Summary
Kinesin-8 motors regulate the size of microtubule structures, using length-dependent accumulation at the plus-end to preferentially disassemble long microtubules. Despite extensive study, the kinesin-8 depolymerase mechanism remains under debate. Here, we provide evidence for an alternative, tubulin curvature-sensing model of microtubule depolymerization by the budding yeast kinesin-8, Kip3. Kinesin-8/Kip3 uses ATP hydrolysis, like other kinesins, for stepping on the microtubule lattice, but at the plus-end, Kip3 undergoes a switch: Its ATPase activity is suppressed when it binds tightly to the curved conformation of tubulin. This prolongs plus-end binding, stabilizes protofilament curvature, and ultimately promotes microtubule disassembly. The tubulin curvature-sensing model is supported by our identification of Kip3 structural elements necessary and sufficient for plus-end binding and depolymerase activity, as well as by the identification of an α-tubulin residue specifically required for the Kip3-curved tubulin interaction. Together, these findings elucidate a major regulatory mechanism controlling the size of cellular microtubule structures.
Introduction
The size of internal cellular structures must be matched to cell size. A well-studied example is the mitotic spindle, whose optimal length is necessary for accurate chromosome segregation and, in metazoans, for the positioning of the cytokinetic apparatus. Spindle size and length are controlled by multiple mechanisms, illustrating principles likely to be relevant to size regulation of other intracellular structures.
Spindle size is controlled globally by the concentration or activity of factors that promote microtubule growth or disassembly (Helmke and Heald, 2014; Reber et al., 2013; Wilbur and Heald, 2013). Consistent with this idea, species differences in the activity of individual microtubule regulators have been shown to explain species-specific differences in spindle length (Loughlin et al., 2011). Thus, the amount or activity of microtubule regulators, or the amount of tubulin itself (Good et al., 2013; Hazel et al., 2013), can serve as a rheostat for spindle size, without requiring direct molecular mechanisms to measure microtubule or spindle length.
In addition to this global regulation, “measuring” mechanisms that mediate length-dependent microtubule assembly or disassembly have also been described. The best-studied length-dependent mechanism occurs through the activity of the kinesin-8 family of microtubule motors. Kinesin-8s are required to maintain normal spindle length in all eukaryotes studied (Gandhi et al., 2004; Goshima et al., 2005; Gupta et al., 2006; Rischitor et al., 2004; Savoian and Glover, 2010; Stumpff et al., 2008; West et al., 2001). They are also implicated in the length-regulation of other microtubule structures such as the cilia (Niwa et al., 2012).
The budding yeast kinesin-8, Kip3, can regulate microtubule dynamics in a length-dependent manner because it combines two biochemical activities: processive motility and microtubule depolymerase activity. Other kinesins typically have a single activity, for example, motile kinesins that are adapted for processive transport of cargo (e.g. kinesin-1), or depolymerases that have lost motility (e.g. kinesin-13s). Kip3 exhibits run lengths significantly longer than the average cellular microtubule length (Su et al., 2011; Varga et al., 2006; Varga et al., 2009). Therefore, all Kip3 molecules that land on the lattice reach the plus-end, where they dwell for extended periods of time (Su et al., 2011; Varga et al., 2006; Varga et al., 2009). Consequently, long microtubules accumulate more Kip3 at their ends than shorter ones. Selective trimming of long microtubules occurs because Kip3 triggers microtubule disassembly in direct proportion to its plus-end concentration (Su et al., 2011; Varga et al., 2006; Varga et al., 2009). A similar combination of motility and microtubule dynamics regulation is the basis for other mechanisms regulating microtubule length, (Marshall, 2015; Subramanian et al., 2013), highlighting the general importance of this class of mechanism for the size regulation of subcellular structures.
Although the length-dependent microtubule regulation by kinesin-8s is well established, the molecular mechanism (s) generating microtubule disassembly, and how they acquired depolymerase activity while preserving and integrating the ancestral motility, has been uncertain and controversial. Indeed, whether all kinesin-8s are active depolymerases has been debated (Du et al., 2010; Mayr et al., 2007; Stumpff et al., 2008). In one model, incoming kinesin-8s “bump-off” end-bound molecules together with the associated terminal tubulin dimer (Varga et al., 2009).In this model, depolymerase activity is thus intrinsically linked to and driven by kinesin-8 motility. Although this model is conceptually appealing, the mechanics of the “bump-off” event are uncertain. For example, it is not known if motility-based forces are sufficient to trigger Kip3-tubulin dissociation from the microtubule end. An alternative proposal, based on studies of human Kif18A, is that kinesin-8s cap the microtubule plus-end, blocking the addition of new tubulin subunits (Du et al., 2010; Stumpff et al., 2008). Such end capping would then promote microtubule catastrophe by slowing microtubule elongation, leading to loss of the stabilizing GTP-tubulin cap due to hydrolysis (Duellberg et al., 2016). Both the bump-off and plus-end capping models of the kinesin-8 mechanism contrast with that proposed for kinesin-13, which is thought to induce microtubule depolymerization by strongly binding to curved tubulin at the ends of microtubules (Ogawa et al., 2004).
Here, we present data supporting a new model for the mechanism of microtubule disassembly by Kip3. We find that the depolymerase activity of Kip3 is independent of its motility. Inconsistent with existing models, a monomeric Kip3 motor construct has depolymerase activity and triggers microtubule catastrophe without slowing microtubule growth. Our experiments suggest that Kip3’s depolymerase activity results from a two-state tubulin-binding switch; Kip3 uses a conventional interaction with the microtubule lattice to arrive at the plus end, and then converts to a mode of interaction characterized by a strong, preferential binding to curved tubulin. Interactions with curved tubulin also suppress Kip3 ATPase activity, further promoting strong binding to tubulin. We identify Kip3’s Loop11 sequence as both necessary and sufficient for its strong interaction with curved tubulin dimers. Indeed, we were able to generate a synthetic depolymerase by grafting Kip3’s Loop11 with two other Kip3 segments onto the backbone of kinesin-1. Finally, we identified an α-tubulin residue required for Kip3 to bind preferentially to curved tubulin dimers. Mutating this residue selectively abrogated Kip3-mediated depolymerization without affecting Kip3 motility or the activity of other depolymerases. Thus, in addition to providing a new model for the mechanism of depolymerase activity, our results elucidate evolutionary adaptations that underlie the acquisition of a new depolymerase activity while retaining motility. The coexistence of motility and microtubule destabilization activity in one molecule have enabled the nanometer-scale kinesin-8 motors to accurately regulate the micron-scale lengths of microtubules.
Results
Microtubule disassembly by Kip3 does not require motility
Kip3 has the strongest depolymerase activity of the kinesin-8s studied to date (Gupta et al., 2006; Mayr et al., 2007; Niwa et al., 2012). To test whether this increased activity depends on Saccharomyces cerevisiae-specific sequence insertions in the motor domain, we deleted the coding sequence for 56 residues from Loop1 and 31 residues from Loop2 (dL1, dL2, Fig. S1A). We purified these constructs (Fig. S2A) and tested their motility and depolymerase activity in vitro. These deletions had no effect on Kip3 motility or depolymerase activity towards GMPCPP-stabilized microtubules in vitro (Fig. S1B–E, Sup. File S1 A–D, P–S). Moreover, a mutant construct containing these deletions also fully rescued the benomyl sensitivity of a kip3Δ null strain (Fig. S4A). Thus, S. cerevisiae-specific insertions into Loop1 and Loop2 do not mediate its robust depolymerase activity and are dispensable for Kip3’s basic function. Because constructs lacking these S. cerevisiae sequences, which are predicted to be disordered, were purified to higher yield (not shown), all of the monomeric Kip3 constructs described below lack these sequences.
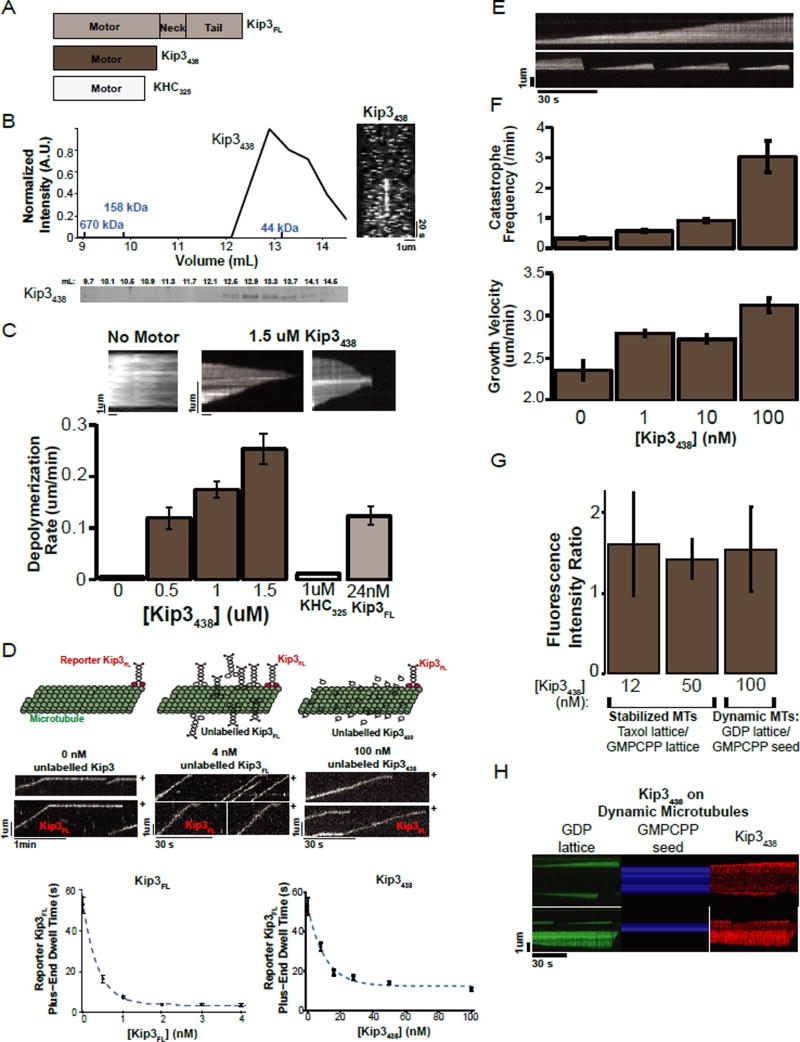
Kip3 microtubule depolymerase activity could be an intrinsic feature of its motor domain or an emergent property of its motility and ability to dwell at the plus-end (Du et al., 2010; Peters et al., 2010; Varga et al., 2009). To distinguish between these possibilities, we tested whether a non-motile, monomeric construct, Kip3438 (Fig. 1A) could depolymerize GMPCPP-stabilized microtubules (MTs). Gel filtration and single molecule motility assays showed that Kip3438 is monomeric and non-motile (Fig. 1B). Surprisingly, the non-motile, monomeric construct, Kip3438, depolymerized both ends of GMPCPP-stabilized MTs in a concentration-dependent manner (Fig. 1C; Sup. File S1 X). This effect is specific to Kip3, because 1uM of kinesin-1 (KHC325) monomer did not depolymerize GMPCPP-stabilized MTs (Fig. 1C; Sup. File S1 AE). Two-ended depolymerization by a monomeric and non-motile Kip3 construct indicates that motility is not required for depolymerase activity. At 0.5uM Kip3438 we observed a depolymerization rate of 0.12 +/− 0.02 um/min equivalent to that observed with 24nM Kip3 dimer. Because Kip3438 lacks motility and cannot concentrate at the plus-end, it is expected that higher concentrations of the monomer would be required to achieve the end concentration required for depolymerization than for the motile dimer. Kip3438 depolymerized microtubules from both ends, in contrast to the plus-end specificity of the Kip3 dimer, which is expected for a monomeric construct that cannot concentrate on one end (Fig. 1C top).
Figure 1. Motility is not required for Kip3 depolymerase activity.
A. Diagrams of the Kip3FL, Kip3438 and KHC325 constructs. Subscripts denote last residue of the protein construct. FL=Full length.
B. Kip3438 is monomeric and non-motile. Representative S200 gel filtration elution profile for Kip3438. The graph shows the intensity of the coomassie-stained elution fractions (normalized to the maximum, n= 3 runs). In blue are molecular weights of protein standards. The molecular weight of Kip3438, calculated from its sequence, is 74.6 KDa. Right: Representative kymograph of fluorescently labelled Kip3438 on taxol-stabilized microtubules showing that Kip3438 lacks motility (n=17 experiments). Scale bar: 1um (horizontal), 20s (vertical).
C. Concentration-dependent depolymerization of GMPCPP-stabilized microtubules by monomeric Kip3438. Top: kymographs showing GMPCPP-stabilized microtubules over time in the absence (control) or in the presence of 1.5 uM Kip3438. Scale bar: 1um (vertical), 1 min (horizontal). Bottom: Quantification of GMPCPP-stabilized microtubule depolymerization rates (mean +/− SEM (standard error of the mean); nMT=151–278) of Kip3438. The depolymerization rate of 1uM monomeric kinesin-1 (KHC325) and 24 nM dimeric Kip3FL are shown as controls (mean +/−SEM; nMT=87, 241).
D. Non-motile monomeric Kip3 can reduce the end-residence time of plus-end-bound dimeric Kip3FL. Top panel: Cartoons illustrating the experimental design. Middle: (left) Control kymograph of fluorescently labelled reporter Kip3FL (<0.05 nM Kip3FL-TMR) on taxol-stabilized microtubules. Unlabeled Kip3FL (middle) or Kip3438 (right) both decrease the plus-end dwell time of the labeled Kip3FL reporter. The microtubule plus-end is marked as (+). Scale bar: 1um (vertical), 1 min (horizontal) left kymograph (0 nM unlabeled Kip3); 30 s (horizontal) for both unlabeled Kip3FL and Kip3438 kymographs. Bottom: Left: The plus-end dwell time (mean +/− SEM; n=50–175 events) of fluorescently labelled Kip3FL is shown in the presence of spiked-in unlabeled Kip3FL (<0.05 nM Kip3FL -TMR, 0 nM to 4 nM unlabeled Kip3FL dimer). Right: The plus-end dwell time (mean +/− SEM; n=149–242 events) of fluorescently labelled Kip3FL is shown in the presence of varying concentrations of Kip3438 (<0.05 nM Kip3FL-TMR, 0 to 100 nM unlabeled Kip3438). The data points were weighted by the SEM and were fit to a single exponential (dotted blue line).
E. Kymographs of dynamic microtubules illustrating Kip3438-induced catastrophes. Microtubules grown in 14 uM tubulin rarely catastrophe (top) whereas 100 nM Kip3438 increases the frequency of catastrophes (bottom. Scale bar: 2 um (vertical), 30s (horizontal).
F. Microtubule catastrophe induced by varying concentrations of Kip3438. Top: The catastrophe frequency of dynamic microtubules grown (14uM tubulin) at the indicated concentrations of Kip3438 (mean +/− SE (standard error); nMT=35–273). Middle: Kip3438 subtly increases, rather than decreases, microtubule growth rates. Microtubule growth velocity as a function of Kip3438 concentration (mean +/− SE; nMT=35–273).
G. No preferential binding of Kip3 to GMPCPP (GTP-like) microtubules; Kip3 instead exhibits a slight preference for the GDP lattice. Ratio of the fluorescence intensity of Kip3438 bound to taxol-stabilized microtubules (GDP-like) relative to GMPCPP-stabilized microtubules (GTP-like) at [Kip3438]: 12 nM Kip3 (N=12 flow-cells) or 50 nM Kip3 (N=6 flow-cells). For dynamic microtubules (GDP lattice), the fluorescence intensity of 100 nM Kip3438 was measured. The ratio of the intensity of Kip3438 on the microtubule lattice (GDP-like), to the GMPCPP-seed (GTP-like) is quantified in the right column (N=12 flow-cells). (Mean+/−SE). In all assays, the ends of microtubules (600 nm from end) were excluded from quantification.
H. Kymographs illustrating the preferential binding of Kip3 to the GDP microtubule lattice. Dynamic microtubules were grown at 14 uM tubulin (green, left) from GMPCPP seeds (blue, middle). The fluorescence intensity of Kip3438 on the GDP-lattice and the GMPCPP seed (red, right) was imaged under TIRF microscopy and quantified as show in G.
A “bump-off” model, where terminal tubulin loss is driven by motility forces, has been proposed to explain the mechanism of Kip3-mediated microtubule disassembly (Varga et al., 2009). The “bump-off” model was inferred from experiments where unlabeled dimeric Kip3FL molecules decrease the residence time of labeled plus-end bound motors [(Varga et al., 2009), Fig. 1D, left]. However, because Kip3438 depolymerized microtubules, we determined whether this non-motile construct (unlabeled Kip3438) could also decrease the end residence time of a labeled, full length motor (Kip3FL). Indeed, just like unlabeled Kip3FL, increasing concentrations of unlabeled Kip3438 decreased the plus-end dwell time of labeled Kip3FL dimers in a dose-dependent manner (Fig. 1D). Thus, motility is not required to decrease the residence time of Kip3 at the plus-end.
We next utilized the Kip3438 construct to test if the depolymerase activity of the motor resulted from an indirect effect of its plus-end accumulation -- capping the microtubule plus-end (Du et al., 2010; Stumpff et al., 2008). Under this model, increasing concentrations of Kip3438, should decrease the microtubule growth velocity in parallel with an increase in the catastrophe frequency. Dynamic microtubules were grown from GMPCPP seeds with varying Kip3438 concentrations and imaged by TIRF microscopy. In the absence of Kip3438, at 14 uM tubulin, catastrophes were rare (0.3 +/− 0.05 /min). Adding monomeric Kip3438 increased the catastrophe frequency in a concentration-dependent manner, with a 10-fold increase in the catastrophe frequency observed at 100 nM Kip3438 (Fig. 1E, F top). However, in these conditions where the frequency of catastrophe was high, we did not observe slowing of microtubule growth and in fact observed a small increase in the growth rate (Fig. 1F, bottom). Thus, slowing of microtubule growth, although it may contribute in vivo, is not essential for Kip3-mediated disassembly, indicating that the monomeric Kip3 motor domain has intrinsic microtubule destabilizing activity.
We also considered the possibility that Kip3 might regulate microtubule plus-end dynamics through a selective high-affinity interaction with plus-end tubulin in a GTP-like or hydrolysis intermediate state, analogous to what has been described for the microtubule plus-end tracking protein, EB1 (Maurer et al., 2014; Zhang et al., 2015). However, the plus-end dwell time of Kip3FL on GDP-like (taxol-stabilized), was 52.9 +/−4.2 s (n=149) as compared with 38.2 +/−6 s (n=206) on GTP-like (GMPCPP-stabilized) microtubules. Furthermore, using a fluorescence assay for Kip3438 binding to a mixed population of microtubules, the relative affinity of Kip3438 for GMPCCP-stabilized microtubules was, in fact, lower (~66%) than for taxol-stabilized microtubules (Fig. 1G) or the lattice of dynamic microtubules (Fig. 1G, H). Thus, the high affinity of Kip3 for the microtubule plus-end cannot be explained by preferential binding to the GTP-cap.
Defining the Kip3 elements required for depolymerase activity
The above results prompted us to consider alternative mechanisms. We speculated that Kip3 depolymerase activity results from a unique binding interaction with curved tubulin subunits at the end of the microtubule. Indeed, a growing number of microtubule plus-end regulators preferentially recognize curved tubulin --either tubulin dimers in solution or tubulin subunits that can exist in a curved conformation at the ends of microtubules (Brouhard and Rice, 2014). As an initial step towards testing this hypothesis, we used domain swaps and mutagenesis to identify regions of Kip3 specifically required for its depolymerase activity. We focused on the Kip3 Loop2, Loop11 and the neck sequences, which each display family-specific sequence conservation that might reflect specialized functions. To determine the role of these regions in Kip3’s depolymerase activity we first created Kip3 chimeras replacing each region of interest with the corresponding segment from kinesin-1, and then tested the chimeras for depolymerase activity, microtubule and tubulin binding.
The first heptad of the coiled-coil neck of Kip3 and other kinesin-8s is predicted to have a low probability of forming a coiled-coil (Fig. S3A). We had previously shown that substitution of the Kip3 neck with a strong coiled-coil –a leucine zipper- abolished Kip3’s depolymerase activity [Kip3FL-CC, Fig. S3B–E; Sup. File S1 V (Su et al., 2013)]. To test the idea that a flexible sequence at the start of the Kip3 neck is required for depolymerase activity, we added three glycine residues to Kip3FL-CC between the leucine zipper and the motor domain (Fig. S3B). Indeed, the flexible glycine linker fully restored depolymerase activity to the leucine zipper containing construct (Fig. S3F; Sup. File S1 W). Using a single molecule competition assay to measure the relative affinity of the constructs for tubulin, we estimated that Kip3FL-CC had a ~25-fold reduced affinity for tubulin dimers [(Su et al., 2013), see schematic, Fig. S3G].
Moreover, the glycine linker also restored tubulin-binding to the leucine zipper construct (Fig. S3G). Together, this suggests that Kip3 depolymerase activity requires a flexible connection between the motor heads at the base of the neck, which itself is necessary for tubulin binding. Moreover, the results are consistent with the idea that strong Kip3 tubulin dimer-binding is fundamental to microtubule depolymerase activity.
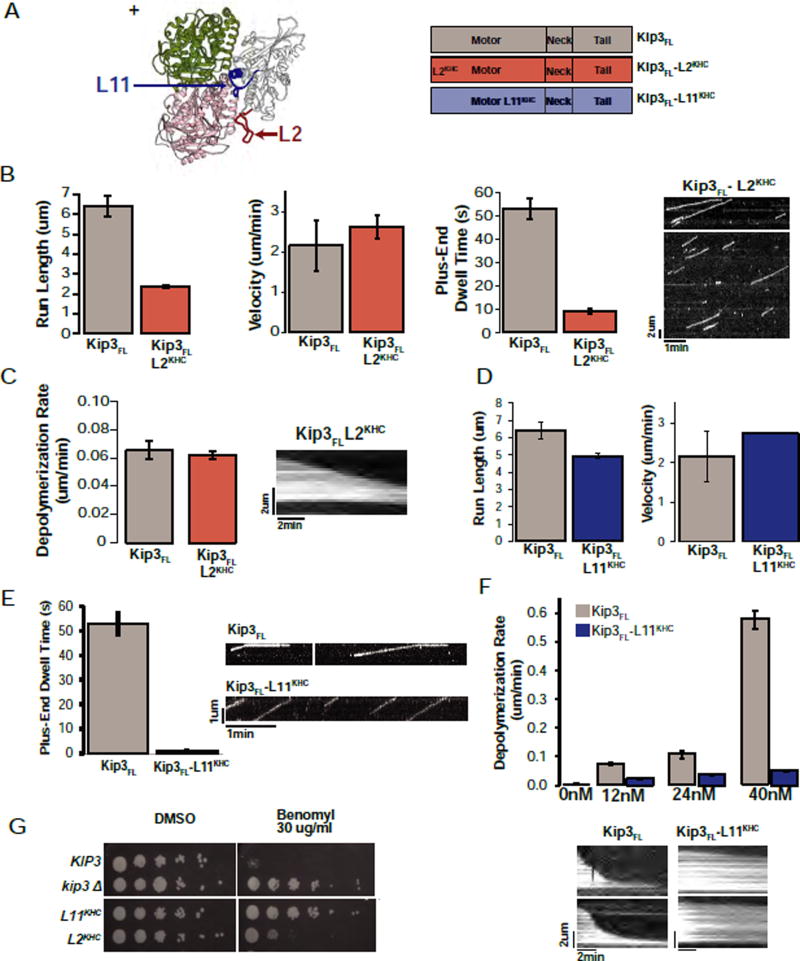
Loop2 plays a critical role in kinesin-13 mediated microtubule depolymerization, and prior structural studies led to the suggestion that Loop2 of kinesin-8s might similarly mediate depolymerase activity (Peters et al., 2010; Kim et al., 2014). To test this proposal, we created a chimera in which Kip3’s entire Loop2 sequence was replaced by the corresponding segment from kinesin-1 (Kip3FL-L2KHC, Fig. 2A). Although Kip3FL-L2KHC exhibited a somewhat reduced run length (2.38 +/−0.08 um, Fig. 2B; Sup. File S1 E) and a shortened plus-end dwell time (9.3 +/− 1.1 s, Fig. 2B; Sup. File S1 E), its depolymerase activity was indistinguishable from the wild-type control, under conditions of similar flux of motors to the microtubule plus-end (0.065 +/− 0.007 um/min vs. 0.062 +/− 0.003 um/min, respectively (p=0.67, Fig. 2C, Fig. S4B–C; Sup. File S1 P,T). Furthermore, although expressed at reduced steady-state levels, Kip3FL-L2KHC partially complemented a kip3Δ null strain (Fig. 2G, S4A). Thus, Loop2 contributes to Kip3’s processivity and plus-end residence time, but, in contrast to kinesin-13, is not required for its depolymerase activity.
Figure 2. Structural elements required for Kip3 microtubule depolymerase activity.
A. Homology model of Kip3 bound to tubulin based on the human Kif18A structure (3LRE). β-tubulin is shown in dark green (marked +) and α-tubulin in pink (4LNU). Loop2 is in red, Loop11 in blue. Right: diagrams of the Kip3FL, Kip3FL-L2KHC and Kip3FL-L11KHC constructs. Superscripts denote the origin of the swapped region.
B. L2 is required for Kip3 processivity. Quantification of the motility (run-length, velocity and plus-end dwell time) of Kip3FL and Kip3FL-L2KHC on taxol-stabilized microtubules imaged by TIRF microscopy (mean +/− SEM; n=75–870 events). The kymograph illustrates the reduced plus-end dwell time and run length of the Kip3FL-L2KHC construct. Scale bar: 1 min (horizontal), 2um (vertical).
C. Kip3 Loop2 is dispensable for microtubule depolymerization. Kip3FL or Kip3FL-L2KHC have comparable depolymerization rates of GMPCPP-stabilized microtubules (mean +/−SEM; nMT=96–407). Depolymerization rates are shown at concentrations of the input motors has been corrected to achieve comparable flux to the plus-end (~170 motors/min; see methods, Fig S4B, input: 12nM and 40nM respectively). Kymograph showing depolymerization of a GMPCPP-stabilized microtubule over time by Kip3FL-L2KHC as quantified above. Scale bar: 2 min (horizontal), 2um (vertical).
D. The motility of Kip3FL-L11KHC is comparable to Kip3FL. Quantification of the run length (left) and velocity (right) of Kip3FL (grey) and Kip3FL -L11KHC (blue) on taxol-stabilized microtubules (mean +/− SEM; n=155–1022 events).
E. Specific requirement of Kip3 Loop11 for prolonged plus-end binding. Plus-end residence time of fluorescently labelled Kip3FL and Kip3FL-L11KHC (mean +/− SEM; n=149–493 events) on taxol-stabilized microtubules. Right: Representative kymographs from single molecule assays quantified in D, E showing the motility and plus-end residence of Kip3FL and Kip3FL-L11KHC. Scale bar: 1 um (vertical), 1 min (horizontal).
F. Kip3 Loop11 is required for depolymerase activity. Depolymerization rate of GMPCPP-stabilized microtubules by the indicated concentrations of Kip3FL (grey) or Kip3FL-L11KHC (dark blue, mean +/− SEM; nMT=69–439 and nMT=121–265 respectively). Bottom: Kymographs illustrating the depolymerization of GMPCPP-stabilized microtubules by Kip3FL or Kip3FL-L11KHC at 40 nM input. Scale bar: 2 um (vertical), 2 min (horizontal).
G. The Loop11 sequence is required for KIP3 function. KIP3 strains are sensitive to benomyl whereas the kip3-L11KHC strain exhibits resistance comparable to the null allele (kip3Δ). kip3-L2KHC strains display intermediate benomyl resistance. L2KHC and L11KHC experiments were performed in a kip3Δ strain background. The indicated strains were spotted at 10-fold dilutions. Partial complementation by L2KHC is likely explained, at least in part, by its low steady state expression (see Fig S4A).
Because the Loop11 (L11) segment of kinesin-8s is highly basic (predicted pI ~ 11.7 for Kip3 as compared to a predicted pI ~4.1 for kinesin-1), we evaluated the importance of Kip3’s Loop11 sequence for depolymerase activity. Replacement of Kip3’s Loop11 with the Loop11 from kinesin-1 (Kip3FL-L11KHC) yielded a motor whose velocity and run length was indistinguishable from that of Kip3 (Fig. 2D; Sup. File S1 F). However, like kinesin-1, Kip3FL-L11KHC failed to dwell at the microtubule plus-end (Fig. 2E; Sup. File S1 F) and had less than 10% depolymerization activity of the Kip3FL control (0.051 +/− 0.003 um/min vs. 0.575 +/1 0.69 um/min at 40nM respectively, Fig. 2F; Sup. File S1 U). Additionally, consistent with the lack of depolymerase activity, Kip3FL-L11KHC failed to complement a kip3Δ null strain (Fig. 2G). Moreover, a complementary loop swap experiment (KHC560-L11Kip3, Fig. 3A, blue) demonstrated that Kip3’s Loop11, unlike the Loop1 or Loop2 segments, was sufficient to enable kinesin-1 to dwell at the microtubule plus-end (Fig. 3B, E; Sup. File S1 K). The combined addition of Loop11 and Loop2 into kinesin-1, further increased its end-residence time to 52.1 +/− 3.7 s, comparable to the Kip3FL control (52.9 +/− 4.3s; Fig. 3B; Sup. File S1 A, L), generating a motor with similar motility to Kip3FL (Fig. 3C,D; Sup. File S1 L). Individually, the Kip3 Loop11, Loop2, or the Kip3 neck sequences are not sufficient for depolymerase activity (Fig. 3F; Sup. File S1 AD, AF–AH). Thus, Kip3’s Loop11 sequence is both necessary and sufficient for plus-end recognition and is required for microtubule depolymerase activity.
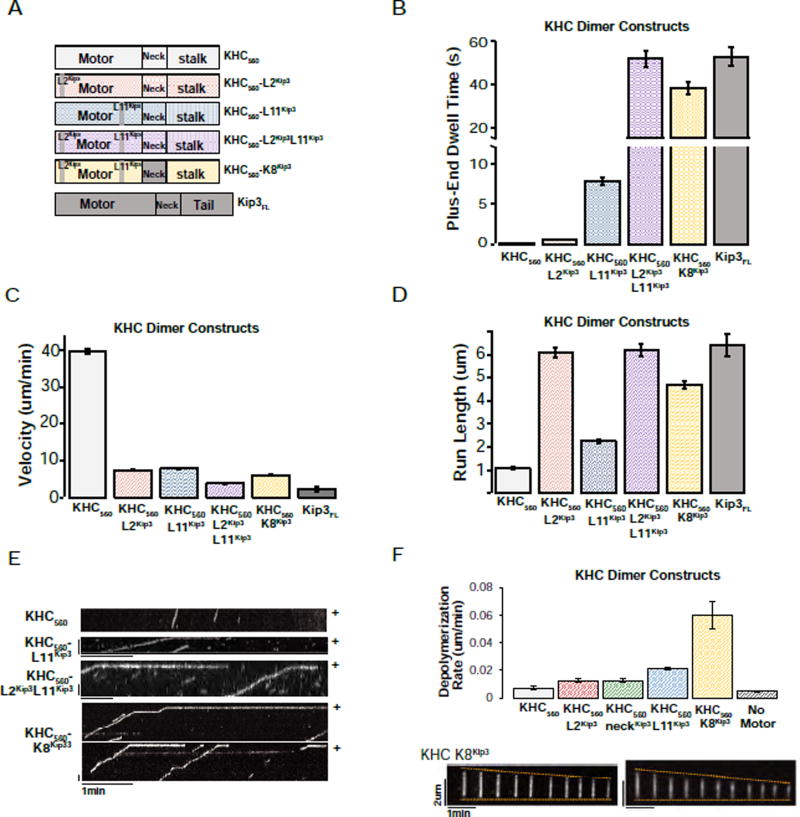
Figure 3. Kip3’s L11 is sufficient to convey plus-end binding to KHC and Kip3’s L2 is sufficient to increase the processivity of KHC.
A. Domain swaps of Kip3 segments into dimeric KHC. Diagrams of the chimeric constructs used. Superscripts denote the origin of the inserted segment.
B. Domain swap showing that Kip3 Loop11 is a transportable segment sufficient for prolonged plus-end binding. Kip3 and a KHC chimera containing Kip3’s L2, L11 and the family-specific neck segment (KHC560-K8Kip3) have comparable plus-end residence times. Shown is the end-residence time (mean +/− SEM; n=149–394 events) of the indicated KHC variant constructs compared to unmodified KHC560 and Kip3FL. The superscript denotes the origin of the indicated loop that was grafted into KHC560.
C. Either Kip3 L2 or L11 reduce the velocity of KHC560. The velocity (mean +/− SEM; n=155–795 events) of KHC560, KHC chimeras and Kip3 on taxol-stabilized microtubules.
D. Kip3 and a KHC chimera containing Kip3’s L2, L11 and family-specific neck segment (KHC560-K8Kip3) have comparable run lengths (mean +/− SEM; n=155–795 events.)
E. Representative kymographs from single molecule assays illustrating the motility and plus-end residence of KHC560 and KHC560 variant motor constructs. Scale bar: 1 min (horizontal), 1 um (vertical).
F. KHC-K8Kip3 is a synthetic microtubule depolymerase. Depolymerization rate of GMPCPP-stabilized microtubules by the indicated kinesin-1 chimeras (120nM, mean +/− SEM; nMT=72–278). The KHC560-NeckKip3 construct has a velocity of 7.4 +/− 0.1 um/min and a run length of 1.2 +/− 0.1 (n=547), similar to KHC. Bottom: GMPCPP-stabilized microtubules shown at 29s intervals after the addition of 120 nM KHC560-K8Kip3 (nMT=84). Scale bars: 2 um (vertical), 1 min (horizontal).
Having identified three structural features of Kip3 (neck, Loop2 and Loop11) that contribute to its distinctive microtubule end-residence, tubulin-binding, and depolymerase activities, we tested whether grafting these three elements together into kinesin-1 (KHC560-K8Kip3 Fig. 3A, yellow) would be sufficient to generate a synthetic microtubule depolymerase. Indeed, KHC560-K8Kip3 was nearly as processive as Kip3FL (Fig. 3D; Sup. File S1 M), had a plus-end dwell time that was comparable to that of Kip3FL (Fig. 3B; Sup. File S1 M), and depolymerized GMPCPP-stabilized microtubules (Fig. 3F; Sup. File S1 AI KHC560-K8Kip3 showed ~10-fold reduced specific activity). The reduced activity of KHC560-K8Kip3 could either be because additional segments of the motor domain also contribute to full activity or because of structural imperfections in the chimeric construct. Thus, the neck, Loop2, and Loop11 elements of Kip3 are necessary and sufficient for depolymerase activity.
Kip3’s Loop11 is necessary and sufficient for preferential binding to unpolymerized tubulin
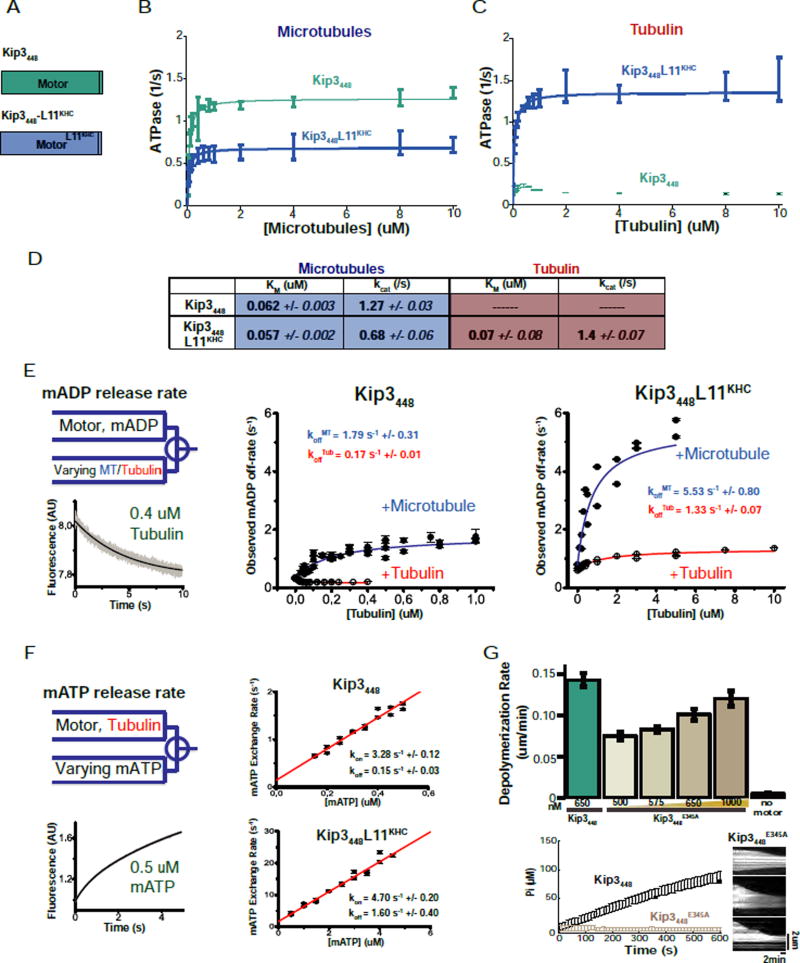
Tubulin dimers in the microtubule lattice lie in a straight conformation, but tubulin dimers in solution and at the plus-end adopt curved conformations (Brouhard and Rice, 2014). Based on the results described above, we first asked whether Kip3’s Loop11 mediated a specific interaction with soluble (curved) tubulin. We measured the affinity of Kip3438 and Kip3438-L11KHC (Fig. 4A) for tubulin using a direct binding assay, biolayer interferometry. Our measurements revealed that the Kip3 Loop11 sequence increased the tubulin binding affinity of the motor by 7-fold (Fig. 4B). These observations are consistent with the inference from the plus-end residence results suggesting that Kip3 L11 mediates the binding of Kip3 to the curved tubulin found at the plus-end and in solution (Fig. 2E, 3B, 4B).
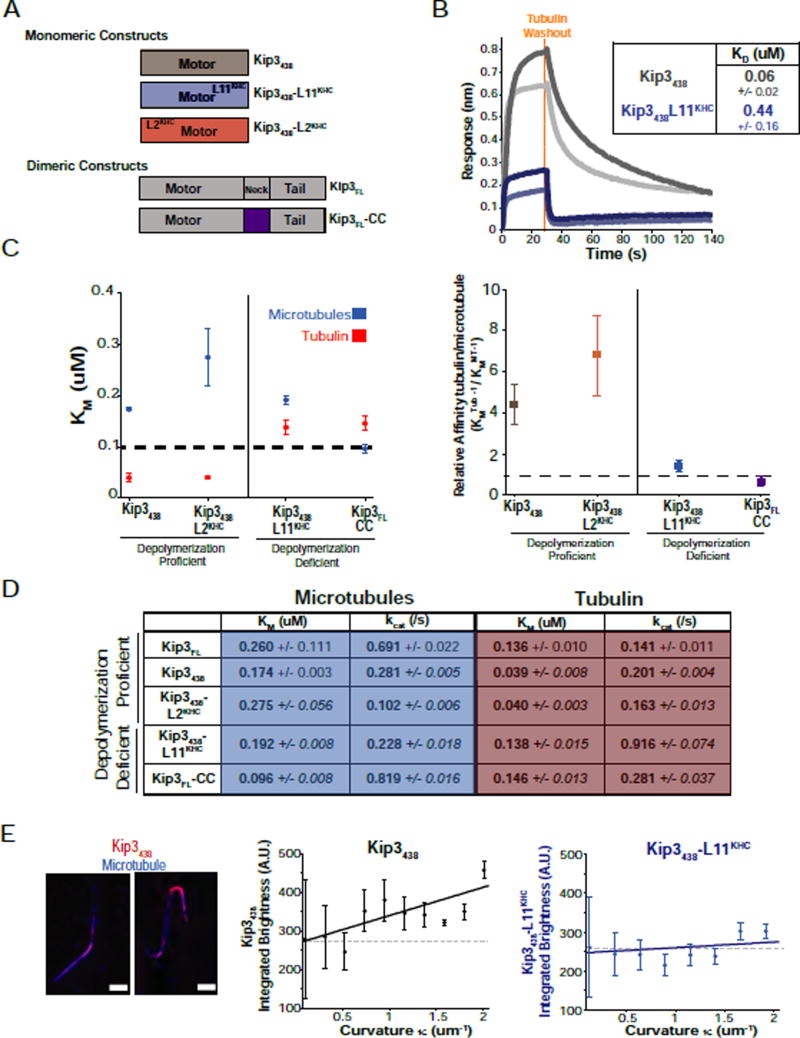
Figure 4. Strong tubulin dimer binding by depolymerase proficient Kip3 variants.
A. Diagram of monomeric and dimeric Kip3 variant constructs. Superscripts denote the origin of the grafted segment. The neck in Kip3FL has been replaced with a leucine zipper (LZ, purple) in Kip3FL-CC.
B. Loop11 is required for strong tubulin binding. Affinity measurements of the binding of tubulin to immobilized Kip3438. Biolayer interferometry example traces of tubulin binding to Kip3438 at 0.5 uM tubulin (light grey), 1uM tubulin (dark grey) versus Kip3438-L11KHC at 1 uM tubulin (light blue) or at 2 uM tubulin (dark blue). Dotted orange line indicates the time point of tubulin washout, allowing measurement of koff values. KD values are listed in the inset (mean +/− SE, nKip3 438=30, nKip3 438-L11KHC=28 concentrations).
C. Depolymerization proficient constructs have a higher relative affinity for tubulin relative to microtubules. Left: Michaelis-Menten constants for microtubules (blue) or tubulin (red) for Kip3438, Kip3438-L2KHC, Kip3438-L11KHC and Kip3-CC (KM, uM, mean +/− SEM, n=3–4 curves). KM values are from steady-state ATPase assays shown in D. ATPase curves are shown in Fig. S5. A line at 100 nM is drawn for visualization of the results. Right: Relative affinity of the constructs for tubulin over the microtubule shown as (KMTubulin)−1 / (KMMicrotubule)−1.
D. Values from steady state ATPase assays of the indicated Kip3 constructs with microtubules or free tubulin as substrate (0–10 uM). Shown are mean +/− SEM; n=3−4 curves. Raw data and fits are presented in Fig. S5.
E. Preferential binding of Kip3438 to regions of higher curvature on bent microtubules. Left: Representative images of fluorescently labelled Kip3438 bound to taxol-stabilized microtubules bent under flow. Scale bar: 2 um. Integrated brightness for Kip3438 (middle, black, nMT=138) or Kip3438-L11KHC (right, blue, nMT=528) plotted against the local curvature (κ) of the microtubule (mean +/− SE). Solid lines are linear fits to the data with slope mKip3438=68.4 AU/um−1 and mKip3438_L11KHC= 27.9 AU/um−1. Dashed lines represent a line with a slope of 0.
We then measured the microtubule and tubulin-stimulated ATPase activity of Kip3 variants as a complementary way to assess their relative affinities for tubulin and microtubules. If Kip3 depolymerizes microtubules by strong binding to curved tubulin at the plus-end, we expected that Kip3 should exhibit a lower Michaelis-Menten constant (KM) for tubulin relative to microtubules, an effect that should depend on its L11 sequence. The Michaelis-Menten constant (KMMT) for microtubule-stimulated ATPase activity was similar for Kip3FL (dimer), Kip3438, Kip3438-L2KHC, Kip3FL-CC (dimer) and Kip3438-L11KHC (Fig. 4C,D). This indicates that Kip3-specific Loop11 and neck sequences do not contribute significantly to its affinity for the microtubule. By contrast, these sequences markedly affected the interaction between Kip3 and tubulin (Fig. 4C–D): depolymerization proficient motors (Kip3438 or Kip3438-L2KHC) had significantly lower Michaelis-Menten constants for tubulin (KMTub) relative to microtubules (4.5-fold or 6.9-fold, respectively; Fig. 4C–D), whereas depolymerase-deficient constructs, Kip3438-L11KHC and Kip3FL-CC had a nearly equivalent KM for tubulin as for microtubules (Fig. 4C–D). Furthermore, swapping Kip3’s Loop11 sequence into kinesin-1 decreased 9.4-fold the KM of the motor for tubulin (Fig. S5B, C), consistent with the biolayer interferometry data. Thus, these experiments reveal that Kip3-specific sequences in Loop11 and the neck allow Kip3 to bind tightly and preferentially to tubulin over microtubules (Fig. 4B–D).
As an additional test of the preferential binding of Kip3 to curved tubulin, we measured the accumulation of fluorescently labeled Kip3438 on regions of increased microtubule curvature that were induced by fluid flow (Bechstedt et al., 2014; Roostalu et al., 2015). Consistent with preferential curved tubulin binding, Kip3438 accumulates at flow-induced microtubule bends (Fig. 4E, S4D). No such enrichment on curved microtubule surfaces was observed for either Kip3438-L11KHC or KHC325 (Fig. 3E right, S4D). Altogether, these results demonstrate that tubulin binding is strongly associated with depolymerase activity and that strong and selective curved tubulin binding is mediated by Kip3’s Loop11 segment.
The Kip3 Loop11 suppresses tubulin-stimulated ATPase activity
In addition to affecting the affinity of Kip3 for tubulin, the Kip3438 Loop11 sequence also selectively suppressed its tubulin-stimulated ATPase activity. Whereas the microtubule stimulated ATPase activity of Kip3438-L11KHC was 80% of the ATPase activity of Kip3438 (Fig. 4D), the tubulin-stimulated ATPase activity (kcat) of Kip3438-L11KHC was nearly 5-fold higher than that of Kip3438 (Fig. 4D). Our Kip3438 construct, modeled on a construct used to determine the crystal structure of human Kif18A (Peters et al., 2010), lacks 10 residues of the neck linker that contribute to maximal ATPase activity in other kinesins (Rice et al., 1999; Tomishige and Vale, 2000). Indeed, a monomer construct containing these residues, Kip3448 (Fig. 5A), had higher microtubule-stimulated ATPase activity (Fig. 5B, D) and also increased depolymerase activity (Fig. S4E). However, just like Kip3438, Kip3448 had markedly reduced tubulin-stimulated ATPase activity (Fig 5C–D). From biolayer interferometry, both monomeric constructs had comparably high affinities for tubulin (Fig. S4F). Moreover, swapping the kinesin-1 Loop11 sequence into Kip3448 (Kip3448-L11KHC), abolished its depolymerase activity (Fig. S4E; Sup. File S1 AB) and selectively reduced its affinity for tubulin without affecting its interaction with microtubules (Fig. 5B–D, S4F). Thus, the analysis of independent monomeric Kip3 constructs shows that Kip3’s Loop11 selectively increases its affinity for tubulin and suppresses its tubulin-stimulated ATPase activity.
Figure 5. Kip3 Loop11 mediates tubulin-specific suppression of ATP turnover.
A. Diagrams of monomeric constructs: Kip3448 and Kip3448-L11KHC.
B. & C. The Kip3 Loop11 segment specifically suppresses tubulin-stimulated ATPase activity but not its microtubule stimulated ATPase activity. Steady state ATPase assays comparing Kip3448 and Kip3448-L11KHC stimulated by: taxol-stabilized microtubules (B) or tubulin (C) (mean +/− SEM; n=3–4 curves). The data was fitted to a Michaelis-Menten curve, except for the tubulin-activated ATPase curve of Kip3448 which showed concentration dependent inhibition of activity.
D. Values from steady state ATPase assays with Kip3448 or Kip3448-L11KHC shown in B–C, using microtubules or free tubulin as substrate (0–10 uM). Shown are mean +/− SEM; n=3–4 curves.
E. Slow tubulin-stimulated ADP release by Kip3448. mantADP-incubated Kip3448 (middle) or Kip3448-L11KHC (right) was rapidly mixed with varying concentrations of microtubules or tubulin in a stopped-flow apparatus. Left, top: scheme of the experiment. Left, bottom: Representative trace of fluorescent signal decay as mantADP unbinds the motor. Observed microtubule- and tubulin-dependent mantADP release rates are plotted for Kip3448 (middle) and Kip3448-L11KHC (right). mantADP release rates (koff) for Kip3448 and Kip3448-L11KHC in saturating microtubule (MT) or tubulin (Tub) are shown in the figure in blue and red, respectively. N=5–6 per point. The x-axis scale ([Tubulin] (uM)) for each construct was chosen as the concentration where the motor reaches a constant maximal mantADP release rate. Each construct shows different kinetics and therefore plateaus at different substrate concentrations. The reported mantADP release rate constants are derived from a fit to the curves shown here as described in the methods section.
F. Tubulin binding to Kip3448 does not inhibit ATP binding. Left, top: scheme of the experiment. Left, bottom: representative trace of the fluorescent signal detected upon mixing of motor-tubulin complex with mantATP. A bi-exponential signal arises with two distinct phases. The initial fast phase corresponds to mantATP exchange to tubulin-bound (APO-state) motor. The slow phase (shown in Fig. S4G) combines ADP dissociation from tubulin-motors with mantATP exchange in the following cycle. The data were fitted to the equation: kobs=kon[mATP]+koff (see methods). The mantATP on-rate (kon) and off-rate (koff) of each construct are shown in the figure. Right, top: Kip3448, in the presence of 2.5 uM tubulin. The slow nucleotide off-rate is consistent with the slow hydrolysis turnover in ATPase assay in (C) and the slow mantADP off-rates in (E). Right, bottom: Kip3448-L11KHC, in the presence of 2.5 uM tubulin. N=5–6 per point.
G. ATP hydrolysis is not required for Kip3 depolymerase activity. Shown are the depolymerization rates (mean +/− SEM; nMT=102–229) for Kip3448 or ATPase deficient Kip3448E345A. Bottom left: Control validating that Kip3448E345A lacks microtubule (1 uM)-stimulated ATPase activity (phosphate produced over time) Kip3448E345A (mean+/− SD; n=3 events per condition comparing Kip3448E345A with Kip3448); input 100 nM motor. Bottom right: Kymographs of GMPCPP-stabilized microtubules showing depolymerization by Kip3448E345A. Scale bar: 2 um (vertical), 2 min (horizontal).
Tuning of the Kip3 catalytic cycle by tubulin-binding
In motile kinesins the release of ADP from the active site is rate limiting and stimulated by microtubule binding (Hackney, 1988). To test whether ADP release or ATP binding were suppressed by tubulin-binding, we performed stopped-flow experiments with fluorescent nucleotide analogues mantADP (mADP) or mantATP (mATP). In solution, Kip3’s basal ADP-release rate (0.33 s−1 +/− 0.01) is considerably higher than other motile kinesins (Alonso et al., 2007; Cross, 2004) and significantly faster than its basal ATP hydrolysis rate (kbasal= 0.13 s−1 +/− 0.01). Therefore, in the absence of tubulin or microtubules, ATP hydrolysis and not ADP-release is rate limiting.
mADP release from both Kip3448 and Kip3448-L11KHC was comparably stimulated by microtubules (Fig. 5E), as expected. The maximal microtubule-stimulated mADP release rate, however, was ~3-fold lower for Kip3448 relative to Kip3448-L11KHC (Fig. 5E, middle). The rate of Kip3448 mADP release (koff=1.79 /s−1 +/− 0.31) was similar to its microtubule-stimulated ATPase activity (kcat=1.27 /s−1 +/− 0.03), suggesting that on microtubules, ADP release by Kip3 is rate limiting as for other motile kinesins. Thus, excepting the high basal ADP-release rate, Kip3 behaves much like other motile kinesins in the presence of microtubules.
Tubulin affected the Kip3 ATPase cycle differently than microtubules. Whereas microtubules stimulated ADP release from Kip3448, tubulin did not stimulate and even slightly inhibited ADP release (Fig. 5E, middle). This contrasts with the stimulation of ADP-release in both translocating and depolymerizing kinesins by tubulin (Alonso et al., 2007; Friel and Howard, 2011). The suppression of Kip3448 ADP release by tubulin required the Kip3 L11 sequence (Fig. 5E, right), consistent with our ATPase results. Altogether, the microtubule and tubulin stimulated ADP release experiments suggest that Kip3’s L11 sequence suppresses ADP release, an effect that is preferentially observed with tubulin dimers.
The slow tubulin stimulated ADP release of Kip3448 suggested that the complex may exist in the ADP state for a relatively long time (koff−1= 6.0 s +/− 0.8, Fig. 5E), which we confirmed by biolayer interferometry measurements of the residence time of Kip3448ADP on tubulin (koff−1= 35.7 s +/− 7.7, Fig. S4F). However, despite the relatively long lifetime of the Kip3448ADP-tubulin complex, ADP reduces the affinity (KD) of Kip3448 for tubulin 4-fold relative to ATP (Fig. S4F). In the presence of ADP, Kip3448ADP is also unable to depolymerize GMPCPP-stabilized microtubules (not shown). Thus, Kip3’s ability to recognize curved tubulin is largely nucleotide independent, however, depolymerization appears to require that the motor be in a relatively strong tubulin binding state.
We then considered the possibility that tubulin binding may inhibit subsequent steps in the catalytic cycle of Kip3. We used mantATP exchange assays to test whether tubulin inhibits ATP binding to the motor which would prolong the lifetime of the strong binding apo state relative to other steps in the catalytic cycle. However, the ATP on-rate for tubulin-bound Kip3448 (kon=3.28 +/− 0.12 uM−1s−1, Fig. 5F) is fast, comparable to that of KHC (Gilbert et al., 1995). We measured a similarly high ATP on-rate for tubulin-bound Kip3448-L11KHC (Fig. 5F), or for Kip3448 bound to microtubules (not shown). Thus, tubulin does not block ATP binding to Kip3 and ATP binding is not influenced by Loop11, arguing against the idea that the relevant strong binding state for Kip3-mediated depolymerization is the apo state. Therefore, we infer that strong tubulin binding is promoted by either ATP or possibly ADP.Pi-liganded Kip3.
ATP hydrolysis is not required for Kip3-mediated MT depolymerization
If strong binding of Kip3 to curved tubulin dimers is the key step for microtubule depolymerization, ATP hydrolysis by Kip3 might not be required for depolymerase activity. Kinesins generally bind strongly to microtubules or tubulin in the ATP or the no nucleotide (apo) states. To test this idea, we mutated the conserved glutamate in the switch II helix (E345A) that is required for ATP hydrolysis in all kinesins, and verified the inactivating effect of the E345A mutation (Fig. 5G, bottom). Indeed, the Kip3448E345A monomer depolymerized GMPCPP-stabilized microtubules in a concentration dependent manner (Fig. 5G, top; Sup. File S1 AC). As expected for a mutant that should poorly recycle because of slow dissociation from tubulin, Kip3448E345A displayed somewhat reduced depolymerase activity relative to Kip3448 (Fig. 5G). Thus, ATP hydrolysis is not required for Kip3-mediated microtubule depolymerization.
An α-tubulin residue required for Kip3 binding to curved but not straight tubulin
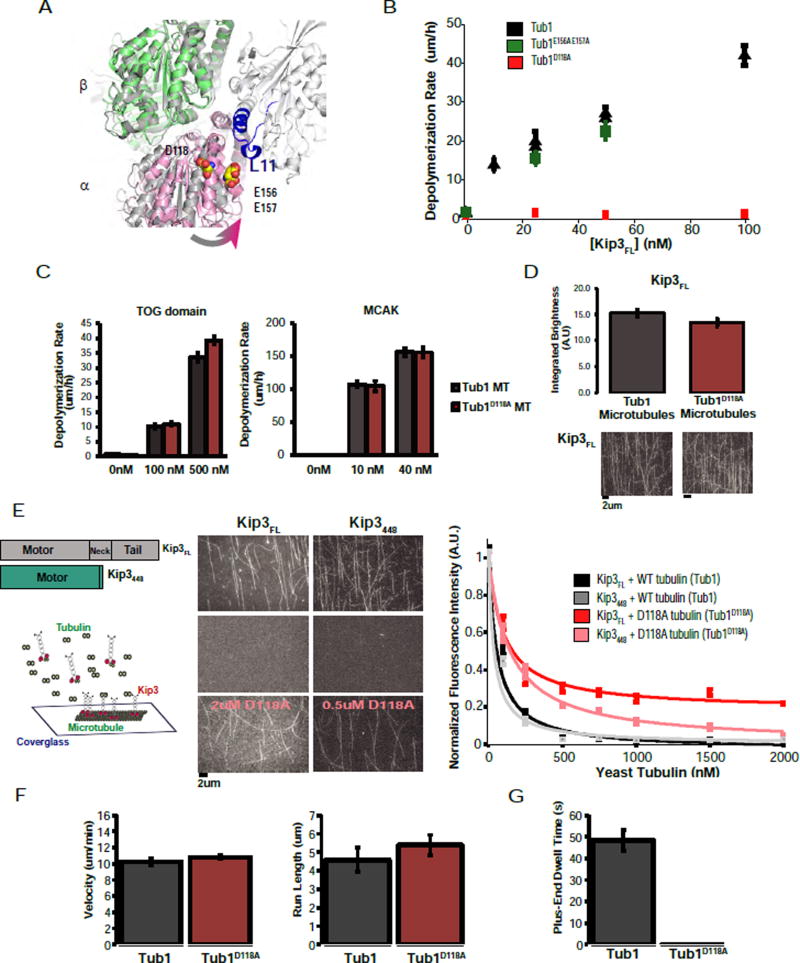
Our data demonstrate that Kip3’s Loop11 sequence is critical for depolymerase activity, for preferential binding to tubulin over microtubules, and for tubulin-specific suppression of ATPase. To find a surface of αβ-tubulin required for these effects, we focused on residues that are likely to move closer to Loop11 in the transition from straight to curved tubulin (see Methods). We used the yeast tubulin structure in this analysis because of the ability to generate and purify yeast tubulin mutants (Ayaz et al., 2012; Johnson et al., 2011). This approach identified a region of α-tubulin that contained a number of acidic residues, we chose to focus on D118 and the E156/E157 pair (Fig. 6A).
Figure 6. An α-tubulin residue specifically required for Kip3 curved tubulin-binding and depolymerase activity.
A. Structure of KHC bound to tubulin (4LNU, light grey) with Loop11 highlighted in blue. Structures of straight αβ-tubulin (3J6G, dark grey) and curved αβ-tubulin (4FFB, α in pink, β in green) are aligned using the β subunit. Selected residues on α-tubulin, D118 and E156/E157 are shown in space filling mode. These residues were chosen from a larger set of α-tubulin residues that are likely to be more proximal to Loop11 in curved tubulin relative to straight tubulin.
B. D118A yeast microtubules are insensitive to depolymerization by Kip3FL. Quantification of the depolymerization rate of the Kip3FL dimer on GTPγS-stabilized yeast microtubules (black), Tub1E156A E157A microtubules (green) and Tub1D118A microtubules (red) (mean +/− SEM; n=10–30).
C. Tub1D118A microtubules are depolymerized by MCAK or TOG domains at the same rate as control microtubules. Depolymerization rate of Tub1 (black) or Tub1D118A (red) GTPyS-stabilized yeast microtubules at various concentrations of Stu2 TOG domains (Left) or MCAK (right). Mean +/− SEM (nMT=20).
D. Tub1D118A does not affect Kip3FL microtubule binding. Integrated fluorescence intensity of Kip3FL binding to either wild-type or Tub1D118A GTPyS-stabilized yeast microtubules; imaged under TIRF microscopy. Shown are mean +/− SEM; N=5 flow cells per condition. Bottom: representative images of Kip3FL bound to Tub1 or Tub1D118A microtubules.
E. Soluble tubulin can compete for Kip3FL binding to the microtubule lattice. Left: Tubulin competition assay schematic. Middle: Representative images of 10nM Kip3 bound to microtubules at 0 nM tubulin, 2 uM tubulin and 0.5 uM, 2 uM D118A tubulin. Scale bar= 2 um. Right: Integrated fluorescence intensity of Kip3FL and Kip3448 on the microtubule lattice with or without competition from soluble tubulin. Data was normalized to fluorescence intensity at 0 nM tubulin and fit to a competition model. Shown are the mean +/−SEM, N=5 flow cells per condition.
F. Kip3FL has comparable motility on control and Tub1D118A microtubules. Shown are velocity and run length measurements (mean +/− SEM; n=100 events).
G. The Tub1D118A mutation abolishes Kip3FL‘s ability to dwell at the microtubule plus-end. Plus-end residence time (mean +/−SEM; n=100 events per condition) of Kip3FL on Tub1 (grey) and Tub1D118A microtubules (red). Scale bar: 5 um (vertical), 1 min (horizontal).
We made charge-neutralizing mutations in yeast α-tubulin to determine if either of these candidates was required for Kip3’s depolymerase activity and preferential binding to curved tubulin. Control, dimeric Kip3FL induced plus-end-specific depolymerization of GTPγS-stabilized yeast microtubules, as expected. By contrast, Tub1D118A yeast microtubules were completely resistant to depolymerization by dimeric Kip3FL (Fig. 6B; Sup. File S1 AK, AM). The absence of Kip3-induced depolymerization of Tub1D118A microtubules cannot be explained by a general stabilizing effect of the mutation because Tub1D118A microtubules were depolymerized at normal rates by the human depolymerizing kinesin-13, MCAK, or by TOG domains from yeast Stu2 (Fig. 6C; Sup. File S1 AN–AQ). Unlike Tub1D118A microtubules, Kip3FL efficiently depolymerized Tub1E156A/E157A yeast microtubules (Fig. 6B; Sup. File S1 AL), providing further evidence of the specificity of the Tub1D118A variant. Thus, the D118 residue of Tub1, which is conserved in other α–tubulins, has a critical and specific role in Kip3-mediated microtubule depolymerization.
Does the D118A mutation also eliminate preferential binding of Kip3 to unpolymerized tubulin, as predicted by our model? Limited quantities of mutant tubulin prevented direct binding measurements. However, we previously reported that soluble tubulin reduces the on-rate of Kip3FL for microtubules (Fig. S3G). Therefore, we performed fluorescence-based competition binding assays using TIRF microscopy. This approach allowed us to compare the affinity of Kip3 for control Tub1 and Tub1D118A tubulin. Consistent with the depolymerase assays, Kip3FL has a 10-fold lower affinity for soluble Tub1D118A tubulin (Fig. 6E) relative to control tubulin. Likewise, the monomeric construct, Kip3448 exhibited a 4-fold lower affinity for soluble Tub1D118A tubulin (Fig. 6E). However, in the absence of soluble tubulin, the binding of Kip3FL for GTPγS-stabilized Tub1D118A and control microtubules was similar (Fig. 6D). Therefore, the D118 residue of α-tubulin is critical for selective binding of Kip3 to tubulin.
We next examined Kip3’s motility and plus-end dwell times on control and Tub1D118A microtubules. We noted some differences in the motility of Kip3FL on GTPγS-stabilized yeast microtubules as compared with taxol-stabilized bovine microtubules: Kip3FL’s velocity was nearly 5-fold faster on yeast GTPγS-stabilized microtubules (10.3 um/min +/− 0.4, versus 2.2 um/min +/− 0.6; Fig. 6F; Sup. File S1 N–O) but its run-length was similar (4.6 um +/− 0.6 vs. 6.4 um +/− 0.5, Fig. 6G; Sup. File S1 N–O). The run-length and velocity of Kip3FL were nearly identical on Tub1D118A as compared to control yeast microtubules (Fig. 6F; Sup. File S1 N–O). This indicates that the lattice interactions of Kip3 were not significantly affected by the Tub1D118A substitution (Fig. 6D). Strikingly, although Kip3FL had a prolonged plus-end dwell time on control microtubules (48.4 s +/− 4.8, Fig 6G; Sup. File S1 N), the Tub1D118A substitution abolished the ability of Kip3FL to dwell at the plus-end (Fig. 6G). Together, these results demonstrate that the D118 residue of α-tubulin is required for Kip3 depolymerase activity because it enables Kip3, in a Loop11-dependent manner, to recognize, a curved conformation of tubulin at the microtubule end.
Discussion
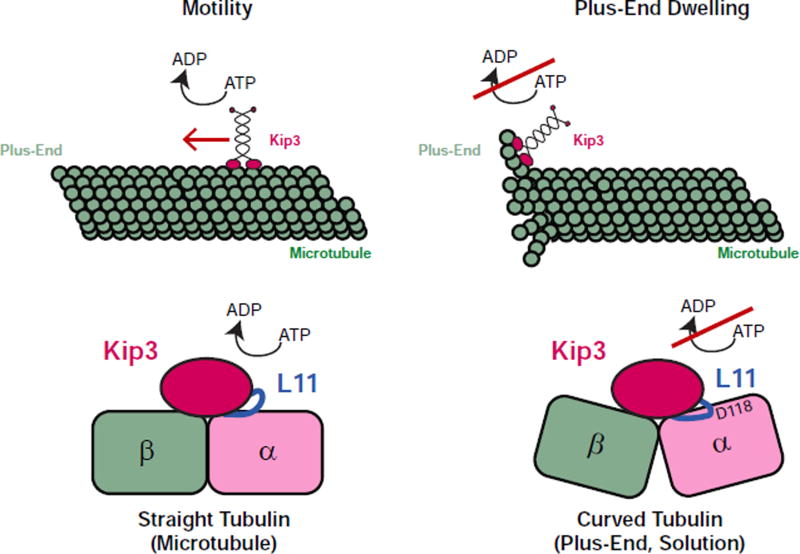
Kinesin-8s play a central role in the regulation of microtubule length and the size of microtubule structures such as the mitotic spindle. Although the high processivity of these motors is known to be essential for length-dependent microtubule disassembly (Varga et al., 2009), the depolymerase mechanism of kinesin-8s has remained unclear. Our experiments suggest that Kip3 undergoes a binding switch at the microtubule plus-end that is critical for depolymerase activity. At the plus-end, Kip3 binds with high affinity to the curved conformation of terminal tubulin subunits. This tight binding is accompanied by suppression of Kip3’s ATPase activity, resulting in a long-lived Kip3-tubulin interaction that stabilizes protofilament curvature, promoting microtubule disassembly (Fig. 7). This model is supported by our identification of Kip3’s Loop11 as an element that is necessary and sufficient for the recognition of curved tubulin. Further support comes from our identification of a residue on α-tubulin that is predicted to move closer to Kip3’s Loop11 when tubulin is in the curved conformation. Mutation of this tubulin residue mimics the Kip3 Loop11 substitution: it prevents the preferential binding of Kip3 to curved tubulin without altering Kip3 motility on the lattice. These findings explain how a motile kinesin acquired depolymerase activity and have general implications for the mechanisms by which conserved proteins can evolve novel activities.
Figure 7. Two-state binding switch model for Kip3 length dependent microtubule regulation.
Schematic of the two modes for Kip3 binding to tubulin: a motile kinesin and a depolymerase, binding straight (left) and curved (right) tubulin respectively. On the microtubule lattice, Kip3 uses ATP-dependent processive stepping to walk to the plus-end. Similar to other kinesins, straight tubulin in the microtubule lattice stimulates ATP hydrolysis. Binding of Kip3 to curved tubulin found at the microtubule plus-end and in solution suppresses ATPase activity, induces high affinity binding, and promotes microtubule catastrophe or the disassembly of stabilized microtubules. In the cartoon, the proximity of Kip3 L11 to the D118 residue on α-tubulin, both of which are required for the curved tubulin-selective interaction, is highlighted.
Kip3 has distinct tubulin conformation-selective binding modes
The observation that monomeric Kip3 can depolymerize microtubules excludes a fundamental role for motility in the depolymerase mechanism (Fig. 1C). Monomeric Kip3 also induces microtubule catastrophe at concentrations where it does not inhibit microtubule growth (Fig. 1F). Likewise, we could exclude a model where Kip3’s depolymerase activity is mediated by preferential binding to the GTP-liganded tubulin at the plus-end (Fig. 1G–H).
We therefore considered the alternative possibility that Kip3 directly recognizes the curved conformation of tubulin at the plus-end. Tubulins on the microtubule plus-end are less constrained by lattice interactions, and can therefore adopt curved states, similar to those observed in solution (Brouhard and Rice, 2014). Consistent with distinct Kip3 interaction modes for curved and straight tubulin, the affinity of Kip3 for curved tubulin is 5-fold stronger than its affinity for the straight tubulin in the lattice (Fig. 4C, D). This strong tubulin binding can explain the long plus-end dwell time of Kip3: the ~48s lifetime of the monomeric Kip3-tubulin complex measured by biolayer interferometry closely corresponds to the ~53s plus-end residence time of native Kip3 (Fig. 2E, 4B, S4F). The relationship between plus-end binding and tubulin binding is also underscored by our analysis of Kip3 and tubulin mutants discussed below. Finally, the high affinity interaction between Kip3 and tubulin markedly suppressed ATP hydrolysis by Kip3 (Fig. 5 B–D).
The strongest support for the idea that Kip3 has two conformation-selective binding modes for tubulin comes from our identification of mutations in both Kip3 and tubulin that selectively disrupt Kip3’s interaction with curved tubulin. Our loop swap experiments established that Kip3’s family-specific Loop11 sequence is required for strong curved tubulin binding: grafting kinesin-1’s Loop11 onto Kip3 abolished preferential binding to tubulin (Fig. 4B), and nearly eliminated its plus-end dwell time without significantly affecting Kip3’s interaction with the lattice (Fig. 2D, E; 4C, D); grafting Kip3’s Loop11 onto kinesin-1 demonstrated sufficiency for tubulin and plus-end binding (Fig. 3B, S5B, C; although for this construct there were effects on velocity and run length). The Loop11 sequence was also required for tubulin suppression of Kip3’s ATPase activity as well as for its depolymerase activity (Fig. 5C, D; 2F). These data are consistent with Kip3’s Loop11 being the major feature of the motor required to recognize the curved conformation of tubulin.
We also identified a residue on yeast α-tubulin that is required for curved tubulin recognition by Kip3. Using rigid body fitting, we identified candidate α -tubulin residues predicted to be closer to Kip3’s Loop11 when tubulin is in the curved conformation. Indeed, Tub1D118A microtubules were completely resistant to depolymerization by Kip3, but were depolymerized normally by the kinesin-13 MCAK, or by the TOG domains of Stu2/XMAP215 (Fig. 6C). The Tub1D118A mutation also abolished the ability of Kip3 to dwell at the plus-end (Fig. 6G), and eliminated the preferential binding of Kip3 for unpolymerized Tub1D118A dimers up to 10-fold (Fig. 6E). Importantly, the velocity and run-length of Kip3 on Tub1 and Tub1D118A microtubules were indistinguishable (Fig. 6F), indicating that Tub1D118A does not affect the interaction of Kip3 with the lattice (Fig. 6D). Thus, the tub1D118A mutation recapitulates the effect of replacing Kip3’s Loop11 sequence with that of kinesin-1. These mutations likely identify reciprocal regions of a Kip3-tubulin binding interface that is specific for the curved conformation of tubulin.
Although Loop11 is a critical element enabling selective binding of Kip3 to curved tubulin, it is not sufficient for depolymerase activity. To create a synthetic depolymerase on the backbone of kinesin-1 also required transplanting Kip3’s Loop2 segment and the Kip3 neck (Fig. 3F). The Loop2 segment contributes to depolymerase activity by enhancing Kip3’s processivity and thus plus-end flux (Fig. 2 B–C, S4C). It also enhances Kip3’s affinity for the plus-end, promoting the prolonged dwell time (Fig. 2B), necessary for depolymerase activity. Our experiments suggest that flexibility of the initial portion of the Kip3 neck is its critical feature contributing to depolymerization (Fig. S3). We speculate that flexibility between the motor heads may prevent steric clashes that might disrupt the productive interaction between a motor head and curved tubulin that favors depolymerization.
Together, our data suggest that strong and preferential tubulin binding is sufficient for Kip3 depolymerase activity. Although Kip3 can bind curved tubulin when it is liganded to either ADP or ATP (Fig. 4B, S4F), it is only in a nucleotide-liganded state with high affinity for tubulin that it is competent for depolymerization (Fig. 4B, 4C, S4F). ATP hydrolysis is not required for depolymerase activity (Fig. 5G), indicating that Kip3ATP can mediate depolymerization. The lack of depolymerase activity from Kip3ADP could either be a simple consequence of reduced affinity for tubulin or reflect more complex, qualitative differences in how Kip3 binds tubulin in its different nucleotide states. Addressing these questions in the future, as well as defining the curved tubulin-Kip3 interface in detail, will require structures of Kip3 bound to tubulin in different nucleotide states.
While this manuscript was in preparation, the structure and characterization of a ciliary kinesin-8, Kif19A, was reported (Wang et al., 2016). Broadly consistent with our conclusions, the switch II cluster of monomeric Kif19A was poorly ordered, leading to the inference of conformational flexibility in the microtubule binding surface that could enable Kif19A to bind both curved and straight tubulin. However, there are significant differences between the mechanistic model of the Wang et al. study and the model described here. Wang et al. propose a model where the Loop2 segment of Kif19A is the key structural element recognizing curved tubulin and triggering depolymerization, analogous to the Loop2-dependent depolymerase mechanism of kinesin-13s. For Kip3, our mutational analysis excludes this model and demonstrates that Loop2 mainly enhances the affinity of Kip3 for tubulin in the straight conformation: Kip3’s Loop2 segment enhances processivity but, after normalization for plus-end flux, is not required for depolymerase activity (Fig. 2B, C; S4B, C). By contrast, our work demonstrates that Kip3’s Loop11 segment (a segment that was not resolved in the Wang et al., structure) is the critical element required for sensing curved tubulin.
It has been unclear and debated whether all kinesin-8s regulate microtubule dynamics through the identical mechanism (Varga et al. 2009, Du et al. 2010; Mayr et al. 2007, Niwa et al. 2012, Walczak et al. 2013). Differing reports on the biochemical properties of different kinesin-8s could reflect differences in constructs or purification schemes. Alternatively, as elucidated here for Kip3, other kinesin-8s could have undergone subtle tuning of their catalytic cycles to optimize them for species-specific functions. It will therefore be interesting to determine the catalytic activity of Kif19A for its native substrate, the triplet microtubules of the centriole (Niwa et al. 2012).
Facile paths to generate evolutionary novelty
Our findings illustrate how small scale tuning of binding affinities and rate constants can generate strikingly different macroscopic properties for otherwise conserved proteins, providing a mechanistic basis for the acquisition of a novel protein function. Kinesin-1 is optimized for transport: its ATPase activity is strongly stimulated by the microtubule lattice and mechanochemical coupling between the motor head promotes the processivity required for transport (Cross, 2004). In keeping with the fact that these motors do not have a major role in regulating microtubule dynamics, kinesin-1 has no special affinity for the plus-end and walks off, rather than dwells, once it reaches the end (Fig. 3 B, E). By contrast, kinesin-13s are specialized, high activity microtubule depolymerases (Walczak et al., 2013). The microtubule binding surface of kinesin-13s (e.g. via Loop2) is specifically adapted to bind curved tubulin, to the extent that steric clashes between the lattice and MCAK prevent motility (Mulder et al., 2009). The ATPase cycle of MCAK/kinesin-13 has also been tuned to maximize the efficiency of microtubule disassembly. Lattice binding of MCAK suppresses its ATPase activity, generating a low affinity interaction that allows MCAK diffusion to the microtubule end. Once at the end, MCAK promotes disassembly through a high affinity interaction with curved tubulin which stimulates its ATPase activity, enabling it to recycle for additional rounds of catalysis. (Hunter et al., 2003; Wagenbach et al., 2008).
Kip3 has acquired yet another set of small scale changes in its tubulin interaction which result in a motor that is uniquely adapted to measure and regulate cellular microtubule length. Like kinesin-1, Kip3 uses ATP hydrolysis to power motility. The unusually high processivity of Kip3, critical to its length-dependent depolymerase activity, is enhanced by interactions mediated by Loop2 (presumably electrostatic interactions) and other elements of the microtubule binding interface. Once it reaches the plus-end, Kip3’s ATPase activity is altered, but differently from MCAK/kinesin-13. By contrast with MCAK, the Kip3 ATPase activity is stimulated by microtubules but suppressed by tubulin. Slowing of Kip3’s ATP turnover rate prolongs strong and preferential binding to curved tubulin and thus microtubule disassembly. Kip3’s strong microtubule end-binding (via Loop11), however, comes at the expense of poor recycling from tubulin, most likely contributing to Kip3’s lower depolymerization rate relative to MCAK. In aggregate, these features enable Kip3 to function as a length-dependent microtubule depolymerase. The emergence of novel molecular behavior from small scale effects on binding affinities and/or catalytic rates appears to be a common theme in evolution (Takeichi, 1990; Thu et al., 2014; Vendome et al., 2014), explaining, at least in part, the expansion of gene families and what can be striking functional diversity of homologous proteins.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by David Pellman (david_pellman@dfci.harvard.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Expression plasmids contained DNA sequences from Saccharomyces cerevisiae the residue numbers of the constructs used in this manuscript are provided in the key resource table. Recombinant proteins were overexpressed in S. cerevisiae (PY7490), a protease-deficient yeast strain, in synthetic complete medium (-Leu). Protein expression was induced by addition of 2% galactose. Cells were grown after induction for 18 hr. at 30°C.
METHOD DETAILS
Strains and coding sequences for recombinant proteins
Yeast strains were in the S288C genetic background except those used for protein purification, which were in the W303-1A background. A protease-deficient yeast strain was used for protein expression (Hovland et al., 1989).
All Kip3 constructs were introduced into a 2µ plasmid with a GAL1,10 promoter for expression (pRS425 backbone). A HALO tag (Promega) for fluorescent labelling was fused to the coding sequence through a GGSGGSLQ linker. The HALO tag enables covalent attachment of fluorescent dyes in a 1:1 stoichiometry. Sequence comparisons were performed using ClustalW and CobaltAlignment (NCBI) using the sequences of known kinesin-8s, kinesin-13s and human kinesin-1 as comparison. Point mutations in Kip3 were performed using QuikChange XL (Stratagene). For large domain swaps (L1, L2, L11) the region of interest was synthesized (Genewiz) with a 20bp region of homology to the recipient plasmid. For other segments (neck, L11), the region was amplified via PCR. The plasmid was then linearized using PCR and the mutated segment was introduced into the vector using In-Fusion (Clontech). The original kinesin-1 plasmid and the KHC349 CysLight plasmids were gifts from the Vale Lab. All residue numbers given in the resource table correspond to Kip3 wild-type (S. cerevisiae) or human kinesin-1, depending on which protein backbone was used for domain swaps. All Kip3438 and Kip3448 constructs harbor the dL1 and dL2 deletions [kip3 Δ(30–85; 106–134)]. All constructs were verified by DNA sequencing (Genewiz).
Plasmids to express wild-type yeast αβ-tubulin were obtained by introducing the S. cerevisiae genes encoding for Tub1p and Tub2p into the galactose-inducible 2um expression plasmids p426Gal1 and p424Gal1, respectively. Both plasmids were co-transformed into the yeast strain JEL1 for expression (Ayaz et al., 2012; Johnson et al., 2011). Plasmids to express the D118A, and E156A/E157A mutations of Tub1p (yeast α-tubulin) were made by QuikChange (Stratagene) mutagenesis, using as template an expression plasmid for the gene encoding Tub1 containing an internal His6 tag (Sirajuddin et al., 2014). A plasmid to express a Tub2p C-terminal Strep-tag II (yeast β-tubulin) was made by QuikChange (Stratagene) mutagenesis, using an expression plasmid encoding wild-type Tub2 as template and with primers designed according to the manufacturer’s instructions. The integrity of all expression constructs was confirmed by DNA sequencing.
Benomyl Resistance Assays
To assay and compare the ability of the KIP3 constructs to complement a kip3Δ strain in vivo, each construct was labelled with GFP and introduced into the KIP3 locus under the native promoter. Each strain was assayed for benomyl sensitivity. Cells were grown in liquid YPD culture at 30°C for one day, measured their OD600, normalized according to the OD600 reading, and then spotted onto YPD plates containing either 30ug/ml benomyl (Sigma) and 1% DMSO or YPD+1%DMSO in a 1:10 series dilution. Plates were placed at 30°C for two days and then imaged. Liquid cultures were aliquoted, lyzed and Kip3 protein expression was assayed by western blot.
Kinesin purification
Protein expression was induced by addition of 2% galactose for 18h at 30°C prior to harvesting cells and freezing in liquid nitrogen. Cells were disrupted by mechanical force using a blender. The yeast powder was thawed and dissolved in cold lysis buffer (50 mM NaPO4, 500 mM NaCl, 10% glycerol, 0.5 mM ATP-Mg, 40 mM imidazole, 2 mM DTT, 1% Triton X-100, protease inhibitor cocktail tablets (Roche), and 2mM PMSF; pH 7.4). Dounce homogenization was used to further lyse cells at 4°C. After centrifugation for 45,000 rpm, 45 min at 4°C, the supernatant was collected and incubated with Ni2+ sepharose (GE Healthcare) for 30 min 4°C. The beads were washed 3 times with wash buffer (50 mM NaPO4, 500 mM NaCl, 10% glycerol, 0.1 mM ATP-Mg, 25 mM imidazole, 0.1 mM DTT, protease inhibitor cocktail tablets, and 1 mM PMSF; pH 7.4). The protein was fluorescently labeled with 10uM TMR (dimer constructs) or 30uM TMR (monomer constructs, HALO-TMR; Promega) on beads for 1 h at 4°C. Elution off the Ni beads was performed at a rate of <1ml/min with elution buffer (50 mM HEPES, 300 mM NaCl, 50 mM EDTA, 5 mM MgCl2, 40mM Imidazole, 50uM ATP-Mg, and 0.1 mM DTT; pH 7.4). The protein eluates were adjusted to 250 mM NaCl and loaded onto a Uno S1 (Bio-rad) ion exchange column, equilibrated with 50 mM HEPES, 250 mM NaCl, and 50 uM ATP-Mg at pH 7.4. The protein was eluted by a linear gradient to 1M NaCl over 10 column volumes. Eluates were supplemented with 10% sucrose and centrifuged at 80,000 rpm for 5 min at 4°C. The supernatant was collected and snap-frozen in liquid nitrogen and stored at −80°C.
Yeast Tubulin Purification
Wild-type yeast αβ-tubulin was purified from conditionally overexpressing strains of S. cerevisiae using Ni-affinity and ion exchange chromatography (Ayaz et al., 2012; Johnson et al., 2011). Strep-tagged constructs were purified from conditionally overexpressing strains of S. cerevisiae using Ni-affinity, followed by affinity chromatography with the use of a StrepTactin affinity chromatography. Protein was eluted from the StrepTactin resin in storage buffer (10 mM PIPES pH 6.9, 1 mM MgCl2, 1 mM EGTA) + 50 µM GTP + 2.5 mM desthiobiotin, according to the manufacturer’s protocols. All tubulin samples were stored in storage buffer (see above) containing 50 µM GTP.
TOG Purification
TOG1(1–317) was expressed and purified as described previously (Ayaz et al., 2014; Ayaz et al., 2012). TOG1317 was overexpressed in E. coli and purified by Ni-affinity and gel filtration chromatography. TOG1 was stored in RB100 (25 mM Tris pH 7.5, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA). MCAK was purchased from Cytoskeleton and resuspended according to the manufacturer’s protocol.
Western blotting
Yeast cells were grown to OD 0.6 and washed with PBS. The cell pellet was lysed by TCA (trichloroacetic acid, MP Biomedicals) and subsequent bead beating: 250ul of 20% TCA was added to the cell pellet. After 5 min on ice, 250ul of glass beads were added and samples were vortexed in bead beater 3 times for 1 minute, with 1 minute breaks at 4C. Samples were then spun at 13,300 rpm for 10 minutes at 4C and the pellet was washed with 750ul of 100% ice cold ethanol. Washed pellets were resuspended in 40ul 1M Tris HCl pH=8 and 80ul 2× SDS sample buffer and boiled for 5 minutes. The samples were diluted 1:3 in 1× SDS sample buffer and 15ul of the sample was loaded in an SDS PAGE gel (BioRad any Kd mini protean TGX). The gel was run at 0.2A in Tris-glycine-SDS buffer for an hour. Dry transfer to a PVDF membrane was carried out in an iBlot 2 (Thermofisher). The membrane was cut horizontally at the 50kD protein standard mark. The PVDF membrane was blocked in 5% milk diluted in TBST for 30 minutes at room temperature and incubated with primary antibody. Rabbit anti-GFP monoclonal antibody (Abcam, E385, ab32146) was used in a 1:3,000 dilution. Mouse anti-actin monoclonal antibody (Abcam, ab824) was used in a 1:3,000 dilution. Membranes were washed 5 times for 5 minutes in TBST and then incubated with secondary antibody for 30 minutes (ECL, HRP linked, GE Healthcare). Membranes were then washed 5 times for 5 minutes with TBST. Autoradiography film (Denville Scientific) was used to image the membranes with an exposure time of 1 minute.
Preparation of stabilized microtubules (Bovine Tubulin)
Tubulin was purified from bovine brains by two rounds of assembly and disassembly as described in (Castoldi and Popov, 2003). HiLyte 488, HiLyte 647, and biotin -labeled tubulin were purchased from Cytoskeleton. Motility assays were carried out using biotinylated, fluorescently labelled, Taxol-stabilized microtubules (10% labelled tubulin). BRB80 (80 mM PIPES-K+, 1 mM EGTA, 1 mM MgCl2, pH 6.9) supplemented with 1mM GTP, 1mM MgCl2, 1mM DTT and 10% DMSO was incubated with a tubulin mix (20 µM tubulin, 2 µM fluorescein-labeled tubulin, 2 µM biotin-labeled tubulin) at 37C for 1hr. Taxol was added to a concentration of 20 uM and was always present in assays with Taxol-stabilized microtubules.
To prepare GMPCPP-stabilized microtubules, microtubule seeds were first grown by incubating seed mix (20 uM tubulin, 0.2 uM biotin-tubulin, 5 uM fluorescein-tubulin, and 1 mM GMPCPP-Mg in BRB80 pH 6.9) at 37°C for 30 min. The elongation mix was made by diluting a seed mix aliquot at 1:10 ratio with BRB80 and 0.5 mM GMPCPP-MgCl. The seeds were added to the elongation mix at a 1:40 ratio. GMPCPP-stabilized microtubules were elongated for 3 h at 37°C before use.
Single Molecule Assays (Motility)
Flow chambers were prepared by taping double sided tape onto glass microscope slides (Fisher Scientific) and placing a microscope cover glass on top creating a channel a volume of ~10–15ul. Chambers were incubated sequentially with the following solutions, interspersed with washes with BRB80 + 20uM Taxol: 0.5mg/ml biotinylated-BSA (Vector Laboratories), 0.5mlg/ml streptavidin (Sigma), a 1:100 dilution of taxol-stabilized microtubules in BRB80+taxol. The last wash was performed with Reaction Buffer (BRB80 pH 7.2 with 0.5 mg/mL k-casein, 5% glycerol, 2 mM ATP-Mg, 20 uM Taxol and 100mM KCl, unless otherwise specified). Kip3 was diluted in reaction buffer at 4C and introduced in the assay chamber in reaction buffer with an oxygen scavenging mix (0.2 mg/mL glucose oxidase, 0.035 mg/mL catalase, 25 mM glucose and 1mM DTT). For end-residence time experiments as a function of total motor concentration, unlabeled motor was diluted in reaction buffer and this was then mixed with TMR-labelled motor(<0.05 nM), introduced into the flow-cell and imaged. The TMR-labeled motors and HiLyte-488-labelled microtubules were imaged at room temperature in a Nikon Ti-E microscope with TIRF-illuminator, perfect-focus system with a 100× 1.49 NA Nikon oil immersion TIRF objective and 488 nm and 561 nm lasers (Agilent module). Images were recorded on an Andor DU-897 (EMCCD) camera at 100ms exposure and EMCCD gain of 150.
Motor velocities and run lengths were analyzed using a software developed in Labview (National Instruments) for selecting microtubules, creating kymographs and tracking the position and brightness of motor tracks. Software was validated using manual analysis of kymographs in ImageJ. Matlab was used to compile the data and perform distribution fittings for velocities (normal distribution), run lengths (exponential distribution) and plus-end dwell times (exponential distribution), using Scott’s rule to determine bin number in the Matlab distribution fitting tool, similarly to Su et al. 2011, 2013.
Kip3 motility assays using microtubule from yeast tubulin
Wild-type or mutant yeast microtubules were stabilized by GTPγS, and flow chambers were prepared with StrepTactin-HRP. Pre-formed, GTPγS-stabilized wild-type or D118A yeast MTs were introduced into the chambers and allowed to incubate for 10 minutes, followed by a wash with BRB80 to remove unbound MTs. Solutions containing 5 pM Kip3 in imaging buffer (BRB80 + 500 µM GTPγS + 0.5 mg/mL k-casein + 100 mM KCl + 2 mM ATP + 1 mM DTT+ antifade reagents) were introduced into the chamber immediately prior to data collection.
Kip3 motility reactions were imaged by total internal reflection fluorescence (TIRF) microscopy using a Nikon Ti Eclipse microscope, with a TIRF Apo 60×/1.49 objective lens and a spectral laser system using a 561 nm, 50mW solid-state laser. Metamorph 7.8 was used to control the microscope. TIRF images of Kip3 signal were recorded every 2 min for 10–20 minutes. Kip3 motility and dwell times were measured manually using ImageJ. Kymographs of Kip3 runs and dwells were taken over the time course of the movie and were used to determine average rate of motility. Kip3 dwell times and run lengths were measured and analyzed using Matlab, similarly to the bovine microtubule data.
Competition Assay to determine the affinity of Kip3 variants for tubulin
Flow-chambers were prepared as described for the motility assays with the exception that the reaction buffer for these experiments was supplemented with 1mM GTP. Bovine tubulin was diluted in reaction buffer+GTP to the desired final concentration. TMR-labelled motors and bovine tubulin were pre-mixed and incubated for 3 minutes on ice. The sample was brought to room temperature prior to introduction into the flow chamber. TMR-labelled motors and fluorescently labelled microtubules were imaged using TIRF microscopy as described in the motility assays. For landing rate quantification, motors were analyzed independently at various tubulin concentrations. Landing events were determined from kymographs and the rate was calculated as the number of events per micron of microtubule per nM motor per second. The data was fit in MATLAB (Levenberg-Marquardt) to the equation k=kmax/([Tubulin]/Ki+1); where Ki represents the relative affinity to the competitor.
Yeast Tubulin Competition Assays
Wild-type or mutant yeast microtubules stabilized by GTPγS were prepared as described above. Samples containing either free wild-type yeast tubulin or free D118A mutant yeast tubulin were prepared at increasing concentrations with a constant concentration of monomeric Kip3-TMR in imaging buffer (see yeast microtubules motility or depolymerization assay). Samples were allowed to incubate on ice for 15 minutes.
Flow chambers containing stabilized microtubules were prepared as described above. Samples containing monomeric Kip3-TMR with variable amounts of free tubulin were flowed into the chamber and reactions were monitored by TIRF, as described above. Multiple images of MTs were recorded by both TIRF conditions (561 laser) and with differential interference contrast microscopy (DIC) using DIC prisms, using illumination at 550 nm by inserting a bandpass filter of 550/100 nm (Olympus) in the light path, to ensure the presence of microtubules in the absence of Kip3-TMR coating. MT fluorescence intensities were measured manually using ImageJ.
TIRF assay for Kip3 Lattice binding
Fluorescence intensity measurements to compare the relative binding of Kip3 to Taxol-stabilized (GDP-like) and GMPCPP-stabilized (GTP-like) microtubules were performed similarly to motility assays using TIRF imaging. Taxol-stabilized microtubules were grown with HiLyte-488 –labelled tubulin and GMPCPP-stabilized microtubules were grown with HiLyte-647-labelled tubulin. Taxol-stabilized and GMPCPP-stabilized microtubules were introduced into the same flow chamber with the presence of 20uM taxol in the reaction buffer. Taxol was added for microtubule stability as Kip3 is unable to depolymerize taxol-stabilized microtubules. Kip3 was introduced at the desired concentrations and imaged as described in the motility assays. Analysis of background-subtracted Kip3 brightness on the microtubule lattices was performed in ImageJ. The ratios of Kip3 fluorescence intensity on the GMPCPP lattice relative to the GDP-like lattice were calculated using all of the microtubules in a field of view. The ends (600 nm from the end) of stabilized or dynamic microtubules were excluded from the analysis to avoid end-specific effects.
The effect of Kip3 variants on dynamic microtubules
Assays were carried out similarly to the experiments described in (Su et al., 2013). HiLyte 647-labeled GMPCPP seeds were immobilized to the coverslips in the flow-cells as described for the motility assay. Tubulin growth mix (14 uM tubulin, 0.7 uM HiLyte 488-labeled tubulin, 1 mM GTP, 2 mM ATP-Mg, 1 mg/ml k-casein, 0.1% methyl cellulose and an oxygen scavenging system described in the motility assay) was introduced into the chamber. Microtubules were allowed to grow for 5 min at room temperature prior to flowing in motors diluted in the same growth mix. Flow cells were imaged in a Nikon Ti-E microscope equipped with a temperature enclosure at 37C, a TIRF-illuminator, perfect-focus system, 100× 1.49 NA Nikon oil immersion TIRF objective and 491 nm, 561 nm and 642 nm lasers (spectral applied research module). Images were recorded on a Hamamatsu ImagEM (EMCCD) camera at 100ms exposure and EMCCD gain of 150. Image analysis was performed as previously described in (Su et al., 2013). Pauses were included in the calculation of growth rates.
Depolymerization Assays
Experimental conditions were similar to those for the motility assay, with the following modifications: GMPCPP-stabilized microtubules were used instead of taxol-stabilized microtubules. Taxol was not included in the reaction mixtures. Wide-field images were acquired every 12 s for 12 min on an upright Zeiss AxioImager M1 microscope using a 63× objective and a Photometrics Cool Snap HQ camera (similar to the conditions used in Su et al., 2013). Images were analyzed first by manually selecting microtubules using the same Labview software described in the motility assays. A custom matlab software was used to automatically measure microtubule length in each frame. The coordinates of the traced microtubules were exported into Matlab. The region of interest was centered at the centroid of the microtubule and expanded to a rectangular region comprising the entire length of the trace with an additional 3 pixels on each side. “Open” and “close” morphological operations and ‘canny’ edge detection were performed using in-built matlab functions in the vision package. The final object was thinned to a line and its length exported. All microtubule images and the final objects were visually inspected. Depolymerization rates were calculated as changes in microtubule length over time.
For experiments that required normalization of motor flux to the plus-end, the on-rate, run-length and velocity were first measured as described in the section: Single Molecule Assays, Motility. The on-rate was calculated as the number of motors landing per micron of measured microtubule lattice per second, per input motor concentration. With these parameters we then calculated an expected flux of motors to the plus-end: Nmotors α kon*RL*v/LMT. Where kon represents the motor on-rate, RL the run length of each construct, v the velocity and LMT the microtubule length. We then directly measured the motor flux-rate by TIRF microscopy and experimentally adjusted the protein input to obtain a given flux-rate, similarly to the assays performed in Su et al. 2011. The protein concentration at which control-level flux-rates were obtained was then used as the protein input in the depolymerase assay.
Yeast Microtubule Depolymerization Assays
Wild-type and mutant yeast αβ-tubulin were polymerized in the presence of 500 µM GTPγS in assembly buffer (100 mM PIPES pH 6.9, 10% glycerol, 2 mM MgSO4, 0.5 mM EGTA). The mixture was incubated for 90 minutes at 30°C.
Flow chambers were prepared as described (Gell et al., 2010). His-Tag Antibody (1:200, Millipore) or StrepTactin-HRP Conjugate (1:200, Biorad) was incubated in the chamber for 10 minutes, blocked by incubation with 1% Pluronic F-127 in BRB80 for 5 minutes, and washed with BRB80. Pre-formed, GTPγS-stabilized wild-type or D118A yeast MTs were then introduced into the chamber and allowed to incubate for 10 minutes, prior to a BRB80 wash to remove unbound MTs. Solutions containing a range of Kip3, MCAK or TOG1 concentrations in imaging buffer (BRB80 + 500 µM GTPγS + 0.1 mg/mL BSA + 100 mM KCl (Kip3) + 2 mM ATP (Kip3) + antifade reagents, without the addition of β-mercaptoethanol (Gell et al., 2010) were introduced into the chamber immediately prior to data collection.
MT depolymerization reactions were imaged by total internal reflection fluorescence (TIRF) microscopy using an Olympus IX81 microscope with a TIRF ApoN 60×/1.49 objective lens, a 561 nm 50 mW solid-state laser and Hamamatsu ORCA-Flash2.8 CMOS camera (Olympus). Temperature was maintained at 30°C using a WeatherStation temperature controller with enclosure fit to the microscope’s body. Micro-Manager 1.4.16 was used to control the microscope. Images of MT shrinking were recorded every 60 seconds for about 10 minutes. MT length was measured manually using ImageJ. Lengths of MT’s were taken over the time course of the movie and were used to determine rate of depolymerization over an hour time span.
Steady State ATPase Assays
ATPase assays were performed using the EnzChek phosphatase assay kit (Molecular probes, ThermoFisher Scientific). Assays with Kip3 were performed in ATPase buffer (50 mM HEPES pH 7.5, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 2mM ATP, 5% sucrose, 0.2mg/ml k-casein, 1mM GDP) with 0–10 uM tubulin or taxol-stabilized microtubules. ATPase assays with KHC were carried out in a modified ATPase buffer (50 mM HEPES pH 7.5, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 2mM ATP, 5% sucrose, 0.2mg/ml k-casein, 1mM GDP). All experiments, with either tubulin or microtubules, were carried out at the same final salt concentration, irrespective of the volume. For assays including taxol-stabilized microtubules, taxol was present in the reaction at 20uM. The absence of tubulin polymerization was verified at the highest tubulin concentration by adding fluorescently-labelled tubulin and imaging the sample with TIRF microscopy. No microtubules were observed in the assay conditions used. Tubulin concentrations were calculated by denaturation with 6M GuHCl and protein absorbance at 280nM and by SDS-PAGE gels run against tubulin standards. A SpectraMax plate reader (Molecular Devices) was used to monitor the reaction (Pi in solution) at OD360 every 12 s for 10 min. Data were weighted by 1/SD (standard deviation) and fitted to the Michaelis Menten equation using Matlab.
Gel Filtration
Gel filtration was performed using purified proteins in a Superdex S200 10/300 GL (GE Healthcare) running on an AKTA P-960 (GE Healthcare). The column was equilibrated using ATPase buffer without GDP, k-casein or added sucrose. Bio-Rad gel filtration protein standards were used as reference. The sample or the protein standards were injected in a volume of 200 ul, run at a flow rate of 0.5 ml/min and collected in 200 ul fractions. The fractions were run on an SDS-PAGE gel (“Any Kd”,Bio-Rad) and coomassie-stained. After destaining, protein bands were scanned, background subtracted and quantified using imageJ.
Tubulin Affinity Measurements (biolayer interferometry)
Tubulin affinity measurements were performed in an Octet Red 384 instrument (ForteBio), all assays were performed in ATPase buffer without added k-casein, with 10% sucrose, in ATP, unless otherwise stated. Monomeric Kip3 was immobilized on an anti-penta-HIS biosensor chip (HIS1K, fortebio) at an input concentration of 100nM. Unbound motor was washed with ATPase buffer. Tubulin was diluted in ATPase buffer and introduced at a range of concentrations (10-fold the expected KD) of the motor construct. Upon reaching steady state, the sensor was automatically switched to a reaction buffer-only condition to measure tubulin off-rates. The data was analyzed using ForteBio software using both the measured kon and koff from a range of concentrations and steady state data to obtain the KD.
Bent microtubule Assay
Taxol-stabilized microtubules were introduced into a flow chamber as described in the motility assay section. Buffer flow (BRB80+Taxol) was maintained and later reversed during the settling of the microtubules onto the cover glass. This resulted in microtubule bending. TMR-labelled motors were introduced into the flow chamber containing bent microtubules and imaged as described above. Microtubules were first manually traced in a custom labview program (National Instruments), the microtubule coordinates were then exported and analyzed in Matlab. Integrated fluorescence intensity, over 30s, of TMR-labelled motors as a function of absolute microtubule curvature was quantified using a custom software in Matlab based on cubic B-splines, similarly to (Bechstedt et al., 2014). The Matlab code created a cubic B-spline onto the trace of the microtubule and calculated the local curvature and the background-subtracted fluorescence intensity. The values were then averaged over a user-defined number of bins. The fluorescence intensity was then plotted as a function of local curvature. Absolute values of intensity are a function of laser power, alignment and camera gain. Camera gain was chosen to maximize the sensitivity and dynamic range of the camera as the experiment was performed across different motor concentrations.
Homology model
Sequence comparisons were performed using ClustalW and Cobalt Alignment (NCBI) using the sequences of known kinesin-8s, kinesin-13s and human conventional kinesin as comparison obtained from the NCBI database. The structural homology model of Kip3 was constructed in the UCSF Chimera program (v. 1.9) interface to Modeler. The Kip3 homology model was constructed using as a reference the structure of Kif18A (3LRE) and KHC on the tubulin dimer (1MKJ, 4LNU, 4HNA).
The comparison between curved and straight tubulin was performed in PyMol by aligning the carbon residues (1:180) on β-tubulin from 4FFB (curved tubulin) and 3J6G (straight tubulin). The crystal structure of kinesin in ADP.AlFx bound to tubulin (4LNU) was then aligned by the carbon residues on β-tubulin. The Loop 11 region from kinesin-1 was replaced in silico with the Loop 11 segment from Kip3. Acidic residues on α-tubulin that were within 15 Å of Loop 11 amino acids when tubulin was in a curved rather than a straight conformation were selected for mutagenesis.
Stopped Flow experiments
Stopped-flow setup
Stopped-flow measurements were carried out on an Applied Physics SX20 spectrofluorometer using 356 nm excitation and a HQ480SP emission filter, as previously described (Chen et al., 2015). For tubulin- and MT-simulated mantADP release assays, 0.1–0.3 µM monomeric motors were first incubated with 0.5 µM mantADP for 5 minutes to exchange nucleotide, and were then flushed against varying concentrations of tubulin or MT plus 2 mM ADP to chase off the bound mant-nucleotide. In MT-induced mantADP release assays for WT motors, the data were fit to a bi-exponential, where the fast phase results from motors binding to the MT and the slow phase results from motors binding to residual tubulin in solution. Otherwise, data were fit to a single exponential. Because the observed mantADP dissociation resulted from both tubulin-bound and free motors, the observed rate constant for tubulin-triggered mantADP release, kobs was fit to the function:
where and correspond to the mADP off-rate on tubulin and in free solution, respectively, and K0.5 denotes the apparent MT affinity. In mantATP exchange assays, 1 µM motors were first incubated with 2.5 µM tubulin, and this complex was flushed against varying concentrations of mantATP. Fluorescence traces were fit to a bi-exponential, with a fast phase corresponding to the mantATP exchange rate for apo motors, and a slow phase corresponding to the cold ADP release proceeding mATP exchange. The observed rates for the fast phase were fit to estimate the nucleotide on- and off-rates, as determined previously (Chen et al., 2015):
QUANTIFICATION AND STATISTICAL ANALYSIS
Motor velocities and run lengths on bovine microtubules were analyzed using a software developed in Labview (National Instruments) for selecting microtubules, creating kymographs and tracking the position and brightness of motor tracks. The software displays the movie using the IMAQ vision package and prompts the user to manually trace the microtubules with a multi segmeneted-line onto the microtubule channel. The trace is used to create a kymograph by summing over a width of 8 pixels, centered on the user-defined line, over all movie frames. Motor tracks are manually selected from the kymographs and the coordinates of these traces are corrected by fitting them to the brightest points within 5 pixels of the region of interest. The brightest points are found using the Ni Vision module “Peak-Valley Detector” using a pre-set threshold of 100 to ensure a selection above background. Since the user has defined the motor tracks a priori, this acts as a refinement step to obtain the trajectory of the motor. Additionally, as an orthogonal approach, we measured the particle velocity and run length directly in the movies. Tracks were automatically traced based on the particle brightness over a 6 pixel square region, with the Ni Vision module “Peak-Valley Detector” (threshold of 100–150). The software was validated using manual analysis of kymographs in ImageJ. For assays with yeast microtubules, Kip3 motility and dwell times were measured manually using ImageJ. Kymographs of Kip3 runs and dwells were taken over the time course of the movie and were used to determine average rate of motility. Matlab was used to compile the data and perform distribution fittings for velocities (normal distribution), run lengths (exponential distribution) and plus-end dwell times (exponential distribution), using Scott’s rule for bin number, (Su et al., 2013). Statistical tests were performed using a two-tailed un-paired t-test. For tubulin competition assays the number of motors landing on a microtubule were manually counted from kymographs and for monomeric Kip3 assays, fluorescence intensities were measured manually from background subtracted images using ImageJ. Fits of ATPase Data to the Michaelis Menten equation were weighted by 1/SD (standard deviation) and were performed using the Levenberg-Marquardt fitting function in Matlab.
Supplementary Material
Related to Figures 1–6, S1, S3, S4.
A–O. Histograms showing the distribution of the motility data: run length (left), velocity (middle) and plus-end dwell time (right) for the indicated constructs, imaged at single molecule conditions on taxol-stabilized microtubules (see methods). Red line represents fits of an exponential (run length, plus-end dwell time) or normal (velocity) distribution to the data.
P-AQ. Histograms showing the distribution of the depolymerization rate of GMPCPP-stabilized microtubules at the indicated concentrations, for each motor construct.
Highlights.
A tubulin binding switch enables length-dependent microtubule disassembly by Kip3
Tubulin binding suppresses Kip3 ATPase activity, promoting depolymerase activity
Loop11 sequence in Kip3 is necessary and sufficient for curved tubulin binding
Kip3 disassembly-resistant microtubules support the two-state binding switch model
Acknowledgments
We are grateful to the Reck-Peterson Laboratory and the NIC@HMS imaging center for the usage of TIRF microscopes. We acknowledge the center for molecular interactions at HMS-BCMP for the use of the Octet system. We acknowledge A. Leschziner and R. Hernandez and VJ for fruitful discussions. D. Pellman is supported by Howard Hughes Medical Institute and a National Institute of Health grant (GM61345). HAS was supported by an international fellowship from the Howard Hughes Medical Institute. LMR is supported by the NIH (GM098543) and by the NSF (MCB 1054947 and 1615938). EAG was supported by NIH T32 GM008297, and by an NSF Graduate Research Fellowship, Grant No. 2014177758. WH is supported by National Institute of Health grant GM076476.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
HAS and D.P. conceived the project and wrote the manuscript, with input from the other authors. LR and HAS designed yeast tubulin experiments. LR performed tubulin structural comparisons. EG performed and optimized the assays with yeast tubulin and analyzed the data with LR. GYC and WH designed and performed nucleotide kinetic experiments, discussed and analyzed the data. ES assisted with cloning and performed in vivo and ATPase experiments. HAS performed the remainder of the experiments and analyzed the data.
References
- Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA. An ATP Gate Controls Tubulin Binding by the Tethered Head of Kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:αβ-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Munyoki S, Geyer EA, Piedra F-A, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, Rice LM. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. eLife. 2014;3:e03069. doi: 10.7554/eLife.03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt S, Lu K, Brouhard Gary J. Doublecortin Recognizes the Longitudinal Curvature of the Microtubule End and Lattice. Current Biology. 2014;24:2366–2375. doi: 10.1016/j.cub.2014.08.039. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM. The contribution of αβ-tubulin curvature to microtubule dynamics. The Journal of Cell Biology. 2014;207:323–334. doi: 10.1083/jcb.201407095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization–depolymerization in a high-molarity buffer. Protein Expression and Purification. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- Cross RA. The kinetic mechanism of kinesin. Trends in Biochemical Sciences. 2004;29:301–309. doi: 10.1016/j.tibs.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Chen G-Y, Arginteanu DFJ, Hancock WO. Processivity of the Kinesin-2 KIF3A Results from Rear Head Gating and Not Front Head Gating. Journal of Biological Chemistry. 2015;290:10274–10294. doi: 10.1074/jbc.M114.628032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The Kinesin-8 Kif18A Dampens Microtubule Plus-End Dynamics. Current Biology. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Duellberg C, Cade NI, Holmes D, Surrey T. The size of the EB cap determines instantaneous microtubule stability. eLife. 2016;5:e13470. doi: 10.7554/eLife.13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel CT, Howard J. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. The EMBO Journal. 2011;30:3928–3939. doi: 10.1038/emboj.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila Kinesin-like Protein KLP67A Is Essential for Mitotic and Male Meiotic Spindle Assembly. Molecular Biology of the Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. Chapter 13 - Microtubule Dynamics Reconstituted In Vitro and Imaged by Single-Molecule Fluorescence Microscopy. In: Leslie W, John JC, editors. Methods in Cell Biology. Academic Press; 2010. pp. 221–245. [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Webb MR, Brune M, Johnson KA. Pre-Steady-State Kinetics of the Microtubule-Kinesin ATPase. Biophysical Journal. 1995;68:357s–357s. [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic Volume Modulates Spindle Size During Embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length Control of the Metaphase Spindle. Current Biology. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hackney DD. Kinesin ATPase: rate-limiting ADP release. Proceedings of the National Academy of Sciences. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in Cytoplasmic Volume Are Sufficient to Drive Spindle Scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. The Journal of Cell Biology. 2014;206:385–393. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland P, Flick J, Johnston M, Sclafani RA. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The Kinesin-Related Protein MCAK Is a Microtubule Depolymerase that Forms an ATP-Hydrolyzing Complex at Microtubule Ends. Molecular Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM. Design, Overexpression, and Purification of Polymerization-Blocked Yeast αβ-Tubulin Mutants. Biochemistry. 2011;50:8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Wilbur Jeremy D, McNally Francis J, Nédélec François J, Heald R. Katanin Contributes to Interspecies Spindle Length Scaling in Xenopus. Cell. 2011;147:1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. Subcellular Size. Cold Spring Harbor Perspectives in Biology. 2015;7:a019059. doi: 10.1101/cshperspect.a019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer Sebastian P, Cade Nicholas I, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 Accelerates Two Conformational Transitions Important for Microtubule Maturation and Dynamics. Current Biology. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU. The Human Kinesin Kif18A Is a Motile Microtubule Depolymerase Essential for Chromosome Congression. Current Biology. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mulder AM, Glavis-Bloom A, Moores CA, Wagenbach M, Carragher B, Wordeman L, Milligan RA. A new model for binding of kinesin 13 to curved microtubule protofilaments. The Journal of Cell Biology. 2009;185:51–57. doi: 10.1083/jcb.200812052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Developmental Cell. 2012;23:1167–1175. doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A Common Mechanism for Microtubule Destabilizers--M Type Kinesins Stabilize Curling of the Protofilament Using the Class-Specific Neck and Loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. The EMBO Journal. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Jülicher F. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol. 2013;15:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rischitor PE, Konzack S, Fischer R. The Kip3-Like Kinesin KipB Moves along Microtubules and Determines Spindle Position during Synchronized Mitoses in Aspergillus nidulans Hyphae. Eukaryotic Cell. 2004;3:632–645. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol. 2015;17:1422–1434. doi: 10.1038/ncb3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. Journal of Cell Science. 2010;123:767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment. Developmental Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta Mohan L, Jr, Pereira-Leal José B, Reck-Peterson Samara L, Pellman D. Mechanisms Underlying the Dual-Mode Regulation of Microtubule Dynamics by Kip3/Kinesin-8. Molecular Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti S-C, Tan L, Darst Seth A, Kapoor Tarun M. Marking and Measuring Single Microtubules by PRC1 and Kinesin-4. Cell. 2013;154:377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: A Molecular Family Important in Selective Cell-Cell Adhesion. Annual Review of Biochemistry. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Thu Chan A, Chen Weisheng V, Rubinstein R, Chevee M, Wolcott Holly N, Felsovalyi Klara O, Tapia Juan C, Shapiro L, Honig B, Maniatis T. Single-Cell Identity Generated by Combinatorial Homophilic Interactions between Protocadherins. Cell. 2014;158:1045–1059. doi: 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Vale RD. Controlling Kinesin by Reversible Disulfide Cross-Linking. J Cell Biol. 2000;151:1081–1092. doi: 10.1083/jcb.151.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 Motors Act Cooperatively to Mediate Length-Dependent Microtubule Depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, Kaczynska A, et al. Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4175–E4184. doi: 10.1073/pnas.1416737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenbach M, Domnitz S, Wordeman L, Cooper J. A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. The Journal of Cell Biology. 2008;183:617–623. doi: 10.1083/jcb.200805145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. Microtubule-Depolymerizing Kinesins. Annual Review of Cell and Developmental Biology. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- Wang D, Nitta R, Morikawa M, Yajima H, Inoue S, Shigematsu H, Kikkawa M, Hirokawa N. Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. eLife. 2016;5:e18101. doi: 10.7554/eLife.18101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, Troxell CL, McIntosh JR. Two Related Kinesins, klp5 and klp6, Foster Microtubule Disassembly and Are Required for Meiosis in Fission Yeast. Molecular Biology of the Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figures 1–6, S1, S3, S4.
A–O. Histograms showing the distribution of the motility data: run length (left), velocity (middle) and plus-end dwell time (right) for the indicated constructs, imaged at single molecule conditions on taxol-stabilized microtubules (see methods). Red line represents fits of an exponential (run length, plus-end dwell time) or normal (velocity) distribution to the data.
P-AQ. Histograms showing the distribution of the depolymerization rate of GMPCPP-stabilized microtubules at the indicated concentrations, for each motor construct.