ABSTRACT
Two-component systems (TCSs), the predominant signal transduction pathways employed by bacteria, play important roles in physiological metabolism in Streptomyces. Here, a novel TCS, GluR-GluK (encoded by SCO5778-SCO5779), which is located divergently from the gluABCD operon encoding a glutamate uptake system, was identified as being involved in glutamate sensing and uptake as well as antibiotic biosynthesis in Streptomyces coelicolor. Under the condition of minimal medium (MM) supplemented with different concentrations of glutamate, deletion of the gluR-gluK operon (gluR-K) resulted in enhanced actinorhodin (ACT) but reduced undecylprodigiosin (RED) and yellow type I polyketide (yCPK) production, suggesting that GluR-GluK plays a differential role in antibiotic biosynthesis. Furthermore, we found that the response regulator GluR directly promotes the expression of gluABCD under the culture condition of MM with a high concentration of glutamate (75 mM). Using the biolayer interferometry assay, we demonstrated that glutamate acts as the direct signal of the histidine kinase GluK. It was therefore suggested that upon sensing high concentrations of glutamate, GluR-GluK would be activated and thereby facilitate glutamate uptake by increasing gluABCD expression. Finally, we demonstrated that the role of GluR-GluK in antibiotic biosynthesis is independent of its function in glutamate uptake. Considering the wide distribution of the glutamate-sensing (GluR-GluK) and uptake (GluABCD) module in actinobacteria, it could be concluded that the GluR-GluK signal transduction pathway involved in secondary metabolism and glutamate uptake should be highly conserved in this bacterial phylum.
IMPORTANCE In this study, a novel two-component system (TCS), GluR-GluK, was identified to be involved in glutamate sensing and uptake as well as antibiotic biosynthesis in Streptomyces coelicolor. A possible GluR-GluK working model was proposed. Upon sensing high glutamate concentrations (such as 75 mM), activated GluR-GluK could regulate both glutamate uptake and antibiotic biosynthesis. However, under a culture condition of MM supplemented with low concentrations of glutamate (such as 10 mM), although GluR-GluK is activated, its activity is sufficient only for the regulation of antibiotic biosynthesis. To the best of our knowledge, this is the first report describing a TCS signal transduction pathway for glutamate sensing and uptake in actinobacteria.
KEYWORDS: glutamate, Streptomyces coelicolor, two-component system, secondary metabolism
INTRODUCTION
Two-component systems (TCSs), typically consisting of a transmembrane histidine kinase (HK) and a cytoplasmic response regulator (RR), are the major signaling pathways employed by microbes to sense and respond to a wide range of environmental stresses or stimuli (1–3). Upon sensing specific signals, HK immediately undergoes autophosphorylation at a conserved His residue and the phosphoryl group is subsequently transferred to a conserved Asp residue of the cognate RR, which then interacts with promoter regions and thereby regulates the transcription of target genes (4, 5). In bacteria, the number of TCSs (especially HKs) encoded within an organism's genome is usually directly linked to the degree of the complexity of the environment in which an organism lives.
Streptomycetes are high-GC-content, Gram-positive soil bacteria that exhibit a complex developmental life cycle, including erection of aerial mycelia and spore formation (6, 7). More importantly, they have strong capabilities to produce numerous chemically distinct secondary metabolites widely used in medicine and agriculture, such as antibiotics, insecticides, and immunosuppressants (8, 9). The coordination of the complex morphological differentiation as well as the production of secondary metabolites requires the existence of different signal transduction systems. Thus, in comparison to other bacteria, streptomycetes harbor a large number of TCS genes contained within the genomes (10). For instance, Streptomyces coelicolor, a model actinomycete strain, which can produce at least four known secondary metabolites, including the blue actinorhodin (ACT), red undecylprodigiosin (RED), yellow type I polyketide (yCPK), and calcium-dependent antibiotic (CDA) (6, 11, 12), possesses up to 84 HK genes and 80 RRs. Among them, at least 67 form paired TCSs; the others include 13 orphan RRs and 17 unpaired HKs (13).
To date, a number of TCS genes have already been identified as closely associated with different physiological processes in S. coelicolor (12–15), including rapA1-rapA2 (16), cutR-cutS (17), absA1-absA2 (18), afsQ1-afsQ2 (19, 20), draR-draK (21), and ohkA (22) being mainly involved in secondary metabolism; ragK-ragG (23), ramR (24), bldM (25), and whiI (26) being closely related to development; phoP-phoR (27) and glnR (28) acting as key regulators for primary metabolism; and vanS-vanR being involved in vancomycin resistance (29). However, the signals directly sensed by TCSs largely remain to be determined. So far, only two direct signals of TCSs were identified experimentally in S. coelicolor, including vancomycin and undecylprodigiosin (RED). They are sensed by the HK VanS and an orphan RR, RedZ, which are involved in vancomycin resistance and RED biosynthesis, respectively (30, 31).
Here, through functional screening of a TCS gene deletion mutant library of S. coelicolor, a typical TCS operon, SCO5778-SCO5779 (called the gluR-gluK operon [gluR-K]), was identified to play a differential role in antibiotic biosynthesis on minimal medium (MM) supplemented with glutamate, positive for RED and yCPK biosynthesis but negative for ACT production. Intriguingly, gluR-K is transcribed divergently from the gluABCD gene cluster (SCO5774 to SCO5777), which is possibly responsible for glutamate uptake (32). Furthermore, we demonstrated that GluR-GluK could directly respond to glutamate and then activate the transcription of gluABCD, thus facilitating glutamate transport into the cell. Finally, bioinformatics analysis revealed that the gluR-K–gluABCD module involved in glutamate sensing and uptake is widely distributed in actinobacteria.
RESULTS
GluR-GluK involved in differential regulation of antibiotic biosynthesis by S. coelicolor under glutamate-supplemented MM culture condition.
Bioinformatics analysis revealed that GluR (encoded by SCO5778) is an OmpR family response regulator (RR) with a winged helix-turn-helix (wHTH) DNA-binding domain, and GluK (encoded by SCO5779) is a typical transmembrane sensor histidine kinase (HK) (see Fig. S1 in the supplemental material). An alignment of the deduced GluR amino acid sequence with its homologs from seven different Streptomyces species revealed that key residues of the phosphorylation pocket were highly conserved and that aspartate (Asp) at position 53 might be the phosphorylation site of GluR (Fig. S2). Similarly, based on the alignment of the deduced GluK amino acid sequence with its seven homologs, we found that histidine (His) at position 261 might be the autophosphorylation site of GluK. In addition, some key residues closely associated with ATP binding were predicted (Fig. S3).
Using the highly efficient clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 genome editing method (33), the ΔgluR-K mutant with in-frame deletion of the putative gluR-K operon was generated on the basis of the model strain S. coelicolor M145 and confirmed by PCR and DNA sequencing (Fig. S4). Then, three often-used media including MS, R2YE, and minimal medium (MM) supplemented with different concentrations of glutamate were used for phenotypic analysis. The results showed that, when grown on the complex medium MS or R2YE, the ΔgluR-K mutant exhibited no obvious phenotypic changes compared with the wild-type M145 (Fig. 1A). However, when grown on MM supplemented with a low concentration of glutamate (10 mM), the ΔgluR-K mutant showed clearly decreased red-pigmented RED but increased blue-pigmented ACT production (Fig. 1B). Meanwhile, when cultured on MM supplemented with a high concentration of glutamate (75 mM), the ΔgluR-K mutant exhibited obviously reduced yellow-pigmented yCPK but increased ACT production (Fig. 1B). In addition, we found that deletion of gluR-K resulted in impaired bacterial growth, especially on MM supplemented with 75 mM glutamate.
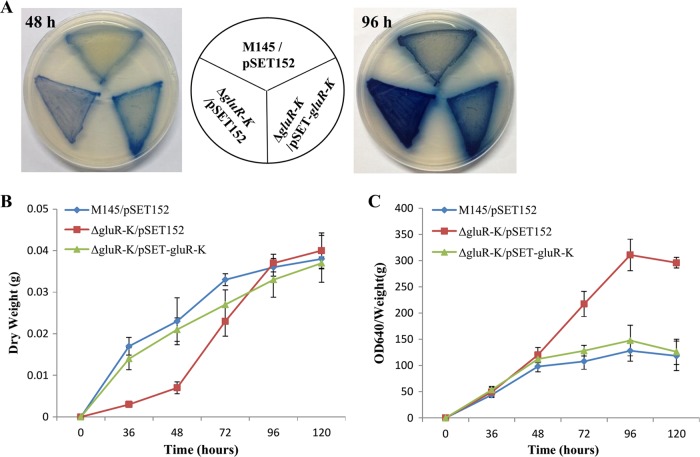
FIG 1.
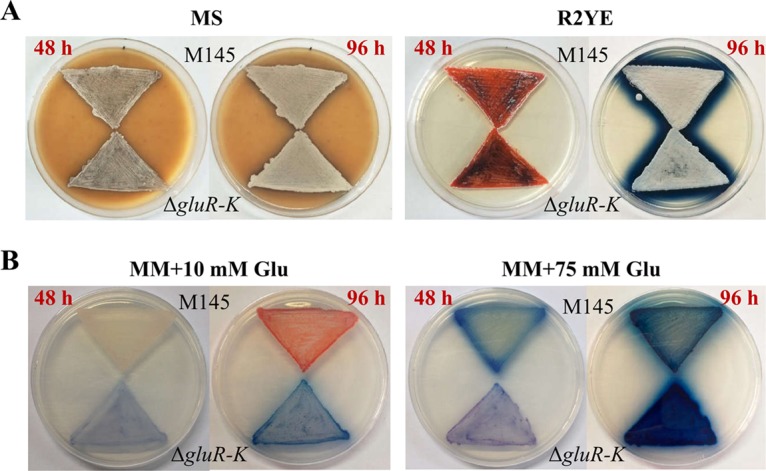
Phenotypes of S. coelicolor M145 and the ΔgluR-K mutant on the complex medium MS or R2YE (A) and MM supplemented with 10 or 75 mM glutamate (Glu) (B). The images for MS or R2YE were photographed from the front, and the images for MM were photographed from the back at 48 and 96 h.
To verify that the phenotypic alterations in the ΔgluR-K mutant were indeed due to the deletion of gluR-K, a genetic complementation assay was performed. The gluR-K operon with the putative promoter region (342 bp) was obtained by PCR and cloned in the integrative vector pSET152 to yield the complemented plasmid pSET-gluR-K. Then, the plasmid pSET-gluR-K was introduced into the ΔgluR-K mutant, generating the ΔgluR-K/pSET-gluR-K complemented strain. Subsequently, quantitative analysis of bacterial growth and ACT production was performed under the culture condition of MM with 75 mM glutamate. It was found that introduction of the complemented plasmid into the ΔgluR-K mutant could easily restore the phenotypic changes of the mutant to the wild-type strain (Fig. 2A). Additionally, the results obtained from quantitative analysis also indicated that deletion of gluR-K resulted in impaired growth only at the early growth stage (before 72 h) (Fig. 2B) and markedly increased ACT production at relatively late stage (after 48 h) (Fig. 2C). Collectively, we demonstrated that GluR-GluK plays a differential role in the regulation of antibiotic biosynthesis in S. coelicolor, positive for yCPK and RED but negative for ACT production.
FIG 2.
Effects of gluR-K deletion on bacterial growth and antibiotic biosynthesis of S. coelicolor. (A) Phenotypic analysis of the ΔgluR-K mutant grown on MM with 75 mM glutamate. Three S. coelicolor strains, including M145/pSET152, the ΔgluR-K mutant with pSET152 (ΔgluR-K/pSET152), and the complemented ΔgluR-K/pSET-gluR-K strain, were cultured for two different time intervals (48 and 96 h) before imaging. (B and C) Growth curves (B) and quantitative analysis of ACT production (C). Three S. coelicolor strains were grown on MM with 75 mM glutamate covered with sterile plastic cellophane. The samples were collected at five time points as indicated.
Genetic organization of the gluR-K–gluABCD gene cluster is widely distributed in actinobacteria.
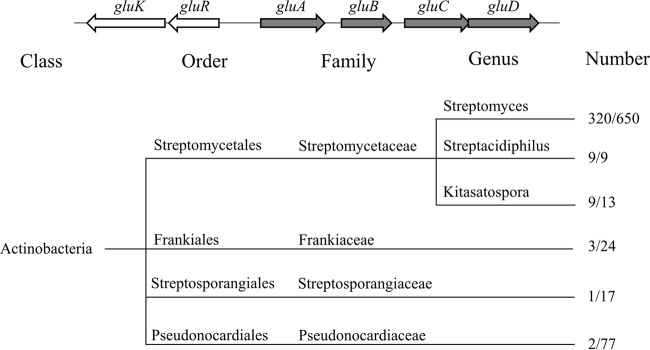
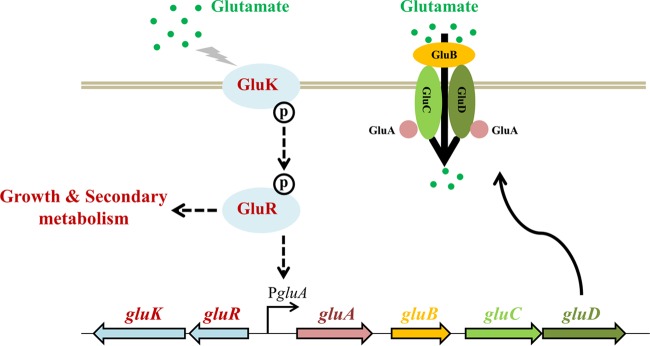
In analysis of the genomic location of gluR-K, we found that these genes are transcribed divergently from the gluABCD gene cluster, which encodes a glutamate uptake system, including an ATP-binding protein (GluA), a glutamate binding protein (GluB), and two glutamate permeases (GluC and GluD) (Fig. 3). GluABCD display 71%, 50%, 59%, and 48% amino acid sequence identities to their counterparts from Corynebacterium glutamicum, respectively. In C. glutamicum, GluABCD has been identified as being involved in glutamate uptake but not export (32). Bioinformatics analysis revealed that the genetic locus, including gluR-K and the gluABCD gene cluster, is widely distributed in actinobacteria with complete genome sequences available, including the orders Streptomycetales, Frankiales, Streptosporangiales, and Pseudonocardiales (8). Among these orders, the gluR-K–gluABCD gene cluster occurs most frequently in the order Streptomycetales, in which this cluster is found in three genera, including Streptomyces, Streptacidiphilus, and Kitasatospora, with rates of approximately 49% (320/650), 100% (9/9), and 69% (9/13), respectively (Fig. 3). Considering that genetic organization of the gluR-K–gluABCD gene cluster is highly conserved in actinobacteria, we speculated that GluR-GluK might directly regulate the expression of the gluABCD gene cluster.
FIG 3.
Distribution of the gluR-K and gluABCD gene cluster in different orders of actinobacteria. The right and left numbers in the ratio represent all the species with the available complete genome sequences and the number of species containing the gluR-K–gluABCD locus, respectively.
GluR-GluK directly activates the transcription of gluABCD.
First, using reverse transcription-PCR (RT-PCR) analysis, we confirmed that four genes, gluA, gluB, gluC, and gluD, were cotranscribed, forming a single operon (Fig. 4A). However, we could not exclude the possibility of the presence of promoters in the intergenic regions of the gluABCD gene cluster. Next, to examine whether GluR-GluK regulates the transcription of gluABCD directly, we performed electrophoretic mobility shift assays (EMSAs) with purified GluR protein. The intergenic region between gluABCD and gluR-K (gluA-Rint) was amplified from the genomic DNA of M145 and labeled by Cy5. We observed that GluR specifically bound to the gluA-Rint probe (Fig. 4B). Furthermore, to assess if phosphorylated GluR has a stronger affinity for gluA-Rint than nonphosphorylated GluR, 5 mM (final concentration) acetyl phosphate was added to phosphorylate GluR in the EMSA. As shown in Fig. S5, phosphorylation of GluR significantly enhanced its binding affinity for gluA-Rint, suggesting that the activity of GluR is dependent on phosphorylation.
FIG 4.
The response regulator GluR directly activates glutamate uptake through binding to the intergenic region between gluABCD and gluR-K. (A) Cotranscriptional analysis of the gluABCD gene cluster. RT-PCR was performed using cDNA as the template and RNA as the negative control. (B) EMSA of GluR binding to the intergenic region between gluABCD and gluR-K (gluA-Rint). The assay was performed using the indicated (micromolar) amounts of purified His6-GluR protein and Cy5 fluorescence-labeled gluA-Rint probe. EMSAs in the presence of 100-fold unlabeled, specific probe (S) or nonspecific (NS) competitor DNA (salmon sperm DNA) were performed as controls. (C) RT-qPCR analysis of the gluABCD gene cluster in the wild-type strain M145 and the ΔgluR-K mutant at 48 and 96 h grown on MM with 75 mM glutamate. Gene expression is represented as fold change after being normalized to M145. (D) Analysis of the gluA promoter activity in M145 and the ΔgluR-K mutant using xylE as a reporter. Promoter activities were assayed after the S. coelicolor strains were grown on MM with 75 mM glutamate for 48 and 96 h, respectively. The error bars indicate the standard deviations from three independent experiments. (E) An illustration of GluR-binding sequence in the intergenic region of gluABCD and gluR-K. The sequence protected by GluR is shown in red. The −10 and −35 elements of the gluA promoter are boxed, and the transcription start site (TSS) is indicated by a bent arrow, which was determined by 5′ RACE.
Subsequently, we compared the expression of gluABCD between the wild-type M145 and the ΔgluR-K mutant by quantitative real-time RT-PCR (RT-qPCR) analysis. Samples were harvested for RNA preparation from these two strains grown on 75 mM glutamate-supplemented MM for 48 and 96 h, respectively. As shown in Fig. 4C, the expression of all four genes was significantly decreased upon deletion of gluR-K at the tested points. Furthermore, using the xylE reporter assay, we found that the gluA promoter activities in M145 were much higher than those in the ΔgluR-K mutant (Fig. 4D). These results indicated that GluR directly activates the transcription of gluABCD in S. coelicolor. Given that gluR-K and gluABCD are located divergently, GluR may be autoregulated by binding to gluA-Rint. To test this possibility, we compared the gluR promoter (PgluR) activities in the M145 and ΔgluR-K strains by using the xylE reporter assay. As shown in Fig. S6, deletion of gluR-K has little effect on xylE activities under the control of PgluR, suggesting that GluR-GluK does not function as an autoregulator.
To determine the precise GluR-binding regions for regulation of gluABCD expression, four small overlapping DNA fragments extending from −352 to +3 with respect to the transcription start site (TSS) of gluA, which was identified by 5′ rapid amplification of cDNA ends (5′ RACE) (Fig. 4E), were generated and EMSAs were conducted. We found that GluR bound to fragments 3 and 4 (covering from −197 to −41 and −117 to +3, respectively) but not to two other fragments (Fig. S7), suggesting that the GluR-binding sequence is possibly located in the overlapping region of fragments 3 and 4, extending from −117 to −41. According to the position of the GluR-binding sequence, it could be suggested that binding of GluR to the upstream region of the gluA promoter (before the −35 element) might help to recruit RNA polymerase and then upregulate the transcription of the gluABCD gene cluster, thereby facilitating glutamate uptake under the glutamate-based MM culture condition.
GluK acts as a glutamate sensor.
As described above, when grown on three different tested media, the ΔgluR-K mutant displayed obvious phenotypic changes only on MM supplemented with 75 mM glutamate (Fig. 1). Here, we compared the transcription levels of gluR-K under the culture conditions of MS, R2YE, and MM with 75 mM glutamate using RT-qPCR to explore whether GluR-GluK was activated at the transcriptional level. We found that gluR-K exhibited similar transcript levels (showing similar threshold cycle [CT] values) under the conditions of these three different media (Fig. S8), indicating that the activity of GluR-GluK is not regulated at the transcriptional level.
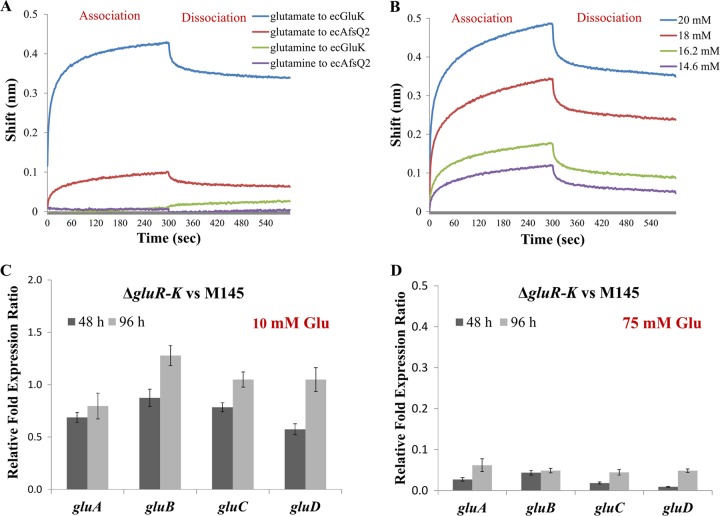
TCS HKs are usually transmembrane proteins, and the extracellular domains of HKs often act as the sensors for environmental signals (4, 34). Bioinformatics analysis using the TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/) showed that GluK harbors two transmembrane domains and that there exists an extracellular domain (ecGluK; total of 145 amino acids from 28 to 172) (Fig. S9A). This prompted us to suggest that the activity of GluR-GluK may be controlled at the posttranslational level and that glutamate is possibly the direct signal of GluK. To address this possibility, we performed biolayer interferometry (BLI) assays to analyze the direct interaction between ecGluK and glutamate. As a label-free technique, BLI was recently developed to assess molecular interactions (35) and involves three main steps including immobilization of His-tagged ecGluK on a nickel-nitrilotriacetic acid (Ni-NTA) sensor, placing the sensor in a solution of glutamate, and finally putting the sensor in a buffer for glutamate dissociation. ecGluK was expressed as a recombinant protein with His6-Trx, resulting in His6-Trx–ecGluK. To exclude possible interaction between His6-Trx tag and glutamate, the extracellular domain of HK AfsQ2 (ecAfsQ2, expressed as the recombinant protein His6-Trx–ecAfsQ2) (Fig. S9B) (19) and His6-Trx tag were also used as negative controls in this assay. In addition, given that the ΔgluR-K mutant showed no obvious phenotypic changes on MM supplemented with 75 mM glutamine (a structural analogue of glutamate) (Fig. S10), glutamine was used as a negative control in the BLI tests. The results showed that ecGluK specifically bound to glutamate with a clear response shift (0.44 nm) but not to glutamine. Although ecAfsQ2 also exhibited a slight response shift (0.1 nm) for glutamate, the background response of Trx tag also reached 0.07 nm (Fig. 5A and S11). These results suggested that ecAfsQ2 cannot bind to both glutamate and glutamine. Furthermore, we also observed a clear dose-dependent effect of the interaction between ecGluK and glutamate, further confirming that the interaction between ecGluK and glutamate was specific (Fig. 5B). Taken together, these findings revealed that GluK directly responds to the glutamate signal.
FIG 5.
The extracellular domain of GluK specifically bound to glutamate. (A) Biolayer interferometry (BLI) analysis of glutamate binding to the extracellular domain of GluK (ecGluK). Glutamine and the extracellular domain of HK AfsQ2 (ecAfsQ2) were used as two negative controls. The concentration of glutamate or glutamine was 20 mM. The concentration of ecGluK or ecAfsQ2 was 2.5 μg/ml. (B) BLI dose-response curves of ecGluK-glutamate interactions. The concentrations of glutamate were 20, 18, 16.2, and 14.6 mM, respectively. (C and D) RT-qPCR analysis of the expression of the gluABCD gene cluster in M145 and the ΔgluR-K mutant on MM supplemented with 10 mM glutamate (C) and 75 mM glutamate (D), respectively. Gene expression is represented as fold change after being normalized to M145.
To explore the physiological significance of glutamate as the signal of GluK, we compared the transcription of gluABCD between the ΔgluR-K mutant and M145 on MM supplemented with two different concentrations of glutamate, 10 mM (low) and 75 mM (high). We found that under the condition of MM with 75 mM glutamate, deletion of gluR-K resulted in significantly decreased expression of gluABCD. However, when grown on MM with 10 mM glutamate, the ΔgluR-K mutant exhibited no significant changes in the expression levels of gluABCD (Fig. 5C and D). It is worth noting that the ΔgluR-K mutant produced markedly reduced RED but enhanced ACT levels on MM with 10 mM glutamate (Fig. 1), suggesting that GluR-GluK was activated under the condition of MM with 10 mM glutamate. Therefore, it could be suggested that upon sensing a high glutamate concentration (75 mM), activated GluR-GluK could regulate both antibiotic biosynthesis and glutamate uptake. However, under low glutamate concentrations (such as 10 mM), although GluR-GluK is activated, its activity is sufficient only for the regulation of antibiotic biosynthesis, which is possibly ascribed to the requirement of higher GluR-GluK activity for activation of glutamate uptake than for regulation of antibiotic biosynthesis.
GluR-GluK indirectly regulates antibiotic biosynthesis in S. coelicolor.
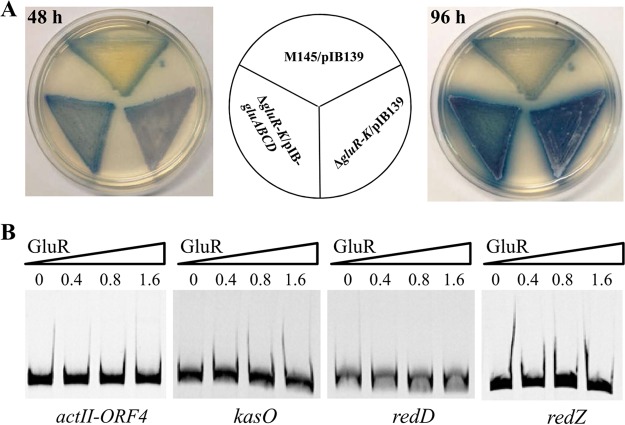
It was previously reported that glutamate addition enhanced yCPK biosynthesis under the MM culture condition (18). Since deletion of gluR-K resulted in markedly reduced gluABCD transcription, glutamate uptake would be impaired in the ΔgluR-K mutant, thereby leading to reduced yCPK biosynthesis. To test this possibility, we introduced the gluABCD gene cluster (cloned in the integrative vector pIB139) under the control of the strong, constitutive promoter ermE*p into the ΔgluR-K mutant, and then phenotypic changes in yCPK production were tested. As shown in Fig. 6A, introduction of the gluABCD gene cluster into the ΔgluR-K mutant has no obvious effect on yCPK. In addition, we found that ACT biosynthesis was not restored as well upon complementation of the gluABCD gene cluster. These results demonstrated that the role of GluR-GluK in antibiotic biosynthesis is independent of its effect on glutamate uptake in S. coelicolor.
FIG 6.
GluR-GluK indirectly regulated antibiotic biosynthesis in S. coelicolor. (A) Phenotypic analysis of overexpression of the gluABCD gene cluster in the ΔgluR-K mutant on MM with 75 mM glutamate. The gluABCD gene cluster was under the control of the strong constitutive promoter ermE*p. M145/pIB139 and ΔgluR-K/pIB139 strains were used as a positive and a negative control, respectively. The images were photographed from the back at 48 and 96 h. (B) EMSA of GluR binding to the respective promoter regions of cluster-situated regulatory genes of these three different antibiotics (ACT, yCPK, and RED). The assays were performed using the indicated amounts of purified His6 tag-GluR protein (micrograms) and Cy5 fluorescence-labeled promoter probes.
To assess whether GluR-GluK exerts its effects on ACT, RED, and yCPK production directly via the respective pathway-specific regulatory genes, EMSAs were performed using purified His6-GluR protein. Four probes containing the respective promoter regions of four pathway-specific regulatory genes, actII-ORF4, redZ/red, and kasO, which are responsible for the biosynthesis of ACT, RED, and yCPK, respectively (14), were amplified from the genomic DNA of M145 and labeled by Cy5. The results showed that GluR could not bind to the promoter regions of these four tested genes (Fig. 6B), suggesting that GluR-GluK regulates antibiotic biosynthesis in an indirect manner.
DISCUSSION
In this study, a novel TCS, GluR-GluK, was identified as being involved in glutamate sensing and uptake as well as antibiotic biosynthesis in S. coelicolor. A possible GluR-GluK working model for glutamate sensing and uptake was proposed (Fig. 7). Upon sensing high concentrations of glutamate (e.g., 75 mM), GluK was activated by autophosphorylation and the phosphoryl group was then transferred to GluR. Phosphorylated GluR controlled the expression of the gluABCD gene cluster via a direct interaction with the promoter region, thereby facilitating glutamate uptake from the environment. In addition, GluR-GluK played a differential role in antibiotic biosynthesis in S. coelicolor, positive for RED and yCPK biosynthesis but negative for ACT production. The role of GluR-GluK in antibiotic biosynthesis is independent of its function in glutamate uptake and is also indirectly mediated by their respective pathway-specific activators. A previous work reported that in Pseudomonas putida, TCS AauRS not only promotes uptake of glutamate by activating the expression of a glutamate ABC transporter but also affects the expression of glutamate dehydrogenase (36). If it is the same case for GluR-GluK, deletion of gluR-K might result in the altered expression of glutamate metabolism-related genes, which would possibly affect the precursor supply of antibiotic biosynthesis. This possibility might partially account for phenotypic changes in antibiotic production of the ΔgluR-K mutant. However, the detailed mechanism for GluR-GluK-mediated regulation of secondary metabolism remains to be investigated.
FIG 7.
Schematic illustration of GluR-GluK-mediated glutamate sensing and uptake in S. coelicolor. Upon receiving the glutamate signal, HK is activated by autophosphorylation and the phosphoryl group is then transferred to GluR. Activated GluR binds to the gluABCD promoter region, resulting in upregulation of gluABCD transcription and thus facilitating glutamate uptake. In addition, GluR-GluK also plays an important role in bacterial growth and secondary metabolism in S. coelicolor.
Previously, knowledge of glutamate uptake in the actinobacteria was mainly obtained from C. glutamicum, which is used for large-scale production of glutamate (32, 37). C. glutamicum possesses a primary glutamate uptake system (GluABCD, an ABC transporter) as well as a secondary transporter, GltS (38). GltS is the sodium-coupled l-glutamate uptake system and can promote reuptake of excreted glutamate (39). However, until now, little was known about the regulation of the two transporters. In addition, unlike the conserved genetic organization of gluR-K–gluABCD in the genus Streptomyces (Fig. 3), there are no GluR-GluK homologs in Corynebacterium. On the other hand, it should be noted that TCS-mediated regulation of amino acid uptake has also been reported in other bacteria, especially in the genus Pseudomonas. For example, in Pseudomonas putida, AauRS could mediate the uptake of several acidic amino acids, such as glutamate and aspartate (36). In Pseudomonas fluorescens, regulation of the histidine uptake process involves two TCSs, CbrAB and NtrAB (40). However, so far, the direct signals answered by these histidine kinases are still to be determined. Taken together, our work is the first report describing a TCS signal transduction pathway for glutamate sensing and uptake in actinobacteria.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. The primers are listed in Table S2 in the supplemental material. S. coelicolor M145 and its derivatives were grown on MS medium (soybean flour, mannitol, and agar, all at 20 g/liter) at 30°C for spore preparation, intergeneric conjugal transfer, and phenotypic observation (41). If the CRISPR-Cas9 genome editing tool was employed for gene deletion, M-Isp4 medium [soybean flour, 5 g/liter; mannitol, 5 g/liter; starch, 5 g/liter; tryptone, 2 g/liter; yeast extract, 1 g/liter; NaCl, 1 g/liter; (NH4)2SO4, 2 g/liter; K2HPO4, 1 g/liter; CaCO3, 2 g/liter; agar, 20 g/liter; trace element solution, 1 ml, pH 7.2] was used for conjugal transfer (33, 41). For phenotypic analysis, two other media, minimal medium (MM) supplemented with different concentrations of glutamate (10 or 75 mM) and R2YE, were also utilized (41). S. coelicolor cultures grown in liquid YEME medium (yeast extract, 3 g/liter; peptone, 5 g/liter; malt extract, 3 g/liter; glucose, 10 g/liter; sucrose, 340 g/liter; MgSO4·7H2O, 1.24 g/liter) on an orbital shaker (200 rpm) at 30°C were harvested for total DNA isolation (41).
Escherichia coli DH5α and BL21(DE3) were used for DNA cloning and protein overexpression, respectively. E. coli ET12567/pUZ8002 was used for conjugal transfer from E. coli to S. coelicolor. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium or on LB agar plates. Antibiotics (50 μg/ml of ampicillin, apramycin, and kanamycin) were added when necessary.
Construction of the S. coelicolor mutant.
The ΔgluR-K mutant was constructed by the CRISPR-Cas9 genome editing method (33). Briefly, the upstream and downstream regions (711 and 758 bp, respectively) of gluR-K were obtained by PCR using the primer pairs Del-gluR-K-up-fw/rev and Del-gluR-K-down-fw/rev, respectively. Using the plasmid pCB003 as the template, the single guide RNA (sgRNA) transcription cassette was obtained by PCR with the primers gluR-K-sgRNA-fw/rev. Then, three DNA fragments were assembled by an overlapping PCR with the primers gluR-K-sgRNA-fw and gluR-K-down-rev, followed by double digestion with HindIII and SpeI. The resulting DNA fragment was cloned into pKCCas9 to yield pKCCas9-gluR-K, which was introduced into the wild-type M145 by conjugal transfer. The resulting strains were subsequently grown on solid MS medium without apramycin at 37°C for two rounds to remove the plasmid pKCCas9-gluR-K. The correct double crossovers were verified by colony PCR with the primers ID-gluR-K-fw/rev, yielding the ΔgluR-K mutant. The primers used are listed in Table S2.
Complementation assays.
Using M145 genomic DNA as the template, the gluR-K operon and the gluABCD gene cluster with their corresponding putative promoter regions were obtained by PCR with the primer pairs Com-gluR-K-fw/rev and Com-gluABCD-fw/rev, respectively. Two PCR products were cloned in the integrative vectors pSET152 and pIB139 to generate the complemented plasmids pSET-gluR-K and pIB-gluABCD, respectively. The obtained plasmids were introduced into the ΔgluR-K mutant, resulting in two complemented strains, the ΔgluR-K/pSET-gluR-K and ΔgluR-K/pIB-gluABCD strains, respectively. The empty vectors, pSET152 and pIB139, were transferred into the M145 and ΔgluR-K strains, respectively, generating two positive controls, M145/pSET152 and M145/pIB139, and two negative controls, the ΔgluR-K/pSET152 and ΔgluR-K/pIB139 strains, respectively.
Analysis of bacterial growth and ACT production.
For the quantitative analysis of bacterial growth and ACT production, S. coelicolor strains with equivalent spore amounts (optical density at 450 nm [OD450] of 0.5) were cultivated on minimal medium (MM) agar plates with 75 mM glutamate (8-cm diameter) covered with sterilized plastic cellophane. The cultures were harvested at five different time points (36, 48, 72, 96, and 120 h) for analysis of both bacterial growth and ACT production. For bacterial growth determination, mycelia from the whole plates were collected, dried, and weighed. Analysis of ACT production was carried out using a previously described method (21).
Protein overexpression and purification.
The gluR open reading frame (ORF) was obtained by PCR amplification from the genomic DNA of M145 using the primers OE-gluR-fw/rev. The PCR product was cloned into the expression vector pET28a between the NdeI and EcoRI sites to generate pET28a-gluR, which was further verified by DNA sequencing. Similarly, the DNA fragments encoding the respective extracellular regions of GluK and AfsQ2 (ecGluK and ecAfsQ2) were obtained using the primer pairs OE-ecGluK-fw/rev and OE-ecAfsQ2-fw/rev and cloned into the expression vector pET32a between the EcoRV and EcoRI sites to yield pET32a-ecGluK and pET32a-ecAfsQ2, respectively. Protein overexpression in E. coli BL21(DE3) and purification were performed as previously described (33). The purities of His6-GluR, His6-Trx–ecGluK, His6-Trx–pAfsQ2, and His6-Trx tag were detected by 10% SDS-PAGE. The concentrations were determined by the Bradford protein assay kit (Sangon, China).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described by Tiffert et al. with some modifications (28). All primers used for EMSAs contain a universal DNA sequence (5′-AGCCAGTGGCGATAAG-3′) for the following Cy5 labeling of the DNA probes (Table S2). The probes harboring the respective promoter regions of tested genes were generated by PCR amplification from the genomic DNA of M145. Then, the PCR products were labeled with the Cy5-labeled universal sequence described above via a second round of PCR. The conditions for EMSA were as previously described (21). To verify the specificity of GluR-probe interaction, 100-fold excess amounts of nonspecific DNA (salmon sperm DNA) or each unlabeled specific probe were first incubated with purified His6-GluR protein for 20 min, followed by the addition of each labeled specific probe. After incubation for 30 min at 25°C, DNA-protein complexes were loaded onto a nondenaturing 6% polyacrylamide gel and electrophoresed in the 0.5× Tris-borate-EDTA (TBE) buffer. Finally, the gels were scanned directly by a FLA-9000 phosphorimager (Fujifilm, Japan). To assess the DNA-binding affinity of GluR after phosphorylation, 5 mM (final concentration) acetyl phosphate was added into the EMSA reaction buffer (42).
Determination of the transcriptional start sites.
5′ rapid amplification of cDNA ends (5′ RACE) analysis was performed to identify the transcription start site (TSS) of the gluA gene using a 5′ full RACE kit with tobacco acid pyrophosphatase (TAP) (TaKaRa, China). This experiment was performed according to the procedure recommended by the manufacturer. The gene-specific primers are listed in Table S2.
RNA preparation, RT-PCR, and RT-qPCR.
RNA preparation, RT-PCR, and quantitative real-time RT-PCR (RT-qPCR) analysis were performed as previously described (19). The primers used are listed in Table S2 in the supplemental material. S. coelicolor strains were grown on three different media including MS, R2YE, and MM with 10 or 75 mM glutamate, covered with sterilized plastic cellophane. Cultures were harvested for RNA preparation at different time points (48 and 96 h). RT-qPCR analysis was performed in the MyiQ2 two-color real-time PCR detection system (Bio-Rad, USA) by using iQ SYBR green Supermix (Bio-Rad, USA). The reactions were conducted in triplicate for each transcript and repeated with three independent samples. The hrdB gene (SCO5820, encoding the principal sigma factor) was used as an internal control. The relative expression levels of tested genes were normalized to those of hrdB. The relative fold changes in the expression of each gene were determined using the 2−ΔΔCT method (43). Error bars indicate the standard deviations from three independent biological replicates.
Determination of the gluA and gluR promoter activities via the xylE reporter assay.
The reporter xylE assay was employed for the analysis of the gluA and gluR promoter activities and was performed as previously described (44). The xylE gene encodes the catechol 2,3-dioxygenase responsible for the conversion of the colorless catechol into the yellow 2-hydroxymuconic semialdehyde (45). Using the primers RE-gluA-fw/rev, the DNA fragment containing the promoter region of the gluABCD operon (PgluA, 151 bp) was obtained by PCR amplification and cloned into the vector pZH-xylE between XbaI and NdeI, resulting in the reporter plasmid pZH-PgluA-xylE. Then, the recombinant plasmid was introduced into M145 and the ΔgluR-K mutant to generate M145/pZH-PgluA-xylE and ΔgluR-K/pZH-PgluA-xylE, respectively. Similarly, the DNA fragment containing the promoter region of the gluR-K operon (PgluR, 164 bp) was obtained by PCR using the primers RE-gluR-fw/rev and cloned into the vector pZH-xylE, resulting in the reporter plasmid pZH-PgluR-xylE. The recombinant plasmid was transferred into M145 and the ΔgluR-K mutant to generate M145/pZH-PgluR-xylE and ΔgluR-K/pZH-PgluR-xylE, respectively.
BLI assay.
The biolayer interferometry (BLI) assay was performed using an Octet Red96 system (Pall FortéBio Corp., Menlo Park, CA) (35). His6-Trx, His6-Trx–ecGluK, and His6-Trx–ecAfsQ2 (2.5 μg/ml) proteins were immobilized on NTA biosensors, followed by incubation with 20 mM glutamate or glutamine in the binding buffer (Tris-HCl, 50 mM; KCl, 50 mM; EDTA, 0.5 mM; pH 8.0). For dose-response analysis of ecGluK-glutamate interactions, His6-Trx–ecGluK (2.5 μg/ml) was incubated with different concentrations of glutamate as indicated. Data were analyzed by the Octet data analysis software 7.0 (Pall FortéBio Corp., Menlo Park, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by the National Natural Science Foundation of China (31430004, 31421061, 31630003, 31370081, and 31570072) and the Science and Technology Commission of Shanghai Municipality (16490712100).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00097-17.
REFERENCES
- 1.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 5.Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 7.Bush MJ, Tschowri N, Schlimpert S, Flardh K, Buttner MJ. 2015. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol 13:749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 8.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clement C, Ouhdouch Y, van Wezel GP. 2016. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler MS, Robertson AAB, Cooper MA. 2014. Natural product and natural product derived drugs in clinical trials. Nat Prod Rep 31:1612–1661. doi: 10.1039/C4NP00064A. [DOI] [PubMed] [Google Scholar]
- 10.Martín JF, Sola-Landa A, Rodriguez-Garcia A. 2012. Two-component systems in Streptomyces, p 315–332. In Gross R, Beier D (ed), Two-component systems in bacteria. Caister Academic Press, Poole, United Kingdom. [Google Scholar]
- 11.Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E. 2010. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156:2343–2353. doi: 10.1099/mic.0.038281-0. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez H, Rico S, Diaz M, Santamaria RI. 2013. Two-component systems in Streptomyces: key regulators of antibiotic complex pathways. Microb Cell Fact 12:127. doi: 10.1186/1475-2859-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. 2004. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150:2795–2806. doi: 10.1099/mic.0.27181-0. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Wang W, Shu D, Zhang W, Chen L, Qin Z, Yang S, Jiang W. 2007. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl Microbiol Biotechnol 77:625–635. doi: 10.1007/s00253-007-1184-5. [DOI] [PubMed] [Google Scholar]
- 17.Chang HM, Chen MY, Shieh YT, Bibb MJ, Chen CW. 1996. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol Microbiol 21:1075–1085. [PubMed] [Google Scholar]
- 18.Brian P, Riggle PJ, Santos RA, Champness WC. 1996. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J Bacteriol 178:3221–3231. doi: 10.1128/jb.178.11.3221-3231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Mast Y, Wang J, Zhang WW, Zhao GP, Wohlleben W, Lu YH, Jiang WH. 2013. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol 87:30–48. doi: 10.1111/mmi.12080. [DOI] [PubMed] [Google Scholar]
- 20.Shu D, Chen L, Wang WH, Yu ZY, Ren C, Zhang WW, Yang S, Lu YH, Jiang WH. 2009. afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl Microbiol Biotechnol 81:1149–1160. doi: 10.1007/s00253-008-1738-1. [DOI] [PubMed] [Google Scholar]
- 21.Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W. 2012. Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol Microbiol 85:535–556. doi: 10.1111/j.1365-2958.2012.08126.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, He J, Zhu H, Yu Z, Wang R, Chen Y, Dang F, Zhang W, Yang S, Jiang W. 2011. An orphan histidine kinase, OhkA, regulates both secondary metabolism and morphological differentiation in Streptomyces coelicolor. J Bacteriol 193:3020–3032. doi: 10.1128/JB.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San Paolo S, Huang J, Cohen SN, Thompson CJ. 2006. rag genes: novel components of the RamR regulon that trigger morphological differentiation in Streptomyces coelicolor. Mol Microbiol 61:1167–1186. doi: 10.1111/j.1365-2958.2006.05304.x. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen KT, Willey JM, Nguyen LD, Nguyen LT, Viollier PH, Thompson CJ. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol Microbiol 46:1223–1238. doi: 10.1046/j.1365-2958.2002.03255.x. [DOI] [PubMed] [Google Scholar]
- 25.Molle V, Buttner MJ. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol Microbiol 36:1265–1278. doi: 10.1046/j.1365-2958.2000.01977.x. [DOI] [PubMed] [Google Scholar]
- 26.Ainsa JA, Parry HD, Chater KF. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 27.Sola-Landa A, Rodriguez-Garcia A, Franco-Dominguez E, Martin JF. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol Microbiol 56:1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x. [DOI] [PubMed] [Google Scholar]
- 28.Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J. 2008. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol Microbiol 67:861–880. doi: 10.1111/j.1365-2958.2007.06092.x. [DOI] [PubMed] [Google Scholar]
- 29.Hutchings MI, Hong HJ, Buttner MJ. 2006. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol Microbiol 59:923–935. doi: 10.1111/j.1365-2958.2005.04953.x. [DOI] [PubMed] [Google Scholar]
- 30.Koteva K, Hong HJ, Wang XD, Nazi I, Hughes D, Naldrett MJ, Buttner MJ, Wright GD. 2010. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat Chem Biol 6:327–329. doi: 10.1038/nchembio.350. [DOI] [PubMed] [Google Scholar]
- 31.Wang WS, Ji JJ, Li X, Wang J, Li SS, Pan GH, Fan KQ, Yang KQ. 2014. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci U S A 111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronemeyer W, Peekhaus N, Kramer R, Sahm H, Eggeling L. 1995. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J Bacteriol 177:1152–1158. doi: 10.1128/jb.177.5.1152-1158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Zheng G, Jiang W, Hu H, Lu Y. 2015. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 34.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekiert DC, Friesen RHE, Bhabha G, Kwaks T, Jongeneelen M, Yu WL, Ophorst C, Cox F, Korse HJWM, Brandenburg B, Vogels R, Brakenhoff JPJ, Kompier R, Koldijk MH, Cornelissen LAHM, Poon LLM, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonawane AM, Singh B, Röhm KH. 2006. The AauR-AauS two-component system regulates uptake and metabolism of acidic amino acids in Pseudomonas putida. Appl Environ Microbiol 72:6569–6577. doi: 10.1128/AEM.00830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JY, Na YA, Kim E, Lee HS, Kim P. 2016. The actinobacterium Corynebacterium glutamicum, an industrial workhorse. J Microbiol Biotechnol 26:807–822. doi: 10.4014/jmb.1601.01053. [DOI] [PubMed] [Google Scholar]
- 38.Burkovski A, Krämer R. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol 58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 39.Trötschel C, Kandirali S, Diaz-Achirica P, Meinhardt A, Morbach S, Krämer R, Burkovski A. 2003. GltS, the sodium-coupled l-glutamate uptake system of Corynebacterium glutamicum: identification of the corresponding gene and impact on l-glutamate production. Appl Microbiol Biotechnol 60:738–742. doi: 10.1007/s00253-002-1170-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XX, Rainey PB. 2008. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 178:185–195. doi: 10.1534/genetics.107.081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 42.McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem 269:31567–31572. [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Luo YZ, Zhang L, Barton KW, Zhao HM. 2015. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth Biol 4:1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 45.Ingram C, Brawner M, Youngman P, Westpheling J. 1989. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol 171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.