ABSTRACT
Progressive T cell depletion during chronic human immunodeficiency virus type 1 (HIV) infection is a key mechanism that leads to the development of AIDS. Recent studies have suggested that most T cells in the tissue die through pyroptosis triggered by abortive infection, i.e., infection of resting T cells in which HIV failed to complete reverse transcription. However, the contribution of abortive infection to T cell loss and how quickly abortively infected cells die in vivo, key parameters for a quantitative understanding of T cell population dynamics, are not clear. Here, we infected rhesus macaques with simian-human immunodeficiency viruses (SHIV) and followed the dynamics of both plasma SHIV RNA and total cell-associated SHIV DNA. Fitting mathematical models to the data, we estimate that upon infection a majority of CD4+ T cells (approximately 65%, on average) become abortively infected and die at a relatively high rate of 0.27 day−1 (half-life, 2.6 days). This confirms the importance of abortive infection in driving T cell depletion. Further, we find evidence suggesting that an immune response may be restricting viral infection 1 to 3 weeks after infection. Our study serves as a step forward toward a quantitative understanding of the mechanisms driving T cell depletion during HIV infection.
IMPORTANCE In HIV-infected patients, progressive CD4+ T cell loss ultimately leads to the development of AIDS. The mechanisms underlying this T cell loss are not clear. Recent experimental data suggest that the majority of CD4+ T cells in tissue die through abortive infection, where the accumulation of incomplete HIV transcripts triggers cell death. To investigate the role of abortive infection in driving CD4+ T cell loss in vivo, we infected macaques with simian-human immunodeficiency viruses (SHIV) and followed the viral kinetics of both plasma RNA and cell-associated DNA during infection. Fitting mathematical models, we estimated that a large fraction of infected cells dies through abortive infection and has a half-life of approximately 2.6 days. Our results provide the first in vivo quantitative estimates of parameters characterizing abortive infection and support the notion that abortive infection represents an important mechanism underlying progressive CD4+ T cell depletion in vivo.
KEYWORDS: abortive infection, SHIV, mathematical modeling, viral kinetics
INTRODUCTION
CD4+ T cells are the primary targets for human immunodeficiency virus type 1 (HIV) infection. The progressive loss of CD4+ T cells in patients chronically infected with HIV is a key mechanism that underlies the development of AIDS (1, 2). Both direct and indirect viral cytopathic effects are involved in CD4+ T cell death. Upon infection, activated CD4+ T cells become productively infected and die rapidly in vivo at a rate of approximately 1 day−1 (3). This form of death is a result of a variety of cytopathic effects, such as viral DNA integration triggering apoptosis (4) and viral peptide presentation on the cell surface, which leads to cytotoxic T lymphocyte-mediated killing (5, 6). In contrast, resting CD4+ T cells are refractory to HIV infection (7). Entry of HIV into these cells often leads to incomplete reverse transcription (8–10). It has been suggested that upon contact with productively infected cells in tissue, cell-to-cell transmission of virus leads to the accumulation of a large amount of incomplete viral DNA transcripts in resting CD4+ T cells, and this accumulation triggers cell death through caspase-1-mediated pyroptosis (11–13). Upon death, these cells further release inflammatory cytokines, including interleukin-1β. The resulting inflammation likely attracts more CD4+ T cells to the site of infection, further fueling HIV infection (13, 14). Ex vivo studies of CD4+ T cells taken from the lymphoid tissue of infected patients suggest that the majority of cells die as a result of abortive infection through cell-to-cell transmission (10, 12, 13). Abortive infection is therefore suggested to be a major mechanism that leads to chronic inflammation, CD4+ T cell depletion, dysregulation of T cell homeostasis, and, ultimately, AIDS (10, 12, 13, 15).
The importance of abortive infection in determining T cell population dynamics raises the question of how quickly abortively infected cells die in vivo and to what extent abortive infection drives T cell depletion. To address these questions, we studied acute viral dynamics in rhesus macaques infected with simian-human immunodeficiency virus (SHIV) 162p3 (SHIV162P3), a chimeric simian-human immunodeficiency virus strain that contains an R5-tropic HIV envelope. SHIV162P3 can be easily transmitted mucosally at low doses, which leads to a dramatic loss of CD4+ T cells in the gut and a gradual loss of peripheral CD4+ T cells (16, 17). This system thus provides a useful model for studying early acute HIV infection. Also, the massive depletion of gut CD4+ T cells provides an opportunity to examine through modeling the impact of abortive infection on viral and cell kinetics. We monitored viral RNA in plasma and total cell-associated SHIV DNA (CA-DNA) in peripheral blood mononuclear cells (PBMCs) in 20 macaques for a period of 15 to 50 weeks and used these data to estimate the rate and fraction of cell death occurring by abortive infection.
RESULTS
Viral RNA and DNA dynamics during SHIV infection.
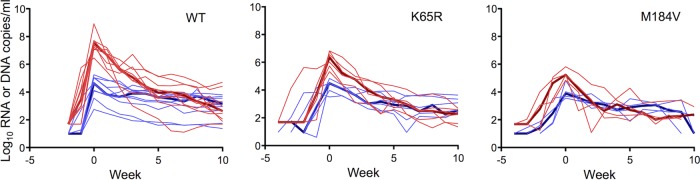
We infected rhesus macaques rectally with wild-type (WT) SHIV162P3 (SHIV162P3WT; n = 10 macaques) or with isogenic SHIV162P3 mutants containing the reverse transcriptase (RT) mutation K65R (SHIV162P3K65R; n = 6) or M184V (SHIV162P3M184V; n = 4). In HIV and simian immunodeficiency virus (SIV), K65R and M184V are associated with resistance to tenofovir and emtricitabine, respectively. To define viral kinetics during acute infection, we measured the levels of SHIV RNA in plasma and total cell-associated SHIV DNA in PBMCs. Figure 1 shows the virus dynamics during the first 10 weeks of acute infection. Overall, SHIV viremia increased rapidly to a high peak (from 105 to 108 copies/ml) soon after infection. Peak plasma RNA levels in macaques infected with the K65R mutant (median = 6.3 log10 copies/ml; minimum and maximum = 5.3 and 6.8 log10 copies/ml, respectively) or the M184V mutant (median = 5.2 log10 copies/ml; minimum and maximum = 4.8 and 5.8 log10 copies/ml, respectively) were significantly lower than those in macaques infected with the WT (P = 0.0017 and P = 0.0020, respectively), likely reflecting the high fitness cost conferred by the K65R and M184V mutations (18–20). After the peak load was reached, the SHIV viral load rapidly decreased to a very low level and stayed roughly constant (at a quasi-steady-state level) in all 20 macaques, irrespective of the strain of the virus (Fig. 1).
FIG 1.
Dynamics of viral RNA (red) and cell-associated SHIV DNA (blue) in plasma measured in 20 rhesus macaques. Measurements are categorized according to the viral strain with which each macaque was challenged. Thin lines, the dynamics in each macaque; thick lines, the median of the dynamics in each category. Note that the x axis, i.e., weeks, is set such that the viral RNA level peaks at 0 week for each macaque.
In contrast to the RNA levels, the CA-DNA levels varied by several orders of magnitude across different macaques, reaching peak levels at times similar to those at which peak viral RNA levels were reached. Peak CA-DNA levels did not differ between WT and drug-resistant virus infections (Fig. 1). After the peak, CA-DNA levels in general exhibited a two-phase decline pattern, where the first phase was rapid and the second phase was very slow.
Construction of a basic mathematical model for SHIV infection dynamics.
To understand the mechanisms that drive the patterns seen in the data, we constructed a viral dynamic model, which we term the basic model. Because SHIV162P3 primarily infects CD4+ T cells in the gut (17), we considered the dynamics of CD4+ T cells in the blood as well as in the gut. The model keeps track of the concentrations of target CD4+ T cells (T1 and T2 in the blood and the gut, respectively), productively infected cells (I1 and I2 in the blood and the gut, respectively), long-lived cells (M1 and M2 in the blood and the gut, respectively), and virus (V). Here, we let the long-lived cell compartment include long-lived infected cells that produce virus (at a rate lower than that for productively infected cells), defectively infected cells, latently infected cells, and cells that have unintegrated DNA. Note that for simplicity we do not explicitly model the transport of virus between the blood and the gut separately; instead, we assume that they reach equilibrium quickly. This is a good assumption, given that the time scale of the infection dynamics that we study here is in weeks, whereas virus is rapidly disseminated into different compartments in hours or days. In this model, we assume that abortively infected cells are at a negligible level, and thus, they are not modeled explicitly. The ordinary differential equations (ODEs) for the model are as follows:
| (1) |
In the model, target cells die at per capita rate d. They move from the blood to the gut at rate kb and from the gut to the blood at rate kg. The scaling parameter r in the model reflects the ratio of the volume of the blood over that of the gut. Since SHIV162P3 primarily infects CD4+ T cells expressing the α4β7 integrin, which are generated and mostly reside in the gut (21), we assume for simplicity that there is a constant generation of target cells in the gut at rate λ but no target cell generation in the blood. Note that the assumption that target cells are generated in the blood instead does not affect the T cell dynamics, since T cells are rapidly transported between the blood and the gut compartment.
Virus infects target cells, which leads to productively infected cells (at rate βI) and long-lived cells (at rate βM). Previous work (22) has suggested that viral infectivity decreases in a time-dependent manner, possibly due to the generation of virus-specific antibodies. Here we use the time (t)-dependent function Γ(t) to describe the decrease in viral infectivity. Specifically, we assume that after an initial delay, τ, virus infectivity decreases exponentially at rate k to a constant level, and γ is the long-term infection constant relative to the time of initial infection.
Productively infected cells and long-lived infected cells die at rates δ and dM, respectively. The total amount of cell-associated SHIV DNA in the blood (which was measured in our experiment) is calculated as the sum of the numbers of productively infected and long-lived cells in the blood. Virus is produced from productively infected cells at per capita rate p and from long-lived infected cells at per capita rate pM. Viruses are cleared at per capita rate c.
We fitted this model to the data from all 20 macaques (see Fig. S1 in the supplemental material; see also Table S1 for best-fit parameter values). In general, this model describes the viral RNA data well: it describes the high peak level of viremia and the rapid and high magnitude of the decrease in the viral load to a low quasi-equilibrium level afterwards. The rapid viral load decrease is driven by a combination of the death of productively infected cells and the lack of newly infected cells. Our model predicts that this lack of newly infected cells is a result of the depletion of target cells and a decrease in viral infection. To confirm the importance of the decrease in viral infectivity to explain the data, we fitted a variation of the model assuming a constant, Γ [i.e., Γ(t) = 1], i.e., constant infectivity, instead and found that this model variation fits the data poorly (Fig. S2 and Table S2).
Despite the agreement of the model with the viral RNA data, the model does not describe the cell-associated proviral DNA data well (Fig. S1). As described above, an interesting feature in the CA-DNA data is that in most of the macaques, the CA-DNA concentration exhibits two-phase declines after the peak viremia, i.e., a fast first-phase decline and a slower second-phase decline. The basic model predicts that the first-phase decline in the levels of both viral DNA and RNA is driven primarily by the death of productively infected cells, and thus, the levels of viral RNA and DNA decrease at the same rate after peak viremia. However, we compared the rate of the first-phase DNA decline with the rate of RNA decline after peak viremia in those rhesus macaques where a two-phase DNA decline was observed (Fig. 2) and found that the rate of DNA decline was lower than the rate of RNA decline in all macaques examined. This strongly suggests that the first-phase decline in CA-DNA levels is mainly reflective of the loss of a cell population other than productively infected cells.
FIG 2.
The level of viral RNA declines quicker than the level of viral CA-DNA after peak viremia in macaques, where the CA-DNA data exhibit a two-phase decline. RNA and DNA data are shown as red and blue dots, respectively. Data are normalized to the peak viral loads, and thus, the blue and red dots at the first time point, i.e., the time of peak viremia, overlap each other. The identifier for each macaque is shown at the top of each panel. Lines show the linear regression of the RNA or DNA data points.
One hypothesis is that the dynamics of latently infected cells drive the first-phase DNA decline after peak viremia. The latently infected cell population is not explicitly modeled in the basic model. To test this hypothesis, we extended the basic model to explicitly model the dynamics of the latently infected cell population using parameter values that were established previously (23) (see Table S3 for the equations defining this extended model). We found that the latently infected cells could not account for the first-phase DNA decline observed in the data (Fig. S3 and Table S4), and to a large extent, the fits resemble those shown in Fig. S1 for the basic model. This is because the frequency of latently infected cells is several orders of magnitude lower than that of long-lived and productively infected cells during the first-phase CA-DNA decline and the fact that latently infected cells are long-lived, reflective of them being resting memory cells. To keep the models simple, we do not explicitly consider this population in the following analysis; rather, we assume that the long-lived cell population includes cells that are latently infected.
Adding abortive infection to describe first-phase CA-DNA decline.
Studies on SIV and HIV infection suggest that a large fraction of CD4+ T cells is abortively infected as a result of cell-to-cell transmission of virus into quiescent cells in tissues and this abortive infection can lead to rapid cell death through pyroptosis (10, 12, 13). We reasoned that if the majority of cells are abortively infected (in contrast to the assumption in the basic model), the first-phase decline in CA-DNA levels seen in the data can be driven by the death of abortively infected cells rather than productively infected cells. To explore this possibility, we added two extra state variables to the ODEs to explicitly track the abortively infected cells in the blood and in the gut compartment (A1 and A2, respectively) and explicitly considered both cell-to-cell viral transmission and cell-free transmission. We assumed that in this model abortive infection occurs only through cell-to-cell transmission in the gut (11, 12). We call this model the abortive infection model, and the equations governing it are as follows:
| (2) |
In this model, cell-free viral transmission leads to productively infected short-lived cells, I, and long-lived infected cells, M, at rates βvI and βvM, respectively; cell-to-cell transmission in the gut leads to productively infected cells, long-lived cells, and abortively infected cells (A) at rates βII, βIM, and βIA, respectively. The available data (i.e., the total concentrations of viral RNA and CA-DNA) prevented us from estimating all 5 infectivity parameters separately. Thus, we first fixed the parameter values for cell-free transmission at low levels (βvI = 5 × 10−9 ml day−1 and βvM = 5 × 10−11 ml day−1), such that most of the infection occurs due to cell-to-cell transmission in the gut, with only a small fraction of infected cells being long-lived cells (24). We then estimated the values of the three infectivity parameters for cell-to-cell transmission together with other parameters. In addition, to keep the model simple, we assumed that the time-dependent decrease in viral infectivity was the same for the two modes of transmission, i.e., Γ1(t) and Γ2(t).
Fitting of this model to the data shows that it describes the two-phase viral DNA decline after peak viremia very well (Fig. 3 and 4; see Table 1 for best-fit parameters). As we have hypothesized, when the majority of cells are abortively infected, the first-phase viral DNA decline is mostly driven by the death of abortively infected cells, whereas the first-phase viral RNA decline is driven by the death of productively infected cells (see Fig. 3 for an example of the fitting results for macaque 34F). To quantify the contribution of abortive infection to CD4+ T cell depletion, we calculated the fraction of abortive infection in each macaque, Ψ, i.e., the ratio of abortive infection events over all infection events during the entire period of data sampling. We found that the majority of cells (for 20 macaques, the mean was 65% and the standard deviation [SD] was 17%, as quantified by the value of Ψ in Table 1) become abortively infected. We estimated that abortively infected cells and productively infected cells die at rates of 0.27 day−1 (SD, 0.14 day−1) and 0.48 day−1 (SD, 0.16 day−1), respectively. This corresponds to a half-life of 2.6 days for abortively infected cells and a half-life of 1.4 days for productively infected cells.
FIG 3.

The abortive infection model predicts that the first-phase DNA decline after peak viremia is driven by the death of abortively infected cells. The simulation results of the abortive infection model were fitted to data from a macaque (animal 34F) challenged with wild-type SHIV (see Table 1 for parameter values). (Top) Fitting of the results for SHIV RNA and DNA data; (bottom) dynamics of target cells, abortively infected cells, productively infected cells, and long-lived infected cells.
FIG 4.
Fitting results of the abortive infection model to data from all 20 macaques. The identifier for each macaque is shown at the top of each panel. Dots show the measured RNA (red) and DNA (blue) levels in the blood, and lines show predictions of the model using the best-fit parameter values reported in Table 1.
TABLE 1.
Best-fit parameter values and predicted fraction of abortive infections derived by fitting the abortive infection model to the data
| Strain | Monkey identifier | dA (day−1) | δ (day−1) | βII (10−8 ml day−1) | βIM (10−8 ml day−1) | βIA (10−8 ml day−1) | kb (day−1) | dM (day−1) | pM (day−1) | γ | k (day−1) | τ (day) | Ψa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 34F | 0.12 | 0.39 | 2.87 | 0.43 | 18.19 | 2.16 | 0.000 | 0.01 | 0.13 | 0.03 | 0.37 | 0.81 |
| AM20 | 0.13 | 0.41 | 6.74 | 0.09 | 8.12 | 0.59 | 0.002 | 0.54 | 0.06 | 0.04 | 0.86 | 0.49 | |
| AO86 | 0.37 | 0.38 | 1.13 | 0.01 | 5.29 | 25.10 | 0.002 | 0.74 | 0.08 | 0.02 | 9.46 | 0.72 | |
| 62P | 0.44 | 0.45 | 1.66 | 0.03 | 2.71 | 0.84 | 0.008 | 0.89 | 0.08 | 0.03 | 8.60 | 0.48 | |
| DM46 | 0.18 | 0.44 | 2.17 | 0.09 | 9.98 | 2.25 | 0.002 | 0.13 | 0.13 | 0.03 | 8.39 | 0.75 | |
| 33160 | 0.12 | 0.52 | 1.80 | 0.16 | 2.14 | 14.76 | 0.009 | 0.03 | 0.15 | 0.50 | 8.48 | 0.43 | |
| AG94 | 0.30 | 0.30 | 1.50 | 1.89 | 3.32 | 8.50 | 0.012 | 0.74 | 0.14 | 0.03 | 10.86 | 0.44 | |
| AI22 | 0.16 | 0.24 | 1.39 | 0.59 | 4.68 | 2.86 | 0.008 | 2.29 | 0.14 | 0.04 | 5.17 | 0.61 | |
| 35720 | 0.53 | 0.54 | 5.39 | 0.27 | 4.87 | 0.55 | 0.016 | 0.05 | 0.14 | 0.04 | 2.00 | 0.40 | |
| 19V | 0.30 | 0.38 | 1.13 | 0.06 | 5.56 | 95.02 | 0.000 | 0.16 | 0.17 | 0.03 | 18.75 | 0.73 | |
| K65R mutant | 9969 | 0.20 | 0.55 | 4.34 | 0.03 | 14.15 | 2.82 | 0.003 | 1.33 | 0.11 | 0.04 | 2.94 | 0.72 |
| RH6635 | 0.41 | 0.51 | 4.93 | 0.48 | 10.39 | 0.06 | 0.014 | 2.33 | 0.13 | 0.50 | 8.46 | 0.39 | |
| 33756 | 0.27 | 0.58 | 2.67 | 0.23 | 14.80 | 0.10 | 0.015 | 7.21 | 0.23 | 0.50 | 4.74 | 0.63 | |
| 33770 | 0.09 | 0.34 | 1.42 | 0.60 | 24.44 | 0.24 | 0.000 | 1.80 | 0.09 | 0.50 | 2.39 | 0.84 | |
| AH8F | 0.36 | 0.37 | 2.24 | 0.09 | 23.44 | 0.54 | 0.006 | 4.14 | 0.13 | 0.20 | 3.16 | 0.86 | |
| AM43 | 0.31 | 0.40 | 2.57 | 0.38 | 20.53 | 0.32 | 0.010 | 0.08 | 0.15 | 0.50 | 8.97 | 0.80 | |
| M184V mutant | 31483 | 0.30 | 0.91 | 3.87 | 0.75 | 7.11 | 0.24 | 0.000 | 1.29 | 0.09 | 0.45 | 2.32 | 0.50 |
| 4284 | 0.14 | 0.56 | 1.59 | 1.39 | 18.46 | 0.34 | 0.000 | 1.20 | 0.13 | 0.50 | 3.54 | 0.79 | |
| 4282 | 0.55 | 0.56 | 2.35 | 0.10 | 24.14 | 0.43 | 0.000 | 4.98 | 0.12 | 0.22 | 2.62 | 0.85 | |
| 33246 | 0.07 | 0.80 | 0.63 | 1.63 | 7.30 | 4.05 | 0.010 | 8.17 | 0.11 | 0.12 | 8.90 | 0.69 | |
| Meanb | 0.27 | 0.48 | 2.62 | 0.46 | 11.48 | 1.27c | 0.01 | 1.91 | 0.13 | 0.22 | 6.05 | 0.65 | |
| SDb | 0.14 | 0.16 | 1.63 | 0.55 | 7.64 | 6.67c | 0.01 | 2.40 | 0.04 | 0.22 | 4.46 | 0.17 |
Ψ, the fraction of abortive infections, which is calculated as the ratio of the number of abortive infection events over the number of all infection events during the entire period of study.
The mean and the standard deviations are calculated for those macaques in which DNA levels exhibited a 2-phase decline, unless indicated otherwise.
The geometric mean and the standard deviations of the estimated values of kb are reported, since the estimated value of kb varies over a wide range.
In the model presented in equations 2, we estimate that all infections are suppressed to low levels due to the time-dependent decrease in the cell-free infection rate. This decrease can be explained by an anti-SHIV antibody response, which typically develops a few weeks after infection (25, 26). We speculate that the decrease in the cell-to-cell infection rate can be a result of both a noncytolytic CD8+ cell-mediated antiviral response (27, 28) and a type I interferon response (29, 30). A type I interferon response can decrease the number of available target cells by generating a protective antiviral state within putative target cells (31). This type of response would also reduce the cell-free virus infection rate. To test the importance of the decrease in the cell-to-cell infection rate in explaining the data, we further fitted a model where the cell-to-cell infection rate is kept at a constant level, i.e., Γ2(t), and found that this model fits the data poorly (Fig. S4 and Table S5). This suggests that both cell-free infection and cell-to-cell infection are partially suppressed a few weeks after infection initiation.
To test the statistical significance of the added complexities, i.e., the time-varying infectivity and abortive infection, we further performed model selection among 4 model variations, the basic model, the basic model assuming constant infectivity, the abortive infection model, and the abortive infection model with constant cell-to-cell infectivity, and computed the corrected Akaike information criterion (AICc) (32). Overall, incorporation of the time-varying infectivity for both cell-free infection and cell-to-cell infection significantly improved the model fit to the data (Table 2). The abortive infection model was statistically significantly better than the other models overall, suggesting that abortive infection is an important mechanism that drives the death of the majority of infected cells. The other models had lower AICc scores than the abortive infection model in some individual monkeys, due to their simplicity compared to the abortive infection model.
TABLE 2.
Model selection using AICc
| Strain | Monkey identifier | ΔAICc scorea |
|||
|---|---|---|---|---|---|
| Basic model | Basic model with constant infectivity | Abortive infection model | Abortive infection model with constant cell-to-cell infectivity | ||
| WT | 34F | 12.2 | 123.7 | 0.0 | 160.1 |
| AM20 | 5.7 | 139.8 | 0.0 | 161.9 | |
| AO86 | 0.0 | 73.9 | 8.7 | 92.8 | |
| 62P | 0.0 | 140.1 | 3.5 | 135.6 | |
| DM46 | 15.0 | 34.6 | 0.0 | 60.1 | |
| 33160 | 26.3 | 39.1 | 0.0 | 46.6 | |
| AG94 | 0.5 | 45.7 | 0.0 | 66.3 | |
| AI22 | 0.0 | 105.9 | 6.9 | 87.9 | |
| 35720 | 17.1 | 45.5 | 0.0 | 51.5 | |
| 19V | 1.0 | 69.4 | 0.0 | 51.8 | |
| K65R | 9969 | 19.6 | 102.8 | 0.0 | 88.6 |
| RH6635 | 24.0 | 76.8 | 0.0 | 26.4 | |
| 33756 | 29.6 | 44.0 | 0.0 | 30.8 | |
| 33770 | 0.0 | 19.4 | 1.4 | 19.2 | |
| AH8F | 23.3 | 154.1 | 0.0 | 97.9 | |
| AM43 | 4.0 | 107.9 | 0.0 | 82.8 | |
| M184V | 31483 | 9.7 | 0.0 | 18.9 | 39.5 |
| 4284 | 3.1 | 0.0 | 8.9 | 41.1 | |
| 4282 | 0.0 | 46.9 | 3.1 | 38.2 | |
| 33246 | 7.9 | 0.0 | 11.6 | 39.7 | |
| Total ΔAICc score | 199.0 | 2,369.5 | 63.1 | 1,418.7 | |
ΔAICc score, change in the AICc score. The lower that the ΔAICc score is, the better that the model is.
To further test the robustness of our conclusion, we performed sensitivity analyses on the 5 fixed parameters in the model, i.e., kg, p, λ, βvI, and βvM (see Materials and Methods). We also tested a model variation where cell-to-cell transmission was not modeled explicitly (Tables S6 and S7 and Fig. S5). In general, the estimated death rates of abortively infected cells and productively infected cells (dA and d, respectively) and the fraction of abortively infected cells were consistent across the fitting results using wide ranges of values for fixed parameters (Fig. S6), and thus, these estimates are robust to the assumptions in our model. Taken together, our sensitivity analyses show that a large fraction of cells is abortively infected and abortively infected cells die at a relatively high rate.
DISCUSSION
Abortive infection has been suggested to be a major pathway that leads to progressive CD4+ T cell depletion and, ultimately, AIDS (10, 13). In this study, by following the dynamics of both the viral RNA level and the total cell-associated SHIV DNA level during SHIV infection in 20 rhesus macaques, we provide a first estimate, to our knowledge, of the death rate of abortively infected cells in an SHIV infection, 0.27 day−1 (SD, 0.14 day−1). This estimated rate in general agrees with that from a recent ex vivo experiment, where more than 80% of human tonsil cells died of abortive infection during a 6-day coculture with HIV-infected cells (10). In addition, we found indirect evidence that innate or adaptive immune responses may play an important role in reducing viral infection by both cell-free transmission and cell-to-cell transmission 1 to 3 weeks after infection initiation.
The estimation of the death rate of abortively infected cells stems from the observation that the first-phase DNA decline after peak viremia is slower than the first-phase RNA decline. Since the first-phase RNA decline is driven mostly by the death of productively infected cells, the slower DNA decline suggests that a mechanism other than the death of productively infected cells underlies the decrease in the amount of viral DNA. It has been shown that the majority of PBMC HIV DNA represents unintegrated HIV genomes (33), suggesting that a major fraction of cells may be abortively infected. Several recent ex vivo studies (10, 13) and an SHIV infection study (34) suggested that the majority of HIV-infected cells die rapidly through abortive infection. Using mathematical models, we showed that the first-phase DNA decline can be a result of the death of abortively infected cells. We estimated that a large fraction (approximately 65%) of infected cells is abortively infected, consistent with the findings of previous studies (10, 13, 34). Alternatively, if the majority of SHIV-infected cells contain integrated proviruses with defective non-replication-competent genomes (35), the estimated death rate would reflect the death of those cells. However, we argue that this is unlikely, given that the half-life of defectively infected cells is expected to be much longer than the 2.6-day half-life that we have estimated. Together, our results support the notion that a large fraction of cells dies through abortive infection at a relatively high rate, suggesting that abortive infection represents an important pathway that leads to CD4+ T cell depletion. Interestingly, a previous study on HIV infection in humans during acute infection showed that in patients treated with highly active antiretroviral therapy, there was a biphasic decline of latently infected cells, with a rapid first phase that was attributed to the loss of cells in preintegration latency followed by a much slower second phase that was attributed to cells with postintegration latency (36). Cells with DNA that has not been integrated can be subject to pyroptosis. Thus, the observed first-phase decline may reflect abortive infection.
Our results also suggest that the macaques in this study developed effective mechanisms to reduce viral infection by both cell-free and cell-to-cell infection mechanisms approximately 1 to 3 weeks after infection. The decrease in the rate of cell-free infection may be the result of an antibody response, which develops within 2 weeks after infection (25, 26, 37, 38). For cell-to-cell infection, we show that its decrease over time is necessary to explain a low level of quasi-steady-state viremia. This decrease may be due to the CD8+ T cell-mediated noncytotoxic antiviral response, which also develops a few weeks after infection (27, 28, 39). Alternatively, it may be due to the innate immune response mediated by interferon signaling, which plays multiple roles in suppressing HIV (30). It inhibits intracellular HIV protein production and thus leads to decreased virion production (31). Furthermore, paracrine signaling of interferon generates an antiviral state in uninfected target cells, which protects them from infection (31). This reduction in the concentration of target cells can lead to decreases in viral infection. We have not explored models with explicit adaptive immune responses, but it is well-known that CD8 depletion leads to increases in the plasma viral load (39–41). Thus, CD8 T cells are likely to be involved in maintaining the low-level quasi-steady-state viremia that we observed. Further experimental studies are needed to investigate the relative importance of the different types of immune responses in suppressing the SHIV162P3 plasma viral load to low levels in macaques. This may have important implications for immunotherapy development.
We used three viral variants to challenge the 20 rhesus macaques in this study: the wild-type virus and two drug-resistant mutant viruses, the K65R and M184V mutants. The viral load exhibited different kinetic patterns in the three macaque groups (Fig. 1). We did not find statistically significant differences in the estimated death rate of abortively infected cells, despite the deleterious effects of these mutations on virus fitness. Although they were beyond the scope of this study, further analyses are ongoing to determine the key parameters that give rise to the differences in the viral kinetic patterns shown in the data and, thus, to understand the impact of the drug-resistant mutants on the SHIV life cycle and infection process.
Our study is also subjected to several limitations. First, we used a less pathogenic SHIV162P3 isolate that may not fully recapitulate the pathogenesis of HIV during the acute or chronic phase of infection. Although our estimates of the fraction of abortive infections and the death rate of abortively infected cells are consistent with those reported in previous studies, additional analyses using more pathogenic SIV or SHIV strains may be needed to further confirm our observations (10, 13, 34). Second, in our mathematical model we assumed that CD4+ T cells are transported between the blood and the gut at constant rates. This assumption allowed us to estimate the dynamics in the gut on the basis of measures of HIV RNA and DNA in the blood. However, further longitudinal studies measuring and sequencing the viral RNA and DNA in both the blood and the gut would provide a more quantitative picture of the mechanistic link between the infection dynamics in the gut and those in the blood.
To conclude, we have studied both the viral RNA and cell-associated DNA dynamics during an SHIV infection. Our study provides a first estimate, to our knowledge, of the death rate of abortively infected cells in vivo. This estimate will be useful in research efforts aimed toward a quantitative understanding of viral and T cell dynamics during an HIV infection and, ultimately, to an understanding of the cause of and the mechanisms that lead to the CD4+ T cell depletion characteristic of AIDS.
MATERIALS AND METHODS
Ethics statement.
All animal procedures were performed according to NIH guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) of the Centers for Disease Control and Prevention (CDC). The CDC IACUC protocol numbers are 2070GARMONC and 2099GARMONC. Macaques were housed at the CDC under the care of CDC veterinarians in accordance with the Guide for the Care and Use of Laboratory Animals, 8th ed. (42). All procedures were performed while the macaques were under anesthesia (10 mg/kg of body weight ketamine or 2 to 6 mg/kg tiletamine-zolazepam [Telazol] given intramuscularly), and all efforts were made to minimize suffering, to improve housing conditions, and to provide enrichment opportunities. For housing, macaques were maintained in cages that met or exceeded the minimum size requirements stipulated in the Guide for the Care and Use of Laboratory Animals (42) (cage dimensions, 30 in. [height] by 30 in. [width] by 30 in. [length]). Steps were taken to reduce animal suffering, which included the provision of enrichment opportunities (e.g., cage features like swings/perches and objects for the macaques to manipulate), an assortment of food selections like fruits, vegetables, or seeds, suitable feeding methods (foraging and task-oriented methods), and humane interactions with caregivers and research staff. All animals had access to clean, fresh water at all times. The commercial diets provided were specifically formulated to meet vitamin C requirements. Prior to the initiation of virus inoculations, compatible macaques were housed in pairs. Once inoculations were initiated, the macaques were separated into single housing (while permitting eye contact) with a cage divider to prevent the possibility of SHIV transmission between the macaques. If one macaque remained uninfected during the course of a study, the animal remained separated from other infected macaques and was not housed in a pair with an infected macaque during the follow-up period. Euthanasia of SHIV-infected macaques (from studies in references 18, 19, 43, and 44) was accomplished in a humane manner (intravenous pentobarbital) by acceptable techniques recommended by the American Veterinary Medical Association Guidelines on Euthanasia (45) and in accordance with the CDC IACUC policy on euthanasia. The senior medical veterinarian and/or trained Comparative Medicine Branch staff verified successful euthanasia by the lack of a heart beat and respiration. Death was not used as a study endpoint.
Quantification of SHIV RNA in plasma.
SHIV162P3 RNA in plasma was quantified using a modification of an existing two-step reverse transcription-PCR (RT-PCR) assay that includes the use of a single RT-PCR (44, 46). Prior to RNA extraction, plasma samples (1 ml) were centrifuged at 43,000 × g for 30 min to concentrate the virus particles. RNA was extracted from 140 μl virus pellets using a QIAamp viral RNA kit (Qiagen) and eluted in 100 μl of elution buffer. To control for the efficiency of extraction, a known amount of virus particles (3 × 105) from an HIV type 1 (HIV-1) strain CM240 (HIV-1CM240) stock (NIH Reference Reagent Program) was added to each sample prior to RNA extraction (44, 46). Reverse transcription and PCR amplification of SHIV (gag) and HIV-1 (env) sequences were done using primers specific for SIVmac239 and HIV-1CM240, respectively (46). The sensitivity of the RT-PCR assay was 50 RNA copies/ml (47, 48).
Quantitation of cell-associated SHIV DNA in PBMCs by a DSP assay.
Rhesus macaque PBMCs were obtained from blood collected in BD Vacutainer CPT cell preparation tubes containing sodium heparin, resuspended in 200 μl of phosphate-buffered saline, and stored at −80°C until use. Genomic DNA was extracted from 200-μl aliquots using a QIAamp DNA and Blood minikit (Qiagen) and resuspended in 100 μl of elution buffer. Total SHIV DNA was quantified using a novel double-stranded primer (DSP) assay coupled with coamplification of the RNase P gene an as internal PBMC control (44). PCR amplification of the SIV long terminal repeat (52 bp) and RNase P (53 bp) was done in an iQ5 thermal cycler (Bio-Rad) using a QuantiFast multiplex kit (Qiagen, Valencia, CA) and included activation of DNA polymerase at 95°C for 90 s, followed by 42 cycles of 94°C for 1 s and 63°C for 20 s. SHIV DNA and cellular RNase P were quantified using standard curves prepared by serially diluting known numbers of copies of plasmid pVP1 containing the 5′ portion of SIVmac239 and known amounts of macaque PBMCs, respectively. The DSP assay is able to consistently detect 10 copies of plasmid pVP1 (44).
Determination of parameter values.
We assumed that the plasma volume of a 7-kg macaque is approximately 300 ml, as described by Vaidya et al. (22), and that the effective volume for the gut compartment was 30 ml, i.e., 10 times smaller than the plasma volume. This gives the ratio of the volume of the blood over the gut (r) of 10. Note that the choice of the value of r does not affect the dynamics of the total concentration of each cell type, because if we sum the ODEs describing the change in the concentration of each type of cell in the two compartments, we find that the parameter r disappears in the ODEs.
We assume that there are a total of 3 × 108 target cells that can be infected by SHIV in an uninfected macaque. This number is consistent with previously estimated numbers in macaques (49, 50). To translate the DNA load measurement (in PBMCs) to a cell concentration, we assumed that there are 1 × 106 PBMCs per ml blood. We set the death rate of uninfected target cells, d, to 0.01 day−1 (22). From the total number of target cells and the volumes of the plasma and the gut, we calculated the influx rate of target cells, λ, to be 105 ml−1 day−1. The production rate of viral particles from productively infected cells and the plasma virus clearance rate were fixed at 4 × 103 virions day−1cell−1 (51) and 23 day−1 (52), respectively.
The distribution of cell populations in the blood and the gut were determined by the ratio of the cell transport rates between the gut and the blood. The ratio was determined by the homing effects of the target cells expressing the α4β7 integrin (21). The values of these two transport rates in rhesus macaques are unknown, to our knowledge. Here, we fixed the transport rate from the gut to the blood, kg, at 0.18 day−1 and estimated the transport rate from the blood to the gut, kb, from the data. The value of kg was approximated roughly as follows. It has been estimated that 2.8 × 107 lymphocytes flow out of the gut of a 35-kg pig per hour (53). Assuming that 40% of the lymphocytes are CD4+ T cells, we estimate that approximately (2.8 × 107) × 40% ×24 ≈ 2.7 × 108 CD4+ T cells leave the gut per day in a pig. If we assume that the number of cells that leave the gut scales linearly with weight, we estimate that roughly (2.7 × 108) × 7/35 ≈ 5.4 × 107 CD4+ T cells leave the gut per day in a macaque. In our model, we assume there are 3 × 108 total target CD4+ T cells. Since the majority of target cells reside in the gut, then the rate of cell transport can be approximated as (5.4 × 107)/(3 × 108) ≈ 0.18 day−1. Note that this estimate is a rough estimate and that the most important quantity that determines the dynamics of infection is the ratio between kg and kb rather than their absolute values, because the time scale of the data is in weeks, which is much longer than the transport rates between the two compartments.
Model fitting and selection.
To estimate the parameter values, we simulated the ODE models (see Text S1 in the supplemental material for determination of initial conditions) and then calculated the residual sum of squares (RSS) between experimentally measured and model-predicted SHIV RNA and DNA levels on a log scale. The RSS for the RNA and DNA data were equally weighted in the calculation of total RSS. We then minimized the total RSS using the Nelder-Mead algorithm (54). To perform model selection, the corrected Akaike information criterion (AICc), which accounts for the low number of data points (range, 18 to 45) for each patient (32), was used. The AICc score is calculated as
where n is the number of data points and K is the number of fitted parameters. When the models were compared, the model with the lowest score was the best model, although a small difference in AICc scores, e.g., ≤2, was not considered significant (32).
Parameter sensitivity analysis.
We tested the robustness of the estimated death rate of abortively infected cells (dA), the death rate of productively infected cells (δ), and the fraction of abortively infected cells against variations in the values of the 5 fixed parameters, i.e., kg, p, λ, βvI, and βvM. Specifically, we changed the value of each of the 3 parameters kg, p, and λ from their baseline values and then reestimated the 11 parameters in the abortive infection model. Since the value of kg is calculated on the basis of estimates from different experimental systems, we increased and decreased the value of kg from its baseline value by 10-fold to reflect the large uncertainty. The value of p was set equal to the rate estimated from human data (51); however, the value of p is estimated to be 10-fold higher in macaques (55). We thus increased the value of p to 2- and 10-fold of its baseline value. The value of λ was calculated roughly on the basis of different estimates of the total number of target CD4+ cells, and thus, we increased or decreased the value of λ from its baseline by 5-fold to reflect the uncertainty. For the parameters βvI and βvM, we increased and decreased their values together by 5-fold.
Supplementary Material
ACKNOWLEDGMENTS
Portions of this work were performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grants R01-AI104373, R01-OD011095, and R01-AI028433 (A.S.P.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00352-17.
REFERENCES
- 1.Haase AT. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol 17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 2.Okoye AA, Picker LJ. 2013. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markowitz M, Louie M, Hurley A, Sun E, Di Mascio M, Perelson AS, Ho DD. 2003. A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J Virol 77:5037–5038. doi: 10.1128/JVI.77.8.5037-5038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. 2013. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 5.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreisberg JF, Yonemoto W, Greene WC. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med 203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swiggard WJ, O'Doherty U, McGain D, Jeyakumar D, Malim MH. 2004. Long HIV type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Res Hum Retroviruses 20:285–295. doi: 10.1089/088922204322996527. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Zhang H, Siliciano JD, Siliciano RF. 2005. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol 79:2199–2210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC. 2010. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz-Arias I, Doitsh G, Yang Z, Sowinski S, Ruelas D, Greene WC. 2015. Blood-derived CD4 T cells naturally resist pyroptosis during abortive HIV-1 infection. Cell Host Microbe 18:463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloway NL, Doitsh G, Monroe KM, Yang Z, Munoz-Arias I, Levy DN, Greene WC. 2015. Cell-to-cell transmission of HIV-1 is required to trigger pyroptotic death of lymphoid-tissue-derived CD4 T cells. Cell Rep 12:1555–1563. doi: 10.1016/j.celrep.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. 2014. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doitsh G, Greene WC. 2016. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe 19:280–291. doi: 10.1016/j.chom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Hottz P, Schechter M, Rong L. 2015. Modeling the slow CD4+ T cell decline in HIV-infected individuals. PLoS Comput Biol 11:e1004665. doi: 10.1371/journal.pcbi.1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harouse JM, Gettie A, Eshetu T, Tan RC, Bohm R, Blanchard J, Baskin G, Cheng-Mayer C. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J Virol 75:1990–1995. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harouse JM, Gettie A, Tan RC, Blanchard J, Cheng-Mayer C. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 18.Cong ME, Youngpairoj AS, Zheng Q, Aung W, Mitchell J, Sweeney E, Hanson DL, Hendry RM, Dobard C, Heneine W, García-Lerma JG. 2011. Protection against rectal transmission of an emtricitabine-resistant simian/human immunodeficiency virus SHIV162p3M184V mutant by intermittent prophylaxis with Truvada. J Virol 85:7933–7936. doi: 10.1128/JVI.00843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong ME, Mitchell J, Sweeney E, Bachman S, Hanson DL, Heneine W, García-Lerma JG. 2013. Prophylactic efficacy of oral emtricitabine and tenofovir disoproxil fumarate combination therapy against a tenofovir-resistant simian/human immunodeficiency virus containing the K65R mutation in macaques. J Infect Dis 208:463–467. doi: 10.1093/infdis/jit189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong M, Heneine W, García-Lerma JG. 2007. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J Virol 81:3037–3041. doi: 10.1128/JVI.02712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 22.Vaidya NK, Ribeiro RM, Miller CJ, Perelson AS. 2010. Viral dynamics during primary simian immunodeficiency virus infection: effect of time-dependent virus infectivity. J Virol 84:4302–4310. doi: 10.1128/JVI.02284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway JM, Perelson AS. 2016. Residual viremia in treated HIV+ individuals. PLoS Comput Biol 12:e1004677. doi: 10.1371/journal.pcbi.1004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 25.Borsetti A, Ferrantelli F, Maggiorella MT, Sernicola L, Bellino S, Gallinaro A, Farcomeni S, Mee ET, Rose NJ, Cafaro A, Titti F, Ensoli B. 2014. Effect of MHC haplotype on immune response upon experimental SHIVSF162P4cy infection of Mauritian cynomolgus macaques. PLoS One 9:e93235. doi: 10.1371/journal.pone.0093235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraft Z, Derby NR, McCaffrey RA, Niec R, Blay WM, Haigwood NL, Moysi E, Saunders CJ, Wrin T, Petropoulos CJ, McElrath MJ, Stamatatos L. 2007. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J Virol 81:6402–6411. doi: 10.1128/JVI.00424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javed A, Leuchte N, Neumann B, Sopper S, Sauermann U. 2015. Noncytolytic CD8+ cell mediated antiviral response represents a strong element in the immune response of simian immunodeficiency virus-infected long-term non-progressing rhesus macaques. PLoS One 10:e0142086. doi: 10.1371/journal.pone.0142086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy JA. 2003. The search for the CD8+ cell anti-HIV factor (CAF). Trends Immunol 24:628–632. doi: 10.1016/j.it.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Coccia EM, Krust B, Hovanessian AG. 1994. Specific inhibition of viral protein synthesis in HIV-infected cells in response to interferon treatment. J Biol Chem 269:23087–23094. [PubMed] [Google Scholar]
- 30.Sivro A, Su RC, Plummer FA, Ball TB. 2014. Interferon responses in HIV infection: from protection to disease. AIDS Rev 16:43–51. [PubMed] [Google Scholar]
- 31.Biron CA. 1998. Role of early cytokines, including alpha and beta interferons (IFN-alpha/beta), in innate and adaptive immune responses to viral infections. Semin Immunol 10:383–390. doi: 10.1006/smim.1998.0138. [DOI] [PubMed] [Google Scholar]
- 32.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach, 2nd ed Springer-Verlag, Berlin, Germany. [Google Scholar]
- 33.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. 2008. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis 197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 34.Matrajt L, Younan PM, Kiem HP, Schiffer JT. 2014. The majority of CD4+ T-cell depletion during acute simian-human immunodeficiency virus SHIV89.6P infection occurs in uninfected cells. J Virol 88:3202–3212. doi: 10.1128/JVI.03428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blankson JN, Finzi D, Pierson TC, Sabundayo BP, Chadwick K, Margolick JB, Quinn TC, Siliciano RF. 2000. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J Infect Dis 182:1636–1642. doi: 10.1086/317615. [DOI] [PubMed] [Google Scholar]
- 37.García-Lerma JG, Otten RA, Qari SH, Jackson E, Cong M, Masciotra S, Luo W, Kim C, Adams DR, Monsour M, Lipscomb J, Johnson JA, Delinsky D, Schinazi RF, Janssen R, Folks TM, Heneine W. 2008. Prevention of rectal SHIV transmission in macaques by daily or intermittent prophylaxis with emtricitabine and tenofovir. PLoS Med 5:e28. doi: 10.1371/journal.pmed.0050028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtis KA, Kennedy MS, Luckay A, Cong ME, Youngpairoj AS, Zheng Q, Smith J, Hanson D, Heneine W, Owen SM, García-Lerma JG. 2011. Delayed maturation of antibody avidity but not seroconversion in rhesus macaques infected with SHIV during oral pre-exposure prophylaxis. J Acquir Immune Defic Syndr 57:355–362. doi: 10.1097/QAI.0b013e3182234a51. [DOI] [PubMed] [Google Scholar]
- 39.Kersh EN, Luo W, Zheng Q, Adams DR, Hanson D, Youngpairoj AS, Cong ME, Butler K, Hendry RM, McNicholl JM, Heneine W, Garcia-Lerma JG. 2012. Reduced inflammation and CD4 loss in acute SHIV infection during oral pre-exposure prophylaxis. J Infect Dis 206:770–779. doi: 10.1093/infdis/jis422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G. 2016. CD8(+) lymphocytes are required for maintaining viral suppression in SIV-infected macaques treated with short-term antiretroviral therapy. Immunity 45:656–668. doi: 10.1016/j.immuni.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 43.Cong ME, Youngpairoj AS, Aung W, Sharma S, Mitchell J, Dobard C, Heneine W, Garcia-Lerma JG. 2011. Generation and mucosal transmissibility of emtricitabine- and tenofovir-resistant SHIV162P3 mutants in macaques. Virology 412:435–440. doi: 10.1016/j.virol.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 44.Cong ME, Pau CP, Heneine W, Garcia-Lerma JG. 2016. Antiretroviral drug activity in macaques infected during pre-exposure prophylaxis has a transient effect on cell-associated SHIV DNA reservoirs. PLoS One 11:e0164821. doi: 10.1371/journal.pone.0164821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.American Veterinary Medical Association. 2013. Guidelines on euthanasia. American Veterinary Medical Association, Schaumburg, IL. [Google Scholar]
- 46.Subbarao S, Otten RA, Ramos A, Kim C, Jackson E, Monsour M, Adams DR, Bashirian S, Johnson J, Soriano V, Rendon A, Hudgens MG, Butera S, Janssen R, Paxton L, Greenberg AE, Folks TM. 2006. Chemoprophylaxis with tenofovir disoproxil fumarate provided partial protection against infection with simian human immunodeficiency virus in macaques given multiple virus challenges. J Infect Dis 194:904–911. doi: 10.1086/507306. [DOI] [PubMed] [Google Scholar]
- 47.Radzio J, Aung W, Holder A, Martin A, Sweeney E, Mitchell J, Bachman S, Pau C, Heneine W, Garcia-Lerma JG. 2012. Prevention of vaginal SHIV transmission in macaques by a coitally-dependent Truvada regimen. PLoS One 7:e50632. doi: 10.1371/journal.pone.0050632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radzio J, Hanley K, Mitchell J, Ellis S, Deyounks F, Jenkins LT, Hanson D, Heneine W, Garcia-Lerma JG. 2014. Physiologic doses of depot-medroxyprogesterone acetate do not increase acute plasma SHIV viremia or mucosal virus shedding in pigtail macaques. AIDS 28:1431–1439. doi: 10.1097/QAD.0000000000000294. [DOI] [PubMed] [Google Scholar]
- 49.Ganusov VV, De Boer RJ. 2007. Do most lymphocytes in humans really reside in the gut? Trends Immunol 28:514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Sopper S, Nierwetberg D, Halbach A, Sauer U, Scheller C, Stahl-Hennig C, Matz-Rensing K, Schafer F, Schneider T, ter Meulen V, Muller JG. 2003. Impact of simian immunodeficiency virus (SIV) infection on lymphocyte numbers and T-cell turnover in different organs of rhesus monkeys. Blood 101:1213–1219. doi: 10.1182/blood-2002-06-1644. [DOI] [PubMed] [Google Scholar]
- 51.Hockett RD, Kilby JM, Derdeyn CA, Saag MS, Sillers M, Squires K, Chiz S, Nowak MA, Shaw GM, Bucy RP. 1999. Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA. J Exp Med 189:1545–1554. doi: 10.1084/jem.189.10.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramratnam B, Bonhoeffer S, Binley J, Hurley A, Zhang L, Mittler JE, Markowitz M, Moore JP, Perelson AS, Ho DD. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354:1782–1785. doi: 10.1016/S0140-6736(99)02035-8. [DOI] [PubMed] [Google Scholar]
- 53.Thielke KH, Pabst R, Rothkotter HJ. 1999. Quantification of proliferating lymphocyte subsets appearing in the intestinal lymph and the blood. Clin Exp Immunol 117:277–284. doi: 10.1046/j.1365-2249.1999.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelder JA, Mead R. 1965. A simplex-method for function minimization. Computer J 7:308–313. doi: 10.1093/comjnl/7.4.308. [DOI] [Google Scholar]
- 55.Chen HY, Di Mascio M, Perelson AS, Ho DD, Zhang L. 2007. Determination of virus burst size in vivo using a single-cycle SIV in rhesus macaques. Proc Natl Acad Sci U S A 104:19079–19084. doi: 10.1073/pnas.0707449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.