Abstract
A universal base analogue forms ‘base pairs’ with each of the natural DNA/RNA bases with little discrimination between them. A number of such analogues have been prepared and their applications as biochemical tools investigated. Most of these analogues are non-hydrogen bonding, hydrophobic, aromatic ‘bases’ which stabilise duplex DNA by stacking interactions. This review of the literature of universal bases (to 2000) details the analogues investigated, and their uses and limitations are discussed.
INTRODUCTION
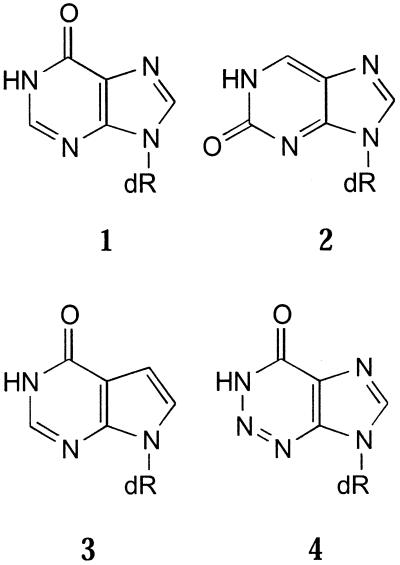
The naturally occurring base hypoxanthine, as its ribo- or 2′-deoxyribonucleoside (1), has been used for many years as an ‘inert’ base (1,2), at least in terms of its ability to form base pairs with the other natural DNA/RNA bases. This has led to a number of applications in primers (3,4) and in probes for hybridisation (1,5,6). Analogues of 1 (Fig. 1), 2′-deoxyisoinosine (2) (7), 7-deaza-2′-deoxyinosine (3) (8) and 2-aza-2′-deoxyinosine (4) (9) have also been investigated and behave in a similar manner. However, 1 is not indiscriminate in its base pairing properties and a wide range of melting temperatures (Tm) are found when it is paired opposite the natural bases in duplexes (5,10). Also, primers containing multiple substitutions by 1 often give rise to non-analysable sequence data. This review will look beyond the use of 1 at the applications of a number of other universal base analogues described during the last few years.
Figure 1.
2′-Deoxyinosine derivatives used as universal DNA analogues.
Almost all of the universal bases that have been described are DNA analogues; very little work has been reported on ribonucleosides. This is an area of potential interest because RNA polymerases are generally less demanding in terms of analogue incorporation and operate at much lower fidelity. Amongst the candidates for universal base analogues a number have been proposed which, in principle, could behave universally but in practice don’t. The main class of such compounds is azole carboxamide derivatives, e.g. 5–8 (11–13; Fig. 2). These compounds could behave ambiguously by rotation around the amide bond, which presents two alternative hydrogen bonding faces, depicted for 5. However, these compounds have proved to be disappointing as universal base analogues because they do not appear to use this potential. They tend to use very specific hydrogen bonding patterns, leading to transition or transversion mutations, but are not able to discriminate between all of the natural DNA/RNA bases. For this reason this class of compound will not be discussed unless there is some specific analogue of interest. Abasic site derivatives, e.g. 9, have also been examined (14–16) and these will also not be discussed here.
Figure 2.
Azole carboxamides designed to be universal base analogues.
The analogues discussed here lack hydrogen bonding sites and are generally hydrophobic aromatic ‘base’ residues. Many of their effects derive from their ability to stack within a duplex and from their hydrophobic character. The applications of universal bases are varied and they are discussed separately. The analogues, incorporated into DNA as their phosphoramidite derivatives, have principally been examined in hybridisation experiments, investigated by Tm values of duplexes containing them. Hybridisation applications include their use as primers for PCR and sequencing, in probes, in ligation and in triplexes. The enzymatic incorporation of 5′-triphosphates of universal bases has so far proven to be of limited utility, but this is also discussed. The main way in which universal base analogues have been shown to be effective is in their increased stacking potential and a number of analogues and applications are discussed separately. Recently there has been an increasing interest in universal base analogues and some of these are detailed towards the end, finishing with what is probably the first in a new series of universal bases, namely ones with hydrogen bonding capability.
The desirable requirements for a universal base have been defined (17). They should: (i) pair with all the natural bases equally when opposite them in an oligonucleotide duplex; (ii) form a duplex which primes DNA synthesis by a polymerase; (iii) direct incorporation of the 5′-triphosphate of each of the natural nucleosides opposite it when copied by a polymerase; (iv) be a substrate for polymerases as the 5′-triphosphate; (v) be recognised by intracellular enzymes such that DNA containing them may be cloned. At present no analogue has been shown to fulfil all these requirements and it may be that a single analogue can’t. However, each of the analogues prepared so far has a set of properties which enable their use for some of these purposes.
APPLICATIONS OF UNIVERSAL BASE ANALOGUES
Hybridisation
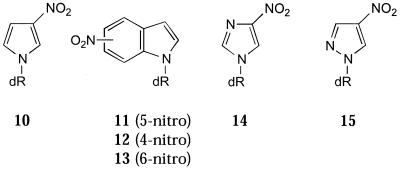
The first universal base analogue to receive prominent attention was 3-nitropyrrole (10) (Fig. 3), described by Bergstrom and co-workers (18–20). The design of this analogue was aimed at maximising base stacking interactions; the presence of the nitro group enhances stacking by polarisation of the π-aromatic system of the pyrrole ring. Modelling indicated that 10 should fit opposite any of the natural bases in a duplex without distorting it. However, whilst 10 was shown to behave indiscriminately in base pairing with the four natural bases, there was a significant destabilisation of duplexes containing it. There is a decrease in Tm value, compared to unmodified duplexes (Tm 57°C), of 11–14°C in a 15mer (20) and even greater if multiple substitutions are made (21). This destabilisation raised questions about how well 10 stacks with the natural bases in a duplex. It was demonstrated that when 10 is incorporated into either strand of an oligopurine–oligopyrimidine duplex it was less indiscriminate. It was shown that A·10 and 10·T base pairs were only destabilising by 3–4°C, whilst in all other cases it destabilised the duplex in the range 7–9°C (22). These data suggest that 10 does not stack well in a duplex. The Tm values of a series of oligonucleotides in which each of the natural bases was successively substituted by 10 has been investigated. The Tm of a natural DNA target showed a U-shaped melting temperature curve as the 3-nitropyrrole residue was moved from each end towards the centre of the duplex (23; see also 24).
Figure 3.
Nitroazole universal base analogues.
A decamer structure containing 10 at the middle opposite adenine has been solved by NMR (25). Two structures (modified and unmodified) were solved and shown to be very similar. However, it was found that stacking of the pyrrole ring with its nearest neighbours was less than had been predicted (19) and that minor perturbations reduced stacking interactions due to the small size of the ring. The bulky nitro group protruded into the major groove, where it could rotate to relieve crowding. It was concluded that the pyrrole ring does not stack as well as expected and the design of future analogues should aim at replacing the nitro group by less bulky substituents.
A related analogue to 10 is 5-nitroindole (11) (21). This analogue has been shown to be generally less destabilising, with only a 2°C decrease in Tm value when incorporated towards the ends of a 17mer duplex and 5°C in the middle (21) (unmodified duplex Tm 72°C). 11 is non-discriminating towards the natural DNA bases, with a Tm range of 3°C, similar to that observed with 10. In addition to 11, the 4-nitroindole (12) and 6-nitroindole (13) derivatives have been studied in melting experiments (21). However, 12 and 13 were more destabilising than 11 (the ribo derivative of 6-nitroindole has also been previously prepared; 27). When contiguous substitutions of 11–13 are made into a duplex the Tm value remains constant, whilst the stacking enthalpy (ΔH) increases. In contrast, 10 does not give increased stacking enthalpy with contiguous substitutions and the Tm value decreases with each additional substitution. This indicates that 11 is much better at stacking within the duplex than 10. The crystal structure of 11 shows that the base residues stack in columns, with overlap between each aromatic base (17).
Whilst it has been shown that duplexes containing 10 have a significantly lower melting temperature than the corresponding natural DNA, Zhang et al. have shown that duplexes in which two 10 residues are opposite each other are more stable (12). They showed that on average the Tm value was raised by 11°C compared to 10 opposite natural DNA bases. This reveals the importance of solvophobic effects, which will be discussed later.
In a study of nitroazole derivatives in the modified Dickerson dodecamer d(CGCXAATTYGCG)2, where X is the nitroazole and Y is A, C, G or T, it was shown that nitroazole residues lower the Tm value compared to the natural sequence (26). Amongst the analogues studied the least discriminating was found to be 10, with a ΔΔG of 0.4 kcal/mol between the least and most stable duplexes. For duplexes containing 11 a ΔΔG of 0.8 kcal/mol was found. With ΔΔG differences as small as this, sequence context can be expected to alter the apparent order of base stability. In a plot of ΔH versus ΔS0 for all the natural and nitroazole- and 2′-deoxyinosine-containing sequences a straight line was obtained. This implies that the correlation between ΔS0 and ΔH is independent of the mode of association of the nucleobases. It is likely that the differences in ΔH and ΔS0 for nitroazole-containing oligonucleotides compared to natural DNA reflects loss of hydrogen bonding interactions with the base opposite the nitroazole (see below). Addition of a ring nitrogen, as a hydrogen bond acceptor, resulted in a preference for base pairing. Thus, nitroimidazole (14) shows a high specificity for pairing with G, whilst the related 4-nitropyrazole (15) shows a preference for pairing with A.
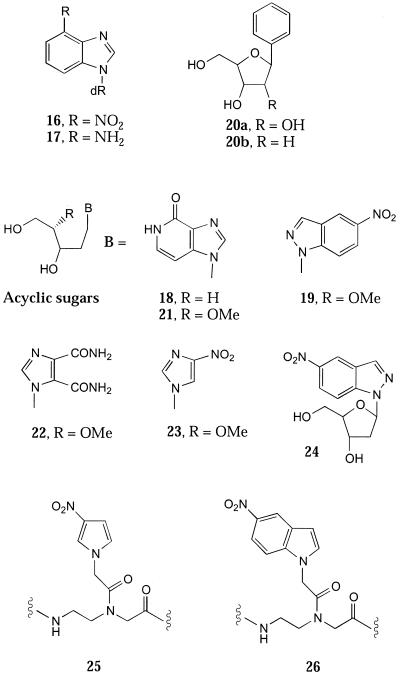
A large number of non-hydrogen bonding base analogues have been investigated, though mainly in hybridisation applications. These include 4-nitrobenzimidazole (16) (28), acyclic sugar analogues of hypoxanthine (18) and 5-nitroindazole (19) (Fig. 4) (29–31). 16 shows a slight preference for base pairing with thymidine, similar to 4-aminobenzimidazole (17) (32). Apart from this bias, 16 behaves indiscriminately towards the other natural bases, but is more destabilising than 11.
Figure 4.
Universal bases have primarily been used in DNA. Other applications include RNA, acrylic nucleotides and PNA.
One of the earliest analogues to be investigated as a universal base was phenyl C-ribonucleoside (20a) (14) [more recently it has been prepared as its 2′-deoxyribosyl nucleoside (20b) to investigate its effect in the hammerhead ribozyme (33)]. These earlier findings of Millican et al. (14) showed that 20a was quite destabilising (ΔTm –16°C) and that it was essentially the same as an abasic site. Thus, the presence of the phenyl ring offers little or no contribution to stacking interactions. Nevertheless, this analogue shows little discrimination towards pairing with each of the natural bases.
A number of acyclic sugar analogues have been prepared by Van Aerschot et al. (29,30) and investigated as potential universal base analogues. The analogues compared are those derived from hypoxanthine (18 and 21), imidazole 4,5-dicarboxamide (22), 3-nitroimidazole (23) and 19 (29,30). Of these, 18 and 21 were the least destabilising but 19 was the least discriminating towards the natural bases (ΔTm range 2°C). The least efficient universal base was 23. The imidazole nitrogen has a significant effect and 23 is most stable when opposite G (29). 22 is less destabilising and shows little discrimination, warranting further investigation as a universal base, particularly as it probably involves formal hydrogen bonding. A detailed comparison of 19 and 11 shows that they are almost indistinguishable (30,31). In fact, the 2′-deoxyribosyl derivative of 5-nitroindazole (24) shows little advantage over 11 in hybridisation terms and in primers for PCR and sequencing (34,35).
The bases 3-nitropyrrole and 5-nitroindole have also been incorporated into peptide nucleic acids (PNA) (36). In PNA–DNA duplexes containing 25 or 26 it was found that the Tm range was narrower than that found for corresponding DNA–DNA duplexes, although significantly higher than the corresponding DNA–DNA duplex containing an A·T base pair.
Primers
The principle application investigated for universal base analogues is their use in primers for PCR and sequencing (37). It has been reported that 10 can be incorporated not only as a string of up to nine substitutions close to the 3′-end of the primer but also at the 3′-end (19,20), although we were unable to reproduce these results with either 10 or 11 (38). Whilst it is reasonable to assume that a universal base may stabilise the primer/template/polymerase structure, there is much evidence that non-hydrogen bonding universal bases are not generally well copied by a polymerase. Various investigators have found primers to be ineffective when a universal base is contained within the first seven or eight bases from, and including, the 3′-end (38–41). Nevertheless, under appropriate conditions a polymerase will add a nucleotide opposite a template universal base, albeit inefficiently.
When amplifying a template 64 nt long containing 11 in the centre, full-length PCR product was obtained (17). When 10 or 11 are copied by Taq polymerase the nucleotide which is preferentially incorporated opposite them is dATP (17,42). In both cases the product yield is low and the incorporation of dATP is possibly due to the polymerase extendase (non-template +1 nt addition) activity (43). However, it has been reported that Taq polymerase incorporates dATP and dTTP (in a 3:1 ratio) opposite 10 (44).
If 11 is incorporated into a primer for PCR or sequencing, with substitutions made at codon third positions, then the effectiveness of the oligomer to prime DNA synthesis is poor when more than two such substitutions are made (45). However, up to four contiguous substitutions of 11 may be made in oligonucleotides, in the middle or 5′-end, and these will prime DNA synthesis in PCR and sequencing (34,38). Longer runs of 11 residues proved to be ineffective, presumably because 11 preferentially forms secondary structures such as hairpin loops (17,46).
As a result of the above observations, universal bases have been used to increase the effective size of short primers without increasing multiplicity. A number of analogues were tested to improve the effectiveness of octamers for cycle sequencing reactions (34). Of the analogues tested, 11 proved to be most effective. Normal octamers are at best poor primers in cycle sequencing, requiring low reaction temperatures. The use of up to four 11 residues at the 5′-end of the primer resulted in much improved performance. The use of other modified bases, such as 5-methylcytosine and diaminopurine (to replace cytosine and adenine), in addition to a tail of 11, resulted in 8mers, now effectively 12mers, that were effective at an extension temperature of 55°C (unpublished data).
Acyclic 19 and 21 have also been used in primers for PCR and sequencing (31). Primers containing up to three consecutive substitutions of either analogue (seven bases from the 3′-terminus) performed at least as well as the unmodified primer of the same length in sequencing reactions. In PCR amplifications they did not perform as well as 11, but full-length product could still be obtained. The least effective analogue for PCR was found to be 21.
During polymerase chain extension reactions it is common for the enzyme to add one or more additional nucleotides to the nascent chain and, in the case of transcription, products of even greater length can be obtained (47). Moran et al. have demonstrated that the use of hydrophobic residues at the 5′-end of the template causes chain termination with a significant decrease in extension beyond the analogue (48). This effect was not observed with an abasic site, showing that the hydrophobic base affects chain termination. Generally, the larger the residue the more effective was termination.
Probes
Oligonucleotide probes have been widely used to target DNA and RNA, for example rRNA. A requirement for such studies is the design of nested probes to target regions of the rRNA. A complete characterisation requires universal probes, which have been difficult to design. Probe design has used a multiplicity of oligomers or base analogues or mismatched probes. The fact that incorporation of 11 destabilises a duplex has been put to advantage. Zheng et al. (49) required probes for rRNA for a variety of species of microorganisms, but the target sequence had a dissociation temperature (Td) value higher than that desired for use in conjunction with other probes. They used probes containing 11 to ensure equal specificity of the probe for all target organisms (49–52). They found that a large spread in Td values decreased when 11 was included in the probe. They also observed a more uniform response from all the members of the complex they were targeting compared to a probe of natural DNA.
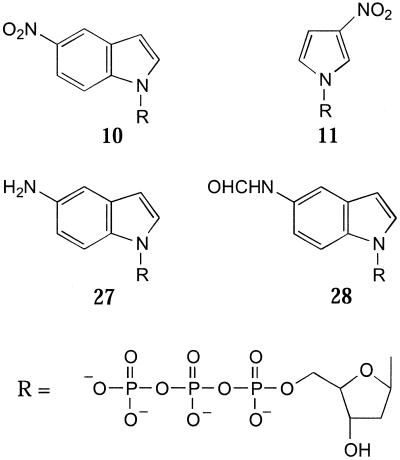
A further application for probing has been described by Smith et al. (53). The –40 M13 forward sequencing primer was 3′-tailed with the 5′-triphosphate derivative of 11 (see below) using terminal deoxynucleotidyl transferase and then hybridised to M13 blotted onto a nylon membrane. The 5-nitroindole tail was then detected using an antibody to the 5-nitroindole base. The limit of detection was not optimised, but detection of 5 fmol of tailed DNA was easily observed.
10 has been used to examine single nucleotide polymorphisms (SNPs) (23,54). A great problem with measuring mismatched oligonucleotide duplexes, particularly for longer oligonucleotides, is that there is a sequence effect that can dramatically alter the Tm value of mismatched oligomers. Many groups have examined this so that mismatched duplexes can be compared more clearly (see 55 and references therein). An alternative approach has been the use of universal bases close to the mismatch site. The incorporation of 10 three or four bases from the mismatch site has a marked effect on the Tm value of the resultant duplex (23; see also 56). An additional artificial mismatch three or four residues from the natural mismatch site destabilises the duplex, but this effect is sequence dependent. Replacement of the artificial mismatch by 10 gave an increase in ΔTm of 8°C (from 3°C). These oligomers were effectively used to probe for SNPs in PCR products (23).
Oligonucleotide chips have been developed for a number of applications: sequencing by hybridisation, mutant diagnostics, gene expression analysis, identification of microorganisms. The size of a library of oligonucleotides increases dramatically with increasing length of oligomer, e.g. a library of all 10mers would require 1 048 576 oligonucleotides. One group has attempted to reduce library size by using 6mers, which only requires 4096 oligonucleotides (57). However, hexamers have a number of shortcomings, namely hybrids have very low stability, difficulties in discriminating terminal mismatches and a wide variation in the stabilities of AT- and GC-rich duplexes. The addition of 11 to one or both ends of the hexamers compensates for these shortcomings, as does a mixture of all four nucleotides. Such additions increase the stabilities of the hexamer duplexes by increasing them to 7mers or 8mers (0.4–0.6 kcal/mol increase in ΔG0 compared to 0.3–0.4 kcal/mol for the four base mixture). They also destabilised terminal mismatches by converting them to internal ones (0.4–1.2 kcal/mol more stable than terminal mismatched oligomers) and equalised the stabilities of AT- and GC-rich duplexes. A 5mer duplex stability can also be significantly increased by addition of 11 at the 5′-terminus, although internal substitutions destabilised the duplex (58).
In a further report (59) the effects of stabilisation by 11 at both the 5′- and 3′-ends of an octanucleotide immobilised on a chip have been investigated. Addition of 11 at the 5′-end of the oligonucleotide enhanced duplex stability by approximately as much as an additional base pair. This stabilisation was not affected by the opposite nucleotide, but there was some effect of the adjacent base. At the 3′-end of the oligomer stabilisation was less marked. Again there was no effect from the opposite base. A theoretical analysis of the use of universal bases in sequencing by hybridisation has also been made, demonstrating the advantage of the use of such analogues (60,61).
Sequencing by MALDI-MS is limited by strand fragmentation and the extent of fragmentation is dependent on sequence. Thymidine-containing oligonucleotide are less susceptible to fragmentation, followed by A, then C/G. Jacutin et al. (62) used 10 as an adenine surrogate in mass spectroscopic analysis of oligonucleotides and found that 10-containing oligonucleotides were less susceptible to fragmentation.
Universal base analogues have also been used to examine protein–DNA interactions. The function of nucleotide excision repair (NER) factors is to protect the genome by removing modified natural bases induced by UV radiation or environmental carcinogens. NER enzymes operate on DNA lesions that may disturb hydrogen bonding between complementary strands and recognise improper base pairing conformations. Destabilisation of Watson–Crick base pairing provides a molecular signal directing the involvement of a NER factor that recognises defective base pairing conformations. 10 and 11 have been used in oligonucleotides to examine the mechanism by which xeroderma pigmentosum group A (XPA) protein discriminates between normal base pairs and non-hybridising DNA constituents (63). With three consecutive 10 or 11 residues in the centre of a 19mer, assembly of XPA–DNA complexes was stimulated more efficiently than by three mismatches. When a duplex containing three 11 residues opposite three 10 residues was examined XPA still retained its strong affinity for the non-hybridising site. This demonstrates that unpaired hydrogen bond acceptors and donors are not required for XPA protein binding to non-hybridising residues. This led the authors to conclude that the affinity of XPA binding is mediated by hydrophobic interactions with aromatic base components that are abnormally exposed to the helical surface of DNA.
11 has also been used to examine the binding of Escherichia coli RNA polymerase with the fork junction DNA containing the –10 region of the promoter (64). It was used to help determine the critical bases in the promoter region for binding by RNA polymerase and to determine the specific nature of the purine requirements at one particular conserved site in that region. Thus the use of 11 helped demonstrate that the adenine at –11 is essential for binding.
Ligation
It has been shown that neither 10 nor 11 are recognised by T4 polynucleotide ligase when present at either the 5′- or 3′-end of an oligonucleotide primer (31). However, Luo et al. showed that when it is near to the 3′-terminus, i.e. still within the active site of the enzyme, 10 causes enhanced fidelity by the thermostable Thermus thermophilus ligase (56). Their approach is based on a method for improving allele-specific PCR using primers with a deliberate mismatch adjacent to the 3′-end (65). Using primers with an A·C mismatch three bases from the 3′-end of a nick site they demonstrated a 4-fold improved fidelity (over a natural G·C base pair) of ligation using Tth DNA ligase. When the A·C mismatch was replaced by a 3-nitropyrrole·A base pair the fidelity of ligation was further improved (9-fold over the G·C primer). If 10 was incorporated at the second site from the nick no ligation was observed.
Triplexes
Triplex formation is generally restricted to oligopurine–oligopyrimidine sites of dsDNA. The introduction of one or two pyrimidine residues into the oligopurine strand leads to substantial destabilisation of triplex formation. The use of non-natural bases might allow for stabilisation of a triplex when the oligopurine strand contains one or two pyrimidine residues, or vice versa. Incorporation of 10, whilst somewhat destabilising in a duplex, was actually extremely destabilising when incorporated into the third strand of a triplex (22). This result was confirmed by another report that examined the effect of a number of non-natural analogues in the third strand of a triple helix (66). However, they found that when the triplex was formed in the presence of the benzopyridoindole triplex-specific intercalator BePI (67), 10 not only discriminated GC from CG and AT from TA base pairs but also the third strand containing 10 gave the most stable triplex. In contrast, the same group found for an alternative third strand sequence that oligonucleotides containing 10 have low stability (68), therefore there is some doubt as to the generality of its use.
10 will stabilise a triplex strand opposite thymidine in a polypurine strand (69). It showed a marked preference for thymidine over any of the other natural nucleotides, 4-fold over cytidine and 6-fold over purines. This effect, though, was found to be sequence specific in that substitutions close to the intercalator end of the triplex strand were deleterious to the binding constant, the stabilising effect only being observed at substitutions further away. This affinity for binding opposite thymidine was explained by molecular modelling, where it was demonstrated that the 3-nitropyrrole·A:T triad is isomorphous to A·A:T.
A series of 5-membered azole nucleosides were investigated in antiparallel triplex formation (70) in non-homopurine sequences. The azoles examined were the 2′-deoxynucleosides derived from pyrazole, imidazole, 1,2,4-triazole and 1,2,3,4-tetrazole. They were incorporated into oligonucleotides for binding to a duplex, with the azole opposite to each of the four natural DNA bases. Each was capable of associating with duplexes containing CG and TA base pairs in antiparallel triplexes with relatively high affinity. The binding in each case was considerably enhanced compared to that found with the natural nucleosides.
Universal nucleoside triphosphates
The practicality of a universal base triphosphate requires that it be randomly incorporated into the growing chain in the presence of the natural triphosphates. This competitive insertion requires that the universal triphosphate be incorporated into DNA with a rate similar to that of the natural triphosphates. Polymerase incorporation of a number of indole derivatives has been demonstrated (53,71), as well as that of 10. Exonuclease-free Klenow fragment inefficiently incorporated both 10 and 11 opposite all the natural bases. However, once incorporated both analogues behaved as chain terminators and no further extension occurred.
Reduction of the nitro group of the 5′-triphosphate of 11 gives the 5-aminoindole nucleotide (27), which in principle can form a single hydrogen bond (Fig. 5). 27 behaves differently from the nitro derivative in that it shows a marked preference for incorporation opposite thymidine, i.e. as A, although it can be inserted opposite the other natural bases to a lesser extent. Once incorporated, the growing chain will only be extended if it was incorporated as T or G. Formylation of 27 gives 28, which behaves more like the 5-nitroindole derivative. The triphosphates of 10 and 11 are inserted relatively efficiently opposite themselves as template bases (unpublished data). In a similar manner, several aromatic, hydrophobic nucleosides also preferentially incorporate opposite themselves, or similar structures, rather than opposite natural nucleotides (72).
Figure 5.
5′-Triphosphates of universal base analogues.
If the 5′-triphosphates of 10 or 11 are used in PCR reactions in the presence of natural triphosphates then with increasing concentration of the analogue triphosphate they progressively inhibit formation of PCR product (71). Given the ability of 11 to enhance stacking interactions, a possible explanation of the inhibition of PCR products is that the universal base is capable of entering the polymerase active site. Once within the active site it forms ‘stable’ interactions with the aromatic amino acid residues present and is therefore not efficiently ejected from the active site.
The 5′-triphosphate of 10 has been incorporated into DNA by the Klenow fragment of DNA polymerase I and avian myeloblastosis virus reverse transcriptase (62,73). Using a protocol that allowed differentiation between no incorporation, chain termination and chain extension it has been tested with a number of commercially available polymerases. It was found that to obtain polymerase incorporation high concentrations of the triphosphate were required and best results were obtained with shorter templates. The only other polymerase that would incorporate 10 was ΔTaq (73).
Whilst these analogues are poor substrates for both Klenow and Taq DNA polymerases, when used with terminal deoxynucleotidyl transferase both analogues are very good substrates (53). 10 gave a tail length similar to that of dCTP, whilst with 11 tailing was better than that obtained with dATP.
Structure/stacking/stabilisation
Duplex stability depends not only on Watson–Crick base pairs but also on geometrical and structural constraints, stacking interactions with adjacent bases and on solvation effects. For RNA, base stacking and hydrogen bonding contribute ∼1 kcal/mol of free energy to the stability of a base pair (74). To understand the stabilisation of duplexes by universal base analogues it is worth examining some of the interactions involved. Much of this literature makes use of analogues that behave universally in hybridisation terms. However, some of the analogues discussed here are not universal bases, but because of their enhanced stacking ability they have been included.
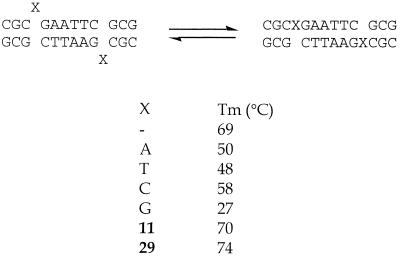
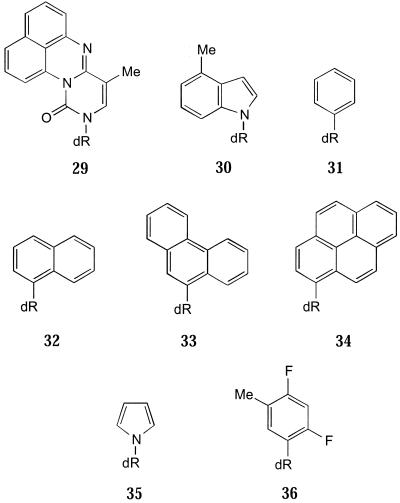
Probably the most important use of universal bases is in the stabilisation of DNA structures. A number of groups have studied the effect of single base bulges in the self-complementary Dickerson dodecamer. In the sequence d(CGCXGAATTCGCG), containing X as the additional base, the additional base does not have a partner to pair with in the classical sense. Thus the stability of such duplexes should reflect the capacity for individual analogues to stack between neighbouring Watson–Crick pairs. When the additional base is A it has been shown that in solution it stacks into the duplex (75), whilst in the solid state it loops out (76). When any of the natural bases are inserted into the Dickerson dodecamer the Tm value is decreased compared to the native dodecamer (17). However, when 11 is added the stability (Tm value) is greater than the parent duplex (Tm 69°C) (Fig. 6). It was suggested that this stabilisation occurs because 11 is sufficiently large to be able to intercalate into the opposite strand. This is further exemplified by incorporating an even larger hydrophobic base, 29 (24), where the melting temperature is raised by a further 4°C.
Figure 6.
Melting temperatures of modified Dickerson dodecamers with an extra internal nucleotide (X).
The native Dickerson dodecamer exists as a B-form DNA duplex. In low salt buffer (<10 mM NaCl) it has been shown to exist in a hairpin structure with a four base hairpin loop. When 10 replaced the two central nucleotides the resulting oligonucleotide was shown to exist as a hairpin structure in salt concentrations of 1 M NaCl (12).
When 11 is incorporated as a 5′-overhang of the dodecamer the effect is to further stabilise the duplex (76 compared with 69°C) (17). The nucleoside derivative of 4-methylindole (30) (77) likewise causes an increase in the melting temperature (72°C). In contrast, when 11 substitutes for the 3′-terminal residue in a flush-ended 20mer duplex the Tm value is considerably depressed (ΔTm 16°C) compared to the unmodified duplex (38). It may be that the additional destabilisation caused by incorporation at the ends of a duplex is due to increased hydrophobic effects without additional stabilisation from stacking interactions.
In a study of hydrophobic isosteres of the natural DNA bases Schweitzer and Kool calculated that hydrophobic nucleosides prefer to self-pair rather than with natural bases, with a selectivity of up to 4–5 kcal/mol (78). When the aromatic analogues benzene (31), naphthalene (32), phenanthrene (33) and pyrene (34) were incorporated as overhanging bases there was an increase in stability from enhanced stacking interactions with increasing size of the overhanging base (77,79). 34 gave an increase of 3.4 kcal/mol (compared to no overhang), whilst 31 gave 1.4 kcal/mol.
Kool and co-workers have studied the effects of aromatic stacking interactions in DNA in great detail (for details see 77–82). They studied the factors affecting aromatic stacking for a series of analogues, which they compared to the natural DNA, by dangling end experiments (79). They calculated various physical parameters (partition coefficient, log P, polarisability, dipole moment, surface area and stacking area) for each analogue and attempted to correlate stacking ability with one of these physical properties. The least polarisable and smallest was pyrrole (35), whilst the most polarisable and largest was 34 (Fig. 7). 34 behaves as a universal base (ΔTm 3°C), though it is somewhat destabilising (80). In the dangling end experiments each analogue was added to the 5′-end of a hexamer of alternating C-G. Of the analogues tested, 35 was the least stabilising (ΔTm 5°C), whilst the most stabilising were 11 (ΔTm 19°C) and 34 (ΔTm 23°C), adding 3.4 kcal/mol stabilisation to the parent duplex. There was no quantitative correlation between any individual physical property and stacking ability, indicating that more than one property is important. A plot of polarisability versus stacking energy correlated qualitatively, with the most (34) and least (35) polarisable bases showing the best and worst stacking potentials, respectively. Probably the best generalised correlation was between stacking surface area and stacking energy. This showed that the compound that had the smallest overlap (35) stacks the least strongly, whilst 34, with the largest area of overlap, stacks the strongest. The exceptions to this are difluorotoluene (36) and 11, which stack more strongly than would be expected from surface area alone. These results show that surface area is not the only factor affecting stacking affinity, but that increased hydrophobicity must also in some way enhance the stabilisation of oligo-nucleotides containing such analogues.
Figure 7.
Hydrophobic, aromatic non-hydrogen bonding bases used to examine base stacking interactions in duplex DNA.
As solvophobic effects are important (79), they examined the effect of base stacking in an organic solvent (77). In a water–ethanol solvent system the greater the effect of solvophobic interactions to molecular interactions the more a co-solvent will weaken the interaction. There is a linear relation of thermal stability with ethanol content. The largest effect found was for 32, demonstrating that it is significantly more sensitive to solvent effects than the natural bases. When a hydrophobic base is in the middle of a DNA strand both faces are removed from water by stacking between adjacent DNA base pairs. At the end of a DNA strand it can still remove one face from water by stacking.
Single-stranded oligo(dA) and oligo(dC) have been shown to be stacked at low temperature, and UV and CD melting experiments can show the effect of ‘unstacking’ (83). When single-stranded d(A21) was made with a single 10 residue in the centre the oligo demonstrated a non-cooperative hyperchromic shift in UV when heated (10–70°C), very similar to d(A21). However, an oligomer of alternating dA and 10 showed no such change in hyperchromicity (22). Thus in this respect 10 is very much like thymine in that it has a weak tendency to stack with its neighbouring bases.
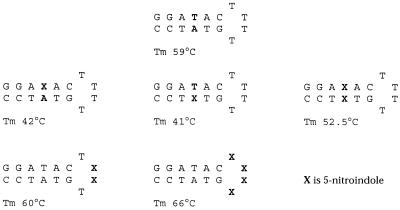
11 has been incorporated in both the stem and bulge of hairpin structures (46,84). When two 11 residues are placed opposite each other in the stem the structure still forms a stable hairpin, with a Tm value lower than that of the unmodified hairpin, but higher than a G:T mismatch. The most stable of the X4 hairpin loops is T4 (85). When this is modified by two or four 11 residues the hairpin has a higher Tm value than the unmodified one (+1.5 and +7°C, respectively; Fig. 8). Four 11 residues in the loop were more stable by up to 1 kcal/mol. It was suggested that the increased stabilisation of hairpins containing 11 residues in the bulge was due to enhanced hydrophobic interactions between two or more consecutive residues. A similar stabilisation of hairpin structures was observed with other hydrophobic analogues (86) in a hairpin loop.
Figure 8.
Effect of substitution by 5-nitroindole (X) on short hairpin structures measured by melting temperature.
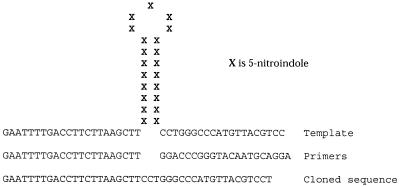
A 64mer oligonucleotide containing 21 consecutive 11 residues has also been shown to adopt a secondary structure, possibly a hairpin (17; Fig. 9). The oligomer exhibited a low hypochromic non-cooperative melting transition monitored at 330 nm by UV, a wavelength at which only 11 absorbs. In addition, melting of the oligomer could be measured by CD. Both UV and CD melting experiments gave a Tm value between 54 and 57°C. The proposed structure of the 64mer was further clarified by PCR, which gave a single product 43 bp long. It was not a primer dimer but corresponded to the polymerase copying across the base of the proposed hairpin structure, probably using its extendase activity to jump the gap, which is good evidence for a helical duplex stem.
Figure 9.
Proposed hairpin structure caused by self-stacking from consecutive 5-nitroindole (X) residues. PCR using the primers gave rise to the product shown corresponding to deletion of the entire 5-nitroindole region.
Other universal base analogues
The two universal base analogues that have received most attention are 10 and 11. However, there are a number of other non-hydrogen bonding base analogues that have been investigated. These include benzimidazole (35,87), 5-fluoroindole (35) and indole (88).
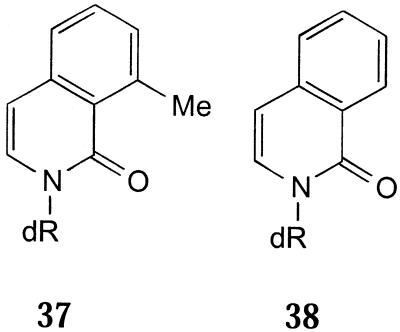
Another class of hydrophobic analogue that is reported to lead to more stable duplexes than 10 and 11 has recently been described (89). Isocarbostyril nucleoside derivatives form stable base pairs opposite each of the natural DNA bases, though the Tm values of duplexes containing them are lower than the corresponding AT duplex. MICS (37) (Fig. 10) directs the insertion of natural triphosphates by Klenow fragment with a 4-fold lower variation in efficiency and, at high dNTP concentration, full-length DNA was synthesised. The 7-propynyl derivative of 37 (90) was examined as its 5′-triphosphate (89). It was inserted more efficiently opposite dA and T, but overall was 40-fold down compared to the natural dNTPs. However, once incorporated, chain termination occurred. Removal of the methyl group of the latter compound gives an analogue that behaves universally in hybridisation experiments (91), though it is less stable than 37 and it forms stable base pairs with itself. Its 5′-triphosphate is incorporated more efficiently, though non-competitively, opposite each of the natural bases. One further analogue, ICS (38), also directs the incorporation of each of the natural triphosphates opposite it with Klenow fragment, but again less efficiently than the natural substrates (72). Whilst some of these compounds appear promising as universal base analogues, no one compound has been shown to fulfil all the criteria outlined earlier. Further work is required for this new class of analogue.
Figure 10.
Universal isocarbostyril nucleoside analogues.
Hydrogen bonding universal base analogues
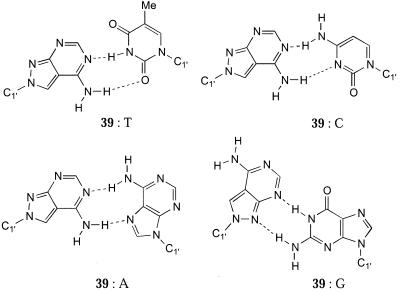
Seela and Debelak (92) recently reported a pyrrolopyrimidine nucleoside (39) in which the sugar is attached to the N8 position. 39 possesses an unexpected feature in that it behaves as a universal base (92). 39 therefore forms a new class of universal base analogues that form bidentate hydrogen bonds with each of the natural DNA bases. Oligonucleotides containing 39 have Tm values within a 2°C range when opposite each of the natural bases. In the absence of structural data the use of bidentate rather than tridentate hydrogen bonds is suggested by the fact that the Tm values of oligonucleotides containing 39 are in the stability range of an A·T base pair rather than C·G. The base pairing motifs probably follow the Watson–Crick or Hoogsteen mode except for the pairing with dG (Fig. 11). Whilst the new base pairs have a different shape compared to those in natural DNA, the new pairs fit well into the DNA duplex. This is clearly a preliminary paper and it will be of considerable interest to know whether the 5′-triphosphate of this analogue behaves as a universal substrate for polymerases.
Figure 11.
Proposed hydrogen bonding motifs for the universal N8-linked analogue 38.
CONCLUSION
As demonstrated, a large number of nucleoside analogues have been examined as universal bases and all fulfil one of the main criteria, in that they do not discriminate between the natural DNA/RNA bases. The first generation universal bases, largely exemplified by 10 and 11, have many demonstrable applications where the only criterion is hybridisation. As substrates for enzymes they have serious limitations. A second class of universal bases is the isocarbostyril nucleosides. These are still hydrophobic analogues and so far little work with them has been reported. Nevertheless, these compounds appear to be improved enzyme substrates, still retaining their hybridisation properties. The final category so far has only one example, which is the N8-pyrrolopyrimidine analogue of adenine. This compound, although still not proven to fulfil other criteria for universal bases, is unique in that it forms bidentate hydrogen bonds with each of the natural bases. If it fulfils the other requirements, then it should find a host of applications in molecular biology.
Acknowledgments
ACKNOWLEDGEMENTS
I am indebted to Dr Dan Brown for his advice and enthusiasm throughout our study of universal base analogues and their applications. I particularly wish to thank him for his critical reading and suggestions for this manuscript. I would likewise thank Drs Dave Earnshaw and Steve Holmes for their comments.
References
- 1.Ohtsuka E., Matsuki,S., Ikehara,M., Takahashi,Y. and Matsubara,K. (1985) An alternative approach to deoxynucleotides as hybridisation probes by insertion of deoxyinosine at ambiguous codon positions. J. Biol. Chem., 260, 2605–2608. [PubMed] [Google Scholar]
- 2.Takahashi Y., Kato,K., Hayashizaki,Y., Wakabayashi,T., Ohtsuka,E., Matsuki,S., Ikehara,M. and Matsubara,K. (1985) Molecular-cloning of the human cholecystokinin gene by use of a synthetic probe containing deoxyinosine. Proc. Natl Acad. Sci. USA, 82, 1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H. and Nichols,R. (1994) PCR amplification using deoxyinosine to replace entire codon and at ambiguous positions. Biotechniques, 16, 24–26. [PubMed] [Google Scholar]
- 4.Kamaya H., Sakaguchi,T., Murata,N., Fujimuro,M., Miura,H., Ishikawa,K., Shimizu,M., Inoue,H., Nishimura,S., Matsukage,A., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) In vitro replication study of modified bases in RAS sequences. Chem. Pharm. Bull., 40, 2792–2795. [DOI] [PubMed] [Google Scholar]
- 5.Martin F.H., Castro,M.M., Aboul-ela,F. and Tinoco,I. (1985) Base-pairing involving deoxyinosine—implications for probe design. Nucleic Acids Res., 13, 8927–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Aerschot A., Peeters,B. and Van Derhaeghe,H. (1987) Hybridisation probes with deoxyinosine, deoxyxanthosine or deoxynebularine at ambiguous codon positions. Nucl. Nucl., 6, 437–439. [Google Scholar]
- 7.Seela F. and Chen,Y. (1995) Oligonucleotides containing fluorescent 2′-deoxyisoinosine: solid phase synthesis and duplex stability. Nucleic Acids Res., 23, 2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seela F. and Mittelbach,C. (1999) 7-Deaza-2′-deoxyinosine: a stable nucleoside with the ambiguous base pairing properties of 2′-deoxyinosine. Nucl. Nucl., 18, 425–441. [Google Scholar]
- 9.Acedo M., De Clercq,E. and Eritja,R. (1995) Synthesis and biophysical and biological properties of oligonucleotides containing 2-aza-2′-deoxyinosine. J. Org. Chem., 60, 6262–6269. [Google Scholar]
- 10.Kawase Y., Iwai,S., Inoue,H., Miura,K. and Ohtsuka,E. (1986) Studies on nucleic-acid interactions 1. Stabilities of mini-duplexes (dG2A4XA4G2.dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC) containing deoxyinosine and other mismatched bases. Nucleic Acids Res., 14, 7727–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergstrom D.E., Zhang,P.M. and Johnson,W.T. (1996) Design and synthesis of heterocyclic carboxamides as natural nucleic-acid base mimics. Nucl. Nucl., 15, 59–68. [Google Scholar]
- 12.Zhang P., Johnson,W.T., Klewer,D., Paul,N., Hoops,G., Davisson,V.J. and Bergstrom,D.E. (1998) Exploratory studies on azole carboxamides as nucleobase analogs: thermal denaturation studies on oligodeoxyribonucleotide duplexes containing pyrrole-3-carboxamide. Nucleic Acids Res., 26, 2208–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pochet S. and Dugue,L. (1998) Imidazole-4-carboxamide and 1,2,4-triazole-2-carboxamide deoxynucleotides as simplified DNA building blocks with ambiguous pairing capacity. Nucl. Nucl., 17, 2003–2009. [Google Scholar]
- 14.Millican T.A., Mock,G.A., Chauncey,M.A., Patel,T.P., Eaton,M.A.W., Gunning,J., Cutbush,S.D., Neidle,S. and Mann,J. (1984) Synthesis and biophysical studies of short oligodeoxynucleotides with novel modifications—a possible approach to the problem of mixed base oligodeoxynucleotide synthesis. Nucleic Acids Res., 12, 7435–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francois P., Perilleux,D., Kempener,Y. and Sonveaux,E. (1990) Flexible aglycone residues in duplex DNA. Tetrahedron Lett., 31, 6347–6350. [Google Scholar]
- 16.Kool E.T. (1998) Replication of non-hydrogen bonded bases by DNA polymerases: a mechanism for steric matching. Biopolymers, 48, 3–17. [DOI] [PubMed] [Google Scholar]
- 17.Loakes D., Hill,F., Brown,D.M. and Salisbury,S.A. (1997) Stability and structure of DNA oligonucleotides containing non-specific base analogues. J. Mol. Biol., 270, 426–435. [DOI] [PubMed] [Google Scholar]
- 18.Bergstrom D.E., Zhang,P.M., Toma,P.H., Andrews,P.C. and Nichols,R. (1993) Synthesis, structure and biochemical applications of a universal nucleic-acid spacer—1-(2′-deoxy-β-d-ribofuranosyl)-3-nitropyrrole. Abstr. Pap. Am. Chem. Soc., 206 (2), 308–ORGN. [Google Scholar]
- 19.Nichols R., Andrews,P.C., Zhang,P. and Bergstrom,D.E. (1994) Synthesis, structure and biochemical applications of a nucleic-acid spacer—1-(2′-deoxy-β-d-ribofuranosyl)-3-nitropyrrole. Nature, 369, 492–493.8202140 [Google Scholar]
- 20.Bergstrom D.E., Zhang,P., Toma,P.H., Andrews,P.C. and Nichols,R. (1995) Synthesis, structure and deoxyribonucleic acid sequencing with a universal base: 1-(2′-deoxy-β-d-ribofuranosyl)-3-nitropyrrole. J. Am. Chem. Soc., 117, 1201–1209. [Google Scholar]
- 21.Loakes D. and Brown,D.M. (1994) 5-Nitroindole as an universal base analogue. Nucleic Acids Res., 22, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amosova O., George,J. and Fresco,J.R. (1997) Effect of the 1-(2′-deoxy-β-d-ribofuranosyl)-3-nitropyrrole residue on the stability of DNA duplexes and triplexes. Nucleic Acids Res., 25, 1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z., Liu,Q. and Smith,L.M. (1997) Enhanced discrimination of single nucleotide polymorphisms by artificial mismatch hybridisation. Nat. Biotechnol., 15, 331–335. [DOI] [PubMed] [Google Scholar]
- 24.Bischofberger N. and Matteucci,M.D. (1989) Synthesis of novel polycyclic nucleoside analogues, incorporation into oligodeoxynucleotides and interaction with complementary sequences. J. Am. Chem. Soc., 111, 3041–3046. [Google Scholar]
- 25.Klewer D.A., Hoskins,A., Zhang,P., Davisson,V.J., Bergstrom,D.E. and Li Wang,A.C. (2000) NMR structure of a DNA duplex containing nucleoside analog 1-(2′-deoxy-β-d-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control. Nucleic Acids Res., 28, 4514–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergstrom D.E., Zhang,P. and Johnson,W.T. (1997) Comparison of the base pairing properties of a series of nitroazole nucleobase analogues in the oligodeoxyribonucleotide sequence 5′-d(CGCXAATTYGCG)-3′. Nucleic Acids Res., 25, 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taktakishvili M.O., Tsitskishvili,T.K., Kikoladze,V.S., Samsonia,S.A. and Preobrazhenskaya,M.N. (1990) Synthesis of 1-β-d-ribopyranosyl-6-nitroindole and ribofuranosyl-6-nitroindole and indoline for phosphotriester oligonucleotide synthesis. Khimiya Geterotsiklicheskikh Soedinenii, 11, 1500–1506. [Google Scholar]
- 28.Seela F., Bourgeois,W., Rosemeyer,H. and Wenzel,T. (1996) Synthesis of 4-substituted 1H benzimidazole 2′-deoxynucleosides and utility of the 4-nitro compound as universal base. Helv. Chim. Acta, 79, 488–498. [Google Scholar]
- 29.Van Aerschot A., Hendrix,C., Schepers,G., Pillet,N. and Herdewijn,P. (1995) In search of acyclic analogues as universal nucleosides in degenerate probes. Nucl. Nucl., 14, 1053–1056. [Google Scholar]
- 30.Van Aerschot A., Rozenski,J., Loakes,D., Pillet,N., Schepers,G. and Herdewijn,P. (1995) An acyclic 5-nitroindazole nucleoside analogue as ambiguous nucleoside. Nucleic Acids Res., 23, 4363–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loakes D., Van Aerschot,A., Brown,D.M. and Hill,F. (1996) Enzymatic recognition of acyclic universal base analogues in oligonucleotides. Nucl. Nucl., 15, 1891–1904. [Google Scholar]
- 32.Seela F. and Wenzel,T. (1995) Oligodeoxyribonucleotides containing 4-aminobenzimidazole in place of adenine: solid-phase synthesis and base pairing. Helv. Chim. Acta, 78, 833–846. [Google Scholar]
- 33.Matulic-Adamic J., Beigelman,L., Portmann,S., Egli,M. and Usman,N. (1996) Synthesis and structure of 1-deoxy-1-phenyl-β-d-ribofuranose and its incorporation into oligonucleotides. J. Org. Chem., 61, 3909–3911. [DOI] [PubMed] [Google Scholar]
- 34.Ball S., Reeve,M.A., Robinson,P.S., Hill,F., Brown,D.M. and Loakes,D. (1998) The use of tailed octamer primers for cycle sequencing. Nucleic Acids Res., 26, 5225–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loakes D., Hill,F., Brown,D.M., Ball,S., Reeve,M.A. and Robinson,P.S. (1999) 5′-Tailed octanucleotide primers for cycle sequencing. Nucl. Nucl., 18, 2685–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Challa H., Styers,M.L. and Woski,S.A. (1999) Nitroazole universal bases in peptide nucleic acids. Org. Lett., 1, 1639–1641. [DOI] [PubMed] [Google Scholar]
- 37.Verma S. and Eckstein,F. (1998) Modified oligonucleotides: synthesis and strategy for users. Annu. Rev. Biochem., 67, 99–134. [DOI] [PubMed] [Google Scholar]
- 38.Loakes D., Brown,D.M., Linde,S. and Hill,F. (1995) 3-Nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res., 23, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith T.H., Latour,J., Leo,C., Muthini,S., Siebert,P. and Nelson,P.S. (1995) Amplification of human transferrin receptor gene using primers containing universal base nucleoside residues at ambiguous positions in degenerate amino-acid codons. Clin. Chem., 41, 10.7813054 [Google Scholar]
- 40.Rohrwild M., Alpan,R.S., Liang,P. and Pardee,A.B. (1995) Inosine containing primers for mRNA differential display. Trends Genet., 11, 300. [DOI] [PubMed] [Google Scholar]
- 41.Yang M., Hayashi,K., Hayashi,M., Fujii,J.T. and Kurkinen,M. (1996) Cloning and developmental expression of a membrane-type matrix metaloproteinase from chicken. J. Biol. Chem., 271, 25548–25554. [DOI] [PubMed] [Google Scholar]
- 42.Day J.P., Bergstrom,D.E., Hammer,R.P. and Barany,F. (1999) Nucleotide analogs facilitate base conversion with 3′ mismatch primers. Nucleic Acids Res., 27, 1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark J.M. (1988) Novel non-templates nucleotide addition-reactions catalysed by prokaryotic and eukaryotic DNA-polymerases. Nucleic Acids Res., 16, 9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoops G.C., Zhang,P., Johnson,W.T., Paul,N., Bergstrom,D.E. and Davisson,V.J. (1997) Template directed incorporation of nucleotide mixtures using azole-nucleobase analogs. Nucleic Acids Res., 25, 4866–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loakes D., Hill,F., Linde,S. and Brown,D.M. (1995) Nitroindoles as universal bases. Nucl. Nucl., 14, 1001–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallone P.M. and Benight,A.S. (1997) Melting behaviour of DNA hairpins containing the universal base 5-nitroindole. Biophys. J., 72, TH426 A421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cazenave C. and Uhlenbeck,O. (1994) RNA template-directed RNA synthesis by T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 91, 6972–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran S., Ren,R.X.F., Sheils,C.J. and Kool,E.T. (1996) Non-hydrogen bonding terminator nucleosides increase the 3′-end homogeneity of enzymatic RNA and DNA synthesis. Nucleic Acids Res., 24, 2044–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng D. and Raskin,L. (2000) Quantification of Methanosaeta species in anaerobic bioreactors using genus- and species-specific hybridisation probes. Microb. Ecol., 39, 246–262. [DOI] [PubMed] [Google Scholar]
- 50.Oerther D.B. and Raskin,L. (1996) In Abstracts of the 96th General Meeting of the American Society for Microbiology. American Society for Microbiology,Washington, DC, Vol. N-97, p. 339.
- 51.De Los Reyes F., Ritter,W. and Raskin,L. (1997) Group-specific small-subunit rRNA hybridisation probes to characterise filamentous foaming in activated sludge systems. Appl. Environ. Microbiol., 63, 1107–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen K.H., Ahring,B.K. and Raskin,L. (1999) Quantification of syntrophic fatty acid-β-oxidising bacteria in a mesophilic biogas reactor by oligonucleotide probe hybridisation. Appl. Environ. Microbiol., 65, 4767–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith C.L., Simmonds,A.C., Hamilton,A.L., Martin,D.L., Lashford,A.G., Loakes,D., Hill,F. and Brown,D.M. (1998) Use of 5-nitroindole-2′-deoxyribose-5′-triphosphate for labelling and detection of oligonucleotides. Nucl. Nucl., 17, 555–564. [DOI] [PubMed] [Google Scholar]
- 54.Baumbach L. (1997) Making the perfect mismatch. Nat. Biotechnol., 15, 318. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen H.-K., Auffray,P., Asseline,U., Dupret,D. and Thuong,N.T. (1997) Modification of DNA duplexes to smooth their thermal stability independently of their base content for DNA sequencing by hybridisation. Nucleic Acids Res., 25, 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo J.Y., Bergstrom,D.E. and Barany,F. (1996) Improving the fidelity of Thermus thermophilus DNA-ligase. Nucleic Acids Res., 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parinov S., Barsky,V., Yershov,G., Kirillov,E., Timofeev,E., Belgovskiy,A. and Mirzabekov,A. (1996) DNA-sequencing by hybridisation to microchip octanucleotide and decanucleotide extended by stacked pentanucleotides. Nucleic Acids Res., 24, 2998–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunitsyn A., Kochetkova,S., Kolganova,N., Tishchenko,E., Gottikh,B. and Florentiev,V. (1997) Stabilisation effect of 5-nitroindole (universal base) on DNA duplexes immobilised on gel matrix. J. Biomol. Struct. Dyn., 15, 597–603. [DOI] [PubMed] [Google Scholar]
- 60.Frieze A.M., Preparata,F.P. and Upfal,E. (1999) Optimal reconstruction of a sequence from its probes. J. Comput. Biol., 6, 361–368. [DOI] [PubMed] [Google Scholar]
- 61.Preparata F.P. and Upfal,E. (2000) Sequencing by hybridisation at the information-theory bound: an optimal algorithm. J. Comput. Biol., 7, 621–630. [DOI] [PubMed] [Google Scholar]
- 62.Jacutin S., Zhang,A.J., Russell,D.H., Gibbs,R.A. and Burgess,K. (1997) Test of the potential of a dATP surrogate for sequencing via MALDI-MS. Nucleic Acids Res., 25, 5072–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buschta-Hedayat N., Buterin,T., Hess,M.T., Missura,M. and Naegeli,H. (1999) Recognition of nonhybridising base pairs during nucleotide excision repair of DNA. Proc. Natl Acad. Sci. USA, 96, 6090–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matlock D.L. and Heyduk,T. (2000) Sequence determinants for the recognition of the fork junction DNA containing the –10 region of promoter DNA by E. coli RNA polymerase. Biochemistry, 39, 12274–12283. [DOI] [PubMed] [Google Scholar]
- 65.Rust S., Funke,H. and Assmann,G. (1993) Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res., 21, 3623–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kukreti S., Sun,J.-S., Loakes,D., Brown,D.M., Nguyen,C.-H., Bisagni,E., Garestier,T. and Helene,C. (1998) Triple helices formed at oligopyrimidine·oligopurine sequences with base pair inversions: effect of a triplex-specific ligand on stability and selectivity. Nucleic Acids Res., 26, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilch D.S., Martin,M.T., Nguyen,C.H., Sun,J.S., Bisagni,E., Garestier,T. and Helene,C. (1993) Self-association and DNA-binding properties of 2 triple helix-specific ligands—comparison of a benzo[e]pyridoindole and a benzo[g]pyridoindole. J. Am. Chem. Soc., 115, 9942–9951. [Google Scholar]
- 68.Kukreti S., Sun,J.-S., Garestier,T. and Helene,C. (1997) Extension of the range of DNA sequences available for triple helix formation: stabilisation of mismatched triplexes by acridine-containing oligonucleotides. Nucleic Acids Res., 25, 4264–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orson F.M., Klysik,J., Bergstrom,D.E., Ward,B., Glass,G.A., Hua,P. and Kinsey,B.M. (1999) Triple helix formation: binding avidity of acridine-conjugated AG motif third strands containing natural, modified and surrogate bases opposed to pyrimidine interruptions in a polypurine target. Nucleic Acids Res., 27, 810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Durland R.H., Rao,T.S., Bodepudi,V., Seth,D.M., Jayaraman,K. and Revanker,G.R. (1995) Azole substituted oligonucleotides promote antiparallel triplex formation at non-homopurine duplex targets. Nucleic Acids Res., 23, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith C.L., Simmonds,A.C., Felix,I.R., Hamilton,A.L., Kumar,S., Nampalli,S., Loakes,D., Hill,F. and Brown,D.M. (1998) DNA polymerase incorporation of universal base triphosphates. Nucl. Nucl., 17, 541–554. [Google Scholar]
- 72.Ogawa A.K., Wu,Y., McMinn,D.L., Liu,J., Schultz,P.G. and Romesberg,F.E. (2000) Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc., 122, 3274–3287. [Google Scholar]
- 73.Martinez C.I., Jacutin,S. and Burgess,K. (1998) Incorporation of unnatural nucleotides by DNA polymerases. Abstr. Pap. Am. Chem. Soc., 215, ORGN, 295. [Google Scholar]
- 74.Petersheim M. and Turner,D.H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp and ACCGGUp. Biochemistry, 22, 256–263. [DOI] [PubMed] [Google Scholar]
- 75.Patel D.J., Kozlowski,S.A., Marky,L.A., Rice,J.A., Broka,C., Itakura,I. and Breslauer,K.J. (1982) Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry, 21, 445–451. [DOI] [PubMed] [Google Scholar]
- 76.Joshua-Tor L., Rabinovich,D., Hope,H., Frolow,F., Appella,E. and Sussmann,J.L. (1988) The three-dimensional structure of a DNA duplex containing looped-out bases. Nature, 334, 82–84. [DOI] [PubMed] [Google Scholar]
- 77.Guckian K.M., Schweitzer,B.A., Ren,R.X.-F., Sheils,C.J., Paris,P.L., Tahmassebi,D.C. and Kool,E.T. (1996) Experimental measurement of aromatic stacking affinities in the context of duplex DNA. J. Am. Chem. Soc., 118, 8182–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schweitzer B. and Kool,E.T. (1995) Hydrophobic, non-hydrogen-bonding bases and base-pairs in DNA. J. Am. Chem. Soc., 117, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guckian K.M., Schweitzer,B.A., Ren,R.X.-F., Sheils,C.J., Tahmassebi,D.C. and Kool,E.T. (2000) Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc., 122, 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matray T.J. and Kool,E.T. (1998) Selective and stable DNA base pairing without hydrogen bonding. J. Am. Chem. Soc., 120, 6191–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schweitzer B.A. and Kool,E.T. (1994) Aromatic nonpolar nucleosides as hydrophobic isosteres of pyrimidine and purine nucleosides. J. Org. Chem., 59, 7238–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kool E.T., Morales,J.C. and Guckian,K.M. (2000) Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed. Engl., 39, 990–1009. [DOI] [PubMed] [Google Scholar]
- 83.Brahms J., Michelson,A.M. and Van Holde,K.E. (1966) Adenylate oligomers in single- and double-stranded conformation. J. Mol. Biol., 15, 467–488. [DOI] [PubMed] [Google Scholar]
- 84.Vallone P.M. and Benight,A.S. (1999) Melting studies of short DNA hairpins containing the universal base 5-nitroindole. Nucleic Acids Res., 27, 3589–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vallone P.M., Paner,T.M., Hilario,J., Lane,M.J., Faldasz,B.D. and Benight,A.S. (1999) Melting studies of short DNA hairpins: influence of loop sequence and adjoining base pair identity on hairpin thermodynamic stability. Biopolymers, 50, 425–442. [DOI] [PubMed] [Google Scholar]
- 86.Ren R.X.F., Schweitzer,B.A., Sheils,C.J. and Kool,E.T. (1996) Formation of stable DNA loops by incorporation of nonpolar, non-hydrogen-bonding nucleoside isosteres. Angew. Chem. Int. Ed. Engl., 35, 743–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Papageorgiou C. and Tamm,C. (1987) Nucleosides and nucleotides. 25. Synthesis of a protected 1-(2′-deoxy-β-d-ribofuranosyl)-1H-benzimidazole 3′-phosphate. Helv. Chim. Acta, 70, 138–141. [Google Scholar]
- 88.Girgis N.S., Cottam,H.B. and Robins,R.K. (1988) Synthesis of 2′-deoxyribofuranosyl indole nucleosides related to the antibiotics SF-2140 and Neosidomycin. J. Heterocycle Chem., 25, 361–366. [Google Scholar]
- 89.Berger M., Wu,Y., Ogawa,A.K., McMinn,D.L., Schultz,P.G. and Romesberg,F.E. (2000) Universal bases for hybridisation, replication and chain termination. Nucleic Acids Res., 28, 2911–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berger M., Ogawa,A.K., McMinn,D.L., Wu,Y., Schultz,P.G. and Romesberg,F.E. (2000) Stable and selective hybridisation of oligonucleotides with unnatural hydrophobic bases. Angew. Chem. Int. Ed. Engl., 39, 2940–2942. [DOI] [PubMed] [Google Scholar]
- 91.McMinn D.L., Ogawa,A.K., Wu,Y., Liu,J., Schultz,P.G. and Romesberg,F.E. (1999) Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc., 121, 11585–11586. [Google Scholar]
- 92.Seela F. and Debelak,H. (2000) The N8-(2′-deoxyribofuranoside) of 8-aza-7-deazaadenine: a universal nucleoside forming specific hydrogen bonds with the four canonical constituents. Nucleic Acids Res., 28, 3224–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]