Abstract
In the current era of big data, the amount of data available is continuously increasing. Both the number and types of samples, or features, are on the rise. The mixing of distinct features often makes interpretation more difficult. However, separate analysis of individual types requires subsequent integration. A tensor is a useful framework to deal with distinct types of features in an integrated manner without mixing them. On the other hand, tensor data is not easy to obtain since it requires the measurements of huge numbers of combinations of distinct features; if there are m kinds of features, each of which has N dimensions, the number of measurements needed are as many as Nm, which is often too large to measure. In this paper, I propose a new method where a tensor is generated from individual features without combinatorial measurements, and the generated tensor was decomposed back to matrices, by which unsupervised feature extraction was performed. In order to demonstrate the usefulness of the proposed strategy, it was applied to synthetic data, as well as three omics datasets. It outperformed other matrix-based methodologies.
Introduction
In the current era of big data, it is often that massive datasets are obtained, including samples with many features. For example, a video dataset can be regarded as time points (samples) vs pixels (features). Audio files consist of time points (samples) vs amplitude (features), and sets of DNA sequences consist of individuals (samples) vs nuclear acid sequences (features). All of these are provided in the form of a matrix, whose rows and columns are features and samples, respectively (of course, rows and columns are exchangeable). Although processing massive datasets is itself problematic, integrating distinct types of datasets is even more difficult. This problem is often annotated as multi-view data processing. For example, an audio visual file can be regarded as time points (samples) vs pixels and amplitudes(both features). In this paper, I consider two types of specific multi-view data processing: one is sharing samples (hereinafter called Case I) and another is sharing features (hereinafter, called Case II). It is formally possible to deal with these two cases as unified; multiple (m > 0) views of data, i.e., X(k), k = 1, … m, each of which is Nk features times M samples shared with multiple views (Case I), can be regarded as a (∑k Nk) × M matrix
| (1) |
where XT is the transposed matrix of X, while, if X(k), k = 1, … m, are Mk samples times N features shared with multiple views (Case II), can be regarded as a N × (∑k Mk) matrix
| (2) |
For both cases, however, we are not sure what will happen by simply merging distinct features as one matrix.
In order to address this problem, a variety of multi-view data processes have been proposed [1, 2]. Independent of the strategy to integrate multi-view datasets, there must be some weights attributed to each view. Since there are no a priori criteria to optimize these weights, some kind of artificial criteria are required. For example, if samples are classified, weights can be optimized so as to discriminate samples coincidentally from classes. Alternatively, if feature extraction is a task, weights can be optimized so as to generate the “best” features regardless of which features are considered good.
The reason weights are required for individual views is that we do not know whether the same weights are acceptable when simply creating new variables by merging or linearly combining them. Suppose are the observed values attributed to the ith feature of the jth sample in the kth view. Generating a merged matrix is to have a matrix where is placed at the (i + ∑k′ < k−1 Nk′)th row and the jth column, as introduced in Eq (1) (Case I). Alternatively, a merged matrix can be generated where is placed at the ith row and the (j + ∑k′ < k−1 Mk′)th column, as introduced in Eq (2) (Case II).
This is not necessarily as simple as it may appear. For example, in Case I, if Nk varies drastically from view to view, the results may be dominated by the views with the maximum number of features. However, it is not clear if views with more features are more important. Alternatively, if the new feature is generated with the linear combination where are coefficient, there are similar problems. If do not vary dependent upon (k), views with more features may dominate the outcome. Reverting row and column, I will discuss Case II as well. In order to compensate for this discrepancy, each view must be weighted based on criteria which are not naturally unique. No previously proposed strategies were free from this problem.
In this paper, I propose a brand new strategy that is free from weighting views; generating tensors whose number of modes is the same as, or one greater than the number of views, and applying tensor decomposition (TD) to them. Using this implementation, I performed feature extraction (FE), which I name TD based unsupervised FE, which is extended from the recently proposed principal component analysis (PCA) based unsupervised FE [3–22].
Materials and methods
Converting multi-view matrices into a tensor with multiplication
If we generate a new feature with neither summation nor merging, the product for Case I——can be regarded as an (m + 1) mode tensor. As each newly generated feature is composed of one feature from individual views, no weight is needed. Similarly, in Case II, , can be regarded as an (m + 1) mode tensor. These tensors are hereinafter called Type 1.
Alternatively, instead of simply multiplying matrix components with the shared columns or rows, they can be summed up as follows: (Case I) and (Case II). These can be regarded as m-mode tensors and are hereinafter called Type II. All variables associated with Type II tensors are written with tildes.
These newly generated m-mode (type II) or (m + 1)-mode (type I) tensors can be processed using any kind of tensor manipulation. For example, for a reduced number of features whose combination can express tensors, TD can be used to gain such features.
In the following subsection, I consider four combinations of types and cases, i.e., type I or II tensors for Case I or II multi-view data, case by case.
Definition and terminology of TD
Since TD is not a popular methodology and the usage of TD for FE is rare, I will briefly introduce TD in this subsection.
TD is the expansion of tensor xn1,n2,…,nm, nk = 1, …, Nk, 1 ≤ k ≤ m in the form
where xnk,ℓk, 1 ≤ k ≤ m, are orthogonal matrices. Since xn1,n2,…,nm is as large as G(n1, n2, …, nm), this formula is clearly overcomplete and does not give unique expansion. In this study, in order to decide G(n1, n2, …, nm), xnk,ℓk, 1 ≤ k ≤ m uniquely, I employ the higher order singular value decomposition (HOSVD) algorithm [23], which has successfully used to analyse microarrays [24] previously. G(n1, n2, …, nm) is a core matrix. xnk,ℓk, 1 ≤ k ≤ m, are singular value matrices and their column vectors are singular value vectors. G(n1, n2, …, nm), having larger absolute values, has more contribution to xn1,n2,…,nm. Since the combination of xnk,ℓk, 1 ≤ k ≤ m, associated with G(n1, n2, …, nm) to which larger absolute values were attributed contributes more collectively to xn1,n2,…,nm, they are more likely to be associated with one another.
For type I tensors this expression is straightforward. (m + 1) modes correspond to m + 1 components, i1, i2, …, im, j (Case I) or j1, j2, …, jm, i (Case II), respectively. On the other hand, for type II tensors, m modes correspond to m components, i1, i2, …, im (Case I) or j1, j2, …, jm (Case II), respectively. Thus, singular value vectors for jth sample (Case I) or ith feature (Case II) are missing. These can be computed via (Case I) or (Case II), k = 1, …, m. Thus, for type II tensor, there are m kinds of sample (Case I) or feature (Case II) singular value vectors in contrast to the type I tensors that have unique sample (Case I) or feature (Case II) singular vale vectors.
The relation to HO GSVD
Higher order generalized singular value decomposition [25] (HO GSVD) is the method that corresponds to singular value vectors when TD is applied to type I tensors. As HO GSVD converts X(k) = U(k)ΛVT where U(k), Λ and V are the Nk × M left singular value matrix, M × M eigenvalue matrix, and the M × M right singular value matrix, U(k) are regarded as feature singular value matrices and V is regarded as a unique (common) sample singular value matrix in the present implementation.
Synthetic dataset
The synthetic dataset used for demonstrating the usefulness of TD based unsupervised FE is defined as:
for 1 ≤ i ≤ N0. for N0 < i ≤ N. 1 ≤ j ≤ M. obeys uniform distribution ∈ [0, 1]. Specifically, c = 0.8, N = 1000, N0 = M = 50.
mRNA and miRNA expression profile
mRNA and microRNA(miRNA) expression profiles of multi-omics data were downloaded from gene expression omnibus (GEO) using GEO ID GSE28884. At first, GSE28884_RAW.tar was downloaded and expanded. For mRNA, 161 files whose names ended by the string “c.txt.gz” were used. Each file was loaded into R by read.csv command and the second column named “M” was employed as mRNA expression values. Probes not associated with Human Genome Organisation (HUGO) gene names were discarded and 13393 probes were remained. For miRNA, 161 files whose names ended by the string “geo.txt.gz” and the corresponding samples of mRNA expression which were measured were used. Each file was loaded into R by read.csv command and the second column (“Count”) was summed using the same third column (“Annotation”) values. Sum totals of less than 10 were discarded. As a result, 755 features remained. Finally, the miRNA expression profile matrix is , and the mRNA expression profile matrix is .
mRNA expressions of epidermal growth factor (EGF) treated breast cancers were downloaded from GEO using GEO ID GSE84096. The file named GSE84096_series_matrix.txt.gz included in “Series Matrix File(s)” was downloaded. Gene expression was divided into 14 control samples and 14 EGF treated samples named and , respectively.
mRNA expressions of vaccination experiments were downloaded from GEO using GEO ID GSE18323. Files named GSE18323-GPL570_series_matrix.txt.gz and GSE18323-GPL571_series_matrix.txt.gz included in “Series Matrix File(s)” were downloaded. As these included two distinct platforms, 22277 commonly included probes were used. Fifty eight samples annotated as “Protected group” (P) at time points T1 to T5, 52 samples annotated as “Delay group” (D) at time points T1 to T5, and 72 samples annotated as “Non-protected group” (NP) at time points T1 to T5, were used. They were named as , and respectively.
All expression profiles were standardized as ∑i xij = 0, and .
PCA-based unsupervised FE
PCA
In contrast to the usual use of PCA, where samples are embedded, the genes were embedded in this implementation.
Suppose xijs satisfy and X is a matrix whose elements are xij. The gram matrix G is defined as G ≡ XXT. Eigenvectors (1 ≤ k ≤ min(M, N)) are then obtained as , where uki is the kth PC score attributed to gene i and λks are the eigenvalues ordered as λk ≥ λk+1. The kth PC loadings attributed to the jth sample vkj are defined as , where because is the eigenvector of the covariance matrix XT X, .
PCA-based unsupervised FE applied to mRNA/miRNA expression
First, the five initial PC loadings vℓ1, js (for mRNA) and (for miRNA), 1 ≤ ℓ1, ℓ2 ≤ 5, were confirmed to have significant sample dependence (P-values < 0.05) with categorical regression,
where and (k = 1, 2) are regression coefficients. δSj takes 1 when jth sample belongs to category S, and isotherwise 0.
Then, assuming that PC scores uℓ1,i1s (for mRNA) and (for miRNA) are normally distributed, the P-values were attributed to the i1th mRNA and i2th miRNA using a χ squared distribution as
where σuℓ1 and is the standard deviation of {uℓ1,i1|1 ≤ i1 ≤ 13393} and , respectively. Pχ2[>x] is the probability that the argument is larger than x under the assumption that the arguments obey a χ squared distribution. The P-values were further adjusted by the Benjamini-Hochberg (BH) criterion [26], and those genes associated with adjusted P-values less than 0.01 were selected as the genes associated with the difference between multiple classes.
PCA-based unsupervised FE applied to vaccination experiments
Although similar to the previous section, some differences are: (i) Only the second PC scores attributed to mRNAs were used for FE. (ii) Instead of mRNA and miRNA, patient category groups, P, D, and NP were used. (iii) mRNAs commonly included in three independent FEs performed considering P, D, and NP, were considered.
TD-based unsupervised FE
For type I tensor generated from multi-omics dataset, in order to identify miRNAs and mRNAs associated with identified sample singular value vectors, it was assumed that and follow multiple normal distributions, and P-values were attributed to the i1th mRNA and the i2th miRNA using χ2 distribution.
where σℓ1(σℓ2) are standard deviations of . Piks were adjusted by using the BH criterion, and mRNAs and miRNAs associated with the adjusted P-value lower than 0.01 for mRNA and 0.05 for miRNA were selected as those associated with identified sample singular value vectors when TD was applied to type I tensor.
When TD was applied to type II tensor, the computations were similar, excluding that adjusted P-value lower than 0.01 and 1 ≤ ℓ2 ≤ 5 were used for miRNA, where and were replaced with and , respectively.
For EGF treated cell lines and vaccination experiments, similar procedures were repeated by replacing gene singular value vectors with xℓ3 = 2,i (EGF, type I) or and (EGF, type II) or , and , (vaccination, type II), respectively.
Where HO GSVD was applied to multi-omics datasets, the feature singular value matrices were replaced with the first five column vectors of U(k) (k = 1 for mRNA and k = 2 for miRNA).
Adjusted P values used as thresholds are always 0.01 for EGF treated cell lines, vaccination, and HO GSVD.
Conversion prob ID to HUGO gene name/ensembl gene ID/GENBANK accession ID
Coincidences between prob ID (mRNA), HUGO gene names, Ensembl gene IDs were downloaded from GEO using GEO ID GPL3676 for multi-omics datasets, and GPL571/570 for vaccination experiments. GENBANK accession ID attributed to probes identified in the EGF treatment were extracted from GPL16686.
Enrichment analysis of g:profiler
Ensembl gene IDs were uploaded to g:profiler [27]. 13393 Ensembl gene IDs were used as background.
Enrichment analysis of genes identified as outliers using each of the first five mRNA singular value vectors obtained by applying TD based unsupervised FE to type I tensor generated from multi-omics datasets
Five distinct P-values were attributed to the i1th mRNA using χ2 distribution:
P-values were adjusted using the BH criterion and mRNAs associated with adjusted P-values less than 0.01 were identified as outliers (see S2 Table). S2 Table was uploaded to g:Cocoa in g:profiler with “Gene Ontology/ Biological Process” specified as the targeted ontology.
Enrichment analysis of MSigDB
HUGO gene IDs or GENBANK accession IDs associated with identified mRNAs were uploaded to http://software.broadinstitute.org/gsea/msigdb/annotate.jsp (registration and login are needed). “CGP: chemical and genetic perturbations” was selected for multi-omics data and EGF treated cell lines, while “C7: immunologic signatures” was selected for vaccination experiments.
Enrichment analysis of DIANA-mirpath
As for TDs applied to type I tensors, the following link was pasted to browser.
As for TDs applied to type II tensors, the following link was pasted to browser.
As for HO GSVD, the following link was pasted to browser.
Categorical regression towards data shown in Fig 1
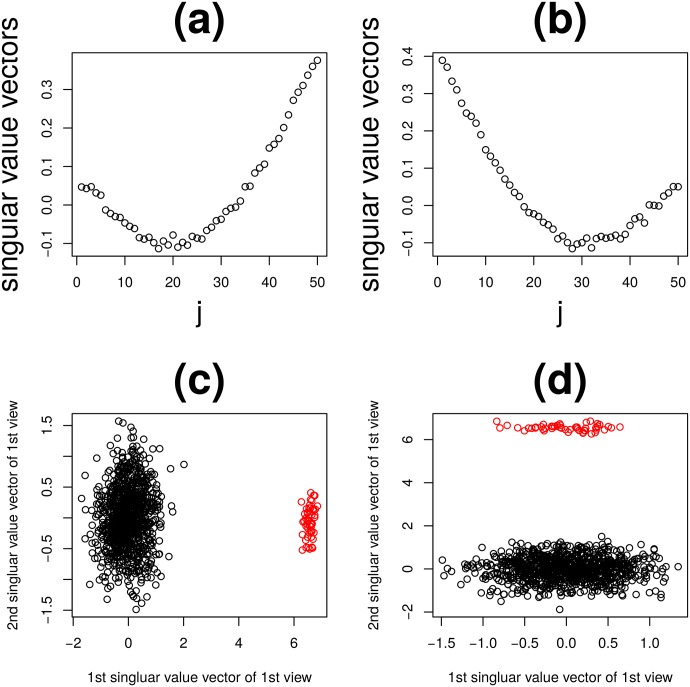
Fig 1.
Boxplots of sample singular value vectors xℓ3,j (a) when TD was applied to the type I tensor and (b), (c), 1 ≤ ℓ3 ≤ 5, when TD was applied to the type II tensor, generated from mRNA and miRNA expression profiles of multi-omics datasets. (d) Sample singular value vectors when HO GSVD was applied to multi-omics datasets. P-values computed by categorical regression attributed to (a) to (d) were below the figures.
For type I tensor (Fig 1(a)),
where and are regression coefficients. δSj takes 1 when jth sample belongs to category S, otherwise 0. The summation is taken over all categories. For type II tensor (Fig 1(b) and 1(c)), xℓ3,j is replaced with or . For HO GSVD (Fig 1(d)), xℓ3,j is replaced with column vectors of V.
Correlations in Fig 2(c) and 2(g)
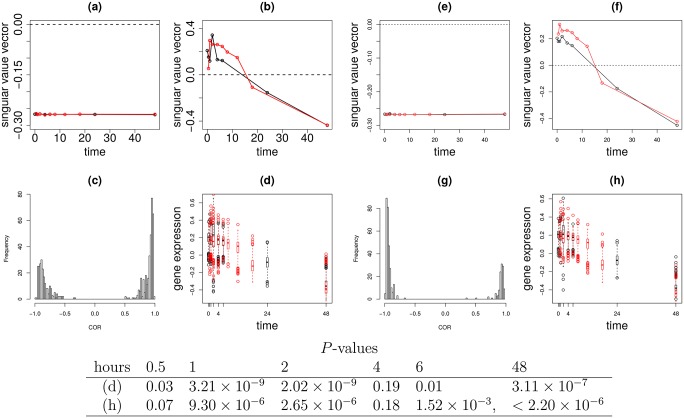
Fig 2. The results of TD applied to type I tensor generated from EGF treatment experiments.
Sample singular value vectors, Black open circle: Red open circle: (a) ℓ1 = 1 (b) ℓ1 = 2. (c) Histogram of the correlation coefficients between sample (time) singular value vectors and selected individual 558 mRNA probes expression profiles. (d) Boxplot of scaled and shifted selected individual 558 mRNA probe expression profiles. Black: control, Red: EGF treated cell lines. The same as (a) to (d), but for type II tensor. Black open circles: Red open circles: (e) ℓ1 = 1 (f) ℓ1 = 2. (g) Histogram of the correlation coefficients between sample (time) singular value vectors and selected individual 398 mRNA probe expression profiles. (h) Boxplot of scaled and shifted selected individual 398 mRNA probe expression profiles. Black: control, Red: EGF treated cell lines. P-values computed by t test of 558 (d) or 398 (h) mRNA probes between with and without EGF treatments are below figures.
The correlations were computed between two vectors of the length Tcontrol + TEGF, and (Fig 2(c)) or between and (Fig 2(g)), where Tcontrol and TEGF are total number of samples in each treatment, respectively. Adjusted P-values attributed to the correlation coefficients were computed via the fdrtool [28] function in the fdrtool package in R [29].
Scaling and shifting prior to plotting Fig 2(d) and 2(h)
As each individual gene expression has its own base line and amplitude, they must be scaled and shifted before being overdrawn. To this end, the linear regression analysis
was employed (Fig 2(c)) where ai and bi are regression coefficients commonly used for control and EGF treated samples. For Fig 2(g), and are replaced with and , respectively. Then, fitted values are used for plots. P-values that exhibit distinction between control and EGF treated sample at time point t were computed by two-sided t test between and within N = 558 (Fig 2(d)) or N = 398 (Fig 2(h)) selected mRNA probes.
Correlations in Fig 3(c)
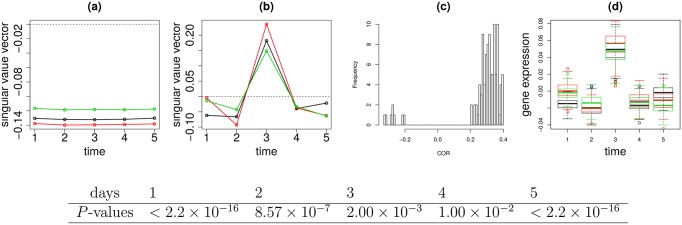
Fig 3. The results of TD applied to type II tensor generated from vaccination.
Sample singular value vectors, Black open circle: Red open circle: Green open circle: (a) ℓ1 = ℓ2 = ℓ3 = 1 (b) ℓ1 = ℓ2 = ℓ3 = 2 (c) Histogram of the correlation coefficients between sample singular value vectors and selected individual 104 mRNA probes expression profiles. (d) Boxplot of scaled and shifted selected individual 104 mRNA probe expression profiles. Black: P, Red:D, green:ND cell lines. P-values computed by categorical regression between P, D, and NP groups are below figures.
Similar to Fig 2, the correlations were computed between two vectors of length TP + TD + TNP, and where TP = 58, TD = 52, and TNP = 72 are the total number of samples in each patient category, respectively. Adjusted P-values were computed via the fdrtool function in the fdrtool package in R [29].
Scaling and shifting prior to plotting Fig 3(d)
The linear regression analysis
was employed, where ai and bi are regression coefficients commonly used for three patient categories (these values differ from those used in Fig 2). Fitted values are used for plots. P-values that exhibit distinction among three patient groups at time point t = 1, 2, 3, 4, 5 days were computed by categorical regression
for 104 commonly selected mRNA probes in the three categories. and are regression coefficients fitted for with fixed t.
Statistical analysis
All statistical analyses were performed in R [29]. HOSVD was performed using the HOSVD function in the rTensor package. PCA was performed using the prcomp function in R. SAM was performed using SAM function in siggenes package. limma was performed in limma function in the limma package. Adjusted P-values were computed by p.adjust function with “BH” options. P-values by χ2 distribution was computed by pchisq function in R. Categorical regression was performed using the lm function in R. RF was performed using randomForest function in randomForest package. KCCA was performed by KCCA function in the kernlab package.
Results
A Work flow chart and list of the variables introduced are in S1 File.
Synthetic dataset
In order to demonstrate the efficacy of our strategy, I applied TD based unsupervised FE to synthetic data. In the following interpretation, I assumed two views of Case I for synthetic dataset, however interpreting it as Case II is straightforward, thus, I do not consider Case II specifically. The method applied to the synthetic dataset, TD based unsupervised FE, is the extension from the recently proposed PCA based unsupervised FE, which has been successfully applied to various bioinformatics problems [5–22].
First, two matrices are generated, each of which is composed of N features times M samples. They are notated as X(k), and k = 1, 2, respectively, whose components are denoted as , and k = 1, 2, respectively. The first N0 row vectors (features), , and 1 ≤ ik ≤ N0, are the noise added linear combination (Fig 4(d) and 4(e)) of constant, linear, and half period sinusoidal function (Fig 4(a) and 4(c)). However, since the coefficients of linear combinations were selected such that the correlation between X(1) and X(2) is negligible, identifying a correlation between two matrix row vectors is usually impossible (Fig 4(f)). Remaining row vectors and N0 < ik ≤ N are simply random number ∈ [0, 1]. The tasks are (i) identify N0 ordered features, and (ii) identify latent correspondence between two views.
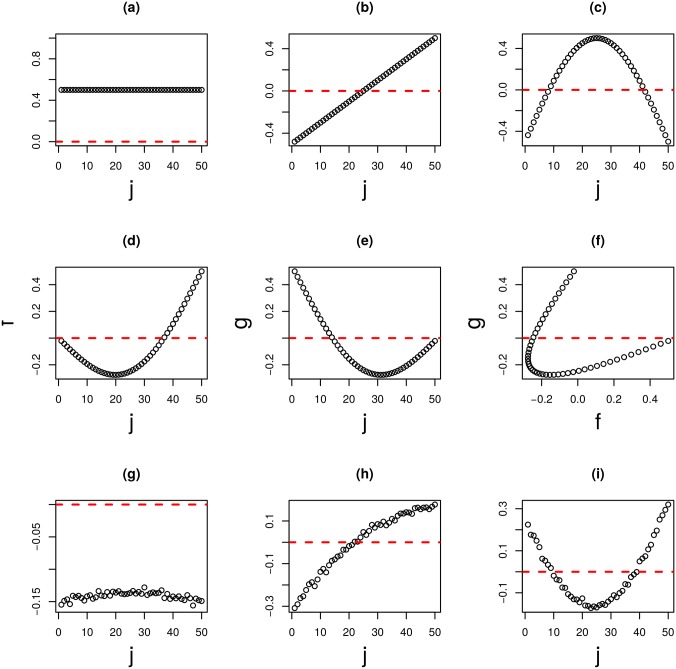
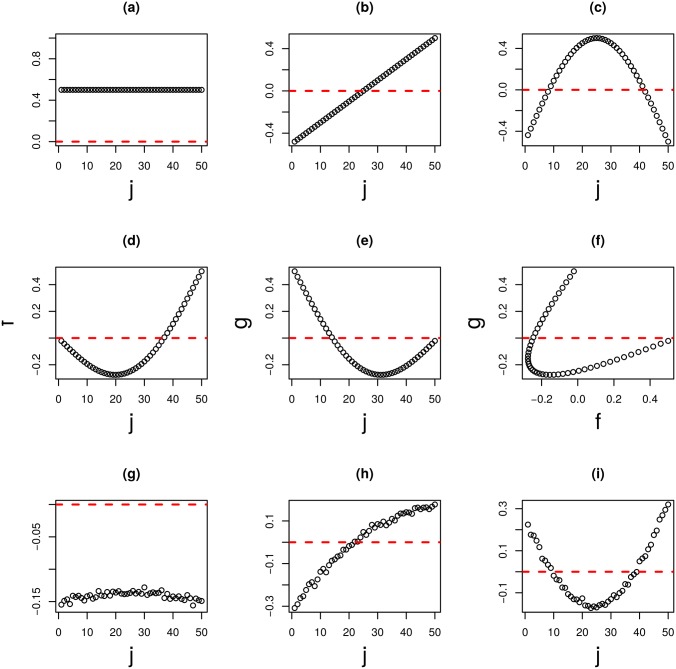
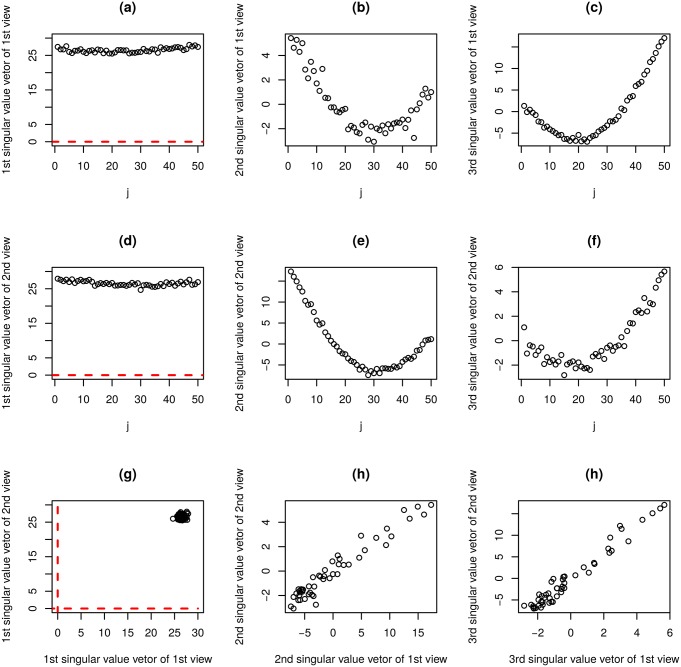
Fig 4. The results of TD applied to the type I tensor generated from a synthetic dataset (M = 50).
(a) to (c) are orthogonal base functions: (a) constant, (b) linear, (c) half period sinusoidal. (d) and (e) base functions used for generating . (d) k = 1, (e) k = 2. (f) is the scatter plot of (d) and (e). (g) to (i) are the first, second, and third sample singular value vectors xℓ3,j and ℓ3 = 1, 2, 3, and are computed by applying TD to synthetic data.
The three mode tensor (type I) was derived from . Fig 4(g)–4(i) shows the first three sample mode singular value vectors, xℓ3,j, ℓ3 = 1, 2, 3, obtained with HOSVD,
| (3) |
It is obvious that Fig 4(g)–4(i) corresponds to Fig 4(a)–4(c), respectively. To my knowledge, it is the only method to decompose linear combinations back into parts in a fully unsupervised manner.
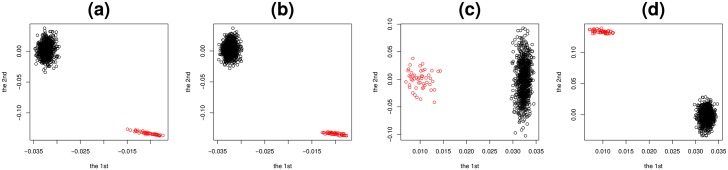
Moreover, as can be seen in Fig 5(a) and 5(b), N0 features are placed as outliers. Thus, TD based unsupervised FE applied to type I tensors generated from matrices’ products can not only decompose linear combinations, but can also identify the limited number of features, 1 ≤ i1, i2, ≤ N0, that contribute to correlations between two matrices, X(k), k = 1, 2.
Fig 5. Feature singular value vectors when TD was applied to type I tensor generated from synthetic data.
(a) and (b) . and type II tensor, (c) and (d) . Red open circles are 1 ≤ i1, i2 ≤ N0 and black open circles are N0 < i1, i2 ≤ N.
Although I could successfully demonstrate that my strategy works well, there is one drawback; it is the computationally extensive method, since its memory as well as computational time are proportional to MN2. In order to reduce computational resources as much as possible, I summed and generated the m(= 2) mode tensor (type II), . Then, is decomposed as
| (4) |
Two sample singular value vectors can be computed as
Since I employed the two views problem, although I occasionally got the two mode tensor, i.e., the matrix, if I consider m(>2) views, I generally get a m mode tensor and the above procedures can be easily extended to m mode tensors.
Fig 6 shows the first three sample singular value vectors, , and (k = 1, 2), obtained with the application of HOSVD to type II tensors. Among these, and k = 1, 2, shown in Fig 6(b) and 6(e) (ℓ3 = 1) correctly reproduce Fig 4(e) while those shown in Fig 6(c) and 6(f) (ℓ3 = 2) does Fig 4(d), respectively. Thus, TD applied to type II tensors also successfully identified latent correlations between X(k), k = 1, 2 (Fig 6(h) and 6(i)). However, it could not depict orthogonal base functions (Fig 4(a)–4(c)) that can be detected by TD applied to type I tensors (Fig 4(g)–4(i)). Additionally, N0 features were successfully identified as outliers (Fig 5(c) and 5(d)). Thus, at the expense of recognition of orthogonal base functions, TD applied to type II tensors successfully reduced the computational resources needed by 1/M and fulfilled tasks (i) and (ii) as defined above.
Fig 6. The results of TD applied to type II tensor generated from synthetic dataset (M = 50).
(a) ℓ3 = 1 (b) ℓ3 = 2 (c) ℓ3 = 3. (d) ℓ3 = 1 (e) ℓ3 = 2 (f) ℓ3 = 3. (g): (a) vs (c), (h): (b) vs (d), γ = 0.97, P = 0, (i): (c) vs (f), γ = 0.97, P = 0. γ: Pearson correlation coefficients. P: associated P-values.
In order to see if these strategies are also useful in practice, integrated analyses of multi-omics datasets were performed with this strategy and are described in the next subsection.
Multi-omics dataset
In the previous subsection, I demonstrated that applying TD based unsupervised FE to tensors generated from matrices’ products could determine latent structures behind pairs of matrices. However, this may only be feasible using synthetic data, as described in the previous subsection. Thus, in order to see if it also works in the situation not prepared specifically fitted to it, I need to show that it works in real situation.
The analysed dataset is composed of two omics profiles. These are mRNA and miRNA profiles which were measured for multi-class breast cancer samples including normal breast tissues [30]. As the samples are shared, the multi-omics data corresponds to Case I data. TD based unsupervised FE was applied to the dataset in order to identify disease critical genes and latent relations between miRNA and mRNA.
At first, TD was applied to the type I tensor generated from the mRNA and miRNA profiles as follows:
where and are expressions of the i1th mRNA and the i2th miRNA from the jth sample. In order to determine whether TD can identify disease critical features, categorical regression analysis was applied to sample singular value vector xℓ3,j, in order to identify coincidences with defined sample classes. If the obtained sample singular value vectors are coincident with sample class labels, it is evidence that TD can process omics profiles properly, as this approach does not employ class labels explicitly. Fig 1 shows the first five sample singular value vectors, xℓ3,j, 1 ≤ ℓ3 ≤ 5, that show significant sample class dependence. Thus, TD could successfully generate disease associated features. This is not a trivial outcome, as sample classification was not used.
Next, I attempted to extract features using the mRNA and miRNA singular value vectors and . To accomplish this, it was necessary to first identify mRNA and miRNA singular value vectors and , associated with sample singular value vectors, xℓ3,j, 1 ≤ ℓ3 ≤ 5 identified above. This can be done by investigating G(ℓ1, ℓ2, ℓ3) since the combinations (ℓ1, ℓ2, ℓ3) associated with larger absolute G values are regarded as more coincident with one another. Table 1 shows the ranking of G for 1 ≤ ℓ1, ℓ2, ℓ3 ≤ 5 (ranked from 1 to 10) based upon their absolute values. The first five sample singular value vectors xℓ3,j, 1 ≤ ℓ3 ≤ 5, are associated with the first two miRNA singular value vectors as well as the first five mRNA singular value vectors, , as only these combinations appear within top ten ranked G(ℓ1, ℓ2, ℓ3)s that represent the amount of coincidence among mRNA and miRNA and samples.
Table 1. Top ranked 10 G(ℓ1, ℓ2, ℓ3)s with larger absolute values among 1 ≤ ℓ1, ℓ2, ℓ3 ≤ 10 when TD was applied to type I tensor generated from and (left) and and (right).
| multi-omics | EGF treatment | ||||||
|---|---|---|---|---|---|---|---|
| ℓ1 | ℓ2 | ℓ3 | G(ℓ1, ℓ2, ℓ3) | ℓ1 | ℓ2 | ℓ3 | G(ℓ1, ℓ2, ℓ3) |
| 1 | 1 | 1 | 1.67 × 105 | 1 | 1 | 1 | −4.03 × 104 |
| 2 | 1 | 2 | −1.03 × 105 | 2 | 1 | 2 | −1.56 × 103 |
| 4 | 1 | 4 | 7.48 × 104 | 1 | 2 | 2 | 1.49 × 103 |
| 3 | 1 | 3 | −6.64 × 104 | 3 | 1 | 3 | 1.05 × 103 |
| 5 | 1 | 5 | 6.23 × 104 | 1 | 3 | 4 | −5.79 × 102 |
| 3 | 2 | 3 | 3.00 × 104 | 4 | 1 | 5 | 4.24 × 102 |
| 1 | 2 | 3 | −2.87 × 104 | 2 | 1 | 3 | 4.16 × 102 |
| 3 | 1 | 5 | −2.33 × 104 | 5 | 1 | 6 | 3.25 × 102 |
| 2 | 2 | 3 | −2.02 × 104 | 1 | 4 | 6 | 3.19 × 102 |
| 1 | 2 | 2 | −1.48 × 104 | 4 | 1 | 4 | −2.62 × 102 |
Four hundred twenty-seven mRNA probes and 7 miRNAs, identified as outliers, using and were selected. The seven selected miRNAs were: hsa-let-7b, hsa-miR-125b, hsa-miR-143, hsa-miR-145, hsa-miR-21, hsa-miR-22, hsa-miR-99a. Remarkably, 143, 145, 21, 22, 99a as well as let-7a, and 125a, which belongs to the same families as 125b and let-7b, were also reported by the original study ([30], Table 1). The mRNAs associated with selected 427 mRNA probes are in S1 Table because of too many numbers.
In order to evaluate obtained mRNAs associated with the 427 selected mRNA probes and 7 miRNAs biologically, the mRNAs were uploaded for enrichment analysis using MSigDB [31] and the miRNAs to DIANA-mirpath [32]. The top 10 enriched gene sets in MSigDB are chiefly related to breast cancer (eight out of ten, see Table 2), while the top ranked pathways identified DIANA-mirpath are “MicroRNAs in cancer” (Table 3). Thus, I concluded that our strategy is successful despite fully unsupervised nature.
Table 2. Overlap between mRNAs identified (S1 Table) and MSigDB.
Top 10 ranked gene sets are presented. Upper rows: type I, lower rows: type II tensors are considered in each gene set name, respectively. The word “BREAST_CANCER/_DUCTAL_CARCINOMA” was presented in bold face in order to emphasize the overlap with breast cancer related gene sets. K: The number of genes in each gene set, k: The number of genes overlapped.
| Gene Set Name | (K) | Description | (k) | k/K | p-value | FDR q-value |
|---|---|---|---|---|---|---|
| SMID_BREAST_CANCER_LUMINAL_B_DN | 564 | Genes down-regulated in the luminal B subtype of breast cancer. | 100 | 0.1773 | 4.00E-105 | 1.36E-101 |
| 88 | 0.1560 | 2.34E-090 | 7.94E-087 | |||
| SMID_BREAST_CANCER_BASAL_DN | 701 | Genes down-regulated in basal subtype of breast cancer samples. | 91 | 0.1298 | 2.06E-082 | 3.50E-079 |
| 86 | 0.1227 | 6.42E-079 | 1.09E-075 | |||
| DOANE_BREAST_CANCER_ESR1_UP | 112 | Genes up-regulated in breast cancer samples positive for ESR1 compared to the ESR1 negative tumors. | 44 | 0.3929 | 5.78E-063 | 6.56E-060 |
| 38 | 0.3393 | 6.29E-053 | 4.28E-050 | |||
| SMID_BREAST_CANCER_RELAPSE_IN_BONE_DN | 315 | Genes down-regulated in bone relapse of breast cancer. | — | — | — | — |
| 51 | 0.1619 | 1.17E-052 | 6.63E-050 | |||
| JAEGER_METASTASIS_DN | 258 | Genes down-regulated in metastases from malignant tumors. melanoma compared to the primar | — | — | — | — |
| 43 | 0.1667 | 5.25E-045 | 2.55E-042 | |||
| WALLACE_PROSTATE_CANCER_RACE_UP | 299 | Genes up-regulated in prostate cancer samples from African-American patients compared to those from the European-American patients. | 55 | 0.1839 | 5.86E-058 | 4.98E-055 |
| — | — | — | — | |||
| SMID_BREAST_CANCER_NORMAL_LIKE_UP | 476 | Genes up-regulated in the normal-like subtype of breast cancer. | 61 | 0.1282 | 2.40E-054 | 1.63E-051 |
| — | — | — | — | |||
| FARMER_BREAST_CANCER_BASAL_VS_LULMINAL | 330 | Genes which best discriminated between two groups of breast cancer according to the status of ESR1 and AR: basal (ESR1- AR-) and luminal (ESR1+ AR+). | 54 | 0.1636 | 5.31E-054 | 3.01E-051 |
| 54 | 0.1636 | 5.03E-056 | 4.27E-053 | |||
| POOLA_INVASIVE_BREAST_CANCER_UP | 288 | Genes up-regulated in atypical ductal hyperplastic tissues from patients with (ADHC) breast cancer vs those without the cancer (ADH). | 51 | 0.1771 | 7.55E-053 | 3.67E-050 |
| — | — | — | — | |||
| MCLACHLAN_DENTAL_CARIES_UP | 254 | Genes up-regulated in pulpal tissue extracted from carious teeth. | 47 | 0.1850 | 1.12E-049 | 4.77E-047 |
| — | — | — | — | |||
| SMID_BREAST_CANCER_BASAL_UP | 648 | Genes up-regulated in basal subtype of breast cancer samples. | 63 | 0.0972 | 1.38E-048 | 5.23E-046 |
| 78 | 0.1204 | 1.18E-070 | 1.34E-067 | |||
| SMID_BREAST_CANCER_LUMINAL_B_UP | 172 | Genes up-regulated in the luminal B subtype of breast cancer. | 38 | 0.2209 | 1.95E-043 | 6.63E-041 |
| 37 | 0.2151 | 3.29E-043 | 1.40E-040 | |||
| DELYS_THYROID_UP_CANCER | 443 | Genes up-regulated in papillary thyroid carcinoma (PTC) compared to normal tissue. | — | — | — | — |
| 48 | 0.1084 | 5.47E-041 | 2.07E-038 | |||
| TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_DUCTAL_NORMAL_DN | 198 | Genes down-regulated in ductal carcinoma vs normal ductal breast cells. | — | — | — | — |
| 36 | 0.1818 | 3.16E-039 | 1.08E-036 |
Table 3. Results of DIANA-mirath using seven miRNAs identified.
Top 10 significant KEGG pathway was presented. gene: number of genes overlapped with miRNAs target genes, miRNA: number of overlapped miRNAs. Numbers both sides of “/” correspond to type I/type II tensors, respectively.
| KEGG pathway | FDR q-value | gene | miRNA |
|---|---|---|---|
| MicroRNAs in cancer | 3.29E-88/4.68E-68 | 115/141 | 7/22 |
| Proteoglycans in cancer | 5.36E-12/9.48E-17 | 116/159 | 7/22 |
| Cell cycle | 1.26E-10/2.61E-12 | 80/104 | 7/22 |
| Renal cell carcinoma | —/2.25E-011 | —/61 | —/22 |
| Protein processing in endoplasmic reticulum | —/3.81E-10 | —/134 | —/22 |
| Hepatitis B | 6.57E-09/5.37E-09 | 79/107 | 7/22 |
| Prion diseases | 5.14E-08/— | 16/— | 7/— |
| Central carbon metabolism in cancer | 2.76E-07/— | 42/— | 7/— |
| Hippo signaling pathway | 3.27E-07/2.40E-07 | 78/109 | 7/22 |
| Chronic myeloid leukemia | —/2.40E-07 | —/62 | —/22 |
| Viral carcinogenesis | —/2.40E-07 | —/158 | —/22 |
| Pancreatic cancer | —/1.84E-06 | —/55 | —/22 |
| Lysine degradation | 1.15E-06/— | 27/— | 6/— |
| FoxO signaling pathway | 2.89E-06/— | 79/— | 7/— |
| Prostate cancer | 4.52E-06/— | 56/— | 7/— |
Next, type II tensor generated from mRNA and miRNA profiles was considered. Applying TD to type II tensor,
gives us two sample singular value vectors
The first five are shown separately in (Fig 1(b) and 1(c)). It is obvious that all of the ten sample singular value vectors are significantly coincident with sample classifications.
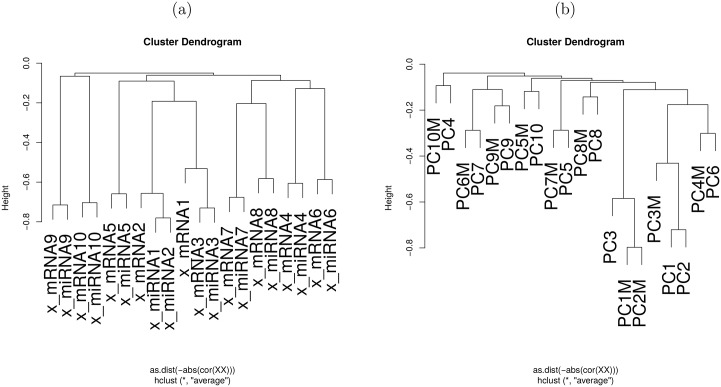
To determine whether TD applied to type II tensors could depict latent correlation between miRNA and mRNA, hierarchical clustering was performed between and (1 ≤ ℓ3 ≤ 10, Fig 7(a)). Here, are always paired with one of (Fig 7(a)). Thus, TD applied to type II tensor could successfully identify latent correlation between two views, i.e., mRNA and miRNA.
Fig 7. Hierarchical clustering of (x_mRNA) and (x_miRNA).
When TD was applied to type II tensor (a) and vℓ3,j (for mRNA, labelled as PC), and (for miRNA, labelled as PCM) when PCA was separately applied to miRNA and mRNA (b) (1 ≤ ℓ3 ≤ 10). Distances were negative signed absolute values of Pearson correlation coefficients. Unweighted Pair Group Method with Arithmetic mean (UPGMA) was employed.
Additionally, I attempted to extract mRNAs and miRNAs using the first five mRNA and miRNA singular value vectors, ,, and 1 ≤ ℓ1, ℓ2 ≤ 5, respectively. In this example two views were emplyed, as the type II tensor is occasionally the matrix . Thus, the core tensor, , is the diagonal matrix. Therefore, the first five feature singular value vectors are automatically associated with the first five corresponding sample singular value vectors. 21 miRNAs (let-7a/b/f, miR-103/125b/141/142-3p/143/145/148a/199a/b-3p/19b/205/21/22/23a/24/26a/30a/451/99a), identified as outliers using the first five miRNA singular value vectors, , were selected. Eight miRNAs (let-7a, miR-21/22/451/142-3p/143/145/99a) were also reported in the original study ([30], Table 1). Three hundred seventy-four mRNA probes, identified as outliers using the first five mRNA singular value vectors , were selected (associated mRNAs are in S1 Table) and were substantially overlapped with those selected when type I tensors were considered (Table 4). In order to evaluate obtained mRNAs and miRNAs biologically, mRNAs associated with 374 probes were uploaded to MSigDB, and miRNAs to DIANA-mirpath. DIANA-mirpath identified “MiRNAs in cancer” as the most significant KEGG pathway (Table 3). Table 2 also shows the overlap with MSigDB. Eight out of ten were breast cancer related, and one of the remaining two is related to metastasis. Thus, these identified mRNAs and miRNAs are biologically reasonable. In conclusion, TD applied to type II tensor works well.
Table 4. Comparison between 426 mRNA probes identified by TD based unsupervised FE applied to type I tensor and 374 mRNA probes identified by TD based unsupervised FE applied to type II tensor, or 427 probes identified by PCA based unsupervised FE separately applied to miRNA/mRNA.
S:selected, NS:not selected.
| TD type II | PCA | ||||
|---|---|---|---|---|---|
| NS | S | NS | S | ||
| TD | NS | 12856 | 110 | 12948 | 19 |
| type I | S | 163 | 264 | 18 | 408 |
Temporally differentially expressed genes
Although the application of TD based unsupervised FE to multi-omics data was successful, one may wonder if the application to muti-omics data is reasonable, as synthetic datasets were more difficult to deal with due to a lack of class labelling. Have I intentionally tried the easiest case? In order to address this possible query, I considered two examples of a more difficult problem: identification of temporally differentially expressed genes. The task is as follows: Given more than one temporal gene expression, identify genes expressed differently among multiple expressions at specific time points. For example, expression of a particular gene obeys f(t) as a function of t under one set of conditions, while it obeys f(t) + C with a constant C under another. Conventionally, this is differentially expressed between the two conditions; however this kind of differential expression is often not of interest when temporal gene expression is considered. One distinction of note would be, for example, differences between expressions at certain time points and not at others (a distinct time dependency between two conditions). However, there are no de facto standard methods that automatically achieve this. In order to address it, TD based unsupervised FE was applied to this problem.
The first example of this application is the comparison of non-small lung cancer cell (NSCLC) line H1975, with and without EGF treatment [33]. Gene expression matrices were divided into two groups (with and without EGF treatment). The type I tensor xt1,t2,i is then generated as
where and are ith gene expressions of cell lines with and without EGF treatment, at time points t1 and t2 after the EGF or control treatments. As they share features (though not samples) in contrast to the previous application which was Case I data, this example uses Case II data. As the samples in this example are divided into two groups based on EGF treatment, it is not fully unsupervised. It is, however, unsupervised in the sense that the type of temporal difference sought is not defined. The tensor was expanded by HOSVD as
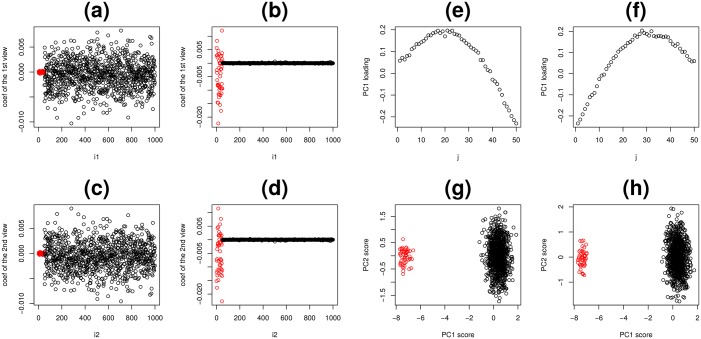
Fig 2(a) and 2(b) shows the and for ℓ1, ℓ2 = 1, 2, respectively. Obviously, those for ℓ1 = ℓ2 = 1 do not have any time dependence while those for ℓ1 = ℓ2 = 2 do. Some temporal difference was observed in the latter, however its significance is unclear. In order to determine said significance, genes identified as outliers had to be selected. This selection process began with identifying the gene singular value vectors associated with and . Table 1 shows the top ranked G(ℓ1, ℓ2, ℓ3)s with larger absolute values. It is obvious that xℓ3 = 2,i is associated with and , as the absolute values of G(2, 2, 1) and G(2, 1, 2) are the second and the third largest in the table. Next, 558 mRNA probes (associated mRNAs are in S1 Table) identified as outliers based on xℓ3 = 2,i were selected. Fig 2(c) shows the histogram of correlation coefficients between the vectors generated by connecting and vs the 558 selected mRNA probes. These are highly correlated (adjusted P-values are less than 0.01). It is remarkable since G(2, 2, 1) and G(2, 1, 2) are smaller than one tenth of G(1, 1, 1) whose absolute value is the largest. This suggests that the amount of contributions of G(2, 2, 1) and G(2, 1, 2) is too little to govern individual gene expression. The high correlation despite this fact speaks to the soundness of our methodology.
The next step was to determine whether the 558 mRNA probes selected exhibit temporal differences. The 558 mRNA probes selected are scaled, shifted, and over drawn as boxplot (Fig 2(d)). Though it is difficult to observe, P-values were computed by two-sided t tests between expressions, with and without EGF treatments at time points, 0.5, 1, 2, 4, 6 and 48 hours (Fig 2). These values are significant only at limited time points with and without EFG treatment. These were merely temporally differently expressed genes. Thus, TD based unsupervised FE applied to type I tensors is effective.
To determine the biological reliability of the selected genes, genes associated with selected 558 mRNA probes were uploaded to MSigDB. The second significant gene set was found to be KOBAYASHI_EGFR_SIGNALING_24HR_DN (the adjusted P value is 1.37 × 10−96), which is reasonable as the genes sought were expressed differently with and without EGF treatments.
The next task was to determine whether type II tensors produce similar results. Type II tensor , which in this example is the matrix since two views are considered, is defined as a summation of a type I tensor over gene index,
that is expanded by HOSVD, which is a simple SVD in the present case
Fig 2(e) and 2(f) shows the and for ℓ1, ℓ2 = 1, 2. Obviously, and do not have any time dependence while and do, as in the case where TD was applied to a type I tensor. Some temporal difference was observed between and . Again, the significance of this temporal difference was unclear. Genes identified as outliers had to be selected to determine the significance of this temporal difference. While there are two gene singular value vectors,
485 and 471 mRNA probes identified as outliers were selected using and , respectively, among which 398 mRNA probes (associated mRNAs are shown in S1 Table) were commonly selected. Fig 2(g) shows the histogram of correlation coefficients between the vector generated by connecting and vs 398 commonly selected mRNA probes. These were highly correlated (adjusted P-values are less than 0.01). This is noteworhty as the smallest contribution from the second singular value vector was 1 × 10−5. This suggests that the amount of contributions of the second singular value vector were too small to govern individual gene expressions. As before, the high correlation despite this fact speaks to the soundness of our methodology.
The next task was to determine whether the genes selected exhibit temporal difference. The genes selected are scaled, shifted, and over drawn as boxplot (Fig 2(h)). Though it is difficult to observe, P-values computed by a two-sided t test between expression at time points, 0.5, 1, 2, 4, 6 and 48 hours (Fig 2) are significant only at limited time points with and without EFG treatment. Although this is of less significance than TD applied to a type I tensor, these are merely temporally differently expressed genes. Thus, TD based unsupervised FE applied to type II tensor is effective.
In order to see the biological reliability of selected genes, the mRNAs associated with commonly selected 398 mRNA probes were uploaded to MSigDB. The second most significant gene set was determined to be KOBAYASHI_EGFR_SIGNALING_24HR_DN (the adjusted P value is 1.37 × 10−128), which, again, was reasonable as the genes sought expressed differently with and without EGF treatments. Although the number of genes selected was less than that by TD applied to type a I tensor, since the P-value is smaller, the significance was greater than that of TD applied to a type I tensor.
As a whole, both TD applied to type I type II tensors is effective.
The next temporally differentially expressed gene detection example is vaccine infection experiment [34]. Patients were divided into three groups, P, D and NP. As sample classification was used, it is also not fully unsupervised. It is, however, unsupervised in the sense that no temporal functional forms are assumed. , , and are the ith gene expressions of protected, delayed, and non-protected, patients at time points t1, t2 and t3 after vaccine treatments, respectively. Type I tensor xt1,t2,t3,i, was defined as
xt1,t2,t3,i would be expanded by HOSVD as
however, the total memory required to store all of this expansion is too large to be prepared. Fortunately, as can be seen in the application to the first temporally differentially expressed gene identification, TD applied to a type II tensor that requires much smaller (1/N) memory can be as effective as TD applied to type I tensor for temporally differentially expressed gene identification. Thus, for this example, I employ only type II tensor , defined as
that can be expanded as
by HOSVD.
Fig 3(a) and 3(b) shows the , and for ℓ1,ℓ2 = 1, 2. Obviously, , and do not have any time dependence while those for ℓ1 = ℓ2 = ℓ3 = 2 do, as in the EGF treated cell line cases. Though , and also seem to have some temporal difference, its significance is again unclear. To determine the significance of this difference, genes identified as outliers had to be selected. There are three gene singular value vectors:
Using these three gene singular value vectors with ℓ1 = ℓ2 = ℓ3 = 2, 104 mRNA probes identified commonly as outliers were selected (S1 Table).
Fig 3(c) shows the histogram of correlation coefficients between the vector generated by connecting , and vs selected 104 mRNA probes. These are highly correlated (adjusted P-values are less than 0.01). This is noteworthy as , being the three largest core tensors associated with these three gene singular value vectors, had the contributions as small as 1 × 10−3 of , the largest one. This suggests that the amount of contributions of the second gene singular value vector is too small to govern individual gene expression. The high correlation despite this fact suggests the soundness of our methodology.
The next task was to determine whether the mRNA probes selected exhibit temporal difference. The mRNA probes selected are scaled, shifted, and overdrawn as boxplot (Fig 3(d)). Though it is difficult to observe, P-values computed by categorical regression assuming three classes (P, D, NP) at time points, 1, 2, 3, 4 and 5 days (Fig 3) are significant at all time points between the three classes. This was clearly not due to simple baseline shifts, as can be seen in Fig 3(d). This is due to as temporally differently expressed genes. Thus, TD based unsupervised FE applied to type II tensor is also effective.
In order to determine the biological reliability of selected genes, the mRNAs associated with the selected 104 mRNA probes were uploaded to MSigDB. The top six significant gene sets were determined to be: GSE13485_X_VS_Y_YF17D_VACCINE_PBMC_DN, GSE10325_MYELOID_VS_LUPUS_MYELOID_DN, GSE13485_X_VS_Y_YF17D_VACCINE_PBMC_DN, GSE13485_X_VS_Y_YF17D_VACCINE_PBMC_DN, GSE13485_X_VS_Y_YF17D_VACCINE_PBMC_DN, and GSE13485_X_VS_Y_YF17D_VACCINATION_PBMC_DN, where (X, Y) = (CTRL, DAY7), (DAY1, DAY7), (CTRL, DAY3), (DAY3, YF17D), and (PRE, POST) in this order, which are associated with adjusted P-values, 2.97 × 10−66, 2.98 × 10−57, 3.69 × 10−55, 4.86 × 10−53, 5.43 × 10−51, and 6.36 × 10−49, respectively. As five out of six are related to vaccination, TD based unsupervised FE selected biologically feasible sets of genes.
In conclusion, TD based unsupervised FE is also effective also for the identification of temporally differentially expressed genes.
Discussion
In the following section I discuss the strategy for applying TD based unsupervised FE to tensors built from matrix products, methodological points of view, and outcomes obtained by applying this strategy to multi-omics datasets from the biological point of views.
Comparisons with various methods applicable to synthetic data
The synthetic datasets that presented in the above sections are very difficult to analyse using standard supervised statistical analysis methods. In the supervised methodology, all background knowledge of given datasets is required, e.g., classification labels or assumed functional forms (for example, monotonic increase/decrease or periodicity). Alternatively, TD applied to type I tensors is Eq (3) and applied to type II tensor is Eq (4), which can be performed in the synthetic dataset prepared without any information in advance. To my knowledge, there are no applicable supervised methodologies for the synthetic datasets presented above. Thus, in the following I discuss only unsupervised methods.
As the first N0 features are derived from the common bases shown in Fig 4(a)–4(c), it is possible to detect them by computing correlations between them. However, as can be seen in Fig 4(d)–4(f), the correlations are highly non-linear, and it is therefore impossible to detect them (in actuality, the Pearson correlation between two variables shown is Fig 4(f) is as small as -0.01).
One may wonder if correlation analysis considering linear combinations, e.g., canonical correlation analysis (CCA), can depict latent correlation. However, in CCA, as M dimensional vectors as numerous as N must be compared, it is an overcomplete problem when M < N (and this is the present case). Thus, canonical correlation coefficients generated from linear combinations are always 1.0. This means that there is no way to detect latent correlation between N0(<N) features.
Similar problems stand for nonlinear correlation analysis like kernel CCA (kCCA). KCCA was applied to matrices X(k), k = 1, 2 in the above synthetic examples. The ten components (this is the kCCA default) generated are classified into two types, each of which are distinct from the first N0 features and remaining (Fig 8(a)–8(d)). Thus, the correlation is apparently successful, however, both results in the correlation coefficients were as large as 1.0 meaning that kCCA evaluated the correlation of two views between the first N0 features and that between remaining features the latter composed of simple random numbers as demonstrated in the above. Thus, kCCA cannot distinguish between latent correlation between the first N0 features and random numbers, and cannot be successfully applied.
Fig 8. Two alternative methods applied to synthetic data.
(a) to (d): The results of kCCA. Vertical axes are the coefficients used for linear combinations of N features, Horizontal axes are i1 and i2, i.e., indices attributed to N features. (a) and (b): the first view, (c) and (d) the second view. (a) and (c): the first type of kCCA results and (b) and (d): the second type of kCCA results. The distinction between the two types of kCCA results is coincident with the distinction between features with latent correlation (1 ≤ j ≤ N0) and those without correlation (N0 < j ≤ N). However, as computed correlation coefficients were as high as 0.99, kCCA failed to identify latent correlation. (e) to (h): PCA separately applied to two views in synthetic data. (e) and (f): the first PC loadings attributed to M samples in each view. (g) and (h) the first and the second PC scores attributed to N features in each view. Red open circles are features with latent correlation (1 ≤ j ≤ N0). Black circles are those composed of random numbers (N0 < j ≤ N).
Finally, PCA based unsupervised FE, which was recently proposed and successfully applied to various integrated analyses of multi-omics datasets, was again applied here. As PCA is equivalent to singular value decomposition (SVD),
Fig 8(e) and 8(f) are vℓ1 = 1, j and , respectively. uℓ1,i1,ℓ1 = 1, 2 is Fig 8(g) and uℓ2,i2, ℓ2 = 1, 2 is Fig 8(h). λℓ2 and λℓ3 are the eigen values computed with PCA. Thus
Compared with Eq (3), if
and PCA is equivalent to TD applied to a a type I tensor. Alternatively, compared with Eq (4), if
| (5) |
then PCA is equivalent to TD applied to a type II tensor. However, HOSVD does not always produce the solution satisfying the above. For example, since Eq (4) computed with HOSVD a standard SVD, is diagonal, while Eq (5) cannot be diagonal since vℓ1,j is not orthogonal to .
Although PCA based unsupervised FE successfully identified the first N0 features associated with latent correlations as outliers (Fig 8(g) and 8(h)), PC loadings attributed to samples (Fig 8(e) and 8(f)) are almost identical to Fig 4(d) and 4(e), which are not correlated (Fig 4(f)). Therefore PCA based unsupervised FE cannot identify latent correlation. In the previous applications of PCA based unsupervised FE aimed at integrated analysis of multi-omics data [14, 22], it was critical to identify pairs of highly correlated PC loadings, otherwise it was not possible to identify which PCs should be used for FE. In this context, PCA based unsupervised FE failed to detect latent correlations among multi-views.
While PCA was unsuccessful, HOSVD was applied to type I tensors in an attempt to decompose vℓ1,j and (Fig 8(e) and 8(f)), which are not orthogonal, into orthogonal bases xℓ3,j as shown in Fig 4(g)–4(i). For this reason, TD applied to type I tensors is superior to PCA when applied to the matrix products of synthetic datasets and can depict latent correlation that PCA failed to identify. As for HOSVD applied to type II tensors, Fig 6(b) and 6(e) correspond to Fig 4(e) while Fig 6(c) and 6(f) correspond to Fig 4(d) shows HOSVD applied to type II tensors can depict the latent correlation that PCA based unsupervised FE failed to detect.
Methodological discussion of TD based unsupervised FE applied to multi-omics data
In contrast to synthetic data (to which no supervised methods apply), the supervised method can be applied to the multi-omics data used in this study can be treated with supervised method as the data uses class labels. Strictly speaking, there are no unsupervised methods applicable to multi-view data processing other than TD based unsupervised FE. We can, therefore, even conclude that is the above strategy is sound. That said, it would be beneficial to demonstrate that the unsupervised methodology is superior to the supervised methodology.
As mentioned in above, most multi-view data processing methodologies require optimization of weights. I do not consider such methodologies, since optimizing weights is a complicated unnecessary process. Therefore, they are not comparable with the unsupervised strategy, which involves no parameter optimization processes.
Thus, the multi-view data processing methodology is considered applicable to merged matrices shown in Eqs (1) or (2), which does not include any weight optimization. Here, I consider three alternative methodologies, Significance Analysis of Microarrays (SAM), [35] Limma [36] and randomforest [37] (RF). As SAM and Limma evaluate features independently, weights attributed to views are not required. Although RF evaluates features in more collective ways, the evaluations are tree based, therefore the absolute values of each feature are not important. Table 5 shows the pertinent results. All three methods are inferior to the methodology presented above, as they failed to identify a significantly small number of features. Limma selected all of 755 miRNAs as significant. Roughly speaking, at least half of mRNAs were identified by these three methods.
Table 5. The numbers of identified mRNAs and miRNAs (multi-omics) and mRNAs (vaccination) using various methodologies.
Multi-omics: Among 427 mRNA probes and 12 miRNAs identified PCA based unsupervised FE, 408 mRNA probes (Table 4) and 9 miRNAs were also identified with TD based unsupervised FE applied to type I tensor.
| SAM | Limma | RF | PCA based unsupervised FE | |
|---|---|---|---|---|
| multi-omics | ||||
| mRNA | 8055 | 6055 | 5079 | 427 |
| miRNA | 148 | 755 | 186 | 12 |
| vaccine | ||||
| mRNA | 11739 | 9445 | 8300 | 18 |
Although these might be enough to demonstrate the superiority of the methodology presented above, measures were also undertaken to reduce the number of mRNAs identified by reducing threshold P-values ensuring that these three methods identify 426 mRNA probes, which is the same number identified by TD applied to type I tensors. As threshold P-values which are too small without any statistical justifications may not be acceptable, in order to evaluate these three methods, unnatural threshold P-values were intentionally used. Top ranked mRNAs selected by using intentionally reduced threshold P-values were uploaded to MSigDB server. However, breast cancer was rarely identified (Table 6). Breast cancer was identified only once by SAM, and was not detected in either limma or RF. These outcomes are in contrast to Table 2, where eight out of ten significant gene sets are breast cancer-related when TD was applied to either type I or type II tensor. Thus, it is obvious that these methodologies are inferior to the methodology presented above in the present study, i.e., TD based unsupervised FE.
Table 6. Top 10 significant overlap gene set in MSigDB with top ranked 400 (approx) mRNA probes identified by alternative methods SAM, limma, and RF, as well as 374 mRNA probes identified by HO GSVD.
“BREAST_CANCER” was presented in bold to emphasize the overlap with breast cancer, whose counts are in parentheses at the right side of method names.
| SAM (1) | limma (0) | RF (0) | HO GSVD (5) | |
|---|---|---|---|---|
| 1 | BLALOCK_ALZHEIMERS_DISEASE_UP | PUJANA_BRCA1_PCC_NETWORK | DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_UP | SMID_BREAST_CANCER_LUMINAL_B_DN |
| 2 | SMID_BREAST_CANCER_NORMAL_LIKE_UP | BLALOCK_ALZHEIMERS_DISEASE_UP | BLALOCK_ALZHEIMERS_DISEASE_UP | SMID_BREAST_CANCER_BASAL_DN |
| 3 | SWEET_LUNG_CANCER_KRAS_DN | KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_UP | HAMAI_APOPTOSIS_VIA_TRAIL_UP | YOSHIMURA_MAPK8_TARGETS_UP |
| 4 | WEST_ADRENOCORTICAL_TUMOR_DN | RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_UP | PUJANA_BRCA1_PCC_NETWORK | SMID_BREAST_CANCER_RELAPSE_IN_BONE_DN |
| 5 | BOQUEST_STEM_CELL_CULTURED_VS_FRESH_UP | PUJANA_CHEK2_PCC_NETWORK | GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_DN | JAEGER_METASTASIS_DN |
| 6 | PUJANA_BRCA1_PCC_NETWORK | DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_UP | MILI_PSEUDOPODIA_HAPTOTAXIS_UP | GOZGIT_ESR1_TARGETS_DN |
| 7 | GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_DN | GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_DN | BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_UP | SMID_BREAST_CANCER_BASAL_UP |
| 8 | HAMAI_APOPTOSIS_VIA_TRAIL_UP | RODRIGUES_THYROID_CARCINOMA_ANAPLASTIC_UP | PUJANA_ATM_PCC_NETWORK | ONDER_CDH1_TARGETS_2_DN |
| 9 | BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_UP | FEVR_CTNNB_TARGETS_DN | PUJANA_CHEK2_PCC_NETWORK | SENGUPTA_NASOPHARYNGEAL_CARCINOMA_WITH_LMP1_DN |
| 10 | PUJANA_CHEK2_PCC_NETWORK | CAIRO_HEPATOBLASTOMA_CLASSES_UP | BLALOCK_ALZHEIMERS_DISEASE_DN | FARMER_BREAST_CANCER_APOCRINE_VS_LUMINAL |
Finally, PCA based unsupervised FE was applied to mRNAs and miRNAs separately (Table 5). PCA based unsupervised FE identified smaller numbers of mRNAs and miRNAs than the above three methodologies. Especially, since mRNAs selected are almost identical to those identified by TD based unsupervised FE applied to type I tensor (Table 4), PCA based unsupervised FE is as effective for the identification of biologically reliable mRNAs. However, it cannot identify latent correlation between miRNAs and mRNAs, as hierarchical clustering of PC loading attributed to samples identified no pairs of miRNAss and mRNA (Fig 7(b)), which were identified when TD applied to type II tensors was considered and without which no integrated analysis of multi-omics datasets by PCA based unsupervised FE were successful. This indicates that the present datasets were more complex and cannot be dealt with using PCA based unsupervised FE in an integrated manner. Thus, I conclude that TD based unsupervised FE applied to type I or type II tensors is the only method for achieving two tasks: (i) identifying sufficiently small numbers of biologically important features, and (ii) identify latent correspondence between multi-omics profiles.
Here, PCA based unsupervised FE is shown to be the only strategy that can compute with the TD based unsupervised FE applied to matrix products. A more detailed comparison of these two strategies may enable us to understand the functionality of TD based unsupervised FE. As can be seen in the application to synthetic data, TD applied to type I tensors attempted to decompose vℓ1,j and v′ℓ2,j into orthogonal bases xℓ3,j. This also occurred in the application to multi-omics datasets. First, I identified that the first miRNA PC loading, , dominates subsequent PC loadings (more than 80%). Next, I also identified that the first mRNA PC loadings attributed to samples vℓ1,j and 1 ≤ ℓ1 ≤ 5, are identical to the first five sample singular value vectors xℓ3,j, 1 ≤ ℓ3 ≤ 5. The Pearson correlations between these five loadings and singular value vectors are −0.94, −0.91, 0.88, −0.97 and −0.97 (Here the signs do not mean anything), respectively. It is also shown the regression analysis of the first miRNA PC loading with the top five mRNA PC loadings, vℓ1,j, 1 ≤ ℓ1 ≤ 5,
covers more than 40% of the first miRNA PC loading . Since the dimension of vector is M = 161, only five components that can cover this amount is highly significant. This suggests that TD can easily decompose single miRNA PC loadings into five mRNA PC loading vℓ1,j, 1 ≤ ℓ1 ≤ 5. The result is that the first five sample singular value vectors (xℓ3,j, 1 ≤ ℓ3 ≤ 5), are almost identical to the first five mRNA PC loadings (vℓ1,j, 1 ≤ ℓ3 ≤ 5). Thus, TD based unsupervised FE can decompose the first dominant miRNA PC loading into five basic (orthogonal) mRNA PC loading. This result is analogous to that seen in the application to synthetic dataset (Fig 4).
It is also important to show that the five mRNA PC loadings vℓ1,j, 1 ≤ ℓ1 ≤ 5 (or the first five sample singular value vectors ) are distinct also from biological point of view. To demonstrate this, five sets of outliner mRNA probes (associated mRNAs are in S2 Table) were selected using each of the first five mRNA singular value vectors () each of which is coincident with the first five sample singular value vectors, ()as G(ℓ1, ℓ2, ℓ3)s with 1 ≤ ℓ1 = ℓ3 ≤ 5 have larger absolute values (Table 1). GO (biological process) BP term enrichments were tested by uploading S2 Table to g:Cocoa in g:profiler [27] (S2 File). It is obvious that five sets of mRNAs identified as outliers using each of the first five mRNA singular value vectors are biologically distinct from one another.
Methodological discussion of TD based unsupervised FE applied to identification of temporally differentially expressed genes
At certain time points in EGF treatment experiments, there is only one measurement, which prevents the application of some statistical methods. Therefore, only vaccination samples were considered. Again, SAM, limma, RF and PCA based unsupervised FE were considered. Here, the samples are assumed to be classified into five time points times three treatments (P, D, NP) equalling 15 classes. Results are shown in Table 5. As with multi-omics data, SAM, limma, and RF failed to identify sufficiently small numbers. This is possibly due to the fact that there are 15 classes. Since even the detection of differences between pairs of any two of the 15 classes can effect results, there are many genes identified as having significant differences. On the other hand, 18 mRNA probes were identified by PCA based unsupervised FE with considering common set when separately applied to three gene expression profiles, and . Thus, relatively successful.
In order to determine biological significance, a reduced number of gene sets was uploaded to MSigDB. Similar to the multi-omics case, SAM indicated too many mRNAs with adjusted P-value = 0, therefore no reduced sets could be generated. Two 300 top ranked mRNA probes sets were generated from limma and RF and associated mRNAs were uploaded to MSigDB together with the genes associated with 18 mRNA probes obtained by PCA based unsupervised FE. Within top 10 ranked significant genes set, no vaccination related genes sets were identified for limma or RF. PCA based supervised FE has only two vaccination related genes sets. GSE13485_X_VS_Y_YF17D_VACCINE_PBMC_DN, (X, Y) = (DAY3, DAY7) and (DAY1, DAY7), were identified as the second (adjusted P = 1.86 × 10−6) and the fourth (adjusted P = 4.30 × 10−5) significant genes sets, which were smaller than those identified when TD was applied to type II tensors, which identified five gene sets associated with vaccination out of six top ranked gene sets.
Thus, one of four tested methods, PCA based unsupervised FE, could produce some significant results which were inferior to those produced by TD based unsupervised FE applied to type II tensors. As a result, TD based unsupervised FE proved more effective than the other methodologies analysed above.
Comparison with HO GSVD
To my knowledge, although there are no methods that comprise a tensor from multiple matrices and applies TD to it, similar trials aiming integration of multiple matrices exist. For example, higher order generalized singular value decomposition (HO GSVD) [25] is one such method. Although HO GSVD does not generate tensor, the outcome is quite similar; a set of feature singular value vectors and a unique (common) sample singular value vector which is equivalent to what TD applied to type I tensors generated from Case I data produces. Although Ponnapalli et al [25] employed the distinct terminology from the present study, I continue to use my own terminology in this subsection to avoid confusion.
First, HO GSVD was applied to synthetic data. Fig 9(a)–9(d) shows the results. Its outcome is close to that when PCA based unsupervised FE was applied to dataset (Fig 8(e)–8(h)). It is in some sense reasonable, since HO GSVD is essentially PCA excluding the fact multiple views share the unique sample singular value matrix, V, where the first the second column vectors correspond to the first PC loading of the first and the second views, respectively.
Fig 9. The results of HO GSVD applied to the synthetic data.
Red open circles are features with latent correlation (1 ≤ j ≤ N0). The first (a) and the second (b) sample singular value vectors and the first vs the second feature singular value vectors of the first (c) and the second views (d).
Next, HO GSVD was applied to multi-omics data. Coincidence between sample singular value vectors and class labeling is shown in Fig 1(d). Although four out of five vectors are significantly coincident with class labeling, significance was substantially less than when TD was applied to type I and II tensors, as the P-values were larger. Thus, HO GSVD can perform well but is less effective than TD applied to type I or II tensors.
Next, 374 mRNA probes identified as outliers using the first five mRNA feature singular value vectors were selected (associated mRNAs are shown in S1 Table). Uploading the mRNAs associated with 374 mRNA probes to MSigDB, I found that only five out of ten top ranked significant genes sets were related to breast cancer (Table 6), while eight out of ten were related to breast cancer when TD was applied to type I or II tensors (Table 2).
Fifteen miRNAs(miR-127-5p/128/181a/190a/301a/30e*/339-5p/340/361-5p/365 /452/454/455-5p/874/135a) identified as outliers using the first and the second miRNA feature singular value vectors were selected. None were reported in the original study ([30], Table 1). Uploading 15 miRNAs to DIANA-mirpath, I found that “MiRNAs in cancer”, which was the top ranked when TD was applied to type I or type II tensors (Table 3), was not included in the top ten ranked KEGG pathways.
HO GSVD cannot be applied to identification of temporally differentially expressed genes, since HO GSVD can be applied only to Case I data where samples are shared between multiple views.
These slightly poorer outcomes of HO GSVD than TD applied to type I or II tensors suggest the usefulness of tensors when analysing multi-view datasets.
Biological validations of mRNAs identified in multi-omics data analysis
In the previous subsection, TD based unsupervised FE applied to product of multi-omica profile matrices was validated chiefly from the methodological perspective, and validated partially from the biological perspective.
In this subsection, I try to validate outcomes biologically in more detail, mainly based upon the consideration from oncology.
The samples analysed are essentially proposing the comparison between tumors with and without metastasis. Thus, it is expected that selected genes are mainly related to cancer oncogenesis related to metastasis.
Since Farazi et al [30] who produced the original study, mainly discuss the aberrant expression of miRNAs among samples, there is no in depth discussion about the role of miRNA/mRNA in metastasis. However, as can be seen in the below, much can be discussed from their dataset.
In order to biologically investigate a set of mRNAs identified when type I tensors were considered, to the mRNAs were uploaded to g:profiler (see S3 Table). A large number of enrichments of biological terms were identified.
For example, in GO BP terms, “leukocyte activation” (GO:0045321) was enriched. It was reported to be related to metastasis. Ihnen et al [38] reported a tumor biological context of activated leukocyte cell adhesion molecules (ALCAM) for the development of metastases in breast cancer. Strell et al [39] concluded that the first two steps of the extravasation of tumor cells and leukocytes, rolling and adhesion, seem to have similarities with regards to the mechanisms and receptors involved. King et al [40] identified ALCAM in metastasis of breast cancer cells to the lung. These suggested that metastasis was mediated by the extravasation similar to that of leukocytes. In relaton to this, “positive regulation of leukocyte chemotaxis” (GO:0002690) was also enriched. Wu [41] reported the role of chemotaxis in cell migration. Gradient of chemotaxis mediates cell migration, and possibly metastasis, too.
In GO (cellular component) CC terms, “extracellular region” (GO:0005576) was enriched. Cho et al [42] reported that Herceptin binds to the juxtamembrane region of HER2, identifying this site as a target for anticancer therapies, while overexpression of HER2 is found in 20-30% of human breast cancers, and correlates with more aggressive tumours and a poorer prognosis. It is also primary biomarker of breast cancer in the original study [30]. More generally, Versteeg et al [43] suggested the importance of extracellular signaling pathway in cancer metastasis. It mediates blood vessel wall damage, which may allow tumours to migrate through blood vessels.
In GO (molecular function) MF terms, “CXCR3 chemokine receptor binding” (GO:0048248) was enriched. CXCR3 was reported as a molecular target in breast cancer metastasis [44]; it inhibits tumor cell migration and promotes of host anti-tumour immunity. As suggested in the above, chemotaxis mediates cell migration and chemokine receptor CXCR3 agonist prevents human T-cell migration [45]. Other than in relation to metastasis, inhibition of CXCR3 was also known to mediate tumor growth [46]. “RAGE receptor binding” (GO:0050786) was also enriched. RAGE was reported to mediate tumor progression and metastasis through binding to S100A7 by modulating the tumor microenvironment [47]. It recruits MMP9-positive tumor-associated macrophages and mediates cell migrations.
Other than in GO terms enrichment, transcription factor (TF) SOX9 target genes were enriched. The relation between SOX9 and metastasis was pointed out by many papers. Got et al [48] reported that co-expression of Slug and Sox9 promotes the tumorigenic and metastasis-seeding abilities of human breast cancer cells. SOX9 protein, which is normally nuclear, was instead localized in the cytoplasm of 25-30% invasive ductal carcinomas (IDCs) and lymph node metastases [49]. Lei et al [50] also suggested that Sox9 expression is related to breast cancer metastasis. Although the above are all related to breast cancer, Sox9 was frequently reported to be related to metastasis in various other cancers.
KEGG pathway “Primary immunodeficiency” (KEGG:05340) was also enriched. Development of cancer in patients with primary immunodeficiencies was reported [51]. Monozygotic twin brothers with primary immunodeficiency presented with metastatic adenocarcinoma of unknown primary [52].
In conclusion, our method and strategy correctly identified many cancer related biological terms/concept enrichments, especially metastasis in breast cancer, which is coincident with the purpose of the original study that did not produce results produced here.
Biological validations of miRNAs identified in multi-omics data analysis
As the relation between mRNAs identified and breast cancer metastasis can be shown, it is necessary to demonstrate the relationship between the miRNAs identified and breast cancer metastasis. Research of the litertature showsthat all seven miRNAs identified when type I tensors were considered(let-7b [53], miR-125b [54–56], miR-143 [57–64], miR-145 [61, 62, 65–68], miR-21 [69–73], miR-22 [74–78] and miR-99a [32, 79]), were reported to be related to metastasis.
Although not all are strictly related to breast cancer, all seven miRNAs identified are frequently reported to be related to metastasis.
Conclusion
In this paper, a new strategy aiming at multi-view data processing that makes use of tensors generated from multi-view matrices products was proposed. As tensors can be generated from individual measurements, observation under combined conditions, which is generally required to produce tensors from datasets, is not necessary. FEs were performed using singular value vectors generated from TD and biological feasibility was confirmed via comparisons with previously generated annotated gene expression profiles. As this strategy is not restricted to gene expression, its application to other datasets is feasible.
Supporting information
A Work flow chart and list of the variables introduced are in S1 File.
(PDF)
BP term enrichments by uploading S2 Table to g:Cocoa in g:profiler.
(PDF)
Full lists of genes selected by various methods.
(XLSX)
Lists of genes associated with five gene singular value vectors.
(CSV)
Enriched terms when list of genes were uploaded to g profiler.
(XLSX)
Data Availability
All data sets from GEO GEO ID: GSE28884, GSE84096, GSE18323.
Funding Statement
This work was supported by Japan Society for the Promotion of Science-17K00417-Prof. Y-h. Taguchi.
References
- 1.Li Y, Wu FX, Ngom A. A review on machine learning principles for multi-view biological data integration. Brief Bioinformatics. 2016;. [DOI] [PubMed]
- 2.Xu C, Tao D, Xu C. A Survey on Multi-view Learning. CoRR. 2013;abs/1304.5634.
- 3.Taguchi YH, Iwadate M, Umeyama H, Murakami Y. Principal component analysis based unsupervised feature extraction applied to bioinformatics analysis. In: Tsai JJP, Ng K-l, editors. Computational Methods with Applications in Bioinformatics Analysis. World Scientific; 2017. p. 153–182. Available from: http://www.worldscientific.com/doi/abs/10.1142/9789813207981_0008
- 4. Taguchi YH. Principal Components Analysis Based Unsupervised Feature Extraction Applied to Gene Expression Analysis of Blood from Dengue Haemorrhagic Fever Patients. Sci Rep. 2017;7:44016 10.1038/srep44016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taguchi YH. Principal component analysis based unsupervised feature extraction applied to publicly available gene expression profiles provides new insights into the mechanisms of action of histone deacetylase inhibitors. Neuroepigenetics. 2016;8:1–18. 10.1016/j.nepig.2016.10.001 [DOI] [Google Scholar]
- 6. Taguchi YH, Iwadate M, Umeyama H. Principal component analysis-based unsupervised feature extraction applied to in silico drug discovery for posttraumatic stress disorder-mediated heart disease. BMC Bioinformatics. 2015;16:139 10.1186/s12859-015-0574-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taguchi YH, Okamoto A. Principal Component Analysis for Bacterial Proteomic Analysis In: Shibuya T, Kashima H, Sese J, Ahmad S, editors. Pattern Recognition in Bioinformatics. vol. 7632 of LNCS. Heidelberg: Springer International Publishing; 2012. p. 141–152. [Google Scholar]
- 8. Ishida S, Umeyama H, Iwadate M, Taguchi YH. Bioinformatic Screening of Autoimmune Disease Genes and Protein Structure Prediction with FAMS for Drug Discovery. Protein Pept Lett. 2014;21(8):828–39. 10.2174/09298665113209990052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinoshita R, Iwadate M, Umeyama H, Taguchi YH. Genes associated with genotype-specific DNA methylation in squamous cell carcinoma as candidate drug targets. BMC Syst Biol. 2014;8 Suppl 1:S4 10.1186/1752-0509-8-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taguchi YH, Murakami Y. Principal component analysis based feature extraction approach to identify circulating microRNA biomarkers. PLoS ONE. 2013;8(6):e66714 10.1371/journal.pone.0066714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taguchi YH, Murakami Y. Universal disease biomarker: can a fixed set of blood microRNAs diagnose multiple diseases? BMC Res Notes. 2014;7:581 10.1186/1756-0500-7-581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE. 2012;7(10):e48366 10.1371/journal.pone.0048366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murakami Y, Tanahashi T, Okada R, Toyoda H, Kumada T, Enomoto M, et al. Comparison of Hepatocellular Carcinoma miRNA Expression Profiling as Evaluated by Next Generation Sequencing and Microarray. PLoS ONE. 2014;9(9):e106314 10.1371/journal.pone.0106314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murakami Y, Kubo S, Tamori A, Itami S, Kawamura E, Iwaisako K, et al. Comprehensive analysis of transcriptome and metabolome analysis in Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Sci Rep. 2015;5:16294 10.1038/srep16294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Umeyama H, Iwadate M, Taguchi YH. TINAGL1 and B3GALNT1 are potential therapy target genes to suppress metastasis in non-small cell lung cancer. BMC Genomics. 2014;15 Suppl 9:S2 10.1186/1471-2164-15-S9-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taguchi YH, Iwadate M, Umeyama H. Heuristic principal component analysis-based unsupervised feature extraction and its application to gene expression analysis of amyotrophic lateral sclerosis data sets. In: Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 2015 IEEE Conference on; 2015. p. 1–10.
- 17. Taguchi YH, Iwadate M, Umeyama H, Murakami Y, Okamoto A. Heuristic principal component analysis-aased unsupervised feature extraction and its application to bioinformatics In: Wang B, Li R, Perrizo W, editors. Big Data Analytics in Bioinformatics and Healthcare; 2015. p. 138–162. [Google Scholar]
- 18. Taguchi YH. Integrative Analysis of Gene Expression and Promoter Methylation during Reprogramming of a Non-Small-Cell Lung Cancer Cell Line Using Principal Component Analysis-Based Unsupervised Feature Extraction In: Huang DS, Han K, Gromiha M, editors. Intelligent Computing in Bioinformatics. vol. 8590 of LNCS. Heidelberg: Springer International Publishing; 2014. p. 445–455. [Google Scholar]
- 19. Taguchi YH. Identification of aberrant gene expression associated with aberrant promoter methylation in primordial germ cells between E13 and E16 rat F3 generation vinclozolin lineage. BMC Bioinformatics. 2015;16 Suppl 18:S16 10.1186/1471-2105-16-S18-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taguchi YH. Identification of More Feasible MicroRNA-mRNA Interactions within Multiple Cancers Using Principal Component Analysis Based Unsupervised Feature Extraction. Int J Mol Sci. 2016;17(5):E696 10.3390/ijms17050696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taguchi YH. Principal component analysis based unsupervised feature extraction applied to budding yeast temporally periodic gene expression. BioData Min. 2016;9:22 10.1186/s13040-016-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taguchi YH, Iwadate M, Umeyama H. SFRP1 is a possible candidate for epigenetic therapy in non-small cell lung cancer. BMC Med Genomics. 2016;9 Suppl 1:28 10.1186/s12920-016-0196-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lathauwer LD, Moor BD, Vandewalle J. A Multilinear Singular Value Decomposition. SIAM Journal on Matrix Analysis and Applications. 2000;21(4):1253–1278. 10.1137/S0895479896305696 [DOI] [Google Scholar]
- 24. Omberg L, Golub GH, Alter O. A tensor higher-order singular value decomposition for integrative analysis of DNA microarray data from different studies. Proc Natl Acad Sci USA. 2007;104(47):18371–18376. 10.1073/pnas.0709146104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ponnapalli SP, Saunders MA, Van Loan CF, Alter O. A higher-order generalized singular value decomposition for comparison of global mRNA expression from multiple organisms. PLoS ONE. 2011;6(12):e28072 10.1371/journal.pone.0028072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 27. Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016;44(W1):W83–89. 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24(12):1461–1462. 10.1093/bioinformatics/btn209 [DOI] [PubMed] [Google Scholar]
- 29.R Core Team. R: A Language and Environment for Statistical Computing; 2015. Available from: https://www.R-project.org/
- 30. Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. 10.1158/0008-5472.CAN-11-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu SH, Zhang CL, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116(2):268–276. 10.1002/jcb.24965 [DOI] [PubMed] [Google Scholar]
- 33. Albrecht M, Stichel D, Muller B, Merkle R, Sticht C, Gretz N, et al. TTCA: an R package for the identification of differentially expressed genes in time course microarray data. BMC Bioinformatics. 2017;18(1):33 10.1186/s12859-016-1440-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vahey MT, Wang Z, Kester KE, Cummings J, Heppner DG, Nau ME, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J Infect Dis. 2010;201(4):580–589. 10.1086/650310 [DOI] [PubMed] [Google Scholar]
- 35. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fortino V, Kinaret P, Fyhrquist N, Alenius H, Greco D. A robust and accurate method for feature selection and prioritization from multi-class OMICs data. PLoS ONE. 2014;9(9):e107801 10.1371/journal.pone.0107801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ihnen M, Kohler N, Kersten JF, Milde-Langosch K, Beck K, Holler S, et al. Expression levels of Activated Leukocyte Cell Adhesion Molecule (ALCAM/CD166) in primary breast carcinoma and distant breast cancer metastases. Dis Markers. 2010;28(2):71–78. 10.3233/DMA-2010-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strell C, Entschladen F. Extravasation of leukocytes in comparison to tumor cells. Cell Commun Signal. 2008;6:10 10.1186/1478-811X-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. King JA, Chambers Z, Kleinfeld S, Chen H, Stevens T, Shevde LA, et al. Potential role of activated leukocyte cell adhesion molecule (ALCAM/CD166) in metastasis of breast cancer cells to the lung. Cancer Research. 2014;66(8 Supplement):654–654. [Google Scholar]
- 41. Wu D. Signaling mechanisms for regulation of chemotaxis. Cell Res. 2005;15(1):52–56. 10.1038/sj.cr.7290265 [DOI] [PubMed] [Google Scholar]
- 42. Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421(6924):756–760. 10.1038/nature01392 [DOI] [PubMed] [Google Scholar]
- 43. Versteeg HH, Spek CA, Peppelenbosch MP, Richel DJ. Tissue factor and cancer metastasis: the role of intracellular and extracellular signaling pathways. Mol Med. 2004;10(1-6):6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu G, Yan HH, Pang Y, Jian J, Achyut BR, Liang X, et al. CXCR3 as a molecular target in breast cancer metastasis: inhibition of tumor cell migration and promotion of host anti-tumor immunity. Oncotarget. 2015;6(41):43408–43419. 10.18632/oncotarget.6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O’Boyle G, Fox CRJ, Walden HR, Willet JDP, Mavin ER, Hine DW, et al. Chemokine receptor CXCR3 agonist prevents human T-cell migration in a humanized model of arthritic inflammation. Proceedings of the National Academy of Sciences. 2012;109(12):4598–4603. 10.1073/pnas.1118104109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oghumu S, Varikuti S, Terrazas C, Kotov D, Nasser MW, Powell CA, et al. CXCR3 deficiency enhances tumor progression by promoting macrophage M2 polarization in a murine breast cancer model. Immunology. 2014;143(1):109–119. 10.1111/imm.12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nasser MW, Ahirwar DK, Ganju RK. RAGE: A novel target for breast cancer growth and metastasis. Oncoscience. 2016;3(2):52–53. 10.18632/oncoscience.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med (Maywood). 2011;236(2):145–155. 10.1258/ebm.2010.010086 [DOI] [PubMed] [Google Scholar]
- 50.Sox9 upregulation in breast cancer is correlated with poor prognosis and the CD44+/CD24-/low phenotype; 2016.
- 51. Salavoura K, Kolialexi A, Tsangaris G, Mavrou A. Development of cancer in patients with primary immunodeficiencies. Anticancer Res. 2008;28(2B):1263–1269. [PubMed] [Google Scholar]
- 52. Wood LA, Venner PM, Pabst HF. Monozygotic twin brothers with primary immunodeficiency presenting with metastatic adenocarcinoma of unknown primary. Acta Oncol. 1998;37(7-8):771–772. 10.1080/028418698430197 [DOI] [PubMed] [Google Scholar]
- 53. Han X, Chen Y, Yao N, Liu H, Wang Z. MicroRNA let-7b suppresses human gastric cancer malignancy by targeting ING1. Cancer Gene Ther. 2015;22(3):122–129. 10.1038/cgt.2014.75 [DOI] [PubMed] [Google Scholar]
- 54. Zhou HC, Fang JH, Shang LR, Zhang ZJ, Sang Y, Xu L, et al. MicroRNAs miR-125b and miR-100 suppress metastasis of hepatocellular carcinoma by disrupting the formation of vessels that encapsulate tumour clusters. J Pathol. 2016;240(4):450–460. 10.1002/path.4804 [DOI] [PubMed] [Google Scholar]
- 55. Glud M, Rossing M, Hother C, Holst L, Hastrup N, Nielsen FC, et al. Downregulation of miR-125b in metastatic cutaneous malignant melanoma. Melanoma Res. 2010;20(6):479–484. 10.1097/CMR.0b013e32833e32a1 [DOI] [PubMed] [Google Scholar]
- 56. Tang F, Zhang R, He Y, Zou M, Guo L, Xi T. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS ONE. 2012;7(5):e35435 10.1371/journal.pone.0035435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. He Z, Yi J, Liu X, Chen J, Han S, Jin L, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15(1):51 10.1186/s12943-016-0533-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58. Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. 2012;33(6):1863–1870. 10.1007/s13277-012-0446-8 [DOI] [PubMed] [Google Scholar]
- 59. Hirahata M, Osaki M, Kanda Y, Sugimoto Y, Yoshioka Y, Kosaka N, et al. PAI-1, a target gene of miR-143, regulates invasion and metastasis by upregulating MMP-13 expression of human osteosarcoma. Cancer Med. 2016;5(5):892–902. 10.1002/cam4.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xia H, Sun S, Wang B, Wang T, Liang C, Li G, et al. miR-143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int J Mol Sci. 2014;15(7):11973–11983. 10.3390/ijms150711973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peng X, Guo W, Liu T, Wang X, Tu X, Xiong D, et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS ONE. 2011;6(5):e20341 10.1371/journal.pone.0020341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang S, Guo W, Tang Y, Ren D, Zou X, Peng X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol Rep. 2012;28(5):1831–1837. [DOI] [PubMed] [Google Scholar]
- 63. Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19(6):1123–1130. 10.1038/mt.2011.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, Wang Y, et al. MicroRNA-143 inhibits migration and invasion of human non-small-cell lung cancer and its relative mechanism. Int J Biol Sci. 2013;9(7):680–692. 10.7150/ijbs.6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang M, Wang J, Deng J, Li X, Long W, Chang Y. MiR-145 acts as a metastasis suppressor by targeting metadherin in lung cancer. Med Oncol. 2015;32(1):344 10.1007/s12032-014-0344-6 [DOI] [PubMed] [Google Scholar]
- 66. Donzelli S, Mori F, Bellissimo T, Sacconi A, Casini B, Frixa T, et al. Epigenetic silencing of miR-145-5p contributes to brain metastasis. Oncotarget. 2015;6(34):35183–35201. 10.18632/oncotarget.5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li YQ, He QM, Ren XY, Tang XR, Xu YF, Wen X, et al. MiR-145 inhibits metastasis by targeting fascin actin-bundling protein 1 in nasopharyngeal carcinoma. PLoS ONE. 2015;10(3):e0122228 10.1371/journal.pone.0122228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei Y, et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget. 2014;5(21):10816–10829. 10.18632/oncotarget.2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372(1-2):35–45. 10.1007/s11010-012-1443-3 [DOI] [PubMed] [Google Scholar]
- 70. Mudduluru G, George-William JN, Muppala S, Asangani IA, Kumarswamy R, Nelson LD, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep. 2011;31(3):185–197. 10.1042/BSR20100065 [DOI] [PubMed] [Google Scholar]
- 71. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. 10.1261/rna.1034808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bornachea O, Santos M, Martinez-Cruz AB, Garcia-Escudero R, Duenas M, Costa C, et al. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci Rep. 2012;2:434 10.1038/srep00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286(45):39172–39178. 10.1074/jbc.M111.285098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7(28):44252–44265. 10.18632/oncotarget.10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei Y, et al. microRNA-22 acts as a metastasis suppressor by targeting metadherin in gastric cancer. Mol Med Rep. 2015;11(1):454–460. [DOI] [PubMed] [Google Scholar]
- 76.Chen M, Hu W, Xiong CL, Qu Z, Yin CQ, Wang YH, et al. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;. [DOI] [PMC free article] [PubMed]
- 77. Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154(2):311–324. 10.1016/j.cell.2013.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wan WN, Zhang YQ, Wang XM, Liu YJ, Zhang YX, Que YH, et al. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol. 2014;9:178 10.1186/s13000-014-0178-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kuo YZ, Tai YH, Lo HI, Chen YL, Cheng HC, Fang WY, et al. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis. 2014;20(3):65–75. 10.1111/odi.12133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Work flow chart and list of the variables introduced are in S1 File.
(PDF)
BP term enrichments by uploading S2 Table to g:Cocoa in g:profiler.
(PDF)
Full lists of genes selected by various methods.
(XLSX)
Lists of genes associated with five gene singular value vectors.
(CSV)
Enriched terms when list of genes were uploaded to g profiler.
(XLSX)
Data Availability Statement
All data sets from GEO GEO ID: GSE28884, GSE84096, GSE18323.