ABSTRACT
A total of 11 multidrug-resistant Pseudomonas aeruginosa clinical isolates were obtained in Nepal. Four of these isolates harbored genes encoding one or more carbapenemases (DIM-1, NDM-1, and/or VIM-2), and five harbored genes encoding a 16S rRNA methyltransferase (RmtB4 or RmtF2). A novel RmtF variant, RmtF2, had a substitution (K65E) compared with the same gene in RmtF. To our knowledge, this is the first report describing carbapenemase- and 16S rRNA methyltransferase-coproducing P. aeruginosa clinical isolates in Nepal.
KEYWORDS: 16S rRNA methylase, Pseudomonas aeruginosa, carbapenemase, multidrug resistance
TEXT
Metallo-β-lactamases (MBLs) confer resistance to all β-lactams, except the monobactams, and are characterized by their efficient hydrolysis of carbapenems (1). The metallo-β-lactamase DIM-1 was first identified in a Pseudomonas stutzeri strain obtained from a Dutch patient in 2007 (2). DIM-1 hydrolyzes broad-spectrum cephalosporins and carbapenems but not monobactams. Since then, DIM-1 producers, including P. stutzeri and Enterobacteriaceae spp., have been isolated in India (3) and Sierra Leone (4), respectively.
Acquired 16S rRNA methyltransferase genes responsible for an extremely high level of resistance against various aminoglycosides are widely distributed among Enterobacteriaceae and glucose-nonfermentative bacteria (5). To date, 10 different 16S rRNA methyltransferases, including ArmA, RmtA, RmtB, RmtC, RmtD, RmtE, RmtF, RmtG, RmtH, and NpmA, have been found in clinical isolates (6–9). One of these, RmtB, was found to have three variants, RmtB2 (accession no. JN968578), RmtB3 (accession no. JN968579), and RmtB4 (accession no. KM999534). The 16S rRNA methyltransferase RmtF was first identified in a clinical isolate of Klebsiella pneumoniae on the island of Réunion in 2011 (7). Since then, RmtF-producing Enterobacteriaceae have been isolated in India, the United Kingdom, the United States, and Nepal (7, 10, 11).
Between 2012 and 2013, 11 multidrug-resistant Pseudomonas aeruginosa clinical isolates were obtained from 11 inpatients treated at a university hospital in Nepal. Multidrug-resistant Pseudomonas aeruginosa isolates are defined as strains showing resistance to carbapenem (MIC ≥ 16 μg/ml), amikacin (MIC ≥ 32 μg/ml), and fluoroquinolone (MIC ≥ 4 μg/ml), as previously described (12). Of these isolates, 7 were from sputum, 3 from urine samples, and 1 from a pus sample. The MICs of various antibiotics were determined using the microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (13). The entire genomes of these isolates were sequenced by MiSeq (Illumina, San Diego, CA). Their genomes were searched for drug resistance genes, including genes encoding β-lactamases (carbapenemases and extended-spectrum β-lactamases), 16S rRNA methyltransferases, and aminoglycoside-acetyl/adenyltransferases, using ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/). Point mutations associated with quinolone resistance were searched in gyrA and parC. Multilocus sequence type (MLST) was deduced, as described by the protocols of the PubMLST (http://pubmlst.org/paeruginosa/) databases. The complete genome of P. aeruginosa IOMTU133 was determined using PacBio RS II (Menlo Park), as described previously (14). The genomic environments surrounding genes encoding carbapenemases and/or 16S rRNA methyltransferases were confirmed by Sanger sequencing. DNA plugs of all isolates tested (digested with I-CeuI or S1 nuclease) were prepared and separated by pulsed-field gel electrophoresis, and Southern hybridization was performed using probes of 16S rRNA, blaDIM-1, blaNDM-1, blaVIM-2, rmtB4, and rmtF2 (15, 16).
All 11 isolates were resistant to meropenem, aztreonam, amikacin, and ciprofloxacin (Table 1), with MICs ≥16 μg/ml. Three isolates showed higher MICs to imipenem or meropenem, ≥64 μg/ml, than the other isolates. Five of the 11 isolates were extremely highly resistant to amikacin and arbekacin, with MICs >1,024 μg/ml, and to ciprofloxacin, with MICs of 32 to 256 μg/ml. All isolates were susceptible to colistin, with MICs ≤0.5 μg/ml.
TABLE 1.
Summary of characteristics of the 11 Pseudomonas aeruginosa strains, including antimicrobial resistance profiles and resistant genes
| Strain | MLST | MICs (μg/ml) fora: |
β-Lactamase(s) | 16S rRNA methylase | Aminoglycoside acetyl/adenylyltransferase(s) | Mutation(s) in DNA gyrase |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ATM | CAZ | AMK | ABK | CIP | CST | GyrA | ParC | |||||
| IOMTU 3 | 654 | 128 | 32 | 16 | 128 | 128 | 1 | 256 | ≤0.5 | VIM-2, PDC-58 | AADB | S83L, D87E | S80L | |
| IOMTU 7 | 244 | 16 | 32 | >1024 | >1024 | 32 | 16 | 32 | ≤0.5 | VEB-Ia, PDC-61 | AAC(6′)-Ib | S83I | S80L | |
| IOMTU 9 | 235 | 512 | >1024 | 32 | >1024 | >1024 | >1024 | 64 | ≤0.5 | NDM-1, VIM-2, PDC-35 | RmtB4 | AACA7, AACC5b | S83I | S80L |
| IOMTU 133 | 1047 | 32 | 64 | 16 | 256 | >1024 | >1024 | 32 | ≤0.5 | DIM-1, PDC-32 | RmtF2 | AAC(6′)-Ib | S83I | S80L |
| IOMTU 143 | 664 | 1 | 4 | 16 | 8 | 128 | 1 | 32 | ≤0.5 | PDC-98 | AAC(6′)-Ib | S83I | S80L | |
| IOMTU 155 | 664 | 1 | 4 | 32 | 8 | 128 | 1 | 32 | ≤0.5 | PDC-98 | AAC(6′)-Ib | S83I | S80L | |
| IOMTU 161 | 664 | 1 | 4 | 16 | 8 | 128 | 1 | 32 | ≤0.5 | PCD-98 | AAC(6′)-Ib | S83I | S80L | |
| IOMTU 179 | 664 | 1 | 4 | 32 | 4 | 128 | 1 | 32 | ≤0.5 | TEM-1, PDC-98 | AAC(6′)-Ib | S83I | S80L | |
| IOMTU 184 | 664 | 8 | 32 | 128 | >1024 | >1024 | >1024 | 64 | ≤0.5 | PSE-2, PDC-98 | RmtF2 | AAC(6′)-Ib | S83I | S80L |
| IOMTU 304 | 235 | 512 | >1024 | 32 | >1024 | >1024 | >1024 | 64 | ≤0.5 | NDM-1, VIM-2, PDC-35 | RmtB4 | AACA7, AACC5b | S83I | S80L |
| IOMTU 487 | 664 | 2 | 32 | 128 | 512 | >1024 | >1024 | 32 | ≤0.5 | PSE-2, PDC-98 | RmtF2 | AADB | S83I | S80L |
IPM, imipenem; MEM, meropenem; ATM, aztreonam; CAZ, ceftazidime; AMK, amikacin; ABK, arbekacin; CIP, ciprofloxacin; CST, colistin.
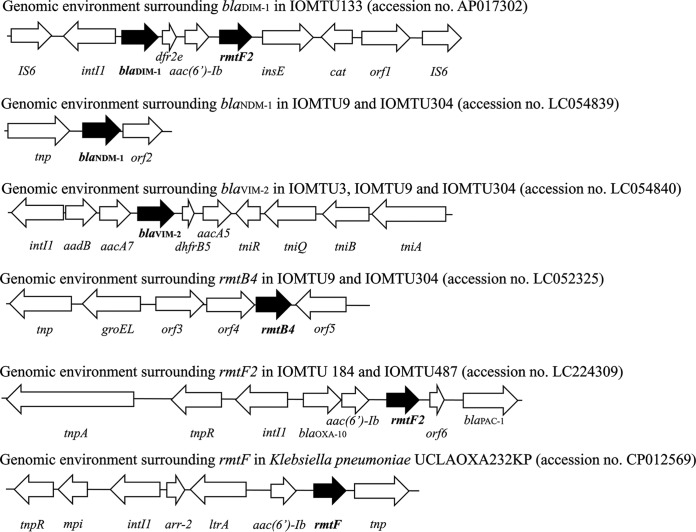
Of the 11 isolates, three had a novel rmtF variant, designated rmtF2 (accession no. LC050387). Analysis of its predicted amino acid sequence revealed a substitution (K65E) compared with the sequence of RmtF. Four isolates had genes encoding one or more metallo-β-lactamases, i.e., blaDIM-1, blaNDM-1, and/or blaVIM-2; and four had genes encoding other β-lactamases, i.e., blaPDCs, blaPSE-2, blaTEM-1, or blaVEB-1a (Table 1). In addition, 5 isolates had a 16S rRNA methyltransferase encoding gene, rmtB4 or rmtF2; and nine had genes encoding an aminoglycoside acetyl- and adenylyl-transferase, including AAC(6′)-Ib, AACA7, AACC5b, and AADB (Table 1). A novel rmtF2 gene was located in the class 1 integron (Fig. 1). All isolates except for IOMTU3 had amino acid substitution point mutations S83I in GyrA and S80L in ParC; IOMTU3 had amino acid substitution point mutations S83L and D87E in GyrA and S80L in ParC.
FIG 1.
Genomic environment surrounding blaDIM-1 in IOMTU133 (accession no. AP017302); blaNDM-1 in IOMTU9 and IOMTU304 (accession no. LC054839); blaVIM-2 in IOMTU3, IOMTU9 and IOMTU304 (accession no. LC054840); rmtB4 in IOMTU9 and IOMTU304 (accession no. LC052325); rmtF2 in IOMTU184 and IOMTU487 (accession no. LC224309); and rmtF in Klebsiella pneumoniae UCLAOXA232KP (accession no. CP012569).
A total of 6 isolates were classified as ST664, two as ST235, and one each as ST244, ST654, and ST1047. The two ST235 isolates harbored blaNDM-1, blaVIM-2, and rmtB4; two of the ST664 isolates harbored rmtF2; the ST654 isolate harbored blaVIM-2; and the ST1047 isolate harbored blaDIM-1 and rmtF2.
Because IOMTU133 belonged to ST1047 and harbored several drug resistance genes, its complete genome was sequenced and deposited in GenBank under accession no. AP017302. This isolate had no plasmids. The complete genome sequence of IOMTU133 had 283-fold coverage for one chromosome, IOMTU133, which consisted of a single circular chromosome of 6,897,018 bp with an average GC content of 65.98%. The chromosome was found to contain 6,245 protein-encoding genes, including 63 tRNA genes and one transfer messenger RNA (tmRNA) gene for all amino acids. IOMTU133 also harbored a carbapenemase-encoding gene, blaDIM-1; a 16S rRNA methyltransferase encoding gene, rmtF2; and an aminoglycoside acetyltransferase encoding gene, aac(6′)-Ib. The blaDIM-1 gene and a novel rmtF2 gene were located within the same integron on the chromosome (Fig. 1).
The genomic environments surrounding blaDIM-1, blaNDM-1, blaVIM-2, rmtB4, and rmtF2 are shown in Fig. 1. The genomic environments of blaDIM-1, blaNDM-1, and blaVIM-2 were IS6-intI1-blaDIM-1-dfr2e-aac(6′)-Ib-rmtF2-insE-cat-orf1 (gene encoding a hypothetical protein)-IS6 (accession no. AP017302), tnp-blaNDM-1-orf2 (gene encoding a hypothetical protein) (accession no. LC054839), and intI1-aadB-aacA7-blaVIM-2-dhfrB5-aacA5-tniR-tniQ-tniB-tniA (accession no. LC054840), respectively. The genomic environments surrounding these carbapenemase-encoding genes were unique to these isolates.
The genomic environment of rmtB4 was tnp-groEL-orf3 (gene encoding queuine tRNA-ribosyltransferase)-orf4 (gene encoding a hypothetical protein)-rmtB4-orf5 (gene encoding a putative Na+/H+ antiporter) (accession no. LC052325), whereas the genomic environment of rmtF2 was tnpA-tnpR-intI1-blaOXA-10-aac(6′)-Ib-rmtF2-orf6 (gene encoding a hypothetical protein)-blaPAC-1 (accession no. LC224309). The rmtF2 in IOMTU133 was located in the same integron as blaDIM-1 (Fig. 1). Compared to the genomic environment surrounding rmtF in K. pneumoniae UCLAOXA232KP plasmid pUCLAOXA232-3.X (accession no. CP012569), both rmtF and rmtF2 were located in class I integron, which contained aac(6′)-Ib in the upstream regions of rmtF and rmtF2; however, the other allelic profiles in each integron were different (Fig. 1). The genomic environments surrounding rmtB4 and rmtF2 were unique to these isolates.
Of all the isolates tested, only IOMTU487 had a 120-kbp plasmid, but the plasmid did not harbor the blaDIM-1, blaNDM-1, blaVIM-2, rmtB4, or rmtF2 genes (see Fig. S1 in the supplemental material). All of these genes were located in the chromosomes (see Fig. S2 in the supplemental material).
The findings of this study indicate that ST664 P. aeruginosa clinical isolates spread in a medical setting in Nepal, because the majority of P. aeruginosa isolates obtained in Nepal were classified as ST664. To date, seven ST664 isolates (PubMLST no. 3401, 3707, 4018, 4033, 4052, 4060, and 4787) have been registered on the PubMLST website (https://pubmlst.org/paeruginosa/). Of these, PubMLST no. 4787 (Pseudomonas aeruginosa VRFPA06) was isolated from human blood in 2012 in India (17), although the details of others were not reported. Of our 11 isolates, only two were classified as ST235, which has been recognized as one of three high-risk clones, i.e., ST235, ST111, and ST175 (18). A P. aeruginosa strain belonging to ST1047, which was originally obtained in Norway and found to produce VIM-type MBLs, was first registered on the PubMLST website in 2011 (PubMLST no. 746).
This is the first report describing carbapenemase- and 16S rRNA methyltransferase-coproducing P. aeruginosa clinical isolates in Nepal. Carbapenemase- and 16S rRNA methyltransferase-coproducing P. aeruginosa was reported in 2007 in Brazil (19) and in 2015 in northeast India (20), which is bordered by Nepal. It is therefore necessary to survey multidrug-resistant P. aeruginosa in medical settings in Nepal.
Accession number(s).
The sequences described were submitted to GenBank under the accession numbers LC050387, LC052325, LC054839, LC054840, LC224309, and AP017302.
Supplementary Material
ACKNOWLEDGMENTS
This study was reviewed and approved by the Institutional Review Board of the Institute of Medicine at Tribhuvan University (reference no. 6-11-E) and the Biosafety Committee at the National Center for Global Health and Medicine (approval no. 28-M-053). The study was supported by grants from International Health Cooperation Research (grant 29-S-5), the Okinawa Communicable Disease Research Hub Formation Promotion Project, Okinawa Prefectural Government Commissioned Projects For Fiscal Year 2016, the Research Program on Emerging and Re-emerging Infectious Diseases from Japan Agency for Medical Research and Development (AMED), and JSPS KAKENHI (grant 16K19133).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00694-17.
REFERENCES
- 1.Bush K. 2001. New β-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis 32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Rodriguez-Martinez JM, Al Naiemi N, Debets-Ossenkopp YJ, Nordmann P. 2010. Characterization of DIM-1, an integron-encoded metallo-β-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob Agents Chemother 54:2420–2424. doi: 10.1128/AAC.01456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshpande LM, Jones RN, Woosley LN, Castanheira M. 2014. Retrospective molecular analysis of DIM-1 metallo-β-lactamase discovered in Pseudomonas stutzeri from India in 2000. Antimicrob Agents Chemother 58:596–598. doi: 10.1128/AAC.01541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, Stenger DA, Taitt CR, Vora GJ. 2013. Identification of blaOXA-51-like, blaOXA-58, blaDIM-1, and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J Clin Microbiol 51:2435–2438. doi: 10.1128/JCM.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother 50:178–184. doi: 10.1128/AAC.50.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Galimand M, Courvalin P, Lambert T. 2012. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother 56:3960–3962. doi: 10.1128/AAC.00660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bueno MF, Francisco GR, O'Hara JA, de Oliveira Garcia D, Doi Y. 2013. Co-production of 16S ribosomal RNA methyltransferase RmtD and RmtG with KPC-2 and CTX-M-group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 57:2397–2400. doi: 10.1128/AAC.02108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Hara JA, McGann P, Snesrud EC, Clifford RJ, Waterman PE, Lesho EP, Doi Y. 2013. Novel 16S ribosomal RNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother 57:2413–2416. doi: 10.1128/AAC.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo L, Hopkins KL, Gutierrez B, Ovejero CM, Shukla S, Douthwaite S, Prasad KN, Woodford N, Gonzalez-Zorn B. 2013. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J Antimicrob Chemother 68:1543–1550. doi: 10.1093/jac/dkt078. [DOI] [PubMed] [Google Scholar]
- 11.Tada T, Miyoshi-Akiyama T, Dahal RK, Mishra SK, Ohara H, Shimada K, Kirikae T, Pokhrel BM. 2013. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int J Antimicrob Agents 42:372–374. doi: 10.1016/j.ijantimicag.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Kirikae T, Mizuguchi Y, Arakawa Y. 2008. Investigation of isolation rates of Pseudomonas aeruginosa with and without multidrug resistance in medical facilities and clinical laboratories in Japan. J Antimicrob Chemother 61:612–615. doi: 10.1093/jac/dkm537. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Tada T, Miyoshi-Akiyama T, Shimada K, Shiroma A, Nakano K, Teruya K, Satou K, Hirano T, Shimojima M, Kirikae T. 2016. A carbapenem-resistant Pseudomonas aeruginosa isolate harboring two copies of blaIMP-34 encoding a metallo-β-lactamase. PLoS One 11:e0149385. doi: 10.1371/journal.pone.0149385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-β-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob Agents Chemother 55:5143–5149. doi: 10.1128/AAC.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. 2014. Biochemical analysis of metallo-β-lactamase NDM-3 from a multidrug-resistant Escherichia coli strain isolated in Japan. Antimicrob Agents Chemother 58:3538–3540. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murugan N, Malathi J, Umashankar V, Madhavan HN. 2014. Comparative genomic analysis of multidrug-resistant Pseudomonas aeruginosa clinical isolates VRFPA06 and VRFPA08 with VRFPA07. Genome Announc 2:e00140-14. doi: 10.1128/genomeA.00140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver A, Mulet X, Lopez-Causape C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother 51:852–856. doi: 10.1128/AAC.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman M, Prasad KN, Pathak A, Pati BK, Singh A, Ovejero CM, Ahmad S, Gonzalez-Zorn B. 2015. RmtC and RmtF 16S rRNA methyltransferase in NDM-1-producing Pseudomonas aeruginosa. Emerg Infect Dis 21:2059–2062. doi: 10.3201/eid2111.150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.