sarcoma showing ossification and monotonous oval to short spindle cell proliferation.(Schoolmeester et al., 2013) PHF1 rearrangements were found in both benign and malignant OFMTs without a conventional typical component (so-called de novo malignant OFMT).(Graham et al., 2013) The identification of recurrent gene rearrangements in OFMT has not only provided a useful diagnostic tool in challenging cases, such as atypical morphology or non-specific immunoprofile, but also helped clarify the dispute over the existence of malignant OFMTs. However, there still remains a small subset of OFMTs lacking known gene rearrangement.(Antonescu et al., 2014) In this study, we take advantage of transcriptome sequencing in two index OFMT cases with frozen material for further fusion gene characterization.
MATERIALS AND METHODS
Index Cases and Patient Selection
The index case 1 was a malignant OFMT arising in a 51 year-old man, as an 11 cm left axillary mass. The case was included in our previous study (Case# 38), lacking gene rearrangements in PHF1, EP400, and BCOR genes by FISH.(Antonescu et al., 2014) The patient subsequently developed a local recurrence 2 years after the initial diagnosis and received radiotherapy followed by resection. One year later he developed bilateral pulmonary metastases, for which he received multiple wedge resections and chemotherapy. Fresh tissue was available from a pulmonary metastasis and submitted for RNA sequencing. Due to subsequent disease progression, the patient succumbed of disease 4 years after diagnosis.
The second index case was a benign OFMT arising in a 36 year-old man presenting with a superficial thigh mass. The tumor was negative for PHF1 and BCOR rearrangements by FISH. Upon re-excision of the primary lesion, fresh frozen tissue was collected for RNA sequencing.
In order to establish the prevalence of the novel gene fusions identified by transcriptome sequencing in the index cases, we searched our files for OFMT lacking known genetic alterations. We selected 4 additional OFMTs which fit the above criteria and had available materials for further molecular testing, 3 of them being included in our previous study (Case No. 34, 35, 37).(Antonescu et al., 2014) The remaining case was a recent case arising in the elbow of a 30 year-old man (Table 1). Similar diagnostic criteria for malignancy were applied as previously described.(Antonescu et al., 2014) Tumors with increased cellularity, mitotic activity more than 2 per 50 high power fields, and/or nuclear pleomorphism or necrosis were classified as malignant OFMT; while cases showing increased cellularity but lacking increased mitotic activity, necrosis or nuclear pleomorphism were defined as atypical OFMTs. Clinical and pathologic information was collected from the electronic medical records. The study was approved by the Institutional Review Board.
Table 1.
Clinicopathologic findings of OFMTs with CREBBP-BCORL1 and KDM2A-WWTR1 fusions
| Case | Age/Sex | Site | Tumor depth | Histology | S100 | Desmin | No. in previous study10 |
|---|---|---|---|---|---|---|---|
| CREBBP-BCORL1 | |||||||
| Index case 1 | 51/M | Axilla | Subcutaneous | Malignant, ossifying | Fpos | Neg | 38 |
| Case 3 | 30/M | Elbow | Subcutaneous | Benign, non-ossifying | Rare | Neg | Not included |
| KDM2A-WWTR1 | |||||||
| Index case 2 | 36/M | Thigh | Dermal-Subcutaneous | Benign, non-ossifying | Neg | Fpos | Not included |
M, male; Fpos, focally positive; Neg, negative.
RNA sequencing and analysis by FusionSeq
Total RNA was extracted using RNeasy Plus Mini (Qiagen), followed by mRNA isolation with oligo(dT) magnetic beads and fragmentation by incubation at 94°C in fragmentation buffer (Illumina) for 2.5 minutes. To reduce the inclusion of artifact chimeric transcripts into the sequencing library due to random priming, an additional gel size-selection (350–400bp) was introduced before the adapter ligation step.(Quail et al., 2008) The adaptor ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2000 (Illumina). All reads were independently aligned with STAR (ver 2.3)(Dobin and Gingeras, 2015) against the human reference genome (hg19). The reads were then analyzed by FusionSeq for potential gene fusions.(Sboner et al., 2010) Paired-end reads mapped to different genes were first used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcript candidates. The mRNA expression levels of the partner genes involved in the translocations were evaluated.
In addition, we also analyzed the gene expression profiles of 5 OFMT cases, each harboring ZC3H7B-BCOR, MEAF6-PHF1, EP400-PHF1, CREBBP-BCORL1, and KDM2A-WWTR1. The first 4 cases were previously published (Case No. 1, 3, 39, 38 in prior report),(Antonescu et al., 2014) while the last one represented the index case 2 from this study. The quantile normalized and log2 transformed RPKM (reads per kilo base per million mapped reads) values were compared to those of a wide array of other sarcomas in our RNA sequencing data to get a differentially expressed gene list of OFMT by a log2 fold change threshold of positive 1 or negative 5 and p-value less than 0.05.
Reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was performed on the index cases to confirm the fusion transcripts identified by RNA sequencing. An aliquot of the RNA material extracted above for RNA sequencing was reverse transcribed by SuperScript III First-Strand Synthesis System (Invitrogen) into complementary DNA. PCR was performed by Advantage 2 PCR kit (Clontech, Mountain View, CA). PCR primers were designed according to the reads from RNA sequencing (Supplementary table 1), using annealing temperatures at 64.5°C for CREBBP-BCORL1 and 66°C for KDM2A-WWTR1. Since WWTR1 was described as a 5′ fusion partner gene in epithelioid hemangioendothelioma,(Errani et al., 2011) we have additionally designed primers to exclude the possibility of WWTR1-KDM2A fusion (Supplementary table 1). The PCR products were then analyzed by agarose gel electrophoresis and purified by QIAquick PCR Purification Kit (Qiagen) for Sanger sequencing.
Fluorescence in situ hybridization (FISH)
FISH was performed on 4 μm-thick formalin-fixed paraffin-embedded (FFPE) tissue sections to confirm the RNA sequencing fusion candidates and subsequently screen the additional cases. Custom probes using bacterial artificial chromosomes (BAC) clones (Supplementary Table 2) were designed flanking the CREBBP, BCORL1, KDM2A and WWTR1 genes, according to UCSC genome browser (http://genome.ucsc.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). DNA from each BACs was isolated according to the manufacturer’s instructions. The BAC clones were labeled with fluorochromes by nick translation and validated on normal metaphase chromosomes. The slides were deparaffinized, pretreated, and hybridized with denatured probes. After overnight incubation, the slides were washed, stained with DAPI, mounted with an antifade solution, and then examined on a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) controlled by Isis 5 software (Metasystems). Fusion FISH assay was also performed on index case 2, combining the BAC clones centromeric to KDM2A and flanking both sides of WWTR1 (Supplementary Table 2).
RESULTS
OFMT with CREBBP-BCORL1 fusion show upregulation of BCORL1
RNA sequencing in case 1 identified a CREBBP-BCORL1 fusion candidate composed of CREBBP (16p13.3) exon 30 fused to BCORL1 (Xq26.1) exon 4 (Fig. 1). The fusion transcript was further confirmed by RT-PCR and break-apart FISH assay of both genes (Fig. 1B,C). The predicted amino acid sequence was in-frame and the protein domains of the chimeric protein are illustrated in Figure 1. The BCORL1 mRNA expression showed up-regulation of the downstream exons distal to the breakpoint (exon 4) (Fig. 1E). In contrast, CREBBP showed no increased expression, either at the whole transcript or individual exon levels (data not shown). Interestingly, investigating a wide array of sarcoma types from our RNA sequencing database, BCORL1 up-regulation was also identified in undifferentiated round cell sarcomas with BCOR genetic alterations, i.e. BCOR internal tandem duplication (ITD) or BCOR-MAML3 gene fusion, which also demonstrated BCOR overexpression.(Kao et al., 2016) In contrast to the OFMT with CREBBP-BCORL1 fusion, tumors with BCOR genetic abnormalities showed overexpression of the entire BCORL1 gene (Fig. 1D,E), suggesting a different underlying molecular mechanism of BCORL1 up-regulation.
Figure 1. CREBBP-BCORL1 fusion in OFMT.
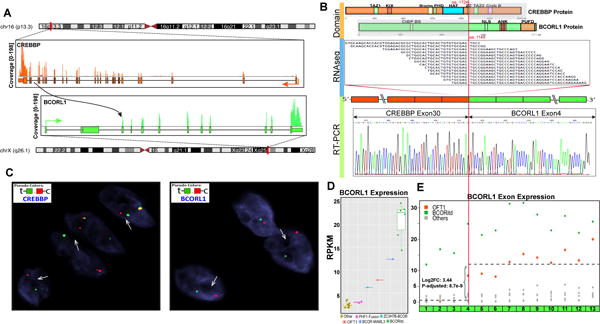
RNA sequencing fusion discovery showed fusion junction reads mapped to the exon 30 of CREBBP (16p13.3) and exon 4 of BCORL1 (Xq26.1); results further confirmed by RT-PCR and Sanger sequencing (A,B). The predicted CREBBP chimeric protein reveals the histone acetyltransferase (HAT) domain being retained in the fusion and joined to the C-terminal part of BCORL1 (B). FISH analyses showed CREBBP and BCORL1 break-apart signals (green, telomeric and red, centromeric, arrows, C). BCORL1 on Xq26 shows split-apart of the only allele in this male patient. High BCORL1 mRNA expression (distal to the exon 4 breakpoint) was noted in the CREBBP-BCORL1 positive OFMT, compared to OFMTs with other gene fusions, including MEAF6-PHF1, EP400-PHF1, and ZC3H7B-BCOR (D,E). Additionally full cDNA BCORL1 overexpression was also identified in round cell sarcomas with BCOR-gene abnormalities (BCOR-MAML3 or BCOR ITD) (D).
CREBBP-BCORL1 occurs in both benign and malignant OFMT
Histologically, the index case 1 showed typical OFMT features, with uniform round cells, arranged in cord-like pattern in a fibromyxoid background (Fig. 3A). Foci of ossification, both in the center and at the periphery of the tumor, were also seen and were accompanied by chondroid metaplasia in areas. The tumor showed variegated cellularity, ranging from hypocellular areas with hyalinized stroma (Fig. 3B) to hypercellular solid areas. The latter showed increased mitotic activity (7/10 high power fields) (Fig. 3C) and focal necrosis, findings in keeping with a malignant OFMT. The tumor wrapped around axillary artery and vein, and lymphovascular invasion was present. Immunohistochemically, the tumor showed very focal S100 positivity, while desmin was negative. The local recurrence showed extensive hyalinization (> 90%) secondary to radiotherapy and the subsequent lung metastases resembled the histology of the primary lesion.
Figure 3. Pathologic findings of OFMTs with CREBBP-BCORL1 (A–F) and KDM2A-WWTR1 fusions (G–I).

Case#1 was composed of uniform evenly-spaced small round cells arranged in cord-like structures in a fibromyxoid to hyalinized stroma (A,B), which based on the increased cellularity and mitotic activity was classified as malignant (arrows, C). Case#3 showed alternating collagenous and myxoid areas with focal microcystic change (D), with bland round cells with uniform nuclei and scant cytoplasm (E) arranged in cord-like to reticular patterns (F). The KDM2A-WWTR1 positive OFMT#2 was located within dermis (G) and superficial subcutis. It was composed of uniform ovoid tumor cells set in a loose fibromyxoid background (H). The tumor showed distinctive giant collagen rosettes throughout (I).
FISH screening of the additional 4 cases identified a second case with similar CREBBP-BCORL1 gene fusion, which occurred in the elbow of a 30 year-old man. Grossly the lesion was well defined and surrounded by a thin fibrous pseudocapsule. Microscopically, the tumor was composed of alternating collagenous and myxoid areas (Fig. 3D). The tumor cells were arranged in cords, reticular pattern, or embedded within the fibrous stroma (Fig. 3E,F). The lesional cells had a round to oval morphology with bland nuclear features and low mitotic activity (<2/50HPFs), in keeping with a benign OFMT. No ossification was present. Immunohistochemically, only rare cells were S100 protein positive and desmin was negative.
The remaining 3 fusion-negative OFMTs included in our previous study were negative for CREBBP, BCORL1, KDM2A, and WWTR1 break-apart by FISH.
KDM2A-WWTR1 was identified in an OFMT with collagenous giant rosettes
In the index case 2, RNA sequencing revealed chimeric reads between KDM2A (11q13.2) exon 14 and WWTR1 (3q23–q24) exon 3. Break-apart FISH showed a typical KDM2A rearrangement, while FISH for WWTR1 showed a constant split, in keeping with an inversion pattern. Fusion FISH assay confirmed the signal association of the 5′-centrometric probe of KDM2A to the 3′-centrometric probe of WWTR1 (Fig. 2A). RT-PCR further demonstrated the in-frame fusion between KDM2A exon 14 to WWTR1 exon 3 (Fig. 2B). No reciprocal WWTR1-KDM2A transcript was amplified by the RT-PCR. As exons 1 and 2 of WWTR1 remain untranslated, the predicted fusion protein would include the entire coding sequence of WWTR1 (Fig. 2B). No up-regulation of KDM2A and WWTR1 mRNA expression levels were noted, compared to other OFMTs or a variety of other sarcomas.
Figure 2. KDM2A-WWTR1 fusion in OFMT.
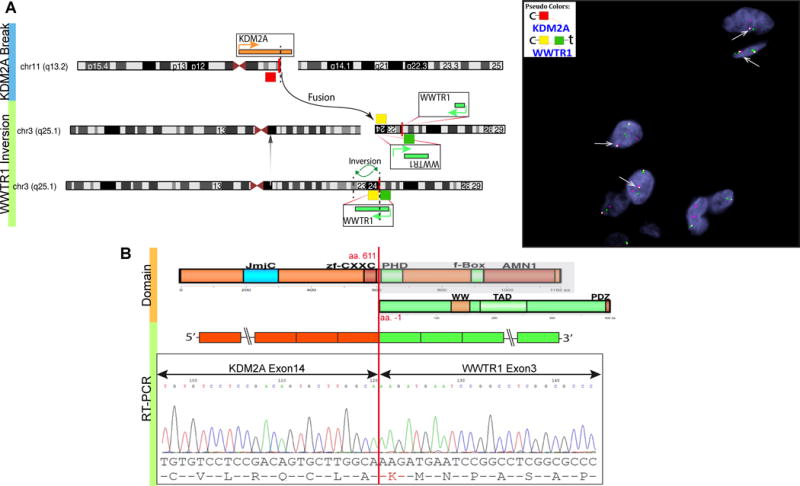
Schematic diagram and fusion FISH assay demonstrate the 5′ KDM2A (centromeric, red) being fused to the inverted 3′ WWTR1 (centromeric, yellow) (A). RT-PCR confirms the KDM2A-WWTR1 in-frame transcript (B). The chimeric protein retains the entire WWTR1 coding region.
The index case 2 occurred in the dermis and superficial subcutis of the thigh in a 36 year-old male (Fig. 3G). The tumor appeared relatively circumscribed and showed a multilobulated growth pattern. The tumor cells had a monomorphic ovoid phenotype, lacking atypia or mitotic activity (Fig. 3H). The lesion showed a distinctive stromal component, with collections of collagenous giant rosettes set in a loose fibromyxoid background (Fig. 3I). There was no ossification noted. Immunohistochemically, tumor was negative for S100, CD34, and only focally positive for desmin. Due to the presence of giant rosettes a diagnosis of low grade fibromyxoid sarcoma was considered in the differential, but MUC4 immunostain was negative and FISH showed no FUS and EWSR1 gene rearrangements.
Most OFMTs form a tight genomic cluster regardless of the fusion type
Using RNAseq data, unsupervised clustering showed a distinct genomic group of the 4 OFMTs harboring EP400-PHF1, MEAF6-PHF1, CREBBP-BCORL1, and ZC3H7B-BCOR, among other sarcomas (Supplem Fig. 1). OFMT with KDM2A-WWTR1 clustered nonspecifically with other tumors types. A list of 257 differentially expressed genes was identified in the 4 OFMTs compared to a large spectrum of sarcomas. The gene list included various genes normally expressed in the central nervous system (i.e. ECEL1, C1QL4, C1QL1, DNER, BAI1, KCNH3, HCN2, NPAS1, KIF1A, GLI1, etc.) and some genes related to bone or cartilage formation (i.e. CRTAC1, PTN, BMP7, PTH1R, TMEM119, etc).
DISCUSSION
In this study we report two novel gene fusions, CREBBP-BCORL1 and KDM2A-WWTR1, expanding the genetic signature of OFMT. Recurrent PHF1 related gene fusions have been identified in the majority of OFMTs, with EP400-PHF1 being the most common variant, but several fusion partners have been reported.(Gebre-Medhin et al., 2012; Graham et al., 2013; Antonescu et al., 2014) A common theme among these fusion partners, including PHF1, EP400, MEAF6, EPC1, and BCOR, is their function implicated in histone modification and epigenetic regulation. Based on the common core function of these genes it appears that epigenetic dysregulation might be a critical event in the pathogenesis of OFMTs. In concordance with this hypothesis, the novel genes identified in this study, including CREBBP, BCORL1 and KDM2A, are also associated with histone modification. CREBBP (CREB binding protein, also known as CBP) encodes an ubiquitously expressed histone acetyltransferase (HAT) and functions as a transcriptional co-activator. CREBBP and its paralog, EP300, are involved in diverse physiological processes, including cell cycle progression, differentiation, apoptosis and DNA repair.(Iyer et al., 2004; Wang et al., 2013) With its HAT activity, CREBBP can acetylate both histones and non-histone proteins, including some transcription factors (e.g. p53, CREB). Germline mutations of CREBBP are associated with Rubinstein-Taybi syndrome, a congenital autosomal dominant disease with a heterogeneous clinical presentation including mental retardation, growth impairment, craniofacial dysmorphism and skeletal abnormalities, as well as an increased predisposition to neoplasms.(Iyer et al., 2004; Roelfsema et al., 2005) Somatic inactivating mutations of CREBBP have been reported in certain types of lymphoma,(Lunning and Green, 2015) while a MYST3-CREBBP fusion was described in acute myeloid leukemia with t(8;16).(Iyer et al., 2004; Diaz-Beya et al., 2013) In our case #1 harboring a CREBBP-BCORL1 fusion, the CREBBP transcript retained most of its coding sequence, except for the last exon. The breakpoint presumably disrupts the zinc finger structure (ZZ domain, Fig. 1) and truncates the C-terminal TAZ2, NCBD and LXXL domains, which are responsible for interaction with many transcription factors (e.g. TFIIB, p53, FOXO3a, STAT1, MEF2, IRF-3), co-activators (e.g. PCAF, GCN5, ACTR), and nuclear receptors (e.g. retinoid X receptor, estrogen receptor, androgen receptor).(Goodman and Smolik, 2000; Vo and Goodman, 2001)
BCORL1 (BCL6 corepressor-like 1), in contrast to CREBBP, encodes a transcriptional corepressor homolog to BCOR, with which shares some degree of amino acid sequences homology, including the ankyrin repeats.(Pagan et al., 2007) However, BCORL1 does not interact with BCL-6, as BCOR does, and has a different mechanism of gene repression by interacting with class II histone deacetylase (HDAC) and C-terminal-binding protein (CtBP).(Pagan et al., 2007) BCORL1 gene rearrangement was previously reported in hepatocellular carcinoma, where BCORL1 exon 11 was fused to ELF4 (E74-like factor 4) exon 8, resulting from an intra-chromosomal inversion of the long arm of chromosome X.(Totoki et al., 2011) Similar to BCOR, inactivating mutations of BCORL1 have been reported in small subsets of acute myeloid leukemia, myelodysplastic syndrome, and chronic myelomonocytic leukemia.(Li et al., 2011; Damm et al., 2013; Yamamoto et al., 2014) The presence of CREBBP-BCORL1 fusion in our OFMT case#1 was associated with mRNA overexpression of the 3′end of BCORL1, which was represented in the fusion transcript. Interestingly, whole transcript BCORL1 up-regulation was also identified in a group of undifferentiated round cell sarcomas harboring BCOR genetic abnormalities, including BCOR ITD and BCOR-MAML3 fusion.(Kao et al., 2016) Additional functional studies are needed to elucidate the mechanism of concurrent BCORL1 upregulation in tumors overexpressing the BCOR homolog, as well as to investigate the biologic consequences of the BCORL1 truncated versus whole transcript upregulation.
KDM2A (lysine demethylase 2, previously known as JHDM1A or FBXL11) is a histone demethylase, which specifically demethylates H3K36me1/me2, affecting chromatin structure and gene transcription.(Cheng et al., 2014) KDM2A plays an important role in cell proliferation of mesenchymal stem cells and can negatively regulate the differentiation toward osteogenic/dentinogenic, adipocytic, and chondrogenic lineages in mesenchymal stem cells. (Dong et al., 2013; Du et al., 2013; Gao et al., 2013) WWTR1 (TAZ) is a transcriptional coactivator of the Hippo pathway and can activate / de-repress the TEAD transcription factor and result in growth limitation.(Mohamed et al., 2015) In epithelioid hemangioendothelioma, WWTR1 is the 5′ gene of the WWTR1-CAMTA1 fusion transcript,(Errani et al., 2011) while in OFMT case 2, WWTR1 was the 3′ partner. In contrast to CREBBP-BCORL1, the KDM2A-WWTR1 fusion did not result in mRNA up-regulation of either constituent genes.
At transcriptional level, OFMTs with various fusion variants, including ZC3H7B-BCOR, MEAF6-PHF1, CREBBP-BCORL1, and EP400-PHF1, clustered tightly together, suggesting that despite different genetic events, they share a common core of downstream targets and gene expression involved in the pathogenesis of this distinct tumor entity. A prior study using gene microarrays showed upregulation of DLK1, EAAT4, HHLA2 and MUC4 genes and downregulation of PMP22 and MYEF2 in OFMT compared to other S100-positive neoplasms, such as schwannoma and nerve sheath myxoma.(Graham et al., 2011) Our RNAseq data could not reproduce these results.
In contrast to most other translocation-associated sarcomas, the fusion transcripts in OFMT do not occur through a simple balanced rearrangement due to the opposite directions of transcription of the gene partners, including EP400-PHF1, EPC1-PHF1, MEAF6-PHF1 and even JAZF1-PHF1.(Antonescu et al., 2014) Thus most OFMTs, including typical and malignant examples, harbor complex structural rearrangements and numerical chromosomal changes by conventional karyotyping.(Sovani et al., 2001; Nishio et al., 2002; Folpe and Weiss, 2003; Kawashima et al., 2007; Gebre-Medhin et al., 2012) Opposite orientation of transcription is also predicted for the KDM2A-WWTR1, but not for CREBBP-BCORL1 fusion.
Quite intriguingly, OFMTs show a significant genetic overlap with low grade endometrial stromal sarcoma (ESS), including identical EPC1-PHF1, MEAF6-PHF1 and ZC3H7-BCOR gene fusions.(Micci et al., 2006; Panagopoulos et al., 2012; Panagopoulos et al., 2013; Micci et al., 2014). More recently, a KDM2B-CREBBP fusion was reported in a low grade ESS.(Micci et al., 2016) However, the CREBBP breakpoint in ESS is located at the beginning of exon 2, while in OFMT at the end of exon 30, although most of the cDNA is retained in both variants. Similar to KDM2A, KDM2B belongs to the same family of F-box proteins, both being involved in differentiation of mesenchymal stem cells via histone modification.(Deng et al., 2015)
In summary, we expand the genetic spectrum of OFMTs with 2 novel gene fusions, CREBBP-BCORL1 and KDM2A-WWTR1. In keeping with other translocations reported in OFMT, these partner genes play a role in histone modification, reinforcing the emerging role of epigenetic deregulation in the oncogenesis of OFMT. This new report further reinforces prior studies suggesting that the presence of gene fusions in OFMTs are not associated with the risk of malignancy, being present in both benign and malignant examples. However, these rare variant fusions appear more prone to occur in lesions with unusual morphology, lack of ossification or non-specific immunoprofile.
Supplementary Material
Supplementary Figure 1. Unsupervised hierarchical clustering of 5 OFMTs among a wide variety of other soft tissue neoplasms. Four OFMTs form a distinct group despite different gene fusions, including EP400-PHF1, MEAF6-PHF1, CREBBP-BCORL1, and ZC3H7B-BCOR. OFMT with KDM2A-WWTR1 did not cluster together.
Acknowledgments
Supported in part by: P50 CA140146-01 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA)
Footnotes
Conflicts of interest: none
References
- Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, Agaram NP, Sboner A, Fletcher CD. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors–molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–193. doi: 10.1002/gcc.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O, Patel DJ. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 2014;28:1758–1771. doi: 10.1101/gad.246561.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y, Itzykson R, Sanada M, Shiraishi Y, Gelsi-Boyer V, Renneville A, Miyano S, Mori H, Shih LY, Park S, Dreyfus F, Guerci-Bresler A, Solary E, Rose C, Cheze S, Prebet T, Vey N, Legentil M, Duffourd Y, de Botton S, Preudhomme C, Birnbaum D, Bernard OA, Ogawa S, Fontenay M, Kosmider O. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169–3177. doi: 10.1182/blood-2012-11-469619. [DOI] [PubMed] [Google Scholar]
- Deng P, Chen QM, Hong C, Wang CY. Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int J Oral Sci. 2015;7:197–204. doi: 10.1038/ijos.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele B, Przybyl J, Quattrone A, Finalet Ferreiro J, Vanspauwen V, Geerdens E, Gianfelici V, Kalender Z, Wozniak A, Moerman P, Sciot R, Croce S, Amant F, Vandenberghe P, Cools J, Debiec-Rychter M. Identification of a novel, recurrent MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int J Cancer. 2014;134:1112–1122. doi: 10.1002/ijc.28440. [DOI] [PubMed] [Google Scholar]
- Diaz-Beya M, Navarro A, Ferrer G, Diaz T, Gel B, Camos M, Pratcorona M, Torrebadell M, Rozman M, Colomer D, Monzo M, Esteve J. Acute myeloid leukemia with translocation (8;16)(p11;p13) and MYST3-CREBBP rearrangement harbors a distinctive microRNA signature targeting RET proto-oncogene. Leukemia. 2013;27:595–603. doi: 10.1038/leu.2012.278. [DOI] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:111411–19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Yao R, Du J, Wang S, Fan Z. Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp Cell Res. 2013;319:2874–2882. doi: 10.1016/j.yexcr.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Du J, Ma Y, Ma P, Wang S, Fan Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells. 2013;31:126–136. doi: 10.1002/stem.1255. [DOI] [PubMed] [Google Scholar]
- Enzinger FM, Weiss SW, Liang CY. Ossifying fibromyxoid tumor of soft parts. A clinicopathological analysis of 59 cases. Am J Surg Pathol. 1989;13:817–827. doi: 10.1097/00000478-198910000-00001. [DOI] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C, Bridge JA, Hogendoorn PC, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. IARC; Lyon: 2013. [Google Scholar]
- Folpe AL, Weiss SW. Ossifying fibromyxoid tumor of soft parts: a clinicopathologic study of 70 cases with emphasis on atypical and malignant variants. Am J Surg Pathol. 2003;27:421–431. doi: 10.1097/00000478-200304000-00001. [DOI] [PubMed] [Google Scholar]
- Gao R, Dong R, Du J, Ma P, Wang S, Fan Z. Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15INK4B and p27Kip1. Mol Cell Biochem. 2013;379:115–122. doi: 10.1007/s11010-013-1633-7. [DOI] [PubMed] [Google Scholar]
- Gebre-Medhin S, Nord KH, Moller E, Mandahl N, Magnusson L, Nilsson J, Jo VY, Vult von Steyern F, Brosjo O, Larsson O, Domanski HA, Sciot R, Debiec-Rychter M, Fletcher CD, Mertens F. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012;181:1069–1077. doi: 10.1016/j.ajpath.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Graham RP, Dry S, Li X, Binder S, Bahrami A, Raimondi SC, Dogan A, Chakraborty S, Souchek JJ, Folpe AL. Ossifying fibromyxoid tumor of soft parts: a clinicopathologic, proteomic, and genomic study. Am J Surg Pathol. 2011;35:1615–1625. doi: 10.1097/PAS.0b013e3182284a3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RP, Weiss SW, Sukov WR, Goldblum JR, Billings SD, Dotlic S, Folpe AL. PHF1 Rearrangements in Ossifying Fibromyxoid Tumors of Soft Parts: A Fluorescence In Situ Hybridization Study of 41 Cases With Emphasis on the Malignant Variant. Am J Surg Pathol. 2013;37:1751–1755. doi: 10.1097/PAS.0b013e31829644b4. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Kao YC, Sung YS, Zhang L, Huang SC, Argani P, Chung CT, Graf NS, Wright DC, Kellie SJ, Agaram NP, Ludwig K, Zin A, Alaggio R, Antonescu CR. Recurrent BCOR Internal Tandem Duplication and YWHAE-NUTM2B Fusions in Soft Tissue Undifferentiated Round Cell Sarcoma of Infancy: Overlapping Genetic Features With Clear Cell Sarcoma of Kidney. Am J Surg Pathol. 2016 doi: 10.1097/PAS.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H, Ogose A, Umezu H, Hotta T, Tohyama T, Tsuchiya M, Endo N. Ossifying fibromyxoid tumor of soft parts with clonal chromosomal aberrations. Cancer Genet Cytogenet. 2007;176:156–160. doi: 10.1016/j.cancergencyto.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Kilpatrick SE, Ward WG, Mozes M, Miettinen M, Fukunaga M, Fletcher CD. Atypical and malignant variants of ossifying fibromyxoid tumor. Clinicopathologic analysis of six cases. Am J Surg Pathol. 1995;19:1039–1046. doi: 10.1097/00000478-199509000-00007. [DOI] [PubMed] [Google Scholar]
- Koontz JISA, Nucci M, Kuo FC, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. PNAS. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Collins R, Jiao Y, Ouillette P, Bixby D, Erba H, Vogelstein B, Kinzler KW, Papadopoulos N, Malek SN. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood. 2011;118:5914–5917. doi: 10.1182/blood-2011-05-356204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361. doi: 10.1038/bcj.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Gorunova L, Agostini A, Johannessen LE, Brunetti M, Davidson B, Heim S, Panagopoulos I. Cytogenetic and Molecular Profile of Endometrial Stromal Sarcoma. Genes Chromosomes Cancer. 2016 doi: 10.1002/gcc.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Gorunova L, Gatius S, Matias-Guiu X, Davidson B, Heim S, Panagopoulos I. MEAF6/PHF1 is a recurrent gene fusion in endometrial stromal sarcoma. Cancer Lett. 2014;347:75–78. doi: 10.1016/j.canlet.2014.01.030. [DOI] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Finnell V, Fetsch JF. Ossifying fibromyxoid tumor of soft parts–a clinicopathologic and immunohistochemical study of 104 cases with long-term follow-up and a critical review of the literature. Am J Surg Pathol. 2008;32:996–1005. doi: 10.1097/PAS.0b013e318160736a. [DOI] [PubMed] [Google Scholar]
- Mohamed AD, Tremblay AM, Murray GI, Wackerhage H. The Hippo signal transduction pathway in soft tissue sarcomas. Biochim Biophys Acta. 2015;1856:121–129. doi: 10.1016/j.bbcan.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Nishio J, Iwasaki H, Ohjimi Y, Ishiguro M, Isayama T, Naito M, Okabayashi H, Kaneko Y, Kikuchi M. Ossifying fibromyxoid tumor of soft parts. Cytogenetic findings. Cancer Genet Cytogenet. 2002;133:124–128. doi: 10.1016/s0165-4608(01)00581-7. [DOI] [PubMed] [Google Scholar]
- Pagan JK, Arnold J, Hanchard KJ, Kumar R, Bruno T, Jones MJ, Richard DJ, Forrest A, Spurdle A, Verdin E, Crossley M, Fanciulli M, Chenevix-Trench G, Young DB, Khanna KK. A novel corepressor, BCoR-L1, represses transcription through an interaction with CtBP. J Biol Chem. 2007;282:15248–15257. doi: 10.1074/jbc.M700246200. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Micci F, Thorsen J, Gorunova L, Eibak AM, Bjerkehagen B, Davidson B, Heim S. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One. 2012;7:e39354. doi: 10.1371/journal.pone.0039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, Heim S, Micci F. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer. 2013;52:610–618. doi: 10.1002/gcc.22057. [DOI] [PubMed] [Google Scholar]
- Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, Hennekam RC, Peters DJ. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider N, Fisher C, Thway K. Ossifying fibromyxoid tumor: morphology, genetics, and differential diagnosis. Ann Diagn Pathol. 2016;20:52–58. doi: 10.1016/j.anndiagpath.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol. 2013;37:938–942. doi: 10.1097/PAS.0b013e318282da9d. [DOI] [PubMed] [Google Scholar]
- Sovani V, Velagaleti GV, Filipowicz E, Gatalica Z, Knisely AS. Ossifying fibromyxoid tumor of soft parts: report of a case with novel cytogenetic findings. Cancer Genet Cytogenet. 2001;127:1–6. doi: 10.1016/s0165-4608(00)00412-x. [DOI] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, Sakamoto H, Wang L, Ojima H, Shimada K, Kosuge T, Okusaka T, Kato K, Kusuda J, Yoshida T, Aburatani H, Shibata T. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet. 2011;43:464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013;70:3989–4008. doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Abe A, Emi N. Clarifying the impact of polycomb complex component disruption in human cancers. Mol Cancer Res. 2014;12:479–484. doi: 10.1158/1541-7786.MCR-13-0596. [DOI] [PubMed] [Google Scholar]
- Zamecnik M, Michal M, Simpson RH, Lamovec J, Hlavcak P, Kinkor Z, Mukensnabl P, Matejovsky Z, Betlach J. Ossifying fibromyxoid tumor of soft parts: a report of 17 cases with emphasis on unusual histological features. Ann Diagn Pathol. 1997;1:73–81. doi: 10.1016/s1092-9134(97)80011-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Unsupervised hierarchical clustering of 5 OFMTs among a wide variety of other soft tissue neoplasms. Four OFMTs form a distinct group despite different gene fusions, including EP400-PHF1, MEAF6-PHF1, CREBBP-BCORL1, and ZC3H7B-BCOR. OFMT with KDM2A-WWTR1 did not cluster together.


