Abstract
Gene flow between species may last a long time in plants. Reticulation inevitably causes difficulties in phylogenetic reconstruction. In this study, we looked into the genetic divergence and phylogeny of 20 Lilium species based on multilocus analyses of 8 genes of chloroplast DNA (cpDNA), the internally transcribed nuclear ribosomal DNA (nrITS) spacer and 20 loci extracted from the expressed sequence tag (EST) libraries of L. longiflorum Thunb. and L. formosanum Wallace. The phylogeny based on the combined data of the maternally inherited cpDNA and nrITS was largely consistent with the taxonomy of Lilium sections. This phylogeny was deemed the hypothetical species tree and uncovered three groups, i.e., Cluster A consisting of 4 taxa from the sections Pseudolirium and Liriotypus, Cluster B consisting of the 4 taxa from the sections Leucolirion, Archelirion and Daurolirion, and Cluster C comprising 10 taxa mostly from the sections Martagon and Sinomartagon. In contrast, systematic inconsistency occurred across the EST loci, with up to 19 genes (95%) displaying tree topologies deviating from the hypothetical species tree. The phylogenetic incongruence was likely attributable to the frequent genetic exchanges between species/sections, as indicated by the high levels of genetic recombination and the IMa analyses with the EST loci. Nevertheless, multilocus analysis could provide complementary information among the loci on the species split and the extent of gene flow between the species. In conclusion, this study not only detected frequent gene flow among Lilium sections that resulted in phylogenetic incongruence but also reconstructed a hypothetical species tree that gave insights into the nature of the complex relationships among Lilium species.
Introduction
Recent progress in molecular technologies has made extensive molecular data available for phylogenetic studies [1]. With advanced techniques and big data, the understanding of the evolutionary relationships between living organisms has increased dramatically. Meanwhile, new challenges in the field of phylogenetics have arisen. Phylogenetic incongruence is a ubiquitous problem in phylogenetic reconstruction [2–4]. To get insights into the incongruence, Rokas et al. [2,5] and Hollingsworth et al. [6] suggest interpreting both the analytical and biological factors involved in the phylogenetic conflict. Though each individual factor has been examined by earlier studies empirically and/or theoretically (e.g., [7–9]), studies rarely inspect both the analytical and biological factors with empirical data. In our study, the commercially and ethnobotanically important genus Lilium was investigated to explore how both factors were responsible for the phylogenetic incongruence.
Analytical factors include the limitations and defects of the methods, while the biological factors comprise several evolutionary forces that are involved in the phylogenetic conflict. Incomplete lineage sorting, gene flow between taxa, horizontal transfer from the organellar genome, natural selection, and polyploidization are recognized as common biological factors that contribute to phylogenetic incongruence [10–13]. Among these factors, interspecific gene flow often leads to a reticulate evolutionary history [14–15]. While the incongruences stemming from gene flow between the populations or the closely related species are broadly addressed [16–18], the incongruences rooted in the genus-wide or higher-level interspecific gene flow are not well explained [19–20]. Because loci may experience different histories with different paces, combining loci inevitably causes systematic conflicts. There has been considerable debates on the combining multilocus sequence data for phylogenetic reconstruction. Most studies suggest that multilocus analysis effectively extends the number of evolutionarily informative characters and thus increases the accuracy of the phylogeny even with incomplete taxon sampling [21–24]. However, some studies indicate that multilocus sequences also unavoidably reduce the discriminatory power of the phylogenetic analysis [2,25].
The genus Lilium L. (Liliaceae), true lilies, is a group of herbs that are important worldwide for medicine, food, and horticulture [26–29]. This genus occurs in Eurasia and North America and is the most abundant in the Hengduan Mountain Region (HMR) of Southwest China and the Himalayan Mountain Ranges [26,30]. China is considered to be the biodiversity center of this genus, especially Sichuan, Yunnan, and Tibet [31–32], with a total of 55 species [29]. In total, approximately 100 Lilium species are classified into 5 to 11 sections [30, 33–35]. Seven sections in the genus sensu Comber [30] were applied in this study. High levels of morphological variation in Lilium lead to difficulties in the section delimitation. Previous phylogenetic studies suggest that most of the sections are polyphyletic, and some of these studies detected systematic incongruence among the loci [36–44]. Nevertheless, further clarifications on the mechanisms resulting in this incongruence were missing in all these studies. The fundamental understanding of the phylogenetic conflicts in Lilium makes the genus an ideal system for testing both the biological and analytical factors that are involved in phylogenetic incongruence.
There are two primary purposes in our study: 1) using multi-locus analyses to reconstruct the phylogeny of Lilium and to provide phylogenetic implications on the current section delimitation [30] and 2) to test whether analytical and/or biological factors caused the phylogenetic incongruence of Lilium. The incongruence caused by polyploidization was not discussed because most of the Lilium species are diploid, except for the triploid L. tigrinum [45–46]. In this study, we discussed how incomplete lineage sorting and interspecific gene flow can result in phylogenetic conflicts.
Materials and methods
Sampling and DNA extraction
In total, 29 Lilium samples, representing 20 species in seven sections, were collected from Taiwan, France, Japan, and China (Sichuan, Shandong, Yunnan, and Hubei Provinces). We confirm that the field studies did not involve endangered or protected species. No permission was needed because the samples were not collected in a protected area. The sample information is shown in Table 1. Leaf materials were dried with silica gel and stored at -80°C for later experiments. Genomic DNA from each sample was extracted using a CTAB method [47], diluted to 2 ng/μL with TE solution, and stored at -20°C.
Table 1. Information of the investigated Lilium samples.
| Species | Sections | Species codes | Number of samples | Localities and altitudes | Voucher information |
|---|---|---|---|---|---|
| L. speciosum var. gloriosoides | Archelirion | spe | 1 | Taiwan: Yilan (24.94N, 121.88E), 442 m | C.-C. Huang 42 |
| L. speciosum var. gloriosoides | Archelirion | gl8 | 1 | China: Fugong, Yunnan (26.53N, 98.89E), 2700 m | X. Gong PD008 |
| L. speciosum var. gloriosoides (cultivated) | Archelirion | spe | 1 | Taiwan: Shenkeng, Taipei (24.99N, 121.61N), 480 m | Y.-W. Ting 1 |
| L. maculatum | Daurolirion | mac | 2 | Japan: Izu, Shizuoka (34.97N, 138.95E), 44 m | N. Osada J1, J2 |
| L. formosanum | Leucolirion | for | 1 | Taiwan: Taoyuan (24.77N, 121.35E), 1491 m | C.-C. Huang Lf51 |
| L. leucanthum | Leucolirion | leu | 1 | China: Yichang, Hubei (30.67N, 111.25E) | X. Gong PD013 |
| L. sargentiae | Leucolirion | sar | 1 | China: Sichuan (32.15N, 107.26E) | X. Gong PD011 |
| L. sulphureum | Leucolirion | sul | 1 | China: Lijiang, Yunnan (26.88N,100.23E) | X. Gong PD010 |
| L. bulbiferum (cultivated) | Liriotypus | bul | 2 | France: Alpine Botanical Garden du Lautaret (45.03N, 6.40E), 2100 m | L. Malikova PD044, PD048 |
| L. pyrenaicum (cultivated) | Liriotypus | pyr | 2 | France: Alpine Botanical Garden du Lautaret (45.03N, 6.40E), 2100 m | L. Malikova PD038, PD039 |
| L. monadelphum (cultivated) | Liriotypus | mon | 2 | France: Alpine Botanical Garden du Lautaret (45.03N, 6.40E), 2100 m | L. Malikova PD050, PD051 |
| L. martagon (cultivated) | Martagon | mar | 1 | France: Alpine Botanical Garden du Lautaret (45.03N, 6.40E), 2100 m | L. Malikova PD034 |
| L. tsingtauense | Martagon | tsi | 2 | China: Laoshan, Shandong (36.18N, 120.58E), 700 m | X. Gong PD020, PD021 |
| L. ‘Casa Blanca’ (cultivated) | Not specified | cas | 1 | Taiwan: Taipei (25.07N, 121.53E), 170 m | Y.-W. Ting P |
| L. parryi (cultivated) | Pseudolirium | ryi | 1 | USA: Native Botanical Garden of Southern California (34.10N, 117.73W), 270 m | PD057 |
| L. pardalinum (cultivated) | Pseudolirium | num | 1 | USA: Native Botanical Garden of Southern California (34.10N, 117.73W), 270 m | PD056 |
| L. davidii (cultivated) | Sinomartagon | dav | 1 | China: Kunming Botanical Garden (25.15N, 102.74E), 1900 m | X. Gong PD012 |
| L. davidii var. willmottiae | Sinomartagon | wil | 1 | China: Sichuan (32.07N, 104.30E) | X. Gong PD001 |
| L. duchartrei | Sinomartagon | duc | 2 | China: Zhongdian, Yunnan (27.88N, 99.66E), 2800 m | X. Gong PD014, PD015 |
| L. leichtlinii (cultivated) | Sinomartagon | lei | 1 | France: Alpine Botanical Garden du Lautaret (45.03N, 6.40E), 2100 m | L. Malikova PD035 |
| L. taliense | Sinomartagon | tal | 2 | China: Zhongdian, Yunnan (28.08N, 99.46E), 3300 m | X. Gong PD003, PD004 |
| L. nepalense | Sinomartagon | nep | 1 | China: Yunnan (25.04N, 102.58) | X. Gong PD009 |
Primer design and selection
In total, 29 loci were investigated in our study. Of these loci, 20 were randomly selected from the expressed sequence tags (EST) of L. formosanum [48] and the NCBI EST database of L. longiflorum (http://www.ncbi.nlm.nih.gov/nucest). Primers for each selected locus were designed using the software Primer 3.0 [49]. Gene codes, primer sequences, and the putative functions for the 20 EST loci are available in Table 2. In addition, the internal transcribed spacer of the nuclear ribosomal DNA (nrITS) [50], as well as the additional eight chloroplast DNA (cpDNA) loci were employed (Table 2).
Table 2. Primers applied in this study.
| Gene codes | Amplicon length (bp) | Forward primer (5'-3') | Reverse primer (5'-3') | Putative functions (for ESTs) | Notes |
|---|---|---|---|---|---|
| ESTs | |||||
| Lf108 | 361–363 | AGGATTTCTCGAGGACCTAC | AATCCCAATTTGTATTGCAC | pectinesterase/pectinesterase inhibitor | GW590174.1 |
| Lf207 | 343–403 | ACCCTTTTGTTCCACACGAG | AGATTCCCACTGTCCCTGTCT | cytochrome P450-like TBP protein | GW591169.1 |
| Lf210 | 302–324 | GGGCATTTCATTTTCCCTTT | GAGTACCCACAGGGGTCAAA | S-adenosyl methionine decarboxylase | GW591158.1 |
| Lf212 | 279–286 | GCTCCCCATCACAGTTTCTC | CCACAGGGGTCAAAGTCAAA | S-adenosyl methionine decarboxylase | GW591146.1 |
| Lf218 | 266 | CTGCTCCTGAAGGAATCCAA | GGCTTCGAGACCAACATCAC | LLP-B3 protein | GW591136.1 |
| Lf219 | 256–299 | CTAGGCATCACACCCAAATG | TCCTGATGAAGTCGCTGATG | isopentenyl diphosphate isomerase II (IPI2) | GW591124.1 |
| Lf224 | 275–299 | CTCCTCAATCGGCACATTCT | CAGGGGACTCTCTCTCAGGA | sugar carrier protein | GW591114.1 |
| Lf229 | 303 | CTGCTCCTGAAGGAATCCAA | CGTCGAGGGTCGTGTCTACT | LLP-B3 protein | GW591080.1 |
| Lf230 | 326 | CCATGAGTGGAGTGACATGC | CACACGCTGGACTTCACATT | beta-tubulin | GW591069.1 |
| LL02 | 526–545 | CTCGAGAGAGAGGCCAGATG | CCTCTCTTCCATGCCCATTA | pentatricopeptide repeat-containing protein | DN985104.1 |
| LL17 | 527 | TCTGGAAGTTCTGCCCCTTA | CATCAAACTCCCTCCTCGAC | leucine-rich repeat receptor protein kinase MSP1-like | DN985157.1 |
| LL19 | 353–496 | CAAACCCTTAAACCGCATTG | CAAACCAGTCATTCCCCTGT | fatty acid desaturase | DN985108.1 |
| LL21 | 539–545 | GAGACCCGGCACTGTAAGAC | TGCTGCCAACAAGATAGCAC | hypothetical protein | DN985137.1 |
| LL22 | 531–536 | ATACCAGCAAACTGGGATGC | TCCTTGAGCACAATGAAATCTG | U-box domain-containing protein | DN985128.1 |
| LL25 | 532–533 | TCCGTCTTCCACTTCAGGAT | CACCAACAGCAACAGTCTGG | elongation factor 1-alpha | DR992640.1 |
| LL39 | 508–539 | GACCACCAAAATGTGCAAAA | AGATCAAGAGTCGCCGAAAC | pollen preferential protein | DN985087.1 |
| LL50 | 493 | AGCCAGCAAAGGAATTCTGA | AATTCTCCTCCCCGACATTT | subtilisin-like protease | DN985130.1 |
| LL89 | 490–491 | TTGTTCCACACGAGATTTCTGTT | GACAAGGGGAATCCGACTGT | cytochrome P450-like TBP protein | GR882236.1 |
| LL106 | 219–220 | CAGACTACAATTGAGATYGACTCC | ATCAGGGTTGATGCTCTTGC | heat shock protein 70 | BP177599.1 |
| LL107 | 320 | AAGGAGCTTGGGACTGTRATGC | CCTCACTTGGCCATCATGAC | calmodulin | FP052101.1 |
| cpDNA | |||||
| rbcL | 500 | CGCGGTGGACTTGATTTTACC | GGCATATGCCAAACATGAATACC | (Arzate-Fernández AM et al., 2005) | |
| psbC-trnS | 1467–1470 | GGTCGTGACCAAGAAACCAC | GGTTCGAATCCCTCTCTCTC | (Arzate-Fernández AM et al., 2005) | |
| trnT-trnL | 785–818 | CAAATGCGATGCTCTAACCT | AGTCCGTAGCGTCTACCGAT | (Nishikawa et al., 2002) | |
| trnL-trnF | 250–251 | TCCGTCGACTTTATAAGTCGTG | TGCCAGGAACCAGATTTGAACT | (Nishikawa et al., 2002) | |
| atpB-rbcL | 637–643 | GAAGTAGTAGGATTGGTTCTCA | CCAACACTTGCTTTAGTTTC | (Nishikawa et al., 2002) | |
| petA | 605–606 | CCCATTTTTGCACAGCAGGGTTATG | CCCTCKGAAACAAGAAGTT | (Grivet et al., 2001) | |
| ycf4 | 391–392 | GGCCYCGGATTTCCATATAAAG | TGGCGATCAGAACAYATATGGATAG | (Grivet et al., 2001) | |
| psbB | 628–629 | GGATTRCGTATGGGMAATATTGAAAC | CCAAAAGTRAACCAACCCCTTGGAC | (Graham and Olmstead, 2000) | |
| nrITS | 694–704 | GGAAGTAAAAGTCGTAACAAGG | TCCTCCGCTTATTGATATGC | (Peterson et al., 2004) |
PCR, cloning, and sequencing
PCR amplification was conducted with a reaction volume of 50 μL, containing 25 μL of 2×Taq polymerase master mix (Ampliqon, Denmark), 5 μL of template DNA (2 ng/μL), 5 μL of each primer (2 pM), and distilled water. The PCR reactions were performed using the MyCycler thermal cycler (Bio-Rad, USA) with 35 cycles. For each cycle, we set an initial denaturation at 94°C for 50 s, annealing at 48–53°C (optimized for each locus, Table 2) for 50 s, and extension at 72°C for 80 s. A final extension at 72°C for 10 min was applied. The PCR products were run on agarose gels, and the targeted DNA fragments were sliced and purified. The purified PCR products were ligated to a pGEM-T Easy vector at 4°C overnight and transformed into E. coli DH5a cells. Positive clones were validated with blue-white screening followed by colony PCR. To ensure that both diploid alleles were sequenced, we randomly selected five to seven clones from each individual, and discarded the low-frequency clones due to possible PCR errors. Sanger sequencing was conducted in both directions with the universal T7P and SP6 primers using the 96-capillary 3730xl DNA Analyzer (Genomics Biotech Co., Ltd.).
Sequence alignments, indel identification, and tree reconstructions
The sequences of the EST, nrITS, and cpDNA loci of the 20 Lilium species were validated using BLAST on NCBI. All sequences were deposited in the NCBI GenBank, with accession numbers of KX863745-KX865072. In addition, the full-length chloroplast genome and nrITS sequences of Cardiocrinum cordatum (KX575837.1 and KP712019.1, respectively) and Fritillaria taipaiensis (KC543997.1 and KT861551.1, respectively) were downloaded from NCBI as outgroups for the Lilium species. Sequences of each locus were then aligned using CLC Free Workbench (http://www.clcbio.com/) with the default settings, and gap sites were manually checked. Indel events for each locus were identified and coded with SeqState [51] to be incorporated in the phylogeny reconstruction. Genetic distances of each alignment were estimated using the two-parameter model implemented in MEGA 6 [52–53].
For the cpDNA markers, the alignments of all the genes representing 5,432 bp were concatenated, the indels were coded according to Simmons and Ochoterena’s simple coding method [54] in SeqState, and a Bayesian inference tree was generated with MrBayes v. 3.2.6 [55]. The best substitution models for the cpDNA were evaluated by MEGA6, and the substitution model used in MrBayes was set accordingly (S1 Table). We performed > 100,000 steps of a Markov chain Monte Carlo (MCMC) for each gene to ensure the average standard deviation of the split frequencies was lower than 0.01, with a sample frequency of 100, print frequency of 100, and diagnosis frequency of 1,000. After summarizing the parameter values, the potential scale reduction factor was confirmed to be approximately 1.0 for all parameters. Finally, the consensus tree was summarized using the default settings. For nrITS and EST markers, tree reconstructions were also conducted with MrBayes for individual loci with the same settings as the cpDNA.
For integrating the information of cpDNA and nrITS genes to reconstruct the phylogeny, the software BEAST version 1.7.5 [56] was used. An uncorrelated relaxed clock model was set for both cpDNA and nrITS loci. The priors of the substitution rate were set as uniform distributions at initial values of 0.000933 and 0.00968, respectively. The prior of the divergence time of Lilium samples was set as a normal distribution with a mean of 13.6 million years and a standard deviation of 1.5 million years based on the estimation of Gao et al. [57]. The length of the MCMC run was set to 10 million, and the parameters were saved every 1,000 steps. The results of the trees for 10 independent runs with different seeds were combined with the program logcombiner [56], with 50% of the trees discarded as burn-in, and subsequently processed by the program treeannotator [56] to generate a consensus tree (hypothetical species tree). The hypothetical species tree was visualized using FigTree v. 1.4.2. (http://tree.bio.ed.ac.uk/software/figtree/), converted to the Newick format, and then annotated in MEGA6.
Intraspecific genetic variations, recombination rates (Rm)
Genetic diversities among the studied taxa were estimated using DnaSP version 5.0 [58]. The nucleotide diversities (π) and the minimum recombination events were calculated. The potential gene flow among the species was inferred from the recombination events and the shared genetic variations among the species [59].
IMa analyses
To sophisticatedly estimate gene flow between species clusters, the Isolation with Migration model that was implemented in program IMa2 [60] was applied, and six model parameters were calculated using coalescence simulations and Bayesian computational procedures: the divergence time (t), the bidirectional migration rates (m1 and m2), and the effective population sizes of the ancestral (θA) and two current populations (θ1 and θ2). Taxa with a possible hybrid origin, L. davidii var. willmotiae, L. bulbiferum, and L. ‘Casa Blanca’, were excluded. Using the coalescence time of 13.6 million years ago for the Lilium crown group [57], we estimated the substitution rates for the 20 EST loci by dividing the root height of the Bayesian trees by the coalescence time (S2 Table). The infinite site (IS) models were applied to all the loci. Because the IMa2 program does not accept genes containing recombinant fragments, the IMGC program was used to extract the largest non-recombinant DNA fragments from the aligned sequences [61]. After processing our data using the IMGC program, the DNA fragments that were shorter than 100 bp were excluded due to a lack of sufficient genetic information. To ensure that there were enough heating steps for obtaining a reliable result, 20 Markov chain Monte Carlo chains were used with the following heating parameters: ha of 0.96 and hb of 0.9. IMa2 runs were performed and saved using at least 3 million burn-in steps followed by at least 5 million steps (50,000 genealogies). All the effective parameter sample sizes were greater than 100. Three independent runs with different random seeds were performed to check for consistency across the results. The final results were calculated by averaging the values estimated in each run. The migration rate per generation (M) was calculated by multiplying the m value by the geometric mean of the substitution rates (μ).
Results
Phylogeny based on eight cpDNA genes
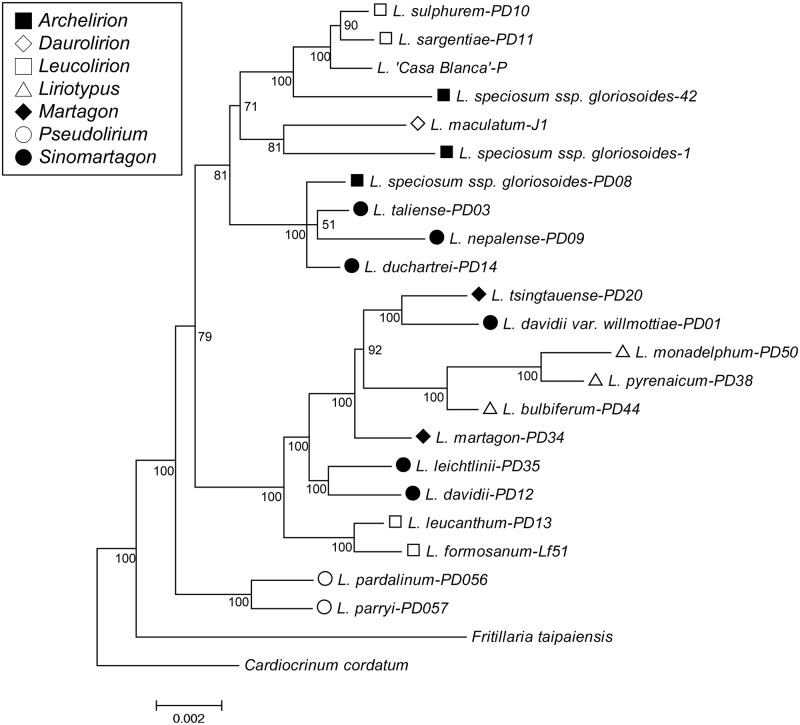
The Bayesian inference phylogram based on the concatenation of eight cpDNA loci showed that the Pseudolirium section was monophyletic and at the basal position, while 18 other species from 5 sections were clustered into two main groups (Fig 1). The Leucolirion, Martagon, and Sinomartagon sections were paraphyletic or polyphyletic, whereas the Liriotypus section was monophyletic. Unexpectedly, three Lilium speciosum var. gloriosoides samples from different populations (two samples from Taiwan and one from Yunnan in China, see Table 1) were not clustered together. Within the Leucolirium section, L. formosanum was sister to L. leucanthum, L. sulphureum was sister to L. sargentiae, but the four taxa were not clustered together. Within the Sinomartagon section, L. leichtlinii was clustered with L. davidii var. davidii, but L. taliense, L. duchartrei, and L. nepalense were clustered with L. speciosum var. gloriosoides from Yunnan (PD08) of the Archelirion section. In addition, L. davidii var. willmottiae of the Sinomartagon section was sister to L. tsingtauense in the Martagon section, suggesting a hybrid origin.
Fig 1. Bayesian inference phylogram reconstructed using the concatenation of eight cpDNA genes with MrBayes.
The tree was rooted at Cardiocrinum cordatum. Sections are coded as follows: section Archelirion, solid square, section Daurolirion, blank diamond, section Leucolirion, blank square, section Liriotypus, blank triangle, section Pseudolirium, blank circle, section Sinomartagon, solid circle, and section Martagon, solid diamond. Posterior probabilities are shown below the branches.
Phylogeny based on nrITS
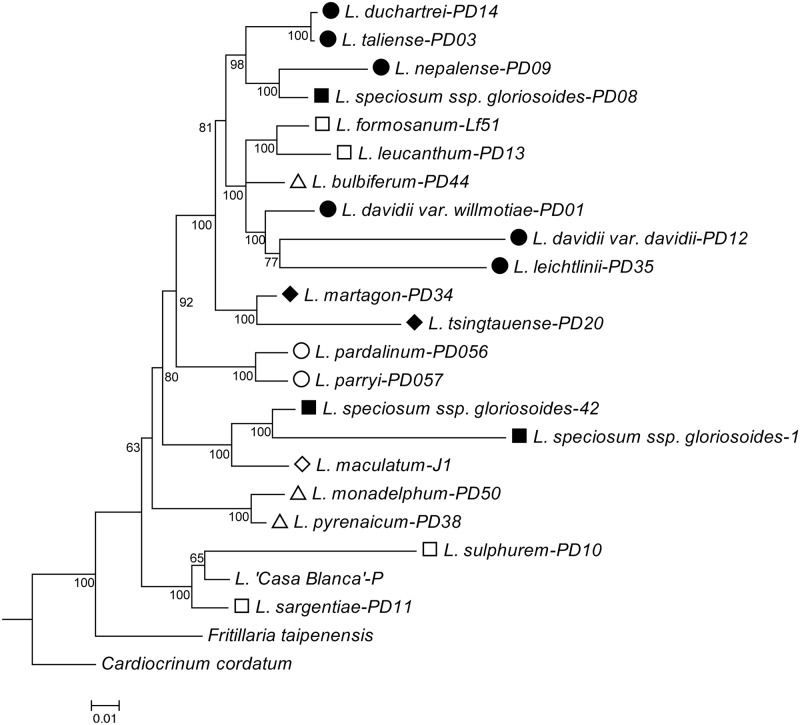
The Bayesian inference tree based on nrITS suggested that 20 Lilium species were divided into one main cluster and several smaller clusters (Fig 2). The main cluster consisted of six species of section Sinomartagon, L. formosanum and L. leucanthum of section Leucolirion, L. bulbiferum of section Liriotypus, and L. speciosum ssp. gloriosoides (section Archelirion) from Yunnan. For the small clusters, two species in the Martagon section were clustered, two species in the Pseudolirium section were grouped together, L. speciosum ssp. gloriosoides (section Archelirion) from Taiwan was clustered with L. maculatum in section Daurolirion, L. monadelphum and L. pyrenaicum in section Liriotypus were clustered, while L. sulphureum and L. sargentiae in section Leucolirion were clustered with the cultivar L. ‘Casa Blanca’. The ITS topology displayed a closer relationship between L. bulbiferum and section Sinomartagon, which has also been revealed in previous studies [38,40,57].
Fig 2. Bayesian inference phylogram reconstructed based on nrITS with MrBayes.
The tree was rooted at Cardiocrinum cordatum. Posterior probabilities are shown below the branches. The sections are coded the same as they are in Fig 1.
Phylogeny based on the combined data of nrITS and cpDNA genes: Hypothetical species tree
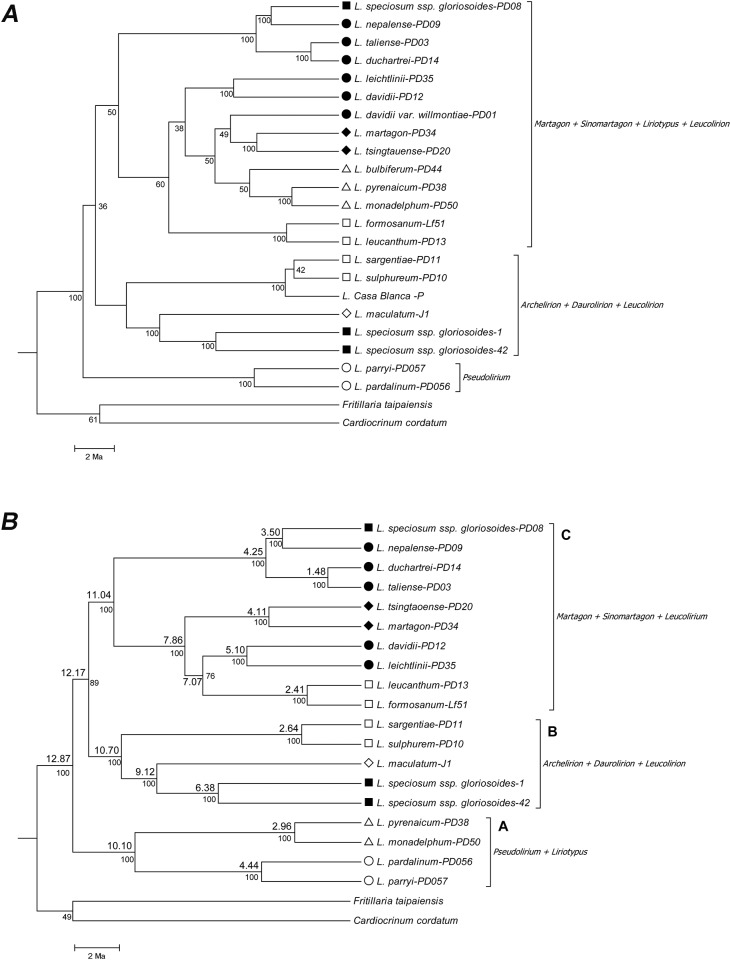
By combining nrITS and cpDNA regions using the BEAST software, the phylogeny showed better resolution on the delimitation of the sections (Fig 3A). This tree uncovered three clusters: Cluster A of L. pardalinum and L. parryi (sect. Pseudolirium); Cluster B of L. sargentiae, L. sulphureum (sect. Leucolirium), L. maculatum (sect. Daurolirion), L. speciosum ssp. gloriosoides in Taiwan (sect. Archelirion), and L. 'Casa Blanca'; and Cluster C comprising L. martagon, L. tsingtauense (sect. Martagon), L. davidii, L. davidii var. willmottiae, L. leichtlinii, L. taliense, L. duchartrei, L. nepalense (sect. Sinomartagon), L. formosanum, L. leucanthum (sect. Leucolirium), L. bulbiferum, L. pyrenaicum, L. monadelphum (sect. Liriotypus), and L. speciosum ssp. gloriosoides in China (sect. Archelirion). When L. davidii var. willmotiae, L. bulbiferum, and L. ‘Casa Blanca’ with a possible hybrid origin were removed from the analysis, the Liriotypus section clustered with the Pseudolirium section of Cluster A instead of the Martagon and Sinomartagon sections of Cluster C, while the other taxa remained in the previous positions, implying an affinity between L. bulbiferum and the Sinomartagon section (Fig 3B). This tree without the hybrid interference better resolved the phylogenetic relationships among the Lilium sections. Here, we identified nine sister groups. They were L. formosanum and L. leucanthum, L. sargentiae and L. sulphureum (sect. Leucolirion), L. davidii var. davidii and L. leichtlinii, L. taliense and L. duchartrei, L. nepalense (sect. Sinomartagon) and L. speciosum var. gloriosoides in China (sect. Archelirion), L. tsingtauense and L. martagon (sect. Martagon), L. pyrenaicum and L. monadelphum (sect. Liriotypus), L. pardalinum and L. parryi (sect. Pseudolirium), as well as L. speciosum var. gloriosoides in Taiwan and L. maculatum (sects. Archelirion and Daurolirion). Hence, the tree was deemed the hypothetical species tree (Fig 3B) in the following discussions. Molecular dating with BEAST revealed that the coalescence time of Lilium was approximately 12.87 million years ago (Ma) (95% CI: 9.88–15.81). The coalescence time of Clusters A, B, and C were dated at 10.10 Ma (95% CI: 6.88–13.33), 10.70 Ma (95% CI: 7.87–13.87), and 11.04 Ma (95% CI: 8.08–14.08), respectively, and the coalescence times of the nine sister groups ranged from 1.48 Ma (L. duchartrei and L. taliense, 95% CI: 0.41–3.16) to 9.12 Ma (L. maculatum and L. speciosum ssp. gloriosoides of Taiwan, 95% CI: 7.87–13.87), suggesting long divergences among these Lilium species (Fig 3B).
Fig 3. The phylogeny reconstructed based on the combined data of nrITS and cpDNA genes with the BEAST analysis.
The tree was rooted at the outgroups of Fritillaria taipaiensis and Cardiocrinum cordatum. The sections are coded as they are in Fig 1. The scale bar denotes 2 million years ago (Ma). Posterior probabilities are shown below the branches. (A) All sampled taxa were analyzed. (B) L. bulbiferum, L. davidii var. willmotiae, L. ‘Casa Blanca’ were removed from the analysis due to their possible hybrid origins. The tree was deemed the hypothetical species tree. Numbers on the branches represent estimated divergence times.
Furthermore, the topology comparisons between the hypothetical species tree (Fig 3B) and the 20 EST trees (S1 Appendix) were conducted focusing on the sister relationships among the groups. Of the gene phylogenies, only the LL22 tree uncovered all the sister groups (S1 Appendix). Of these sister groups, L. pardalinum and L. parryi showed the highest supporting rate (55%), followed by L. formosanum and L. leucanthum (40%) as well as L. sargentiae and L. sulphureum (40%) (S3 Table). Unexpectedly, the sister relationship between L. speciosum ssp. gloriosoides in Taiwan and L. maculatum was not supported by any EST tree.
Intraspecific genetic variations, recombination events, and IMa analyses
The nucleotide diversities of the chloroplast genes among the 20 Lilium species were generally lower than those of the nuclear loci. Locus Lf207 had the lowest nucleotide diversities (π = 0.01527 ± 0.00322), while the nucleotide diversities of Lf210, Lf224, LL19, LL25, LL89, LL106, and nrITS were relatively higher (Table 3). Moreover, fewer recombination events were detected in the chloroplast loci (Table 3). It is likely attributable to maternal inheritance of the chloroplast DNA, which reserved the primordial genetic variation of the ancestral genotypes. In contrast, high recombination rates were detected in most of the EST loci, i.e., Lf108, Lf207, Lf210, Lf212, Lf224, Lf229, LL02, LL17, LL19, LL22, LL25, LL39, LL50, LL89, LL106, and LL107.
Table 3. Recombination rates and nucleotide diversities of the 20 Lilium species.
| Gene codes | Minimum numbers of recombination events (Rm) | Nucleotide diversity (π) |
|---|---|---|
| Lf108 | 16 | 0.05714±0.00213 |
| Lf207 | 12 | 0.01527±0.00322 |
| Lf210 | 23 | 0.05868±0.00626 |
| Lf212 | 15 | 0.05205±0.00456 |
| Lf218 | 9 | 0.03955±0.00265 |
| Lf219 | 7 | 0.03979±0.00171 |
| Lf224 | 28 | 0.14724±0.01027 |
| Lf229 | 11 | 0.03906±0.00233 |
| Lf230 | 6 | 0.02865±0.00279 |
| LL02 | 14 | 0.04163±0.00193 |
| LL17 | 15 | 0.03498±0.00151 |
| LL19 | 20 | 0.07564±0.00829 |
| LL21 | 3 | 0.01854±0.00095 |
| LL22 | 13 | 0.03825±0.0014 |
| LL25 | 23 | 0.07758±0.00654 |
| LL39 | 17 | 0.05378±0.00144 |
| LL50 | 19 | 0.05061±0.00125 |
| LL89 | 28 | 0.0212±0.00486 |
| LL106 | 30 | 0.17965±0.00755 |
| LL107 | 13 | 0.03966±0.00674 |
| ITS | 52 | 0.08574±0.0089 |
| rbcL | 1 | 0.00623±0.00058 |
| psbC-trnS | 3 | 0.00824±0.00055 |
| trnT-trnL | 3 | 0.01754±0.00115 |
| trnL-trnF | 0 | 0.00691±0.00137 |
| atpB-rbcL | 0 | 0.00671±0.00096 |
| petA | 0 | 0.00678±0.00078 |
| ycf4 | 0 | 0.00865±0.00318 |
| psbB | 0 | 0.00586±0.00068 |
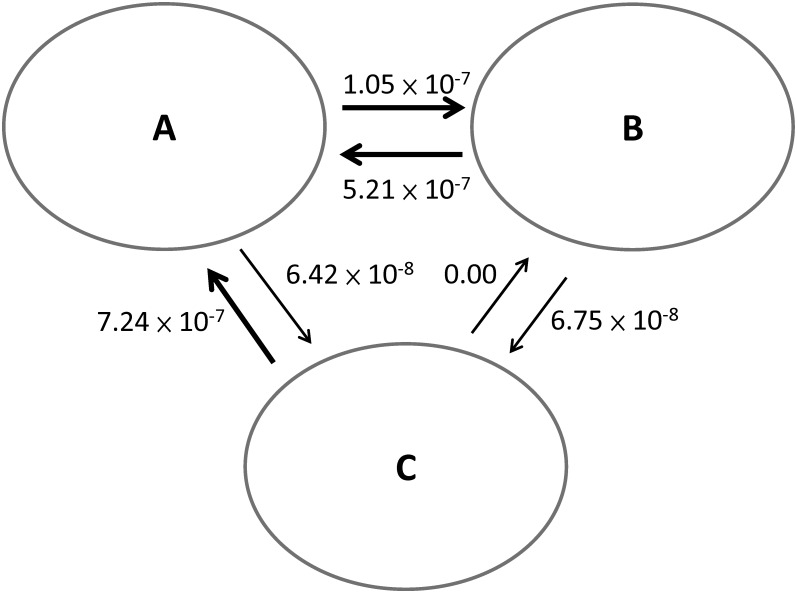
We used IMa2 to evaluate the levels of gene flow among the three Lilium clusters (Fig 4). The taxa that had a possible hybrid origin were excluded from this analysis. The level of gene flow was estimated by the migration rates per generation (M). In the pairwise comparisons between Clusters A, B, and C, the highest level of gene flow occurred from Clusters C to A (M = 7.43 × 10−7), followed by the gene flow from Clusters B to A (M = 5.21 × 10−7) and the gene flow from Clusters A to B (M = 1.03 × 10−7). All the above directions showed significant gene flow, as was suggested by the likelihood ratio test (p < 0.05) (S4 Table). These results suggested that hybridization across sections may have occurred frequently in Lilium.
Fig 4. Gene flow among the three clusters suggested by the hypothetical species tree.
Numbers denote the extent of the gene flow estimated by the IMa2 analysis. The thick arrow indicates significant gene flow as suggested by the likelihood ratio tests.
Indel events
Our results revealed that indel distributions varied among the Lilium taxa in the nuclear loci. The indel events in cpDNA and nrITS were identified and were included in the phylogeny reconstruction, and some informative indels in the EST loci were shown in the S5 Table. For example, a 12-bp insertion at site 20 on LL39 was found in L. tsingtauense and L. martagon (sect. Martagon), and L. taliense and L. duchartrei (sect. Sinomartagon) shared a 1-bp deletion at site 458 on LL22. Some indels were shared by multiple sections. For instance, a 13-bp deletion at site 519 on LL39 occurred in the Pseudolirium (L. pardalinum and L. parryi) and Leucolirion sections (L. sulphureum and L. sargentiae), and a 3-bp deletion at site 264 of LL39 was found in L. speciosum var. gloriosoides (sect. Archelirion) and L. nepalense (sect. Sinomartagon).
Discussion
This study unraveled the factors that contribute to the phylogenetic conflicts by exploring multilocus phylogenies of Lilium. Although phylogenetic conflict can be rampant across loci, only a few studies have addressed the causal factors comprehensively [3–4]. For Lilium, several phylogenetic studies reflected its phylogenetic conflict but failed in illustrating the mechanism or factors resulting in the conflict [41–42]. Here, we evaluated the influences of the analytical and biological factors that led to the phylogenetic conflict in Lilium.
Analytical and biological factors causing phylogenetic conflicts
Multilocus approach
While the debate on the advantages and disadvantages of combining data in reconstructing multilocus phylogeny continues, many recent studies suggested that combining all the available data is feasible and reliable and that elucidating incongruence provides hints into the evolutionary history [21,23,25]. Unfortunately, most of the phylogenetic studies that combined sequences across loci provided very few explanations regarding the reliability of the combined trees (e.g., [62–63]). In our study, with a wide locus sampling, predominant phylogenetic incongruences across different loci were revealed (e.g., S3 Table and S1 Appendix). By combining the cpDNA and nrITS loci, species from the same sections were mostly clustered and the topology of the 17 Lilium species substantially agreed with the taxonomic sections by Comber [30] (Fig 3B). The hypothetical species tree (Fig 3B) uncovered three clusters: Cluster A (sect. Pseudolirium and Liriotypus), Cluster B (Leucolirion, Daurolirion, and Archelirion), and Cluster C (Leucolirion, Martagon and Sinomartagon). The multilocus analyses not only gave an insight into the Lilium phylogeny but also provided opportunities to uncover the taxa that had a hybrid origin as shown earlier and revealed the interspecific gene flow, which is addressed in the following paragraphs.
Interspecific gene flow
Intraspecific gene flow could provide concordance in the species genome. It would homogenize the genomes and thereby block the genome divergence of the isolated populations [64]. However, when gene flow among taxa occurs, phylogenetic incongruence ineluctably arises [14–15]. Interspecific gene flow is not rare in plants [65,66]. Examples include Howea belmoreana and H. forsteriana [17], and Arabidopsis halleri and A. lyrata [18], all revealing uninterrupted gene flow after speciation [18]. Interspecific gene flow causes extreme difficulty with regard to the phylogenetic reconstruction.
The divergence of the crown group of Lilium can be dated back to 13.6 million years ago [57]. All the examined taxa in our study diverged a very long time, even the closest sister pair, L. taliense and L. duchartrei, for whom the divergence was more than 1.4 million years. Of the three clusters identified in the hypothetical species tree (Fig 3B), long divergences between the clusters (more than 10 million years) tended to reject incomplete lineage sorting, which blurs the species delimitation in Lilium. Given the rampant phylogenetic incongruence across the loci, interspecific gene flow may have been largely involved in the evolution of the Lilium species. Here, three inspections were proposed to elucidate the possibility and strength of interspecific gene flow.
First, as demonstrated earlier, L. bulbiferum and L. davidii var. willmottiae were recognized as hybrids based on their inconsistent placements on the cpDNA and nrITS trees (Figs 1 and 2). Second, in the DnaSP analysis, 17 out of 20 (85%) EST loci appeared to have high number of recombination events (Table 3). Third, the result of the IMa2 analyses suggested that historical gene flow likely occurred between the three clusters of the hypothetical species tree, especially the genetic exchanges with Cluster A (Fig 4). It is noticeable that all the samples in Cluster A were cultivars, implying the possibility of artificial hybridization, whether intentional or not. It has been shown that the artificial crosses between the different Lilium sections were common and not difficult [67]. As the strongest gene flow occurred from Cluster C, which predominately originated from China, to Cluster A of America or Europe, the gene flow across continents also implied that artificial hybridization may have blurred the species/section boundaries of the Lilium species.
Overall, even though our inspections have limited power in surveying the quantity and direction of gene flow due to the small sample size of each taxon, our results suggested that extensive gene flow among taxa had occurred in Lilium. The plentiful interspecific gene flow apparently contributed to the difficulties in section delimitation. It was likely that interspecific gene flow arose from artificial hybridizations, and thereby caution ought to be exercised in using cultivated samples for phylogeny reconstruction.
Phylogenetic implications
Our results reflected adequate resolution on the phylogenies, despite a small sample size, reflecting the power of incorporating multiple loci in the phylogenetic reconstruction. Some taxa that were assigned to the same section appeared to be sister groups in the phylogenies of cpDNA and nrITS, thanks to the low genetic recombination and lower substitution rate of the maternally inherited cpDNA (Table 3 and S2 Table; [68]) and/or the concerted evolution of nrITS [69–71].
Our phylogenies affirmed the previous studies that identified the Martagon section as a monophyletic group [37–38,43–44] with close relationships to the Leucolirion and Sinomartagon sections [36] (Fig 3B). Moreover, the Liriotypus section was determined to be polyphyletic in previous studies that include L. bulbiferum [38,40,43], whereas it was determined to be monophyletic in the studies that excluded L. bulbiferum from the analysis [37,44]. Our study revealed similar results, with the cpDNA phylogeny uncovering monophyly of the Liriotypus section, whereas the nrITS phylogeny showed that L. bulbiferum clustered with the Sinomartagon section. Apparently, the hybrid origin of L. bulbiferum caused noise in the phylogenetic inference. Likewise, the BEAST analysis, which was based on the combined data of cpDNA and nrITS, further supported the monophyly of the Liriotypus section and its close affinity to the Sinomartagon section. Interestingly, when L. bulbiferum was removed from the phylogenetic analysis, the Liriotypus section became the neighbor of the Pseudolirium section (Fig 3B). Furthermore, phylogenies of the EST loci revealed that L. bulbiferum was clustered with L. davidii, L. leichtlinii (sect. Sinomartagon), L. monadelphum, or L. pyrenaicum (sect. Liriotypus) (S1 Appendix). These results suggested that L. bulbiferum did show affinity to the Sinomartagon section, as suggested by other studies based on nrITS [38;40;57] and a maternal background from the Liriotypus section based on the chloroplast DNA. Altogether, we suggested that L. bulbiferum is likely a hybrid between the Sinomartagon and Liriotypus section, with the latter as the maternal parent. Furthermore, the Pseudolirium section appeared to be basal in Lilium, both in the phylogenies of cpDNA and the combined data, a finding consistent with the matK gene phylogeny [57].
Our study also revealed that the Leucolirion and Sinomartagon sections were polyphyletic, which largely corroborated earlier phylogenies [36–38,43–44, 57]. In the Leucolirion section, 40% of the EST loci supported that L. formosanum and L. leucanthum were sisters, and 40% of nuclear loci showed that L. sargentiae was related to L. sulphureum, while no data collected here indicated the clustering of these four species (S3 Table, S1 Appendix). The close relationship between L. sargentiae and L. sulphureum also corresponded to the geographic regions where they grow. In contrast, no overlap in the distribution of L. formosanum and L. leucanthum has been reported. Although the allopatric distribution of this sister group might imply a longer divergence between L. formosanum and L. leucanthum, the close phylogenetic relationship indicated their affinity. However, the possibility of a sampling bias that contributes to this allopatric match relationship cannot be ruled out. The cpDNA, nrITS, and the combined data all suggested that L. ‘Casa Blanca’ was clustered with L. sargentiae and L. sulphureum (Figs 1–3). This may infer that part of the parental species of L. Casa Blanca was from the Leucolirion section. The largest section, Sinomartagon, contains 22 taxa, which are morphologically distinguishable from each other [30,72]. Nishikawa et al. [38] divided this section into five groups according to the phylogeny based on the nrITS data. Our results on the Sinomartagon section generally agreed with Nishikawa et al.’s work [38].
Even though our sampling on the Archelirion section was restricted to two populations of L. speciosum ssp. gloriosoides, the hypothetical species tree and most of the EST trees revealed genetic dissimilarities between the populations (Fig 3B and S1 Appendix). The individuals in Taiwan were closely related to the Daurolirion section, while the individual isolated from China was related to L. nepalense of the Sinomartagon section. Accordingly, we suggest assigning L. speciosum ssp. gloriosoides of China to the Sinomartagon section instead of the Archelirion section.
Conclusions
In summary, multilocus analyses enabled us to uncover interspecific gene flow, identify the taxa with hybrid origins, and comprehensively reconstruct the evolutionary history of Lilium. The hypothetical species tree better resolved the section classification of the Lilium species. Our study suggested that future studies exploring both analytical and biological factors that cause phylogenetic conflicts would provide a better understanding of the evolutionary relationship among plant species.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
Data Availability
All sequences were deposited in NCBI GenBank, and the accession numbers were KX863745-KX865072.
Funding Statement
This work was supported by Ministry of Science and Technology: 104-2621-B-006 -002, https://www.most.gov.tw/en/public. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gojobori T, Chiang TY. Opening a new era of “Ecological Genetics and Genomics”. Ecological Genetics and Genomics. 2016;1:8. [Google Scholar]
- 2.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- 3.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends in Genetics. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003 [DOI] [PubMed] [Google Scholar]
- 4.Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology. 2011;9:e1000602 doi: 10.1371/journal.pbio.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rokas A, King N, Finnerty J, Carroll SB. Conflicting phylogenetic signals at the base of the metazoan tree. Evolution and Development. 2003;5:346–359. [DOI] [PubMed] [Google Scholar]
- 6.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254 doi: 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ennos RA, French GC, Hollingsworth PM. Conserving taxonomic complexity. Trends in Ecology and Evolution. 2005;20:164–168. doi: 10.1016/j.tree.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 8.Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences USA. 2009;106: 18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingsworth ML, Clark A, Forrest LL, Richardson J, Pennington RT, Long DG, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Molecular Ecology Resources. 2009;9:439–457. doi: 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 10.Wendel JF, Doyle JJ. Phylogenetic incongruence: window into genome history and molecular evolution In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular Systematics of Plants II: DNA Sequencing. Boston: Kluwer; 1998. p. 26 [Google Scholar]
- 11.Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. Phylogeographic studies in plants: problems and prospects. Molecular Ecology. 1998;7:465–474. [Google Scholar]
- 12.Syvanen M. Evolutionary implications of horizontal gene transfer. Annual Review of Genetics. 2012;46:341–358. doi: 10.1146/annurev-genet-110711-155529 [DOI] [PubMed] [Google Scholar]
- 13.Naciri Y, Linder HP. Species delimitation and relationships: The dance of the seven veils. Taxon. 2015;64:3–16. [Google Scholar]
- 14.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 15.Gauthier O, Lapointe FJ. Hybrids and phylogenetics revisited: a statistical test of hybridization using quartets. Systematic Botany. 2007;32:8–15. [Google Scholar]
- 16.Arriola PE, Ellstrand NC. Crop-to-weed gene flow in the genus Sorghum (Poaceae): spontaneous interspecific hybridization between johnsongrass, Sorghum halepense, and crop sorghum, S. bicolor. American Journal of Botany. 1996: 1153–1159. [Google Scholar]
- 17.Savolainen V, Anstett MC, Lexer C, Hutton I, Clarkson JJ, Norup MV, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441:210–213. doi: 10.1038/nature04566 [DOI] [PubMed] [Google Scholar]
- 18.Wang WK, Ho CW, Hung KH, Wang KH, Huang CC, Araki H, et al. Multilocus analysis of genetic divergence between outcrossing Arabidopsis species: evidence of genome-wide admixture. New Phytologist. 2010;188:488–500. doi: 10.1111/j.1469-8137.2010.03383.x [DOI] [PubMed] [Google Scholar]
- 19.Sang T, Crawford D, Stuessy T. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany. 1997;84:1120 [PubMed] [Google Scholar]
- 20.Ji Y, Fritsch PW, Li H, Xiao T, Zhou Z. Phylogeny and classification of Paris (Melanthiaceae) inferred from DNA sequence data. Annals of Botany. 2006;98:245–256. doi: 10.1093/aob/mcl095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan J. Combining data with different distributions of among-site rate variation. Systematic Biology. 1996;45:375–380. [Google Scholar]
- 22.Maddison WP. Gene trees in species trees. Systematic biology. 1997;46:523–536. [Google Scholar]
- 23.Comas I, Moya A, González-Candelas F. Phylogenetic signal and functional categories in Proteobacteria genomes. BMC evolutionary biology. 2007;7(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanderson MJ, McMahon MM, Steel M. Phylogenomics with incomplete taxon coverage: the limits to inference. BMC Evolutionary Biology. 2010;10:155 doi: 10.1186/1471-2148-10-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soltis ED, Soltis PS. Contributions of plant molecular systematics to studies of molecular evolution. Plant Molecular Biology. 2000;42:45–75. [PubMed] [Google Scholar]
- 26.Woodcock H, Stearn W. Lilies of the world, their cultivation and classification. London: Country Life Limited; 1950. [Google Scholar]
- 27.MacRae E. Lilies: A guide for growers and collectors. Oregon:Timber Press; 1998. [Google Scholar]
- 28.Patterson TB, Givnish TJ. Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: insights from rbcL and ndhF sequence data. Evolution. 2002;56:233–252. [DOI] [PubMed] [Google Scholar]
- 29.Rong L, Lei J, Wang C. Collection and evaluation of the genus Lilium resources in Northeast China. Genetic Resources and Crop Evolution. 2011;58:115–123 [Google Scholar]
- 30.Comber HF. A new classification of the genus Lilium. Lily Yearbook. 1949;13:86–105. [Google Scholar]
- 31.Bao LY, Zhou J, Liu YJ. The wild Lilium resources in Tibet and its development and use. Forest By-Product and Speciality in China. 2004;69:54–55. [Google Scholar]
- 32.Wu XW, Li SF, Xiong L, Qu YH, Zhang YP, Fan MT. Distribution situation and suggestion on protecting wild lilies in Yunnan Province. Journal of Plant Genetic Resources. 2006;7:327–330. [Google Scholar]
- 33.Endlicher S. Genera plantarum secundum ordines naturales disposita. Vindobonae: Apud F. Beck; 1836–1840. [Google Scholar]
- 34.Wilson E. The lilies of eastern Asia: a monograph. London: Dulau & Company Ltd; 1925. [Google Scholar]
- 35.Baranova M. A synopsis of the system of the genus Lilium (Liliaceae). Botanicheskii Zhurnal. 1988;73:1319–1329. [Google Scholar]
- 36.Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T. A molecular phylogeny of Lilium in the internal transcribed spacer region. Journal of Molecular Evolution. 1999;49:238–249. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi K, Kawano S. Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Species Biology. 2000;15:73–93 [Google Scholar]
- 38.Nishikawa T, Okazaki K, Arakawa K,Nagamine T. Phylogenetic analysis of section Sinomartagon in genus Lilium using sequences of the internal transcribed pacer region in nuclear ribosomal DNA. Breeding Science. 2001;51:39–46. [Google Scholar]
- 39.Hayashi K, Kawano S. Bulbous monocots native to Japan and adjacent areas-their habitats, life histories and phylogeny. Acta Horticulturae. 2005;673:43–58 [Google Scholar]
- 40.İkinci N, Oberprieler C, Güner A. On the origin of European lilies: phylogenetic analysis of Lilium section Liriotypus (Liliaceae) using sequences of the nuclear ribosomal transcribed spacers. Willdenowia. 2006;36:647–656. [Google Scholar]
- 41.Nishikawa T. Molecular phylogeny of the genus Lilium and its methodical application to other taxon. Miscellaneous Publication of the National Institute of Agrobiological Sciences; (Japan: ); 2008. [Google Scholar]
- 42.Muratović E, Hidalgo O, Garnatje T, Siljak-Yakovlev S. Molecular phylogeny and genome size in European lilies (genus Lilium, Liliaceae). Advanced Science Letters. 2010;3:180–189. [Google Scholar]
- 43.Lee CS, Kim SC, Yeau SH, Lee NS. Major lineages of the genus Lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. Journal of Plant Biology. 2011;54: 159–171. [Google Scholar]
- 44.Gao YD, Hohenegger M, Harris AJ, Zhou SD, He XJ, Wan J. A new species in the genus Nomocharis Franchet (Liliaceae): evidence that brings the genus Nomocharis into Lilium. Plant Systematics and Evolution. 2012;298: 69–85. [Google Scholar]
- 45.Moens P The fine structure of meiotic chromosome pairing in natural and artificial Lilium polyploids. Journal of Cell Science. 1970;7: 55–63. [DOI] [PubMed] [Google Scholar]
- 46.Peruzzi L, Leitch IJ, and Caparelli KF. Chromosome diversity and evolution in Liliaceae. Annals of Botany. 2009;103: 459–475. doi: 10.1093/aob/mcn230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang WK, Liu CC, Chiang TY, Chen MT, Chou CH, Yeh CH. Characterization of expressed sequence tags from flower buds of alpine Lilium formosanum using a subtractive cDNA library. Plant Molecular Biology Reporter. 2011; 29:88–97 [Google Scholar]
- 49.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. [DOI] [PubMed] [Google Scholar]
- 50.Peterson A, John H, Koch E, Peterson J. A molecular phylogeny of the genus Gagea (Liliaceae) in Germany inferred from non-coding chloroplast and nuclear DNA sequences. Plant Systematics and Evolution. 2004;245:145–162. [Google Scholar]
- 51.Müller K. SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Applied Bioinformatics. 2005;4:65–69 [DOI] [PubMed] [Google Scholar]
- 52.Kimura M. A simple method for estimating evolutionary rates of base substitutions throughcomparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120 [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729 doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381 [PubMed] [Google Scholar]
- 55.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542 doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao YD, Harris AJ, Zhou SD, He XJ. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q-T plateau and the Hengduan Mountains. Molecular Phylogenetics and Evolution. 2013;68:443–460. doi: 10.1016/j.ympev.2013.04.026 [DOI] [PubMed] [Google Scholar]
- 58.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 59.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of dna sequences. Genetics 1985;111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proceedings of the National Academy of Sciences USA. 2007;104: 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woerner AE, Cox MP, Hammer MF. Recombination-filtered genomic datasets by information maximization. Bioinformatics. 2007;23:1851–1853. doi: 10.1093/bioinformatics/btm253 [DOI] [PubMed] [Google Scholar]
- 62.Xu B, Wu N, Gao XF, Zhang LB. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Molecular Phylogenetics and Evolution. 2012;62:346–358. doi: 10.1016/j.ympev.2011.10.007 [DOI] [PubMed] [Google Scholar]
- 63.Lu-Irving P, Olmstead RG. Investigating the evolution of Lantaneae (Verbenaceae) using multiple loci. Botanical Journal of the Linnean Society. 2013;171:103–119. [Google Scholar]
- 64.Kane NC, King MG, Barker MS, Raduski A, Karrenberg S, Yatabe Y, et al. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 2009;63:2061–2075 doi: 10.1111/j.1558-5646.2009.00703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minder AM, Widmer A. A population genomic analysis of species boundaries: neutral processes, adaptive divergence and introgression between two hybridizing plant species. Molecular Ecology. 2008;17:1552–1563. doi: 10.1111/j.1365-294X.2008.03709.x [DOI] [PubMed] [Google Scholar]
- 66.Ellstrand NC. Is gene flow the most important evolutionary force in plants? American Journal of Botany.2014;101:737–753. doi: 10.3732/ajb.1400024 [DOI] [PubMed] [Google Scholar]
- 67.van Tuyl JM, Arens P. Lilium: Breeding history of the modern cultivar assortment. Acta Horticulture. 2011;900:223–230 [Google Scholar]
- 68.Byrne M, Moran GF. Population divergence in the chloroplast genome of Eucalyptus nitens. Heredity. 1994;73:18–28. [Google Scholar]
- 69.Zimmer EA, Martin SL, Beverley SM, Kan YW, and Wilson AC. Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proceedings of the National Academy of Sciences. 1980; 77:2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117 [DOI] [PubMed] [Google Scholar]
- 71.Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- 72.Jefferson-Brown MJ, Howland H. The gardener's guide to growing lilies. Portland: Timber Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
Data Availability Statement
All sequences were deposited in NCBI GenBank, and the accession numbers were KX863745-KX865072.