ABSTRACT
Thirty-nine fosfomycin-resistant Escherichia coli isolates carrying fosA3 were obtained from pigs, chickens, dairy cows, and staff in four northeastern provinces of China between June 2015 and April 2016. The fosA3 gene was colocated with blaCTX-M genes on conjugative plasmids of the incompatibility groups IncN (n = 12), IncN-F33:A−:B−(n = 2), IncF33:A−:B−(n = 14), IncF14:A−:B−(n = 2), and IncI1/sequence type 136 (ST136) (n = 9). Four different genetic contexts of fosA3 were detected among the 39 E. coli isolates. Three potential epidemic plasmids circulated among E. coli strains from this region.
KEYWORDS: CTX-M, antibiotic resistance, resistance genes, horizontal dissemination
TEXT
Due to the increasing prevalence of bacterial infections caused by multidrug-resistant (MDR) or extensively drug-resistant (XDR) Gram-negative pathogens (1, 2), one of the older antibiotics, fosfomycin, has been reintroduced into clinical use (3). While resistance to fosfomycin is mainly due to chromosomal mutations (4), two plasmid-borne fosfomycin resistance genes, fosA3 and fosC2, were first identified in human Escherichia coli in Japan in 2010 (5). Thereafter, a relatively high prevalence of fosfomycin-resistant E. coli isolates carrying fosA3 was found in food animals, pets, and wild rodents in China, although fosfomycin has not been approved for veterinary use in China (6–8).
So far, information on the distribution of fosfomycin-resistant E. coli isolates of animal origin in China has been limited to the southern and central parts of China. However, available information on the occurrence and epidemiological characteristics of fosfomycin-resistant E. coli in other regions of China is scarce. In this study, a total of 370 E. coli isolates were collected from diseased animals and humans (pigs, n = 115; chickens, n = 95; dairy cows, n = 98; dairy farm staff, n = 1) and healthy animals and humans (pigs, n = 13; chickens, n = 19; dairy cows, n = 24; dairy farm staff, n = 5) in northeast China between June 2015 and April 2016.
The MICs of fosfomycin for 370 isolates, determined as previously described (6), revealed that 39 isolates (10.5%) were resistant to fosfomycin (>512 mg/liter) (see Table S1 in the supplemental information). PCR assays for plasmid-mediated fosfomycin resistance genes fosA3, fosA, and fosC2 (7) showed that all 39 isolates were positive only for the fosA3 gene. Additional antimicrobial susceptibility testing by broth microdilution (9, 10) revealed that all 39 fosA3-positive isolates were also resistant to florfenicol, cefotaxime, gentamicin, and tetracycline but susceptible to colistin (see Table S1). PCR amplification and sequencing (11, 12) revealed that all of the fosA3-positive isolates also harbored blaCTX-M genes, and 15 (38.5%) of them produced CTX-M-55 (Table 1). In addition, 29 (74.4%), 22 (56.4%), 20 (51.3%), and 20 (51.3%) isolates carried floR, tet(A), blaTEM-1, and rmtB genes, respectively (Table 1). Pulsed-field gel electrophoresis (PFGE) analysis of the original 39 fosA3-positive E. coli isolates revealed great genetic diversity of the isolates, with 29 major XbaI patterns (see Table S2 in the supplemental material). The results of multilocus sequence typing (MLST) and phylogenetic analysis also revealed a great diversity. Thirty-seven different sequence types (STs) were distributed among 39 fosA3-positive E. coli isolates (Table 1). Most of these strains belonged to the phylogenetic grouping B2 (60.0%), followed by B1 (25.6%), A1 (7.7%), and D (7.7%).
TABLE 1.
Characterization of fosA3-positive E. coli isolates and their fosA3-carrying plasmids
| Isolatea | Host | MLST | Phylogroup | Other cotransferred resistance genesb | Context of fosA3 |
fosA3-carrying plasmids |
|||
|---|---|---|---|---|---|---|---|---|---|
| Size | Replicon type | EcoRI RFLPc | Addiction system(s) | ||||||
| SY301B | Pig | ST761 | B2 | blaCTX-M-65, floR, cfr, tet(A) | III | pECXH3 (∼97 kb) | N (ST7) | A1 | pemKI, hok-mok-sok, srnB-srnC |
| SY301F | Pig | ST3315 | B2 | blaCTX-M-55, floR, cfr, rmtB, strA, strB, tet(A) | I | ∼105 kb | N (ST7) | A2 | pemKI, hok-mok-sok, srnB-srnC |
| SY303P | Pig | ST5714 | B2 | blaCTX-M-65, floR, rmtB, tet(A) | III | ∼100 kb | N-F33:A−:B− | NT | pemKI, hok-mok-sok, srnB-srnC |
| SY301L | Pig | ST5442 | B2 | blaCTX-M-55, floR, oqxA, oqxB, blaTEM-1, tet(A) | II | ∼97 kb | F33:A−:B− | C1 | pemKI, hok-mok-sok, srnB-srnC |
| SY303M | Pig | ST3944 | B2 | blaCTX-M-14, floR, cfr, rmtB, aadA2, blaTEM-1 | IV | ∼97 kb | N | A1 | pemKI, hok-mok-sok, srnB-srnC |
| SY305M | Pig | ST3944 | B2 | blaCTX-M-14, floR, cfr, rmtB, aadA2, blaTEM-1 | IV | ∼97 kb | N | A1 | pemKI, hok-mok-sok, srnB-srnC |
| SY286M | Cow | ST93 | B2 | blaCTX-M-14, rmtB, aadA2, blaTEM-1 | IV | pECM13 (113,006 bp) | IncI1 (ST136) | E1 | pemKI, hok-mok-sok |
| SY287M | Cow | ST98 | B2 | blaCTX-M-14, floR, rmtB, aadA2, blaTEM-1 | IV | pECM13-like | IncI1 (ST136) | E1 | pemKI, hok-mok-sok |
| SY284B | Cow | ST4683 | B1 | blaCTX-M-14, floR, rmtB, aadA2, blaTEM-1 | IV | pECM13-like | IncI1 (ST136) | E1 | pemKI, hok-mok-sok |
| CH293B | Chicken | ST10 | B2 | blaCTX-M-55, floR, strA, strB, tet(A) | I | ∼97 kb | F33:A−:B− | C1 | pemKI, hok-mok-sok, srnB-srnC |
| CH292B | Chicken | ST10 | B2 | blaCTX-M-55, floR, cfr, blaTEM-1, strA, strB, tet(A) | II | pECB11 (92,545 bp) | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| CH291M | Chicken | ST410 | B2 | blaCTX-M-65, floR, rmtB | III | ∼100 kb | N-F33:A−:B− | C3 | pemKI, hok-mok-sok, srnB-srnC |
| CH281F | Chicken | ST5889 | A | blaCTX-M-65, floR, rmtB | III | ∼100 kb | N (ST7) | A2 | pemKI, hok-mok-sok, srnB-srnC |
| CH282M | Chicken | ST1437 | A | blaCTX-M-65, floR, rmtB, oqxA, oqxB | III | ∼100 kb | N (ST7) | A2 | pemKI, hok-mok-sok, srnB-srnC |
| CH285F | Chicken | ST448 | B2 | blaCTX-M-3, floR, strA, strB, tet(A) | I | ∼105 kb | F14:A−:B− | NT | pemKI, hok-mok-sok |
| DH286F | Chicken | ST2518 | B2 | blaCTX-M-3, rmtB, strA, strB, tet(A) | I | pECF12 (77,822 bp) | F33:A−:B− | D1 | pemKI |
| DH286M | Chicken | ST617 | B2 | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | ∼120 kb | F33:A−:B− | NT | pemKI, hok-mok-sok, srnB-srnC |
| JL12G | Cow | ST559 | B2 | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| JL15P | Cow | ST209 | B2 | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| JL17P | Cow | ST359 | A | blaCTX-M-65, tet(A) | III | ∼100 kb | N (ST7) | NT | pemKI, hok-mok-sok, srnB-srnC |
| JT14G | Cow | ST5689 | B1 | blaCTX-M-14, floR, rmtB, aadA2, blaTEM-1 | IV | ∼95 kb | N | A3 | pemKI, hok-mok-sok, srnB-srnC |
| JT14P | Cow | ST5714 | B1 | blaCTX-M-14, floR, rmtB, aadA2, blaTEM-1 | IV | ∼95 kb | N | A4 | pemKI, hok-mok-sok, srnB-srnC |
| HL12L | Cow | ST195 | D | blaCTX-M-55, floR, strA, strB, tet(A) | I | ∼97 kb | F33:A−:B− | C1 | pemKI, hok-mok-sok, srnB-srnC |
| HL36B | Cow | ST4680 | B1 | blaCTX-M-55, strA, strB, tet(A) | I | ∼100 kb | N | A3 | pemKI |
| HL40C | Farmer | ST4680 | B1 | blaCTX-M-14, floR, aadA2, blaTEM-1 | IV | ∼105 kb | IncI1 (ST136) | E2 | None |
| HB37B | Chicken | ST1725 | B1 | blaCTX-M-55, floR, rmtB, strA, strB, tet(A) | I | pECF12-like | F33:A−:B− | D1 | pemKI |
| HB35D | Chicken | ST48 | B2 | blaCTX-M-123 | III | ∼90 kb | F14:A−:B− | NT | pemKI, hok-mok-sok, srnB-srnC |
| HB38B | Cow | ST2055 | B1 | blaCTX-M-14, rmtB, aadA2, blaTEM-1 | IV | ∼140 kb | IncI1 (ST136) | E3 | pemKI |
| HB38L | Cow | ST5693 | B1 | blaCTX-M-14, rmtB, aadA2, blaTEM-1 | IV | ∼120 kb | IncI1 (ST136) | E4 | None |
| HB18F | Cow | ST1081 | B1 | blaCTX-M-14, floR, rmtB, aadA2, blaTEM-1 | IV | ∼140 kb | IncI1 (ST136) | E3 | pemKI |
| HB13B | Chicken | ST167 | B2 | blaCTX-M-55, rmtB, strA, strB, tet(A) | I | pECF12-like | F33:A−:B− | D2 | pemKI |
| HB13M | Cow | ST4463 | B2 | blaCTX-M-65, rmtB, tet(A) | III | ∼120 kb | N | NT | None |
| HB312G | Cow | ST685 | B2 | blaCTX-M-65, floR, rmtB, strA, strB, tet(A) | III | ∼120 kb | IncI1 (ST136) | E4 | None |
| SH312M | Pig | ST1488 | B2 | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| SH312N | Pig | ST1313 | B2 | blaCTX-M-65, floR, oqxA, oqxB | III | ∼120 kb | N | B | pemKI, hok-mok-sok, srnB-srnC |
| SH21F | Pig | ST209 | B2 | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| SH21G | Pig | ST354 | D | blaCTX-M-55, floR, cfr, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| SH21M | Pig | ST648 | D | blaCTX-M-55, floR, blaTEM-1, strA, strB, tet(A) | II | pECB11-like | F33:A−:B− | C2 | pemKI, hok-mok-sok, srnB-srnC |
| SH33L | Cow | ST3743 | B1 | blaCTX-M-65, tet(A) | III | ∼100 kb | IncI1 (ST136) | E5 | None |
The first two capital letters in each strain name represent the city in which the isolate was obtained: SY, Shenyang (Liaoning Province); CH, Changchun (Jilin Province); DL, Dehui (Jilin Province); JL, Jilin (Jilin Province); JT, Jiutai (Jilin Province); HL, Hailaer (Inner Mongolia autonomous region); HB, Harbin (Heilongjiang Province); SH, Suihua (Heilongjiang Province). Isolates from healthy animals are underlined.
Resistance genes were transferred by conjugation and determined by PCR.
RFLP patterns were assigned to the same major RFLP profiles (A–E) when they differed in ≤3 bands. The numbers (e.g., A1, A2 etc.) indicate minor variations within the RFLP patterns. NT, not typeable.
Conjugation by filter mating was conducted as previously described (13) to determine the transferability of fosA3-carrying plasmids. Plasmids of the transconjugants were analyzed by S1 nuclease PFGE and Southern blot hybridization using a fosA3-specific probe. Each transconjugant carried a single fosA3-positive plasmid, which ranged in size between 70 and 140 kb and was assigned to IncFII (n = 18), IncN (n = 12), or IncI1 (n = 9) replicon type (14–16) (Table 1). Restriction fragment length polymorphism (RFLP) analysis was successfully performed on 34 fosA3-carrying plasmids, but only five EcoRI RFLP profiles were detected, suggesting the presence of several epidemic plasmids that may be responsible for the dissemination of fosA3 in northeast China.
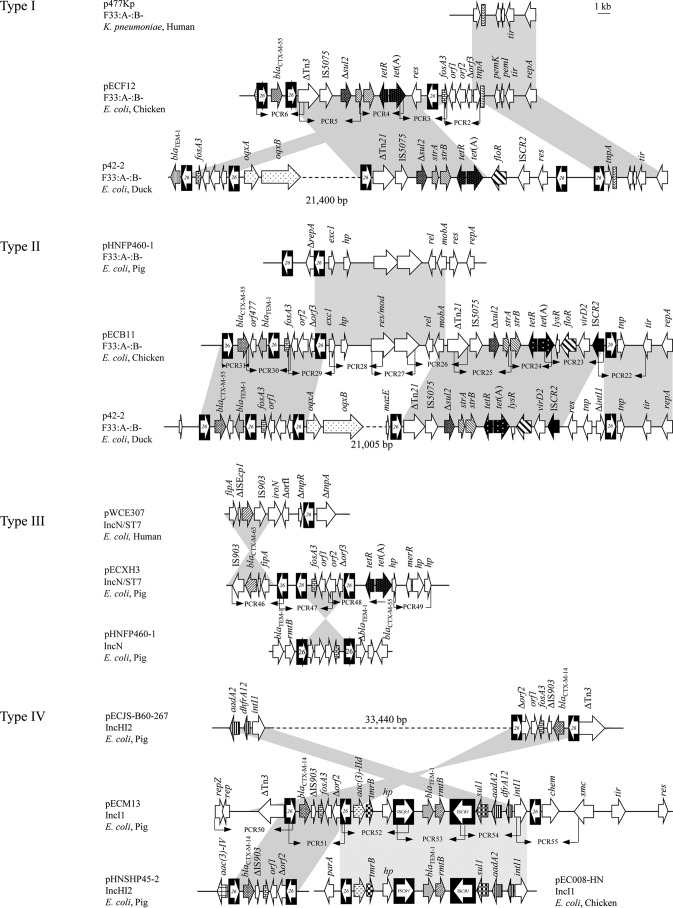
Based on several parameters (i.e., size, EcoRI RFLP profile, and replicon type), four representative fosA3-positive plasmids, namely, pECF12 (F33:A–:B–), pECB11 (F33:A–:B–), pECM13 (IncI1), and pECXH3 (IncN), were subjected to complete plasmid sequencing using the next-generation Illumina MiSeq system. The draft sequences of the plasmids were assembled by the GS de novo Assembler (version 2.8), and gap closure of the plasmids was done by PCR and Sanger sequencing. Except for pECXH3, from which only a fragment of 16,293 bp carrying the fosA3 gene was obtained, the remaining three plasmids were successfully sequenced and assembled. A PCR mapping approach served to determine the genetic context of the fosA3 gene in the remaining fosA3-positive plasmids. Based on the obtained sequences harboring fosA3 in this study, a series of primers (Fig. 1; see also Table S3 in the supplemental material) were designed to amplify several overlapping PCR fragments containing fosA3. Subsequently, all relevant PCR amplicons were cloned into pEASY-T1 and sequenced by primer walking (Invitrogen, Beijing, China). Four genetic environments of fosA3 were identified among the 39 plasmids and designated types I through IV (Fig. 1).
FIG 1.
Schematic presentation and comparison of the genetic environment of fosA3 in four plasmids with other related plasmids. The arrows indicate the positions and directions of transcription of the genes. Gray shading indicates >99% nucleotide sequence identity. PCR2-PCR6, PCR22-PCR31, PCR46-PCR49, and PCR50-PCR55 used to investigate the type I, II, III, and IV structures of the fosA3 environment are indicated by black arrowheads. Δ indicates a truncated gene. A 1-kb distance scale is displayed in the upper right corner. Diagrams were drawn from sequences deposited in the GenBank database under accession numbers LN897475 (p477kp), KT990220 (p42-2), KJ020575 (pHNFP460-1), HM440049 (pWCE307), KX254341 (pECJS-B60-267), KU341381 (pHNSHP45-2), and KP010147 (pEC008-HN).
The type I genetic environment is associated with the F33:A–:B– plasmid pECF12, which is 77,822 bp in length, has a mean G+C content of 53.1%, and contains 52 open reading frames (ORFs) with known functions (see Table S4 in the supplemental material). The pECF12 backbone of 59,497 bp showed high homology with F33:A–:B– plasmids, such as p477kp from human Klebsiella pneumoniae (17) and p42-2 (GenBank accession number KT990220, duck E. coli, China). The multidrug resistance region (MDRR) of 18,325 bp in pECF12 is bracketed on the left-hand side by the IS26 element and on the right-hand side by the ΔIS1 element. It contains three resistance genes, blaCTX-M-55, tet(A), and fosA3. The segment of 3,270 bp (IS26-fosA3-orf1-orf2-Δorf3) represents one of the most common genetic environments of fosA3 which was detected among eight fosA3-positive E. coli isolates from different cities and hosts in the present study (Table 1).
Plasmid pECB11, which harbors the type II structure of fosA3 environments, has a size of 92,545 bp, of which 61,191 bp represents the IncFII typical backbone segment encoding genes for plasmid replication, maintenance, conjugative transfer, and stability functions (see Table S5 in the supplemental material). The 31,354-bp MDRR of pECB11 has a mosaic structure, scattering four IS26 elements flanking two different fragments that harbor several resistance genes. The first segment of 7,724 bp corresponds to the IS26-formed composite transposon carrying the blaCTX-M-55, blaTEM-1, and fosA3 genes, which has been seen in several plasmids (e.g., p42-2). The second segment of 13,378 bp, containing the resistance genes floR, tet(A), strA, strB, and Δsul2, shared high nucleotide sequence identity with that found in p42-2 except for a 1,966-bp insertion between the 5′ end of the ISCR2 and upstream of the IS26 element (Fig. 1).
A 16,293-bp fosA3-carrying segment containing 17 ORFs was identified in the IncN/ST9 plasmid pECXH3 (see Table S6 in the supplemental material). This segment contains the type III structure of fosA3 genetic environments. In pECXH3, the fosA3 gene was located in an IS26-formed composite transposon (IS26-fosA3-orf1-orf2-Δorf3-IS26), which has been seen in plasmids of many replicon types found among isolates of diverse animal and human origins, such as pHNFP460-1 (KJ020575, pig E. coli, China, 2016), pEC012 (KT282968, chicken E. coli, China, 2016), and pSLK172-2 (CP017633, human E. coli, China, 2017) (Fig. 1).
Plasmid pECM13 has a size of 113,006 bp and an average G+C content of 50.6% and comprises 58 putative ORFs (see Table S7 in the supplemental material). This plasmid was assigned to IncI1/ST136 and harbored a novel fosA3 genetic environment, designated type IV. This genetic environment was detected in plasmids of two different replicon types among 11 fosA3-carrying E. coli isolates in this study, including the single isolate from a dairy farm worker. Like other resistance plasmids, pECM13 was divided into two parts, including the 90,949-bp plasmid backbone segment and a 22,057-bp MDRR. The pECM13 backbone is highly similar to that of other IncI1 plasmids, such as pEC15I_1 (KU932029, human E. coli, Finland, 2016) and pFAM22871_2 (KU355874, dairy cow E. coli, Switzerland, 2016). The MDRR of pECM13 also had a mosaic structure and contained five intact mobile elements (three IS26 elements, ISCfr1, and ISCR1) and nine resistance genes. Based on sequence comparisons, two transferable units existed in the MDRR of pECM13. The first transferable unit corresponded to the IS26-formed fosA3-carrying composite transposon, which has been found so far in the mcr-1-harboring plasmid pHNSHP45-2 from a pig E. coli isolate in China (18). The segment IS26-tmrB-ISCfr1-sul2-aadA2-dfrA12-intI1 was the second transferable unit, which has also been seen in another multidrug-resistant plasmid pEC088-HN (GenBank accession number KP010147) from a chicken E. coli isolate in China. Within this segment, the tunicamycin resistance gene, tmrB, coding for a protein of 180 amino acids (aa) was detected downstream of the aac(3)-IId gene.
Based on the known three complete sequences, a total of 73 overlapping PCRs were designed (see Table S3) to amplify 20, 25, and 27 partly overlapping regions covering the whole sequences of pECF12, pECB11, and pECM13, respectively. All replicons were cloned and sequenced. The results revealed that two, six, and two E. coli strains carried virtually the same pECF12-like, pECB11-like, and pECM13-like plasmids, respectively, except for a difference in a few nucleotides compared to the corresponding completely sequenced plasmids. These findings further demonstrate that three epidemic plasmids are maintained in northeast China and appear to be highly efficient vectors in the spreading of fosA3 and several other resistance genes (e.g., blaCTX-M variants, floR, and rmtB).
Accession number(s).
The complete sequences of three plasmids and a partial sequence from pECXH3 have been submitted to the NCBI database with accession numbers KY865321 (pECB11), KY865322 (pECF12), KY865323 (pECM13), and KY865324 (pECXH3).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (grant 2016YFD0501304).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00859-17.
REFERENCES
- 1.Poulakou G, Bassetti M, Righi E, Dimopoulos G. 2014. Current and future treatment options for infections caused by multidrug-resistant Gram-negative pathogens. Future Microbiol 9:1053–1069. doi: 10.2217/fmb.14.58. [DOI] [PubMed] [Google Scholar]
- 2.Sastry S, Doi Y. 2016. Fosfomycin: resurgence of an old companion. J Infect Chemother 22:273–280. doi: 10.1016/j.jiac.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raz R. 2012. Fosfomycin: an old–new antibiotic. Clin Microbiol Infect 18:4–7. doi: 10.1111/j.1469-0691.2011.03636.x. [DOI] [PubMed] [Google Scholar]
- 4.Castaneda-Garcia A, Blazquez J, Rodriguez-Rojas A. 2013. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel) 2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, Chen Z, Liu JH. 2012. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother 56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou J, Yang X, Zeng Z, Lv L, Yang T, Lin D, Liu JH. 2013. Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J Antimicrob Chemother 68:766–770. doi: 10.1093/jac/dks465. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Liu W, Liu Y, Wang J, Lv L, Chen X, He D, Yang T, Hou J, Tan Y, Xing L, Zeng Z, Liu JH. 2014. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front Microbiol 5:688. doi: 10.3389/fmicb.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals; third informational supplement. CLSI VET01-S, 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement CLSI M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M. 2010. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis 29:1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhang WJ, Xu XR, Schwarz S, Wang XM, Dai L, Zheng HJ, Liu S. 2014. Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J Antimicrob Chemother 69:385–389. doi: 10.1093/jac/dkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Fernandez A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 17.Sennati S, Riccobono E, Di Pilato V, Villagran AL, Pallecchi L, Bartoloni A, Rossolini GM. 2016. pHN7A8-related multiresistance plasmids (blaCTX-M-65, fosA3 and rmtB) detected in clinical isolates of Klebsiella pneumoniae from Bolivia: intercontinental plasmid dissemination? J Antimicrob Chemother 71:1732–1734. doi: 10.1093/jac/dkv506. [DOI] [PubMed] [Google Scholar]
- 18.Zhi C, Lv L, Yu LF, Doi Y, Liu JH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.