Abstract
Understanding abnormal resting-state functional connectivity of distributed brain networks may aid in probing and targeting mechanisms involved in major depressive disorder (MDD). To date, few studies have used resting state functional magnetic resonance imaging (rs-fMRI) to attempt to discriminate individuals with MDD from individuals without MDD, and to our knowledge no investigations have examined a remitted (r) population. In this study, we examined the efficiency of support vector machine (SVM) classifier to successfully discriminate rMDD individuals from healthy controls (HCs) in a narrow early-adult age range. We empirically evaluated four feature selection methods including multivariate Least Absolute Shrinkage and Selection Operator (LASSO) and Elastic Net feature selection algorithms. Our results showed that SVM classification with Elastic Net feature selection achieved the highest classification accuracy of 76.1% (sensitivity of 81.5% and specificity of 68.9%) by leave-one-out cross-validation across subjects from a dataset consisting of 38 rMDD individuals and 29 healthy controls. The highest discriminating functional connections were between the left amygdala, left posterior cingulate cortex, bilateral dorso-lateral prefrontal cortex, and right ventral striatum. These appear to be key nodes in the etiopathophysiology of MDD, within and between default mode, salience and cognitive control networks. This technique demonstrates early promise for using rs-fMRI connectivity as a putative neurobiological marker capable of distinguishing between individuals with and without rMDD. These methods may be extended to periods of risk prior to illness onset, thereby allowing for earlier diagnosis, prevention, and intervention.
Keywords: Resting state fMRI, Major depressive disorder, Machine learning, MVPA
Highlights
-
•
Multivariate pattern analysis (MVPA) techniques can be used in prediction.
-
•
Resting state fMRI is a stable and effective method to determine network function.
-
•
Detection of clinical conditions, such as major depressive disorder (MDD), is challenging.
-
•
MVPA with resting state fMRI was successful in distinguishing remitted MDD.
1. Introduction
Major depressive disorder (MDD) is a heterogeneous disorder characterized by variable patterns of nine key symptoms, often in episodic patterns across an individual's lifetime, parallel to the waxing and waning observed in chronic illnesses such as multiple sclerosis (APA, 1994) with relapsing–remitting and relapsing–progressive patterns. Despite awareness that symptom profiles and illness course patterns vary widely across (and perhaps even within) individuals with MDD, prior studies have typically used the full diagnostic spectrum (studies of those with any and all patterns of MDD) and the broadest range in age and experience of illness in cross sectional studies. These prior, broad-stroke, heterogeneous studies may have led to increased type I error with relatively small samples and publication bias or diffusion of important, specific effects, increasing type II error. Together, these would dilute both inferential capability and replication. More recently, subtypes of MDD have been pursued with the emergence of some larger studies including more clinically and demographically homogeneous samples (Korgaonkar et al., 2014). To this end, the present study has strived to constrain a number of features with known impact on brain function in MDD varying from small (e.g., medications, subtypes of MDD) to medium (active symptoms) to very large (age, development) effect sizes. As such, we studied early course MDD in the remitted state among a sample of late-adolescents who were medication-free at the time of scan in order to reduce sources of heterogeneity for between group comparisons. These methodological controls increased homogeneity and have resulted in an emerging model for how mood disorders might have distinct trait/risk features, scar patterns, symptom perturbations, and chronic burden/scar components (Weisenbach et al., 2014, Peters et al., 2016, Votruba and Langenecker, 2013).
Studying individuals with rMDD enables a unique examination of potential trait-based mechanisms of depression and depression relapse (e.g. Marchetti et al., 2012). One method for understanding trait-based markers for MDD involves studying network function through measurements of network connectivity. Disrupted network connectivity has been documented among individuals within a major depressive episode (Greicius et al., 2007, Connolly et al., 2013), including within and between key nodes of posterior cingulate cortex (PCC), medial prefrontal cortex (mPFC), inferior parietal cortex (IPC, Hamilton et al., 2012), amygdala (Siegle et al., 2007), anterior insula, dorsal ACC (Strigo et al., 2008, Briceño et al., 2013), pregenual ACC (Horn et al., 2010), DLPFC (Bench et al., 1992, Mayberg et al., 1999, Jacobs et al., 2014), and hippocampus (Cao et al., 2012, Sambataro et al., 2013).
Although alterations in brain functional connectivity have been demonstrated in MDD, most of these studies have focused on group level analysis. However, there is substantial interest in identifying single subject biomarkers that are clinically applicable as diagnostic or prognostic tools (Mossner et al., 2007, Atluri et al., 2013, Schneider and Prvulovic, 2013). Recent studies (Klöppel et al., 2011, Mourao-Miranda et al., 2012, Orru et al., 2012, Zarogianni et al., 2013, Haller et al., 2014 May, Sundermann et al., 2014) have begun to investigate diagnostic classification of mental disorders using rs-fMRI and multivariate pattern analysis (MVPA). Particularly, pioneering work has examined the clinical applicability of examining resting state data using MVPA (Craddock et al., 2009). MVPA algorithms provide a framework for disease state prediction in which the final goal is to predict the presence or absence of a disease based on observed functional connections. The patterns are learned from multivariate data given predetermined categories, and performance is measured by the prediction accuracy obtained when classifying a new case. Support Vector Machine (SVM) has been used as a classification model often in fMRI research that may offer better prediction accuracy and is less sensitive to noise than alternative MVPA approaches (Mitchell et al., 2004, Chen et al., 2006, LaConte et al., 2007, Mourao-Miranda et al., 2005). In practice, building a robust, generalizable classification model can be challenging because the number of features far exceeds the number of data observations. To avoid model overfitting, there is a need to select informative features before building a classification model. In the context of rs-FMRI research, the features are the functional connections between two regions (voxels). The objective of feature selection include the reduction of prediction error and the improved interpretability of a MVPA model (Guyon and Elisseeff, 2003a, De Martino et al., 2008, Mourao-Miranda et al., 2005). Several filter and wrapper approaches (Craddock et al., 2009, Zeng et al., 2014, Cao et al., 2014) have been used based on univariate t-test, probability density function and recursive feature elimination (Guyon and Elisseeff, 2003b) in fMRI research. Embedded methods learn which features best contribute to the accuracy of the model while the model is being created. The most common type of embedded feature selection methods are regularization methods. In recent years, regularized embedded methods such as LASSO and Elastic Net have demonstrated good effectiveness and sensitivity (Mwangi et al., 2013) in neuroimaging machine learning tasks such as Alzheimer's disease (AD) classification (Casanova et al., 2011, Rao et al., 2011, Shen et al., 2010), treatment response predictions in Attention Deficit Hyperactivity Disorder (Marquand et al., 2012) and Autism Spectrum Disorder classification (Duchesnay et al., 2011). In the context of rs-fMRI, a few studies have applied SVM (Craddock et al., 2009, Cao et al., 2014) in identifying patients with MDD. With task-based data, machine learning using a Gaussian Process Classifier has been used to discriminate adolescents at high risk for mood disorders from healthy adolescents (Mourao-Miranda et al., 2012). However, to our knowledge, rs-fMRI connectivity data have not been explored using machine learning among patients in the remitted state of MDD.
In this research, we built a sparse MVPA framework combining regularized Elastic Net feature selection algorithm and a linear SVM. The advantage of Elastic Net regularization penalty over filter approaches is that it conducts automatic variable selection and continuous shrinkage simultaneously, and selects a group of correlated variables. This feature selection strategy is a state-of-the-art representative of recent advances in L1/L2-constraint based methods. We compared Elastic Net with two filter approaches t-test and Wilcoxon rank sum and performed these evaluations using leave-one-out cross validation in the context of a study of resting state functional connectivity in remitted major depressive disorder (rMDD). This design allowed us to examine network differences in the absence of the state effects of active illness. We hypothesized that building a MVPA framework by employing feature selection strategies combined with SVM would be successful in identifying the discriminant functional connections that predicted prior history of rMDD.
2. Methods
2.1. Participants
Participants were recruited from the University of Michigan (UM) and the University of Illinois at Chicago (UIC) using flyers and multiple forms of posting on the internet. All participants completed an identical assessment protocol, including the Diagnostic Interview for Genetic Studies (DIGS; Nurnberger et al., 1994), the Hamilton Depression Scale (Ham-D; 34 Hamilton, 1960), and a targeted neuropsychological and fMRI battery (not reported here). Participants were considered remitted from MDD if they previously met criteria for at least one major depressive episode (MDE), did not meet current criteria for an MDE in the last three months (Mean 2.5 years well), and currently scored below a 7 on the Ham-D (administered during the phone screen and during the initial diagnostic interview). HCs could not meet current or past criteria (Never Mentally Ill, NMI) for MDD or any other Axis I or II psychiatric disorder and had no first degree relatives with a history of psychiatric illness. In addition, participants were required to be medication free for a period of 30 days prior to the scan and those with substance abuse or dependence within the past six months were excluded.
Diagnosis of past MDD or NMI was confirmed using a modified Family Interview for Genetic Studies completed with a parent, guardian, or older sibling (Nurnberger et al., 1994). The final sample included 38 rMDD (17 UM, 21 UIC) and 29 NMI (16 UM, 13 UIC) between the ages of 18–23 years (66% Female). None of the remitted (r)MDD were taking medications at the time of scan or for the past 30 days (and 21 had never taken any psychotropic medication, 33 had a history of psychotherapy). The rMDD (aged 18–23, modal depressive episodes = 1, modal years well = 4) were compared with data from 29 NMI. Participant demographics and clinical characteristics are presented in Table 1.
Table 1.
Sample demographics and clinical characteristics.
| rMDD | NMI | |
|---|---|---|
| N | 38 | 29 |
| Site | 17 UM/21 UIC | 16 UM/13 UIC |
| Sex | 29 F/9 M | 16 F/13 M |
| Age | 20.97 (SD = 1.53) | 20.97 (SD = 1.55) |
| Years Education | 14.34 (SD = 1.40) | 14.90 (SD = 1.21) |
| Psychoactive medications taken for 3 consecutive months (in the past)? | 13 Yes/25 No | NA |
| Depressive eps. | 1.92 (SD = 1.25) | NA |
| Age of onset | 14.84 (SD = 4.91) | NA |
| Ham-Da | 2.39 (SD = 3.01) | 0.48 (SD = 1.15) |
Indicates that there are significant differences in HAM-D. No participants had any current medication use for at least the past 30 days.
2.2. rs-fMRI data
rs-fMRI data from two 3.0 Tesla GE scanners were collected using eight bilateral seeds in the default mode network (DMN), salience network (SN) and cognitive control network (CCN). Table 2 provides the Montreal Neurological Institute (MNI) coordinates of the ROI seeds. Seeds were derived based on previous literature examining resting state connectivity of the amygdala (Fox et al., 2009, Pannekoek et al., 2013), PCC (McCabe and Mishor, 2011, Bluhm et al., 2011), sgACC (Alexopoulos et al., 2012, Kelly et al., 2009), and anterior superior insula (Margulies et al., 2007, Siegle et al., 2006) in depression. All seeds were verified visually on an average anatomy of the first 55 subjects that participated in the study, and used 19 contiguous voxels (radius 2.9 mm).
Table 2.
Names, abbreviations and MNI coordinates of the ROIs.
| Network/regions | MNI coordinates |
||
|---|---|---|---|
| x | y | z | |
| Default mode | |||
| Posterior cingulate cortex (PCC) | − 5/5 | 50 | 36 |
| Subgenual anterior cingulate (sgACC) | − 4/4 | 21 | − 8 |
| Hippocampal formation (HPF) | − 30/30 | − 12 | − 18 |
| Emotion/salience | |||
| Amygdala (AMYG) | − 23/23 | − 5 | − 19 |
| Anterior insula (INS) | − 36/36 | 13 | 5 |
| Ventral striatum–superior (VSs) | − 10/10 | 15 | 0 |
| Ventral striatum–inferior (VSi) | − 9/9 | 9 | − 8 |
| Cognitive control | |||
| Dorsolateral prefrontal cortex (DLPFC) | − 46/46 | 46 | 14 |
2.3. rs-fMRI preprocessing
Data preprocessing occurred as follows: Slice timing was completed with SPM8 (http://www.fil.ion.ucl.ac.uk/spm/doc/) and motion detection algorithms were applied using FSL (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). Coregistration of structural images to functional images was followed by spatial normalization of the coregistered T1-spgr to the Montreal Neurological Institute (MNI) template. The resulting normalization matrix was then applied to the slice-time-corrected, time series data. These normalized T2* time-series data were spatially smoothed with a 5 mm Gaussian kernel resulting in T2* images with isotropic voxels, 2 mm a side.
2.4. Cross-correlation analysis
The rs-fMRI time series was detrended and mean centered. Physiologic correction was performed by regressing out white matter and cerebral spinal fluid signals (Tabachnick and Fidell, 2007). Motion parameters were regressed out (Behzadi et al., 2007). Based upon the recent literature (Behzadi et al., 2007, Jo et al., 2013), motion volumes were identified based on any TR to TR movement exceeding 1.5 mm and did not differ between groups. Those with significant movement were not included in this final reported sample. Global signal was not regressed due to collinearity violations with gray matter signal, problematic mis-estimates of anticorrelations (Power et al., 2012), and because it does not affect distance–micromovement relationships (Behzadi et al., 2007). Finally time-series were band-pass filtered over 0.01–0.10 Hz. Regions of Interest (ROIs; 2.9 mm radius) were defined in MNI space and spatially averaged time course data were extracted from these regions for each participant. Correlation coefficients were calculated between seed regions (Table 2) and transformed to z scores using a Fisher transformation.
2.5. Feature extraction
The rs-fMRI network was captured by a 16 × 16 symmetric matrix of nodes. We extracted the upper triangle elements of the functional connectivity matrix as classification features, i.e. the feature space for classification was spanned by the (16 × 15) / 2 = 120 dimensional feature vectors.
2.6. Classification algorithm
SVM classification (Vapnik, 1995) is a widely used method for binary classification in fMRI studies. SVMs are supervised learners that work in two steps. In the training step, a subset of the available data points as well as their associated classes is used to iteratively find a linear boundary or hyperplane that separates the two classes optimally. In the testing step, new, previously unobserved data points in the same space as the training points are classified depending on their position relative to the boundary (often the “case left out” in leave one out, small n analyses). For two classes, the SVM algorithm attempts to find a linear decision boundary (separating hyper plane) using the decision function, where defines the linear decision boundary, and is chosen to optimize the boundaries defined by D = + 1 and D = − 1 (known as the margin) between the two class distribution. The decision function that is learned by a SVM is a linear combination of feature values in a particular feature space.
There are differences between different types of SVM, notably in how the relationship between the feature space and the original features (functional connections, in our case) is determined; a given choice of kernel function determines an (implicit) feature space in which a decision takes place.
2.7. Feature selection algorithms
In neuroimaging studies, the number of features are often more than the number of observations (often less than 100), which causes curse-of-dimensionality and small-n-large-p effects. The unimportant features may result in an over fitting problem in machine learning, and therefore reduces model prediction accuracy and generalization ability. We considered two embedded-methods: LASSO and the Elastic Net (Tibshirani, 1996, Zou and Hastie, 2005). The popular LASSO regression (Tibshirani, 1996) method minimizes the Residual Sum of Squares (RSS), similar to Ordinary Least Squares (OLS) regression, but poses a constraint to the sum of the absolute values of the coefficients being less than a constant. This additional constraint is similar to that introduced in Ridge regression, where the constraint is to the sum of the squared values of the coefficients. This simple modification allows LASSO to perform also variable selection because the shrinkage of the coefficients is such that some coefficients can be shrunk exactly to zero. The LASSO computes model coefficient by minimizing the following function R(β) + λ || β ||1, where R(β) is the mean square error on the training set and || β ||1 = . λ controls the degree of sparsity of the solution, i.e. the number of features selected.
Elastic Net (Zou and Hastie, 2005) is similar to LASSO. It differs in that the l1 norm of β is replaced by a combination of l1 and l2 norms. In this case, we minimize R(β) + λ Pα(β), where, for α strictly between 0 and 1, and a nonnegative λ. The λ parameter can be tuned in order to set the shrinkage level, and the higher the λ is, and the more coefficients are shrunk to 0. Elastic Net is the same as LASSO when α = 1. As α shrinks toward 0, Elastic Net approaches ridge regression. For other values of α, the penalty term Pα(β) interpolates between the L1 norm of β and the squared L2 norm of β. The advantage of Elastic Net over LASSO is that the Elastic Net penalty completes automatic variable selection and continuous shrinkage simultaneously, and it can select from a group of correlated variables. It is especially useful for large p small n problems where the grouped variables situation is a particularly important concern (Hastie et al., 2000, Hastie et al., 2001).
2.8. Feature selection and classification method
Due to our limited number of samples, we used a leave-one-out cross-validation (LOOCV) strategy to estimate the generalization ability of our classifier. We used SVM classifier with three strategies: 1) No feature selection, which acted as a baseline to focus the specific contributions of feature selection. 2) Multivariate LASSO and Elastic Net based feature subset ranking. 3) Univariate t-test and Wilcoxon feature subset ranking.
Feature selection algorithms were implemented in MATLAB 2012b and were done in each cross validation fold. For Elastic Net, we have evaluated classification performances for different values of alpha, ranging from .1 to 1.0. When alpha = 1, it is Lasso and when alpha is between 0 and 1, it is Elastic net. Inside each LOOCV fold, 10-fold CV was used to select the best elastic net regularization parameter lambda (λ).
We also evaluated two filter-based algorithms such as Student's t-test and Wilcoxon sum-rank test to rank the features which evaluate if each feature is significantly different between the two classes; these ranking algorithms were employed in each cross validation fold. We used the MATLAB Bioinformatics toolbox to compute these scoring functions.
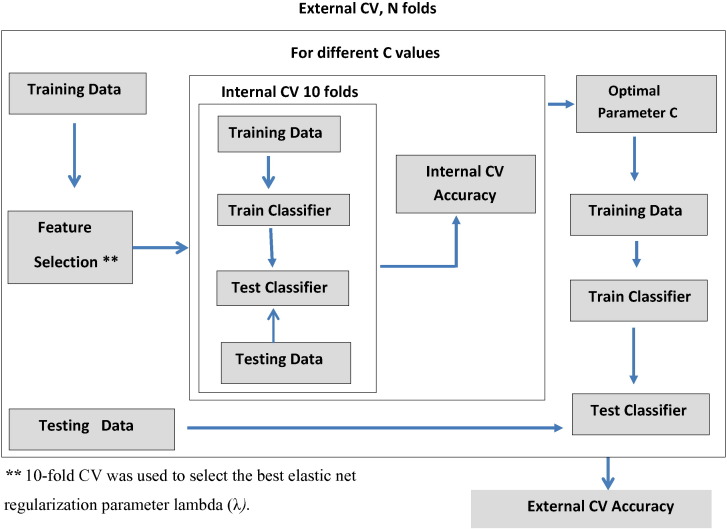
Inside the LOOCV (n = 67), the features were selected based on a feature selection strategy and used as the final feature set for SVM classification. We used default parameter setting in Matlab SVM function for kernel and optimization method. For the selection of soft margin parameter C, we performed another 10-fold CV strategy for different values of C using SVM classifier inside the LOOCV. We selected the parameter value C which produces highest accuracy in 10-fold CV and used in the final SVM model. The classification framework is shown in Fig. 1.
Fig. 1.
The Classification Framework.
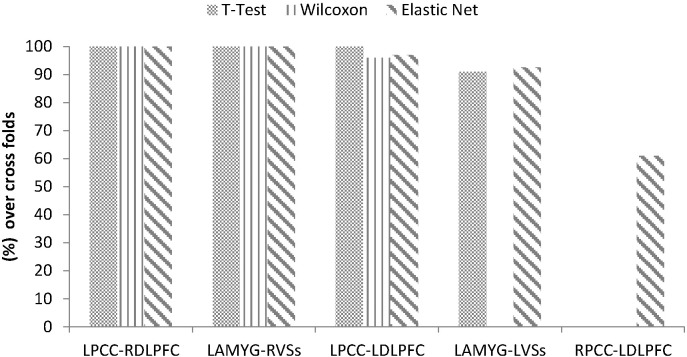
Since we used a LOOCV strategy, the feature ranking was based on different training dataset in each cross validation (CV) fold. Therefore the feature (functional connections) contributions to classification were not evenly distributed. In this study we adopted the concept of consensus functional connectivity (Fair et al., 2012), which is defined as the functional connectivity feature appearing in the final feature set of each CV iteration. We computed the percentages of occurrences of features that contributed to identification of depressed patients across all iterations of the cross validation. The functional connectivities which appeared in the leave-one-out process more than half of the times were shown in Fig. 5, and indicated the most discriminative features between those with rMDD and HC.
Fig. 5.
The contributing functional connections detection of rMDD selected by different feature selection algorithms. Note. The cell values represent the percentages of training folds in which a given connection was selected during the classification (more than 50% were selected as consensus functional connectivities).
We also performed a hold-out validation test by randomly selected nine samples (5 rMDD patients, 4 controls) as testing data set and remaining 58 samples as the raining dataset. We obtained the best alpha and k parameters by re-running the LOOCV using only the training 58 subjects. Once we found the connectivities using 58 subjects with best alpha and k parameters, we used these connections as our final model and tested with the hold-out 9 subjects for prediction. The results are shown in Table 4.
Table 4.
Results of holdout validation in prediction of rMDD.
| Methods | Accuracy (%) | Sensitivity (%) | Specificity (%) | Permutation test* p-Value |
|---|---|---|---|---|
| All Features | 44.4 | 60.0 | 25.0 | >.05 |
| Elastic Net | 77.8 | 80.0 | 75.0 | <.05 |
| t-Test | 77.8 | 80.0 | 75.0 | <.05 |
| Wilcoxon | 66.7 | 80.0 | 50.0 | >.05 |
2.9. Permutation test
The performance of the SVM classifier was evaluated using accuracy, sensitivity and specificity measures. To determine whether classification accuracy exceeded chance levels (50%), we performed permutation testing and derived a p-value. We permuted the class labels 1000 times (each time randomly assigning rMDD and HC labels to each pattern of functional connectivity values) and repeated the entire feature selection algorithm. We then counted the number of times the permuted test accuracy was higher than the one obtained for the true labels. Finally, we divided this number by 1000 and obtained a p-value for classification accuracies. For the permutation test the 10-fold CV and LOOCV structures were maintained.
3. Results
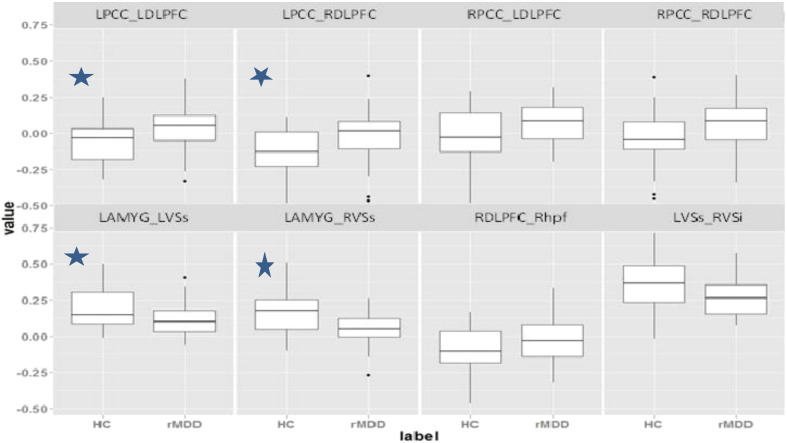
First we employed a two sample t-test using our data set to find any significant functional connectivity differences between healthy controls and rMDD. There were eight connections found to be significantly different among these two groups (Fig. 2). At a more conservative test, we also performed a FDR corrected t-test (q = .2), which found four significant group differences in connections (Fig. 2, left four panels).
Fig. 2.
Significant functional connections based on two sample unadjusted t-test (p < /05). Note: * represents the FCs based on t-test with FDR.
It should be noted that univariate filter methods ranked all variables in terms of relevance, as measured by a score. In each CV fold, we selected the top k FCs according to the score and reported the average accuracy over all CV folds. Fig. 3 illustrates the performance of t-test and Wilcoxon methods for different values of k. As the chart reveals, the highest accuracy was obtained when k = 4 for t-test and k = 3 for Wilcoxon test. With higher k after that, accuracy declined. In contrast, LASSO and Elastic Net regressions derived only important FCs, with a different number of important FCs in each CV fold. As such, the SVM classifier was trained with only the important predictors found in each CV. The average accuracy for different values of α parameters is reported. As illustrated in Fig. 4, the best accuracy (76.1%) was obtained when α = .5. At α = 1, Elastic Net becomes LASSO, and very few variables (mostly two features in each CV fold), and the accuracy is 67.1%.
Fig. 3.
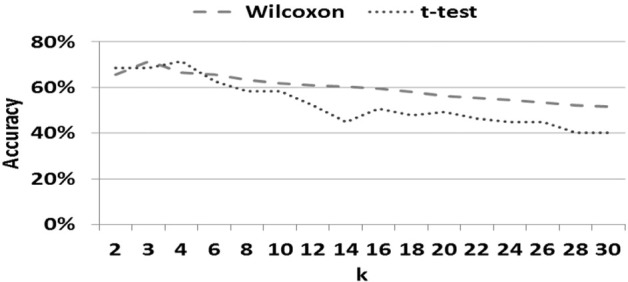
Classification accuracy for rMDD across top connections for two filter methods.
Fig. 4.
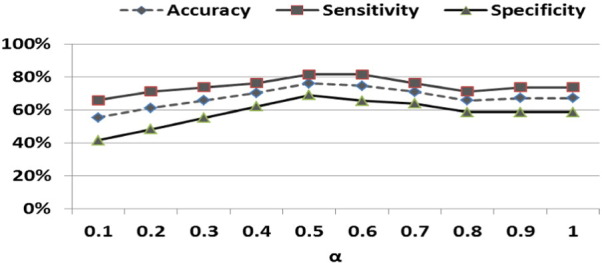
Classification results for rMDD varying by alpha parameter (α) in Elastic Net method. Note. At α = 1, Elastic Net becomes LASSO.
Table 3 summarizes the performance of t-test, Wilcoxon, LASSO and Elastic Net using SVM classifier across 67 folds. Without any feature selection algorithm, the SVM classifier using LOOCV obtained overall accuracy of 45.1% (57.9% for patients and 28.3% for controls). In contrast, the embedded methods LASSO and Elastic Net algorithms discard the unimportant features by forcing them to zero, so there is no ranking of features. The highest classification accuracy of 76.1% was obtained by choosing the Elastic Net feature selection algorithm (81.5% for patients and 68.9% for healthy controls). t-Test and Wilcoxon methods also performed well, both with 71.6% accuracy (76.3% for patients and 65.5% for healthy controls).
Table 3.
SVM Classification accuracies in discriminating between rMDD patients and controls using LOOCV.
| Methods | Accuracy |
Sensitivity |
Specificity |
Permutation testa |
|---|---|---|---|---|
| (%) | (%) | (%) | p-Value | |
| All features | 45.1 | 57.9 | 28.3 | > .05 |
| Elastic Net (α = .2) | 61.2 | 71.1 | 48.3 | < .05 |
| Elastic Net (α = .5) | 76.1 | 81.5 | 68.9 | < .05 |
| LASSO (α = 1) | 67.1 | 73.6 | 58.6 | > .05 |
| t-Test (k = 2) | 68.6 | 73.7 | 62.1 | < .05 |
| t-Test (k = 4) | 71.6 | 76.3 | 65.5 | < .05 |
| t-Test (k = 6) | 64.2 | 68.4 | 58.6 | > .05 |
| t-Test (k = 10) | 56.7 | 57.9 | 55.2 | > .05 |
| Wilcoxon (k = 2) | 68.6 | 73.7 | 62.1 | < .05 |
| Wilcoxon (k = 3) | 71.6 | 76.3 | 65.5 | < .05 |
| Wilcoxon (k = 6) | 64.20 | 71.0 | 55.2 | > .05 |
| Wilcoxon (k = 10) | 58.2 | 65.8 | 48.3 | > .05 |
Permutation test indicates whether the accuracy exceeds chance levels (50%).
The classification accuracies from holdout validation test are shown in Table 4. We found the consensus connectivities using 58 subjects with best alpha and k parameters by carrying out a LOOCV. We used these connections as our final model and tested with the remaining 9 subjects for prediction. Our results showed that the both filter-based t-test and Elastic Net obtained accuracy of 77.8%.
The percentages of functional connections in all folds that contributed to identification of rMDD patients are shown in Fig. 5. We showed only those functional connections for the final model which appeared more than 50% inside LOOCV using 58 subjects. The most discriminative connections based on consensus functional connections were between the left PCC and right DLPFC and between the left amygdala and right VSs among all feature selection algorithms. These connections were selected in all cross validation folds. The connections between left PCC and left DLPFC were selected more than 95% of the time using Elastic Net, Wilcoxon and t-test ranking algorithms. The connections between left amygdala and left VSs were observed in Elastic Net and t-test algorithms in more than 92% of cross validation folds. In addition, Elastic Net identified the connection between right PCC and left DLPFC in 61% of cross validation folds.
4. Discussion
The present study aimed to classify participants into the clinical psychiatric diagnostic categories for rMDD or NMI using SVM of rs-fMRI data. Sixteen nodes from the DMN, SN and CCN were chosen a priori based on previous research indicating their importance in MDD and rMDD (Jacobs et al., 2014, Tremblay et al., 2005, Kaiser et al., 2015) and theories show that these three networks comprise the core neurocognitive networks (Raichle et al., 2001, Seeley et al., 2007, Menon, 2011). We then applied SVM, using four different feature selection algorithms, to identify the top ranked features (connections between two nodes) that distinguished the rMDD from the HC group. In support of our hypothesis, we were able to use SVM of rs-fMRI data to discriminate these participants, even in the remitted state. We found that the most discriminative connection was between the left PCC and the right DLPFC, followed by the left amygdala and the right superior ventral striatum connection. These two connections were selected in 100% of training folds for all feature selection methods.
Our results provide new support in discrimination of rMDD, extending beyond previous studies that have been able to use machine learning classification, including SVM, to discriminate active MDD (aMDD) from HC using rs-fMRI data (Guo et al., 2012, Guo et al., 2014). For example, using SVM with leave-one-out cross validation applied to whole-brain rsfMRI data, active MDD were distinguished from HCs with about 84% classification accuracy (Cao et al., 2014). A similar result (83% accuracy) was obtained in predicting active MDD using 15 pre-determined regions of interest (Craddock et al., 2009). SVM classification performance here (76.1% accuracy) was similar, but not as high as reported in the above mentioned studies. However, it is important to note that we did not employ separation of active MDD from NMI. The results of prior studies of aMDD could capture features related to active illness state, trait features, and potential scar/repetitive scar features, strengthening results. Here, our results could only be related to trait and early scar features, weakening the number of predictors, but strengthening the meaning and actual practical use. In addition, our feature selection algorithms are different and we used a larger sample which can result in more conservative estimates. In the future, we intend to explore other feature selection strategies to use strategies like recursive feature elimination and sparse logistic regression. It is worthwhile to note that, in a clinical setting, when our goal is to find the pattern of functional connectivity that accurately predicts whether a subject suffers from rMDD, we expect a classification performance to be 95–100% accuracy (Orru et al., 2012). This is a promising initial step that can be followed up using additional predictors and moving to at-risk samples.
4.1. DMN–CCN connectivity
One of our main discriminant connections for rMDD was between the left PCC and right DLPFC. These regions are part of the DMN and the CCN, respectively, and reflect meta-analytic findings of hyperconnectivity/hyperactivity of posterior default network components including the PCC with lateral prefrontal areas (Sundermann et al., 2014, Kaiser et al., 2015). Our results also lend support to previous research that found disturbances in the PCC and DMN, also in people with active MDD (Hamilton et al., 2012, Pizzagalli, 2011), including increased connectivity in the PCC in patients with active MDD (Sambataro et al., 2013). As we found these patterns for the first time in young adults with rMDD, it lends credence to the argument that this dysfunctional connection is not related to active symptoms, and it is present early in the course of the disease. A dysfunctional DLPFC may represent a precursor to the hyperactive midline activity reported in MDD, and connections to the DMN may represent a vulnerability marker in people at-risk for MDD (Marchetti et al., 2012). It is possible that cognitive risk factors, including rumination and poor attentional control, are a direct byproduct of DMN hyperconnectivity, and are associated with maladaptive self-focus. Consistent with this argument, individuals with MDD have a tendency to attend to internal at the expense of external stimuli (Greicius et al., 2007, Hamilton et al., 2011, Broyd et al., 2009), and aberrant connectivity within the DMN has been associated with increased rumination and brooding in depression (Hamilton et al., 2011, Berman et al., 2010, Jacobs et al., 2014, Zhu et al., 2012 Apr 1) and decreased sustained attention (Jacobs et al., 2014).
4.2. SN connectivity
Our second main discriminant connection for rMDD was between the left amygdala and right ventral striatum. Within the three core network model, both the ventral striatum and the amygdala are considered part of the salience (and emotion) network. Connections between limbic structures such as the amygdala and the VS highlight how and where affective processes could influence action (Mogenson et al., 1980, Mogenson and Nielsen, 1984). The ventral striatum includes the nucleus accumbens, which is a critical part of the mesotelencephalic dopaminergic reward system. Dopamine function in reward-related processes, and in particular disruption of these in animal models, led to the development of the anhedonia hypothesis by Wise and colleagues (Wise, 1982, Wise, 1985). Subsequent observations of VS dysfunction patients with MDD are interpreted in light of this hypothesis (Epstein et al., 2006, Keedwell et al., 2005, Pizzagalli et al., 2009), including emergence early in the course of the illness.
In contrast, the amygdala is important for the detection of salient stimuli in the environment, including facial expressions, often of a negative or threatening valence (Adolphs et al., 1994, Fu et al., 2008). It functions as a low signal detection system to orient the organism toward such potentially important stimuli (Holland and Gallagher, 1999, Whalen, 1998) so that the correct behavior can be enacted. Amygdala activity has been shown to be heightened in active MDD (Fu et al., 2008, Sheline et al., 2010, Surguladze et al., 2005). SN dysfunction and disrupted connectivity could thus contribute to emotion regulation difficulties in MDD via biasing attention to emotional stimuli, even those below the level of conscious awareness (Victor et al., 2010), and via connections to autonomic regions involved in the psychophysiological emotional response (Drevets et al., 2008, Price and Drevets, 2010).
Given previous findings indicating the importance of sgACC connectivity in active MDD as a predictor of depression refractoriness and severity (Greicius et al., 2007, Connolly et al., 2013, Mayberg et al., 1999), it may be surprising that the current study did not identify sgACC connectivity as a top feature. It is possible that sgACC activity is a state effect of MDD, as sgACC hyperactivity has been reported to normalize following treatment of MDD, and is even present in sad mood induction (Fu et al., 2008, Mayberg et al., 1999, Sheline et al., 2010, Victor et al., 2013). In terms of connectivity, our results failed to support previous findings of studies using machine learning that the connectivity of the sgACC is able to differentiate active MDD patients from HCs with a high degree of accuracy (e.g. Zeng et al., 2014). However our results do support findings that the amygdala is hyperconnected in MDD (e.g. Jin et al., 2011), and suggest that this may be either a risk factor or trait effect, as it persists into the remitted state.
4.3. Lack of SN–CCN connectivity predictor
Studies that have investigated abnormalities in regional functional connectivity in active MDD have reported reduced connectivity between cortical regulatory areas and the amygdala during task performance (Carballedo et al., 2011, Dannlowski et al., 2009, Siegle et al., 2007, Moses-Kolko et al., 2010), and activation in the nucleus accumbens as well as the amygdala during successful inhibition on a cognitive control task has been found to predict post-treatment improvement in depressive symptoms (Langenecker et al., 2007). Anomalous functional connections may be reflective of disrupted interactions between the SN and CCN (Dannlowski et al., 2009). The DLPFC and VLPFC/insula are important for both explicit and implicit cognitive control of limbic regions and the correlated emotional responses (Langenecker et al., 2014). As such, we were surprised that there was not a significant discriminant feature connection between a CCN region and a SN region, such as the amygdala. Thus, anomalous functional connectivity between regions of the salience and cognitive control networks in MDD may be a state effect, or may emerge with longer standing illness course (more episodes).
4.4. Limitations
Multivariate pattern classification of resting state functional MRI data is a challenging task due to small samples with expensive data collection, noisy and high dimensionality of the data, and individual variability. We note several limitations in the current study. The first is a lack of an evaluation data set with which to test our methods and confirm the findings. Future research can confirm classification results with larger sample sizes and/or multicenter imaging data. The second limitation is that we used seed-based functional connectivity patterns. Using a whole brain functional connectivity measure may improve our classification accuracy. There is need for acquiring additional neuroimaging evidence of brain abnormalities including differences in structure that may discriminate individuals with MDD (active or remitted) from HCs. Using the current method, we cannot infer the directionality of connections, and therefore future research could use measures of effective connectivity such as Granger causality (Van den Heuvel and Hulshoff Pol, 2010). Another limitation is that the present study was cross-sectional, therefore we cannot dissociate the scar effects of illness from the risk factors for MDD. We will be following this sample longitudinally to determine which participants relapse, and which features of brain connectivity during remission can be used to classify individuals into relapsing or resilient groups, similar to what has been attempted using structural MRI to predict treatment response in MDD using SVM (Gong et al., 2011). Knowing which individuals are more likely to relapse will allow for more intensive therapies to be targeted to this group in order to avoid the chronic scarring effects of illness. It should be noted that while all of our sample were medication free for the previous month, only some of our sample were medication naïve, so we were unable to dissociate the effects of previous medication use. Future research can combine resting state functional connectivity and structural abnormalities to obtain more reliable clinical diagnosis of MDD. Finally, it is unfortunate that none of the connections observed with the SVM approach were unique from those captured in traditional univariate analyses.
5. Conclusions and future directions
The present study is the first to demonstrate that new SVM techniques can be used with rs-fMRI data to distinguish individuals with a history of MDD from controls with reasonably good accuracy. Efficient information and emotion processing requires coordination between a series of networks, even in the resting state (Di and Biswal, 2013, Spreng et al., 2013). Impaired internetwork connectivity at rest, even for individuals in the remitted state of MDD, were strong predictors of illness history. Our data suggests that there is remarkable potential in combining rs-fMRI data with machine learning-based techniques for advancing our understanding of network-level differences in MDD. It is hoped that machine learning techniques such as those used in the present study will develop further to be able to identify biomarkers that can inform us about individual prognosis such as relapse and thus guide therapeutic decisions and assist in psychiatric research into MDD.
Acknowledgements
This work was supported by NIMH BRAINS RO1 to SAL (MH081911). We thank the staff and faculty at the University of Michigan fMRI Laboratory and the Center for Magnetic Resonance Research at the University of Illinois at Chicago. We thank Laura Gabriel, Michelle Kassel, Anne Weldon, Amanda Baker, Bethany Pester, and Kaley Angers for assistance in data collection for this study.
References
- Adolphs R., Tranel D., Damasio H., Damasio A.R. Impaired recognition of emotion in facial expressions following bilateral damage to the human amygdala. Nature. 1994;372(6507):669–672. doi: 10.1038/372669a0. [DOI] [PubMed] [Google Scholar]
- Alexopoulos G.S., Hoptman M.J., Kanellopoulos D., Murphy C.F., Lim K.O. Functional connectivity in the cognitive control network and the default mode network in late-life depression. J. Affect. Disord. 2012;139:56–65. doi: 10.1016/j.jad.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (APA) 4th ed. American Psychiatric Association; Washington (DC): 1994. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- Atluri G., Padmanabhan K., Fang G., Steinbach M., Petrella J.R., Lim K. Complex biomarker discovery in neuroimaging data: finding a needle in a haystack. Neuroimage Clin. 2013;3:123–131. doi: 10.1016/j.nicl.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y., Restom K., Liau J., Liu T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench, Friston, Brown, Scott, Frackowiak, Dolan The anatomy of melancholia: focal abnormalities of cerebral blood flow in major depression. Psychol. Med. 1992;22(3):607–615. doi: 10.1017/s003329170003806x. [DOI] [PubMed] [Google Scholar]
- Berman M., Nee D., Peltier S., Kross E., Deldin P., Jonides J. Depression, rumination, and the default network. Soc. Cogn. Affect. Neurosci. 2010;6(5):548–555. doi: 10.1093/scan/nsq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm R.L., Clark C.R., McFarlane A.C., Moores K.A., Shaw M.E. Default network connectivity during a working memory task. Hum. Brain Mapp. 2011;32:1029–1035. doi: 10.1002/hbm.21090. (brain/awn23918835868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briceño E.M., Weisenback S.L., Rapport L.J., Hazlett K.E., Bieliauskas L.A., Haase B.D., Ransom M.T., Brinkman M.L., Peciña M., Schteingart D.S., Starkman M.N., Giordani B., Welsh R.C., Noll R.C., Zubieta J.-K., Langenecker S.A. Shifted inferior laterality in women with major depressive disorder is related to emotion-processing deficits. Psychol. Med. 2013;43(7):1433–1445. doi: 10.1017/S0033291712002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyd S.J., Demanuele C., Debener S., Helps S.K., James C.J. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci. Biobehav. Rev. 2009;33:279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Cao X., Liu Z., Xu C., Li J., Gao Q., Sun N. Disrupted resting-state functional connectivity of the hippocampus in medication-naive patients with major depressive disorder. J. Affect. Disord. 2012;141:194–203. doi: 10.1016/j.jad.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Cao L., Guo S., Xue Z., Hu Y., Liu H., Mwansisya T.E., Pu W., Yang B., Liu C., Feng J., Chen E.Y.H., Liu Z. Aberrant functional connectivity for diagnosis of major depressive disorder: A discriminant analysis. Psychiatry Clin. Neurosci. 2014;68:110–119. doi: 10.1111/pcn.12106. [DOI] [PubMed] [Google Scholar]
- Carballedo A., Scheuerecker J., Meisenzahl E., Schoepf V., Bokde A., Moller H.J., Frodl T. Functional connectivity of emotional processing in depression. J. Affect. Disord. 2011;134(1–3):272279. doi: 10.1016/j.jad.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Casanova Ramon, Whitlow C.T., Wagner Benjamin, Williamson Jeff, Shumaker S.A., Maldiian J.A., Espeland M.A. High dimensional classification of structural MRI Alzheimer's disease data based on large scale regularization. Front Neuroinform. 2011;5:22. doi: 10.3389/fninf.2011.00022. (Published online 2011 October 14, PMCID: PMC3193072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Pereira F., Lee W., Strother S., Mitchell T. Exploring predictive and reproducible modeling with the single-subject FIAC dataset. Hum. Brain Mapp. 2006;27(5):452–461. doi: 10.1002/hbm.20243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly C.G., Wu J., Ho T.C., Hoeft F., Wolkowitz O. Resting-state functional connectivity of subgenual anterior cingulate cortex in depressed adolescents. Biol. Psychiatry. 2013;12:898–907. doi: 10.1016/j.biopsych.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock R.C., Holtzheimer P.E., III, Hu X.P., Mayberg H.S. Disease state prediction from resting state functional connectivity. Magn. Reson. Med. 2009;62:1619–1628. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannlowski, Ohrmann, Konrad, Domschke, Bauer, Kugel Reduced amygdala-prefrontal coupling in major depression: association with MAOA genotype and illness severity. Int. J. Neuropsychopharmacol. 2009;12(1):11–22. doi: 10.1017/S1461145708008973. [DOI] [PubMed] [Google Scholar]
- De Martino F., Valente G., Staeren N., Ashburner J., Goebel R., Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. Neuroimage. 2008;43(1):44–58. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- Di X., Biswal B.B. Modulatory interactions of resting state brain functional connectivity. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets W.C., Price J.L., Furey M.L. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct. Funct. 2008;213(1–2):93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesnay E., Cachia A., Boddaert N., Chabane N., Mangin J.-F., Martinot J.-L., Brunelle F., Zilbovicius M. Feature selection and classification of imbalanced datasets: application to PET images of children with autistic spectrum disorders. Neuroimage. 2011;57:1003–1014. doi: 10.1016/j.neuroimage.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Epstein J., Pan H., Kocsis J.H., Yang Y.H., Butler T., Chusid J.…Silbersweig D.A. Lack of ventral striatal response to positive stimuli in depressed versus normal subjects. Am. J. Psychiatry. 2006;163(10):1784–1790. doi: 10.1176/ajp.2006.163.10.1784. [DOI] [PubMed] [Google Scholar]
- Fair D.A., Nigg J.T., Iyer S. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Frontiers in Systems Neuroscience. 2012;6:80. doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M.D., Zhang D., Snyder A.Z., Raichle M.E. The global signal and observed anticorrelated resting state brain networks. J. Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C.H.Y., Mourao-Miranda J., Costafrecla S.G., Khanna A., Marquand A.F., Williams S.C.R., Brammer M.J. Pattern classification of sad facial processing: toward the development of neurobiological markers in depression. Biol. Psychiatry. 2008;63(7):656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Gong, Wu, Scarpazza, Lui, Jia Prognostic prediction of therapeutic response in depression using high-field MR imaging. Neuroimage. 2011;55(4):1497–1503. doi: 10.1016/j.neuroimage.2010.11.079. [DOI] [PubMed] [Google Scholar]
- Greicius M.D., Flores B.H., Menon V., Glover G.H., Solvason H.B. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol. Psychiatry. 2007;62:429–437. doi: 10.1016/j.biopsych.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Cao, Liu, Li, Chen, Zhang Machine learning classifier using abnormal brain network topological metrics in major depressive disorder. Neuroreport. 2012;23(17):1006–1011. doi: 10.1097/WNR.0b013e32835a650c. [DOI] [PubMed] [Google Scholar]
- Guo, Cheng, Cao, Xiang, Chen, Zhang Resting state functional connectivity abnormalities in first-onset unmedicated depression. Neural Regeneration Research. 2014;9(2):153–163. doi: 10.4103/1673-5374.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I., Elisseeff A.E. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003;3:1157–1182. [Google Scholar]
- Guyon I., Elisseeff A.E. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003;3:1157–1182. [Google Scholar]
- Haller S., Lovblad K.O., Giannakopoulos P., Van De Ville D. Multivariate pattern recognition for diagnosis and prognosis in clinical neuroimaging: state of the art, current challenges and future trends. Brain Topogr. 2014 May;27(3):329–337. doi: 10.1007/s10548-014-0360-z. Epub 2014 Mar 28. [DOI] [PubMed] [Google Scholar]
- Hamilton M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, Furman, Chang, Thomason, Dennis, Gotlib Default-mode and task positive network activity in major depressive disorder: Implications for adaptive and maladaptive rumination. Biol. Psychiatry. 2011;70(4):327–337. doi: 10.1016/j.biopsych.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.P., Etkin A., Furman D.J., Lemus M.G., Johnson R.F. Functional neuroimaging of major depressive disorder: a meta-analysis and new integration of base line activation and neural response data. Am. J. Psychiatry. 2012;169:693–703. doi: 10.1176/appi.ajp.2012.11071105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Eisen M.B., Alizadeh A., Levy R., Staudt L., Chan W.C., Botstein D., Brown P. Gene shaving as a method for identifying distinct sets of genes with similar expression patterns. Genome Biol. 2000;1(2):1–0003. doi: 10.1186/gb-2000-1-2-research0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Botstein D., Brown P. Supervised harvesting of expression trees. Genome Biol. 2001;2(1) doi: 10.1186/gb-2001-2-1-research0003. research0003.1–0003.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P.C., Gallagher M. Amygdala circuitry in attentional and representational processes. Trends Cogn. Sci. 1999;3(2):65–73. doi: 10.1016/s1364-6613(98)01271-6. [DOI] [PubMed] [Google Scholar]
- Horn D.I., Yu C., Steiner J., Buchmann J., Kaufmann J., Osoba A., Eckert U., Zierhut K.C., Schiltz K., He H., Biswal B., Bogerts B., Walter M. Glutamatergic and resting-state functional connectivity correlates of severity in major depression - the role of pregenual anterior cingulate cortex and anterior insula. Front. Syst. Neurosci. 2010 Jul 15;4 doi: 10.3389/fnsys.2010.00033. pii: 33. eCollection 2010. PMID: 20700385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.H., Jenkins L.M., Gabriel L.B., Barba A., Ryan K.A., Weisenbach S.L. Increased coupling of intrinsic networks in remitted depressed youth predicts rumination and cognitive control. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Gao, Chen, Ma, Netra, Wang A preliminary study of the dysregulation of the resting networks in first-episode medication naïve adolescent depression. Neurosci. Lett. 2011;503:105–109. doi: 10.1016/j.neulet.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Jo H.J., Gotts S.J., Reynolds R.C., Bandettini P.A., Martin A. Effective preprocessing procedures virtually eliminate distance-dependent motion artifacts in resting state FMRI. J. Appl. Math. 2013;2013:1–9. doi: 10.1155/2013/935154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser R.H., Andrews J.R., Wager T.D., Pizzagalli D.A. Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry. 2015 doi: 10.1001/jamapsychiatry.2015.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedwell, Andrew, Williams, Brammer, Phillips The neural correlates of anhedonia in major depressive disorder. Biol. Psychiatry. 2005;58(11):843–853. doi: 10.1016/j.biopsych.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Kelly A.M., Di Martino A., Uddin L.Q., Shehzad Z., Gee D.G. Development of anterior cingulate functional connectivity from late childhood to early adulthood. Cereb. Cortex. 2009;19:640–657. doi: 10.1093/cercor/bhn117. [DOI] [PubMed] [Google Scholar]
- Klöppel S., Abdulkadir A., Jack C.R., Jr., Koutsouleris N., Mourao-Miranda J., Vemuri P. Diagnostic neuroimaging across diseases. Neuroimage. 2011;61:457–463. doi: 10.1016/j.neuroimage.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar M., Ram K., Williams L., Gatt J., Grieve S. Establishing the resting state default mode network derived from functional magnetic resonance imaging tasks as an endophenotype: a twins study. Hum. Brain Mapp. 2014;35(8):3893–3902. doi: 10.1002/hbm.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConte S.M., Peltier S.J., Hu X.P. Real-time fMRI using brain-state classification. Hum. Brain Mapp. 2007;28(10):1033–1044. doi: 10.1002/hbm.20326. (PubMed: 17133383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenecker S.A., Kennedy S.E., Guidotti L.M., Briceno E.M., Own L.S. Frontal and limbic activation during inhibitory control predicts treatment response in major depressive disorder. Biol. Psychiatry. 2007;62:1272–1280. doi: 10.1016/j.biopsych.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenecker SA, Dawson EL, Mickey BJ, Ransom MT, Walker SJ, Meyers KK, Haxlett KE, Weldon AL, Giordani B, Zubieta JK (2014), Cognitive control and neuronal functioning biomarkers as predictors of treatment response in major depression. Biol. Psychiatry volume: 7 Issue: 9.
- Marchetti I., Koster E.H., Sonuga-Barke E.J., De Raedt R. The default mode network and recurrent depression: a neurobiological model of cognitive risk factors. Neuropsychol. Rev. 2012;22:229–251. doi: 10.1007/s11065-012-9199-9. [DOI] [PubMed] [Google Scholar]
- Margulies D.S., Kelly A.M., Uddin L.Q., Biswal B.B., Castellanos F.X. Mapping the functional connectivity of anterior cingulate cortex. Neuroimage. 2007;37:579–588. doi: 10.1016/j.neuroimage.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Marquand A.F., O'Daly O.G., De Simoni S., Alsop D.C., Maguire R.P., Williams S.C.R., Zelaya F.O., Mehta M.A. Dissociable effects of methylphenidate, atomoxetine and placebo on regional cerebral blood flow in healthy volunteers at rest: a multi-class pattern recognition approach. Neuroimage. 2012;60:1015–1024. doi: 10.1016/j.neuroimage.2012.01.058. (PubMed: 22266414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg, Liotti, Brannan, McGinnis, Mahurin, Jerabek Reciprocal limbic-cortical function and negative mood: converging PET findings in depression and normal sadness. Am. J. Psychiatry. 1999;156(5):675–682. doi: 10.1176/ajp.156.5.675. [DOI] [PubMed] [Google Scholar]
- McCabe C., Mishor Z. Antidepressant medications reduce subcortical resting-state functional connectivity in healthy volunteers. Neuroimage. 2011;57:1317–1323. doi: 10.1016/j.neuroimage.2011.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn. Sci. 2011;15:483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Mitchell T.M., Hutchinson R., Niculescu R.S., Pereira F., Wang X., Just M., Newman S. Learning to decode cognitive states from brain images. Mach. Learn. 2004;57(1–2):145–175. [Google Scholar]
- Mogenson G.J., Nielsen M. Neuropharmacological evidence to suggest that the nucleus accumbens and subpallidal region contribute to exploratory locomotion. Behav. Neural Biol. 1984;42(1):52–60. doi: 10.1016/s0163-1047(84)90424-2. [DOI] [PubMed] [Google Scholar]
- Mogenson G.J., Jones D.L., Yim C.Y. From motivation to action: functional interface between the limbic system and the motor system. Prog. Neurobiol. 1980;14(2–3):69–97. doi: 10.1016/0301-0082(80)90018-0. [DOI] [PubMed] [Google Scholar]
- Moses-Kolko E.L., Perlman S.B., Wisner K.L., James J., Saul A.T., Phillips M.L. Abnormally reduced dorsomedial prefrontal cortical activity and effective connectivity with amygdala in response to negative emotional faces in postpartum depression. Am. J. Psychiatry. 2010;167(11):1373–1380. doi: 10.1176/appi.ajp.2010.09081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner R., Mikova O., Koutsilieri E., Saoud M., Ehlis A.C., Muller N. Consensus paper of the WFSBP task force on biological markers: biological markers in depression. World J. Biol. Psychiatry. 2007;8:141–174. doi: 10.1080/15622970701263303. [DOI] [PubMed] [Google Scholar]
- Mourao-Miranda J, Bokde AL, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: support Vector Machine on functional MRI data. Neuroimage Volume 28, Issue 4, December 2005, Pages 980–995 Special Section: Social Cognitive Neuroscience. [DOI] [PubMed]
- Mourao-Miranda J., de Oliveira L., Ladouceur C.D., Marchand A., Birmaher B., Axelson D.A., Phillips M.L. Pattern recognition and functional neuroimaging help to discriminate healthy adolescents at risk for mood disorders from low risk adolescents. PLoS One. 2012;7(2):e29482. doi: 10.1371/journal.pone.0029482. 10.1371/journal.pone.0029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi B., Tian S., Soares Jair C. Springer Science + Business Media; New York: 2013. A Review of Feature Reduction Techniques in Neuroimaging Neuroinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger J.I., Jr., Blehar M.C., Kaufmann C.A., York-Cooler C., Simpson S.G. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch. Gen. Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Orru G., Pettersson-Yeo W., Marquand A.F., Sartori G., Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci. Biobehav. Rev. 2012;36:1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Pannekoek J.N., Veer I.M., van Tol M.J., van der Werff S.J., Demenescu L.R. Aberrant limbic and salience network resting-state functional connectivity in panic disorder without comorbidity. J. Affect. Disord. 2013;145:29–35. doi: 10.1016/j.jad.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Peters E.M., Balbuena L., Marwaha S., Baetz M., Bowen R. Mood instability and impulsivity as trait predictors of suicidal thoughts. Psychol Psychother Theory Res Pract. 2016;89:435–444. doi: 10.1111/papt.12088. [DOI] [PubMed] [Google Scholar]
- Pizzagalli D.A. Frontocingulate dysfunction in depression: toward biomarkers of treatment response. Neuropsychopharmacology. 2011;36:183–206. doi: 10.1038/npp.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli Holmes, Dillon Goetz, Birk Bogdan. Reduced caudate and nucleus accumbens response to rewards in unmedicated subjects with major depressive disorder. Am. J. Psychiatry. 2009;166(6):702–710. doi: 10.1176/appi.ajp.2008.08081201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Drevets W.C. Neurocircuitry of mood disorders. Neuropsychopharmacology. 2010;35(1):192–216. doi: 10.1038/npp.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle, MacLeod, Snyder, Powers, Gusnard, Shulman A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A., Lee Y., Gass A., Monsch A. Paper Presented at: Engineering in Medicine and Biology Society, EMBC, 2011 Annual International Conference of the IEEE (IEEE) 2011. Classification of Alzheimer's Disease from structural MRI using sparse logistic regression with optional spatial regularization. (PubMed) [DOI] [PubMed] [Google Scholar]
- Sambataro F., Wolf N.D., Pennuto M., Vasic N., Wolf R.C. Revisiting default mode network function in major depression: evidence for disrupted subsystem connectivity. Psychol. Med. 2013;44:2041–2051. doi: 10.1017/S0033291713002596. [DOI] [PubMed] [Google Scholar]
- Schneider B., Prvulovic D. Novel biomarkers in major depression. Curr. Opin. Psychiatry. 2013;26:47–53. doi: 10.1097/YCO.0b013e32835a5947. [DOI] [PubMed] [Google Scholar]
- Seeley W.W., Menon V., Schatzberg A.F., Keller J., Glover G.H., Kenna H. Dissociable intrinsic connectivity networks for salience processing and executive control. J. Neurosci. 2007;27(9):2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline Y.I., Price J.L., Yan Z.Z., Mintun M.A. Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11020–11025. doi: 10.1073/pnas.1000446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Wang L., Liu Y., Hu D. Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. Neuroimage. 2010;49:3110–3121. doi: 10.1016/j.neuroimage.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Siegle G.J., Carter C.S., Thase M.E. Use of FMRI to predict recovery from unipolar depression with cognitive behavior therapy. Am. J. Psychiatry. 2006;163:735–738. doi: 10.1176/ajp.2006.163.4.735. [DOI] [PubMed] [Google Scholar]
- Siegle, Thompson, Carter, Steinhauer, Thase Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: related and independent features. Biol. Psychiatry. 2007;61(2):198–209. doi: 10.1016/j.biopsych.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Spreng RN, Sepulcre J, Turner, GR, Stevens, WD, Schacter DL, (2013) Intrinsic architecture underlying the relations among the default, dorsal attention, and frontoparietal control networks of the human brain J. Cogn. Neurosci. Volume: 25 Issue: 1 Pages: 74–86. [DOI] [PMC free article] [PubMed]
- Strigo I.A., Simmons A.N., Matthews S.C., Craig A.D., Paulus M.P. Association of major depressive disorder with altered functional brain response during anticipation and processing of heat pain. Arch. Gen. Psychiatry. 2008;65(11):1275–1284. doi: 10.1001/archpsyc.65.11.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermann B., Beverborg O.L., Pfleiderer B. Toward literature-based feature selection for diagnostic classification: a meta-analysis of resting state fMRI in depression. Front. Hum. Neurosci. 2014;8(692):1–12. doi: 10.3389/fnhum.2014.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surguladze S., Brammer M.J., Keedwell P., Giampietro V., Young A.W., Travis M.J., Williams S.C., Phillips M.L. A differential pattern of neural response toward sad versus happy facial expressions in major depressive disorder. Biol. Psychiatry. 2005;57(3):201–209. doi: 10.1016/j.biopsych.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Tabachnick B.G., Fidell L.S. Pearson Education; Boston, MA: 2007. Using Multivariate Statistics. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B. 1996;58:267–288. [Google Scholar]
- Tremblay L.K., Naranjo C.A., Graham S.J. Functional neuroanatomical substrates of altered reward processing in major depressive disorder revealed by a dopaminergic probe. Arch. Gen. Psychiatry. 2005;62:1228–1236. doi: 10.1001/archpsyc.62.11.1228. [DOI] [PubMed] [Google Scholar]
- Van den Heuvel M.P., Hulshoff Pol H.E. Exploring the brain network a review on resting state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Vapnik V. Springer-Verlag; New York: 1995. The Natures of Statistical Learning Theory. [Google Scholar]
- Victor T.A., Furey M.L., Fromm S.J., Ohman A., Drevets W.C. Relationship between amygdala responses to masked faces and mood state and treatment in major depressive disorder. Arch. Gen. Psychiatry. 2010;67(11):1128–1138. doi: 10.1001/archgenpsychiatry.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor T.A., Furey M.L., Fromm S.J., Ohman A., Drevets W.C. Changes in the neural correlates of implicit emotional face processing during antidepressant treatment in major depressive disorder. Int. J. Neuropsychopharmacol. 2013;16(10):2195–2208. doi: 10.1017/S146114571300062X. [DOI] [PubMed] [Google Scholar]
- Votruba K.L., Langenecker S.A. Factor structure, construct validity, and age- and education-based normative data for the Parametric Go/No-Go Test. J. Clin. Exp. Neuropsychol. 2013;35:132–146. doi: 10.1080/13803395.2012.758239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenbach S.L., Marshall D., Weldon A.L., Ryan K.A., Vederman A.C., Kamali M., Zubieta J.-K., McInnis M.G., Langenecker S.A. The double burden of age and disease on cognition and quality of life in bipolar disorder. International Journal of Geriatric Psychiatry. 2014;29:952–961. doi: 10.1002/gps.4084. [DOI] [PubMed] [Google Scholar]
- Whalen P.J. Fear, vigilance, and ambiguity: Initial neuroimaging studies of the human amygdala. Curr. Dir. Psychol. Sci. 1998;7(6):177–188. [Google Scholar]
- Wise R.A. Neuroleptics and operant behavior: the anhedonia hypothesis. Behav. Brain Sci. 1982;5(1):39–53. [Google Scholar]
- Wise R.A. The anhedonia hypothesis: mark III. Behav. Brain Sci. 1985;8(1):178–184. [Google Scholar]
- Zarogianni E., Moorhead T.W., Lawrie S.M. Towards the identification of imaging biomarkers in schizophrenia, using multivariate pattern classification at a single-subject level. Neuroimage Clin. 2013;3:279–289. doi: 10.1016/j.nicl.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Shen, Liu, Hu Unsupervised classification of major depression using functional connectivity MRI. Hum. Brain Mapp. 2014;35:1630–1641. doi: 10.1002/hbm.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Xiao J., Liao J., Zhong M., Wang W. (2012). Evidence of a dissociation pattern in resting-state default mode network connectivity in first- episode, treatment-naive major depression patients. Biol. Psychiatry. 2012 Apr 1;71(7):611–617. doi: 10.1016/j.biopsych.2011.10.035. Epub 2011 Dec 15. [DOI] [PubMed] [Google Scholar]
- Zou H., Hastie T. Regularization and variable selection via the Elastic Net. J. R. Stat. Soc. Ser. B. 2005;67:301–320. [Google Scholar]