Abstract
Life as we know it, simply would not exist without DNA replication. All living organisms utilize a complex machinery to duplicate their genomes and the central role in this machinery belongs to replicative DNA polymerases, enzymes that are specifically designed to copy DNA. “Hassle-free” DNA duplication exists only in an ideal world, while in real life, it is constantly threatened by a myriad of diverse challenges. Among the most pressing obstacles that replicative polymerases often cannot overcome by themselves, are lesions that distort the structure of DNA. Despite elaborate systems that cells utilize to cleanse their genomes of damaged DNA, repair is often incomplete. The persistence of DNA lesions obstructing the cellular replicases can have deleterious consequences. One of the mechanisms allowing cells to complete replication is “Translesion DNA Synthesis (TLS). TLS is intrinsically error-prone, but apparently, the potential downside of increased mutagenesis is a healthier outcome for the cell than incomplete replication. Although most of the currently identified eukaryotic DNA polymerases have been implicated in TLS, the best characterized are those belonging to the “Y-family” of DNA polymerases (pols η, ι, κ, and Rev1), which are thought to play major roles in the TLS of persisting DNA lesions in coordination with the B-family polymerase, pol ζ. In this review, we summarize the unique features of these DNA polymerases by mainly focusing on their biochemical and structural characteristics, as well as potential protein-protein interactions with other critical factors affecting TLS regulation.
Keywords: translesion synthesis, replicative bypass, DNA polymerase families, PCNA, ubiquitin, protein interaction network, mutagenesis
Graphical Abstract

Introduction
According to a very simplistic perception of life, the proper assembly of each living organism from a set of molecules relies on an instruction manual encoded in DNA, a double-stranded helix composed of a string of nucleotides. The information written in this manual must be accurately copied before being transferred from generation to generation. The mechanism that ensures the preservation of genetic information is based on the strict specificity of nucleotide base pairing (A with T and G with C) across the shared axis of the double helix. Watson and Crick, who are credited with discovering the molecular structure of DNA, mentioned that specific pairing itself “immediately suggests a possible copying mechanism for the genetic material” (Watson and Crick, 1953). Moreover, they even assumed that base complementarity is the driving force strong enough to guide incoming nucleotides for pairing with the parental DNA strand without the assistance of any enzyme. However, as with any other chemical reaction within a cell, DNA replication depends on the help from an extra set of hands. In fact, an elaborate machinery is required to accomplish the duplication of genomic DNA and the role of a “right-hand” in this process belongs to replicative DNA polymerases, the enzymes specifically designed to copy DNA with high efficiency and accuracy. This task is challenged by a large variety of external and internal damaging agents that constantly threaten DNA stability and obscure the ability of DNA polymerases to read the information written in DNA. In order to overcome such obstacles, cells rely on multiple repair mechanisms that have evolved to restore the integrity of damaged sequences. When repair is incomplete, i.e., when lesions are refractory to repair, or their number is simply overwhelming, cells utilize damage tolerance mechanisms in order to survive. Similar to DNA replication, both DNA repair and damage tolerance depends on DNA polymerases each playing a specific role in copying or replacing modified genomic sequences.
Virtually all of the eukaryotic DNA polymerases characterized in vitro appear able to traverse past damaged DNA sites in a process called translesion DNA synthesis (TLS), although with vastly different efficiencies, and fidelities, and often with the help of protein partners. High fidelity replicative polymerases are the least adept to perform this function. In contrast, reasonably efficient bypass synthesis in eukaryotes has been demonstrated for the A-family polymerases (pols θ and ν), X-family polymerases (pols β, λ, and μ) and the newest addition to the TLS polymerase family, PrimPol. However, within the TLS polymerase superfamily, there is a group of enzymes that appear to be specifically designed to accomplish the lesion bypass task most effectively. The majority of these enzymes comprise the Y-family of DNA polymerases (Ohmori et al., 2001). Mammalian and yeast representatives of the eukaryotic Y-family DNA polymerases, i.e., pols η, ι, κ and Rev1 are the main subject matter of the current review. In addition, when describing specialized TLS polymerases, one cannot avoid inclusion of the phylogenetically unrelated B-family DNA polymerase, pol ζ, which is an indispensable component of the DNA damage tolerance machinery in eukaryotic cells.
In the past two decades, we have witnessed a rapid expansion of TLS research and multiple comprehensive review papers focusing on the various aspects of the TLS mechanism and biological functions of the TLS polymerases have been published [relevant reviews published during the past five last years include: (Boiteux and Jinks-Robertson, 2013; Fuchs and Fujii, 2013; Goodman and Woodgate, 2013; Guengerich et al., 2015; Hedglin and Benkovic, 2015; Ishino and Ishino, 2014; Jansen et al., 2015; Lambert and Lambert, 2015; Malkova and Haber, 2012; Maxwell and Suo, 2014; McIntyre and Woodgate, 2015; Nicolay et al., 2012b; Sale, 2013; Salehan and Morse, 2013; Sharma and Canman, 2012; Sharma et al., 2013; Tomasso et al., 2014; Yang et al., 2013; Yang, 2014)]. In the current review, we intend to present the major recent findings by putting them in an historical context and by focusing on the development of the ideas leading to the current understanding of TLS. We will mainly discuss biochemical and structural studies of the TLS polymerases themselves, as well as their interactions with major regulatory proteins affecting their activation of TLS [for the reviews on a growing body of literature regarding other means of their regulation, such as transcription, inactivation, or proteasomal degradation see (McIntyre and Woodgate, 2015; Sale et al., 2012)].
Historic overview
Before we begin our journey into the exciting world of translesion DNA polymerases, we wanted to take the liberty of presenting a very brief overview of the discoveries of all types of prokaryotic and eukaryotic DNA polymerases, which will illustrate why detection of the TLS polymerases was a paradigm-changing event for scientists working in the field.
The existence of a substance in a cell’s nucleus which we now call DNA, was first revealed in 1869 by Friedrich Miescher [for an excellent review see (Hübscher et al., 2010)]. It took about eight decades to unequivocally prove that this polynucleotide serves as the storage material for genetic information in all living organisms (Avery et al., 1944; Hershey and Chase, 1952). By the early 1950s, the chemical (Chargaff, 1951) and molecular (Watson and Crick, 1953) structure of DNA was uncovered and consequently a possible mechanism for copying the genetic material was proposed. Just three years after Watson and Crick reported the double-helical structure of DNA, the first enzyme able to catalyse replication of a DNA chain was identified (Kornberg et al., 1956) (Figure 1). This discovery was made by a group of scientists led by Arthur Kornberg, who believed that “a biochemist devoted to enzymes could reconstitute any metabolic event in the test tube as well as, or even better than, the cell does it”. Indeed, Kornberg and his colleagues succeeded in the reconstitution of the DNA synthesis reaction with purified components, which besides the DNA polymerase included a primed DNA template, Mg2+ and four deoxynucleotides. This approach is still widely used not only for the direct identification of the enzymes with the ability to synthesize DNA, but also to study the biochemical properties of DNA polymerases and to define the conditions that are required to ensure their optimal performance.
Figure 1. DNA polymerases: 60 years of discoveries.
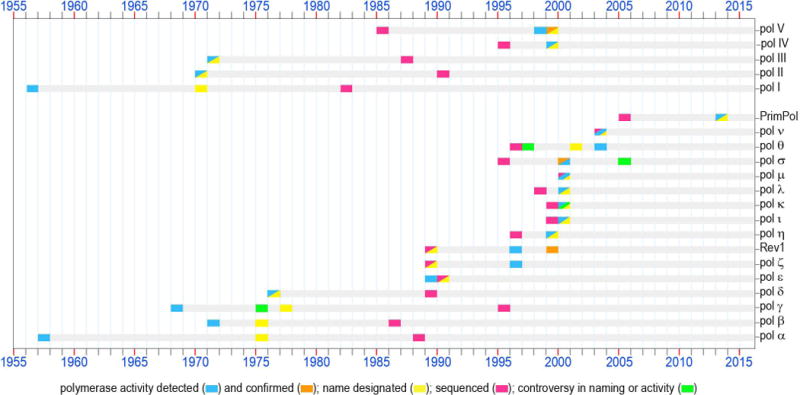
1956 – The first enzyme capable of copying DNA was discovered in E.coli extracts and was assumed at that time to be the only bacterial DNA polymerase (Kornberg et al., 1956). Later, when a second E. coli DNA polymerase was purified, this enzyme which plays an important role in prokaryotic DNA replication and repair was named pol I. The polA gene was sequenced in 1982 (Joyce et al., 1982) (accession P00582a).
1957 – The first eukaryotic DNA polymerase was identified (Bollum and Potter, 1957). When the uniform nomenclature was adopted in 1975, this enzyme was appropriately designated as pol α. Originally assumed to bear the sole responsibility for DNA synthesis in mammalian cells, this polymerase instead plays a key role in the initiation of chromosomal replication. The POLA gene was sequenced in 1988 (Wong et al., 1988) (accession CAA29920).
1968 – An enzyme with DNA polymerase activity was isolated from the rat liver mitochondria (Kalf and Ch’ih, 1968; Meyer and Simpson, 1968). This enzyme, known since 1977 as pol γ (Bolden et al., 1977) is a major polymerase dealing with all transactions involving mitochondrial DNA in mammalian cells. The POLG gene was sequenced in 1995 (Ropp and Copeland, 1995) (accession CAA88012).
1970 – A second DNA polymerase was discovered in E.coli by several groups in the USA and Germany (Knippers, 1970; Kornberg and Gefter, 1970; Moses and Richardson, 1970). According to the chronological order of discovery, it was named pol II. The sequencing of the polB gene was accomplished in 1990 (Chen et al., 1990) (accession P21189).
1971 – During the course of purification of E.coli pol II, a third prokaryotic DNA polymerase was detected (Kornberg and Gefter, 1971). DNA polymerase III is now known to be the main prokaryotic replicative polymerase. The dnaE gene encoding the catalytic α-subunit of pol III was sequenced in 1987 (Tomasiewicz and McHenry, 1987) (accession P10443).
- A second eukaryotic nuclear DNA polymerase later named pol β, was identified in mammalian cells and tissues practically simultaneously by several laboratories in the USA and England (Baril et al., 1971; Berger et al., 1971; Chang and Bollum, 1971; Haines et al., 1971; Weissbach et al., 1971). Very soon, it became apparent that this polymerase does not play a direct role in DNA replication. Instead, extensive research conducted by various groups revealed a major role for DNA pol β in base-excision repair. The POLB gene was sequenced in 1986 (Zmudzka et al., 1986) (accession P06766).
1974 – A formal nomenclature designating each mammalian DNA polymerase with a Greek symbol was proposed in 1974 and accepted by attendees of the international conference on eukaryotic DNA polymerases in 1975 (Weissbach et al., 1975). According to the established nomenclature, the first two mammalian DNA polymerases were designated pols α and β. It is worth noting that the third DNA polymerase which was given the name pol γ was and a forth DNA polymerase found in mitochondria (designated DNA polymerase-mt) were later shown to be identical and the name pol γ has been retained for this mitochondrial polymerase (Bolden et al., 1977).
1976 – The first nuclear polymerase containing an associated 3′→5′ exonuclease activity was purified and called pol δ (Byrnes et al., 1976). Later, this polymerase was shown to be an essential component of the eukaryotic replication machinery. The sequencing of the POLD1 gene was accomplished in 1989 (Boulet et al., 1989) (accession P15436).
1987 – It was proposed that DNA polymerases should be classified into discrete families based on their evolutionary relatedness. The first two evolutionary groups of DNA polymerases were designated as polymerase families A- and B- according to the amino acid homology to E coli pols I and II, respectively (Jung et al., 1987). In 1991 two additional groups typified by the catalytic subunit of E coli pols III and eukaryotic pol β were designated as families C- and X-, respectively (Ito and Braithwaite, 1991). In 1999 family D- was proposed to group polymerases involved in the DNA replication machinery of the Euryarchaeota (Cann and Ishino, 1999). In 2001, proteins originally defined as belonging to the UmuC/DinB/Rev1/Rad30 superfamily and involved in mutagenesis and TLS DNA synthesis were designated as Y-family polymerases (Ohmori et al., 2001).
1989 – The fourth nuclear DNA polymerase in mammalian cells, pol ε, was first reported as a PCNA-independent form of pol δ (Focher et al., 1989). Subsequently, this enzyme was recognized as a distinct DNA polymerase and accordingly it was named pol ε. Similar to pol δ, pol ε is equipped with 3′→5′ exonuclease proofreading activity and is essential for replication of the eukaryotic genome. The POLE1 gene was sequenced in 1990 (Morrison et al., 1990) (accession P21951).
1996 – Saccharomyces cerevisiae DNA pol ζ was characterized as a complex of Rev3 and Rev7 proteins (Nelson et al., 1996b). These studies confirmed the hypothesis that the Rev3 gene long known to be involved in damage-induced and spontaneous mutagenesis, encodes the first DNA polymerase specializing in TLS (Morrison et al., 1989) (accession P14284). This prediction was made based on the homology of Rev3 to other genes encoding B-family DNA polymerases.
- In the same year, the same group discovered dCMP transferase activity for the S. cerevisiae REV1 protein (Nelson et al., 1996a) that was known at the time to be required for the damage-induced mutagenesis and having ~25% identity with the E.coli UmuC protein (Larimer et al., 1989) (accession P12689). It was proposed that the CMP transferase function was important for mutagenic TLS involving pol ζ. However, Rev1 was not recognized as belonging to the broad superfamily of DNA-dependent DNA polymerases until 1999, when deoxynucleotidyl transferase activity was detected in several enzymes homologous to REV1.
1998 – The first evidence suggesting that the E. coli UmuD’2C complex consisting of the umuDC gene products (Perry et al., 1985; Kitagawa et al., 1985) (accession P04152), is a DNA polymerase was demonstrated (Tang et al., 1998). At that time, it was shown that in vitro the UmuD’2C complex could copy an abasic site-containing DNA template without the assistance of any other polymerase, although the possibility of contamination with trace amounts of other DNA polymerase were not entirely ruled out. A year later, additional biochemical studies unequivocally confirmed that the UmuD’2C complex is a bona fide DNA polymerase designated as E.coli pol V (Tang et al., 1999).
1999 –The S.cerevisiae RAD30 gene which was previously identified by sequence homology to prokaryotic UmuC and DinB in 1996 (Kulaeva et al., 1996; McDonald et al., 1997) was shown to encode DNA polymerase η (Johnson et al., 1999b). Shortly thereafter, human polymerase η was characterized (Masutani et al., 1999b). Polη became one of the founding members of the new Y-family of DNA polymerases (Ohmori et al., 2001). Nonsense, or frameshift mutations in the gene (RAD30A, POLH, XPV) encoding pol η are responsible for the Xeroderm Pigmentosum Variant syndrome in humans (Johnson et al., 1999a; Masutani et al., 1999b).
- The same week that the DNA polymerase activity of the UmuD’2C encoded pol V was confirmed, a manuscript describing the TLS activity of E.coli DinB was published (Wagner et al., 1999). This polymerase became known as E.coli DNA pol IV. The dinB gene was originally identified as dinP in 1995 (Ohmori et al., 1995) (accession BAA07593) and despite being shown to be allelic with dinB in 1999 the name of dinP, rather than the correct name of dinB, is still often used in Genbank data files describing related proteins.
2000 –The TLS activity of two eukaryotic Y-family polymerases ι (Tissier et al., 2000b) and κ (Ohashi et al., 2000a) (products of the POLI (RAD30B) and POLK (DINB1) genes, respectively) was demonstrated a year after they were cloned (Gerlach et al., 1999; McDonald et al., 1999) [accession numbers AAD50381 (pol ι) and AAF02541 pol κ)].
- Three X-family polymerases implicated in participating in different types of DNA transactions (such as BER, non-homologous end joining repair, V(D)J recombination, TLS, and sister chromatid cohesion) were discovered. This includes pol λ (Garcia-Diaz et al., 2000) (the sequence was first submitted in 1998 (Blanco, 1998) [accession number CAB65241]), pol μ (Dominguez et al., 2000) (accession CAB65075), and pol σ (Wang et al., 2000) (first sequenced in 1995 (Sadoff et al., 1995) accession P53632). It should be noted that the DNA polymerase activity of pol σ has been contested by Haracska et al., (Haracska et al., 2005b), who suggest that the protein is actually a poly-A RNA polymerase, rather than a bona fide DNA polymerase.
2003 – Two mammalian A-family DNA polymerases that are homologous to the DNA cross-link sensitivity protein Mus308 and implicated in different defense pathways against DNA damage were identified and characterized; pol θ (Seki et al., 2003) and pol ν (Marini et al., 2003). It worth mentioning that pol θ, the only polymerase known to contain a helicase domain, was first identified in 1997 in the genomes of a variety of eukaryotic organisms based on sequence homology to E. coli DNA pol I (Harris et al., 1996; Sharief et al., 1999; Sonnhammer and Wootton, 1997) (accession numbers AAB67306 and AAC33565). The polymerase was originally named pol η (Burtis and Harris, 1997), but was later renamed pol θ (Burgers et al., 2001). Pol ν was sequenced in 2003 (Marini et al., 2003) (accession NP_861524).
2013 – An ability to replicate both damaged and undamaged DNA templates was detected in PrimPol, an enzyme belonging to the archaeal-eukaryotic primase superfamily (Bianchi et al., 2013; García-Gómez et al., 2013). The gene encoding PrimPol was first sequenced in 2005 (Iyer et al., 2005) (accession NP_689896). The ability to catalyze TLS is only one of the broad enzymatic activities of the PrimPol enzymes that have been implicated in a large variety of cellular functions.
a GeneBank Sequence identifiers are listed (Clark et al., 2016).
Soon after Kornberg’s group isolated what is now known as DNA polymerase I from Escherichia coli, a protein with corresponding functions was identified in eukaryotes (Bollum and Potter, 1957). During the following 40 years of painstaking work that was conducted by multiple laboratories around the world, many more enzymes responsible for copying DNA in different organisms were discovered (Figure 1). According to the proposed classification, based upon similarities of the amino acid sequences, all known DNA-dependent DNA polymerases were originally grouped into four families; A-, B-, C-, or X- [such classification was first proposed in 1987 (Jung et al., 1987) and then updated in 1991 (Ito and Braithwaite, 1991)]. In eukaryotes, these families were represented by five DNA polymerases designated by Greek letters given to these proteins in order of their historical discovery [the formal nomenclature was adopted in 1975 (Weissbach et al., 1975)]. These five polymerases, later to be known as “classical” DNA polymerases, that were identified within the 40 years of Kornberg’s initial discovery, included components of the chromosomal replisome; pols α, δ and ε (family B); mitochondrial pol γ (family A); and the DNA repair polymerase, pol β (family X). The latest addition to the eukaryotic DNA polymerase superfamily made at the end of 40-year time span was pol ζ (family B) (Nelson et al., 1996b), a polymerase which appeared to be specifically dedicated to copying damaged genomic and mitochondrial DNA. This discovery can be considered as marking the end of the first “slow” stage in the timeline of DNA polymerase discovery and the beginning of the polymerase “boom era” when enzymes functionally similar to pol ζ, i.e. specializing in translesion DNA synthesis, came to the forefront.
In 1956, Kornberg and his colleagues assumed that the enzyme they found was the only DNA polymerase required for replication of the Escherichia coli (E. coli) genome, just as pol α was for several years considered to be the only polymerase involved in DNA replication in eukaryotic cells. By the mid-1990s, it seemed reasonable that the six eukaryotic polymerases that had been identified by that time were sufficient to fulfil all the needs related to DNA synthesis. Indeed, many DNA polymerases were known to play roles in more than one pathway related to DNA metabolism. Thus, replicative pols δ and ε are also involved in all major DNA excision-repair pathways, where they act to maintain the integrity of the genome by restoring the DNA sequence after damaged bases, or nucleotides, are removed. Both, replicative pol δ (Choi et al., 2010; Daube et al., 2000; Hirota et al., 2016; Meng et al., 2009; Narita et al., 2010; O’Day et al., 1992; Schmitt et al., 2009) and the base excision repair polymerase, pol β (Jiang et al., 2015; Vaisman and Woodgate, 2004; Villani et al., 2011; Villani et al., 2014; Xu et al., 2014) have been implicated in TLS. Even though the discovery of pol ζ hinted that cells might contain more enzymes specializing in the replication of damaged DNA, a prevailing idea was that replicative polymerases could perform this function themselves with the assistance of accessory proteins. In agreement with this idea, it was demonstrated that pol δ-catalysed replication staled at UV-induced cyclobutane thymine dimers (CPDs) can be rescued by addition of the proliferating cell nuclear antigen (PCNA), a co-factor of pol δ (Daube et al., 2000). The working hypothesis was that an interaction with accessory proteins relaxes the normally stringent fidelity of replicative polymerases, thus enabling them to insert nucleotides opposite the damaged bases in an error-prone manner and to continue DNA replication past the lesion. Among the proteins under investigation for such conduct were the products of the E. coli umuD and umuC genes, which were known to be responsible for the majority of damage-induced mutagenesis in bacterial cells (Kato and Shinoura, 1977). By the late 1990s, proteins sharing sequence homology with UmuC had been identified in various organisms from all kingdoms of life. These include DinB (Kulaeva et al., 1996; Ohmori et al., 1995), Rad30 (McDonald et al., 1997) and Rev1 (Larimer et al., 1989), which together with UmuC were proposed to comprise a superfamily of enzymes involved in both error-prone and error-free DNA damage tolerance pathway. Extensive studies aimed at uncovering the mechanism by which these supposed accessory proteins facilitated TLS culminated in the unexpected discovery that the super family of enzymes homologous to UmuC, DinB, Rad30 and Rev1 although lacking sequence similarity to any of the previously identified polymerases, all possess DNA replication activity. The first clue to such activity came from the study of the Saccharomyces cerevisiae Rev1 protein, which was shown to exhibit deoxycytidyl transferase activity in vitro (Nelson et al., 1996a). The idea that the phylogenetically related proteins might possess a similar activity, or even an expanded ability to incorporate all four deoxynucleotides, went largely unexplored until the late 1990s when the difficulties with the purification of these proteins were finally overcome.
In 1998, the first report describing DNA synthesizing activity of a heterotrimer consisting of the UmuC and two UmuD’ subunits was published (Tang et al., 1998). Although the possibility of contamination of the purified complex by trace amounts of other polymerase had not been entirely ruled out at the time the paper was published, subsequent studies unequivocally proved that the UmuD’2C complex which was subsequently named pol V, is able to copy undamaged and abasic site-containing DNA templates without the assistance of any other polymerase (Reuven et al., 1999; Tang et al., 1999). Practically simultaneously, TLS polymerase activity was reported for E.coli DinB (pol IV) (Wagner et al., 1999).
At the same time, watershed discoveries were also made in the field of eukaryotic polymerases. In 1999 the S.cerevisiae Rad30 protein was identified as a bona fide DNA polymerase, pol η which could bypass thymine-thymine cyclobutane dimers efficiently (Johnson et al., 1999b). Soon thereafter, TLS activity was shown for human DNA polymerases η (Johnson et al., 2000c; Masutani et al., 1999b; Zhang et al., 2000a), ι (Johnson et al., 2000b; Tissier et al., 2000b; Zhang et al., 2000c) and κ (Johnson et al., 2000a; Ohashi et al., 2000b; Zhang et al., 2000b)
Importantly, human pol η was shown to be defective in humans with the variant form of Xeroderma Pigmentosum (XPV) (Johnson et al., 1999a; Masutani et al., 1999b), a cancer-prone syndrome characterized by an increased incidence of skin malignancies and sensitivity to sunlight (Lehmann et al., 1975; Maher et al., 1976). This discovery led to the realization that pol η normally protects humans from the deleterious effects of excessive exposure to sunlight through its extraordinary ability to facilitate TLS past UV-induced CPDs very efficiently and accurately (Masutani et al., 1999a).
By the turn of the twenty first century, scientists were armed with new and powerful tools. Rapid progress in genomic sequencing and advanced search mechanisms opened the door for the identification of a whole class of protein related to the UmuC/DinB/Rad30/Rev1-like proteins, which were subsequently named the “Y-family” of DNA polymerases (Ohmori et al., 2001).
In the space of three years, additional members of the A- and X -families were also identified (Figure 1). Indeed, it was an extremely exciting time for scientists working in the DNA polymerase field. While it took four decades to identify the first six eukaryotic DNA polymerases, the same number of polymerases had been discovered in just two years, 1999–2000. In the field of prokaryotic polymerases, the situation was similar – within one year, the number of known polymerases grew from three to five.
During the early years of DNA polymerase research, new enzymes were often given their consensus names many years after their catalytic activity had been demonstrated. In modern times, naming a new polymerase sometimes preceded their biochemical characterization. Thus, yeast pol ζ was purified 7 years after the prediction that the Rev3 gene encodes a DNA polymerase based on the sequence homology to several known polymerases (Morrison et al., 1989; Nelson et al., 1996b). Such predictions occasionally even caused confusion regarding polymerase nomenclature. For example, a putative DNA polymerase homologous to E. coli DNA pol I was identified in 1996 in the genomes of several eukaryotic organisms through the analysis of sequence databases (Harris et al., 1996; Sonnhammer and Wootton, 1997). This enzyme, originally called pol η (Burtis and Harris, 1997) was later renamed pol θ (Burgers et al., 2001) and its intrinsic polymerase activity was not experimentally verified until 2003 (Seki et al., 2003). Interestingly, the same Greek letter designation (θ) was originally given to the product of the human DINB1 gene (Johnson et al., 2000a), which is now known as pol κ. In its turn, pol κ was originally assigned to the product of the TRF4 (Topoisomerase one-requiring function 4) gene (Wang et al., 2000), and later re-designated as pol σ. This enzyme related to the X-family DNA polymerases has been shown to be involved in sister chromatid cohesion (Wang et al., 2000), although another study suggested that instead of possessing DNA polymerase activity, TRF4 protein might only exhibit poly(A) RNA polymerase activity (Haracska et al., 2005b).
The rapid pace with which DNA polymerases were identified at the turn of the century slowed down after 2000 and only one new eukaryotic polymerase, pol ν was discovered in the next decade (Marini et al., 2003). However, there is a good chance that identification of enzymes belonging to the DNA polymerase superfamily is still incomplete. Thus, interest in this research field was recently refueled by the discovery of a whole new class of DNA polymerases possessing both primase and DNA polymerase activities and accordingly named PrimPol (Bianchi et al., 2013; García-Gómez et al., 2013). These enzymes belong to a large functionally diverse superfamily of ancient enzymes equipped with broad enzymatic capabilities related to DNA metabolism and are present in all domains of life. There is a great deal of evidence suggesting that among a large variety of cellular functions fulfilled by PrimPols a very prominent role is that of TLS past various replicase stalling lesions [recently reviewed in (Guilliam et al., 2015; Helleday, 2013)].
Today, we know that the human genome encodes at least 15 DNA template-dependent DNA polymerases (terminal deoxynucleotidyl transferase (TdT), the unique template-independent DNA polymerase, is not included in this review). Only a handful of these enzymes are responsible for the bulk of nuclear and mitochondrial DNA replication and for the significant part of repair DNA synthesis (pols δ, ε, and γ), while all other polymerases are required for the fine-tuning of multiple diverse DNA transactions in cells. Most of these polymerases have been implicated in TLS one way or another. Why do cells need so many enzymes capable of copying damaged DNA? It has been estimated that between 1017–1019 DNA damaging events occur in a human body every day (Lindahl and Barnes, 2000). Even though the majority of modified bases are removed through multiple elaborate repair mechanisms, many lesions nevertheless persist during S-phase, when DNA is being replicated, and this represents a serious challenge for replication fork progression. In some cases, replicative polymerases can overcome such obstacles themselves, but much more often they are forced to give way to specialized enzymes that are better equipped to extend the replication fork past damaged sites. The large amount of overall damage does not, however, justify the necessity to possess such a wide variety of DNA polymerases until one considers the extraordinary variety of DNA lesions and differences in structural DNA distortions. Accordingly, it makes perfect sense that a wide assortment of polymerases with different affinities for various types of damaged DNA are required to perform highly specialized functions, especially since each enzyme is involved in more than one pathway (Lange et al., 2013; Sale et al., 2012; Sharma et al., 2013), including those which are not directly related to replication of damaged DNA (Pillaire et al., 2015). As previously mentioned, in this this review, we will focus on DNA polymerases whose primary functions are related to replication of damaged DNA.
Major tools in studies of eukaryotic TLS
The first insights into error-prone damage tolerance mechanisms were gained through genetic experiments using bacterial and yeast systems. By analyzing mutant strains, multiple genes involved in damage-induced mutagenesis have been identified. It is through these studies that UmuC and Rev1 were first implicated in DNA synthesis, even though they were originally hypothesized to function simply as “mutagenesis proteins”, or accessory factors of the cells DNA replicase (Bridges and Woodgate, 1985). An understanding of how these proteins and their orthologs in other organisms actually function came from biochemical studies in which DNA replication reactions were reconstituted in vitro. This approach is still widely used to study the biochemical properties of DNA polymerases such as fidelity, processivity, and ability to copy damaged DNA. In vitro studies not only provide a direct proof that the protein in question is indeed a TLS polymerase, but also specify the type of DNA lesion each polymerase is able to tolerate and allows for the identification of protein partners, along with the determination of optimal reaction conditions for most efficient and accurate replicative bypass. Biochemical characterization therefore plays an important role in predicting which polymerase is best suited to bypass a specific lesion, as well as in identifying candidates for a backup role. While these studies provide valuable information, they nevertheless have to be substantiated by in vivo evidence and represent only the first step in elucidating the mechanism of translesion replication.
Mechanistic insights into many aspects of TLS polymerases functions have been gained through structure-function studies. Thus, crystallographic studies provided the molecular basis for understanding the common biochemical properties of TLS polymerases in general and the unique features of each polymerase in particular. Such studies also allows one to analyze the structural determinants underlying the recruitment of TLS polymerases to the replication fork and mechanism by which there is a switch between high-fidelity and specialized DNA polymerases is achieved.
An important assay used to not only confirm the participation of a particular TLS polymerase in the cellular response to a specific type of DNA damage, but also to characterize translesion replication in a living cell, is fluorescent microscopy. This method is based on immunological detection of overexpressed proteins, or of a polymerase fused to a fluorescent protein. It provides direct evidence of the localization of TLS polymerases at replication factories and allows one to monitor their interaction with protein partners.
In recent years, the biological role of TLS polymerases has been extensively investigated using knockout mouse models. While homozygous inactivation of murine Rev3 (catalytic subunit of pol ζ) leads to embryonic lethality, mice defective one or more Y-family DNA polymerase have been generated (Esposito et al., 2000; Jansen et al., 2006; Jansen et al., 2014a; Lin et al., 2006; McDonald et al., 2003; Ohkumo et al., 2006; Schenten et al., 2002) [and reviewed in (Goodman and Woodgate, 2013; Guo et al., 2009; Jansen et al., 2015; Lange et al., 2011; Lehmann et al., 2007; Loeb and Monnat, 2008; Makarova and Burgers, 2015; Pillaire et al., 2014; Sale et al., 2012; Seki et al., 2005)]. These studies, together with analysis of epidemiological data in humans with altered expression of TLS polymerases provide invaluable information about the connection between TLS and human health, especially with regard to cancer risk.
Biochemical properties of TLS polymerases
Extensive investigations during the first decade following the discovery of TLS DNA polymerases mainly focused on their biochemical properties [reviewed in (Boiteux and Jinks-Robertson, 2013; Choi et al., 2010; Goodman and Woodgate, 2013; Guengerich et al., 2015; Guo et al., 2009; Katafuchi and Nohmi, 2010; Lehmann, 2005; Makarova and Burgers, 2015; Maxwell and Suo, 2014; Prakash and Prakash, 2002; Shcherbakova and Fijalkowska, 2006; Vaisman et al., 2002; Vaisman et al., 2004; Vaisman and Woodgate, 2004; Waters et al., 2009; Yang et al., 2013; Yang, 2005; Yang and Woodgate, 2007)]. Here, we will summarize the accumulated data and present them in the context of the in vivo observations emphasizing the biological functions of the enzymes. In general, translesion polymerases are characterized by low catalytic efficiency, fidelity and processivity. This set of properties is not surprising taking into account that the main task of TLS polymerases is to help the main replicase to overcome obstacles and keep replication going. The fact that the active site of TLS polymerases is flexible enough to accommodate damaged nucleotides suggests that it is not stringent enough to maintain a tight interaction with DNA so as to prevent the polymerase from dissociating, to ensure the high efficiency of the correct base incorporation, and to exclude binding of non-complementary bases. In fact, the right balance between these biochemical characteristics is very important in maintaining overall genomic integrity: low catalytic activity and a distributive mode of DNA synthesis prevents highly inaccurate polymerases from replicating long stretches of DNA and generating large number of mutations. In all TLS polymerases, low-fidelity results from not only the intrinsic reduced capacity to discriminate between right and wrong nucleotides, but also by the lack of 3′–5′ exonucleolytic proofreading that allows these enzymes to avoid an additional kinetic barrier to translesion synthesis (Khare and Eckert, 2002). It is important to mention that low fidelity, which at first blush seemed to be an inevitable side effect of the necessity to rescue arrested replication, in many cases is actually beneficial for the living organisms (see below). One example illustrating the usefulness of erroneous DNA synthesis is somatic hypermutation, a mechanism by which the immune system adapts to foreign antigens. In order to accomplish this task, targeted mutations in the variable regions of immunoglobulin genes must be generated. One of the pathways by which mutations can be created involves DNA synthesis by error-prone DNA polymerases. While one way or another, all TLS polymerases have been considered as potential suspects responsible for the mutagenesis of antibody genes, only Rev1 and pol η have been unequivocally proven to play a central role in somatic hypermutation (Jansen et al., 2006; Krijger et al., 2013; Seki et al., 2005; Yavuz et al., 2002; Zhao et al., 2013).
B-family pol ζ
Pol ζ has been extensively studied since its discovery as the first DNA polymerase specializing in TLS. But even today, it does not appear that we have a complete picture reflecting the involvement of the enzyme in various cellular processes. On one hand, characterization of a vertebrate homologue has been hindered by the difficulty of purifying the large multi-subunit complex, while investigation of the biological roles of mammalian pol ζ have been hampered by the embryonic lethality of mice with a disruption of the rev3L gene and inviability of murine rev3L−/− mouse embryonic fibroblasts (Mefs). For more than 15 years following the first purification of the overexpressed yeast pol ζ as a heterodimeric complex (Nelson et al., 1996b), pol ζ was believed to consist of a large catalytic Rev3 subunit and much smaller regulatory subunit, Rev7, which stabilizes and increases the activity of the polymerase, as well as mediates the interaction of pol ζ with Rev1 (see below). However, in 2012, several groups independently demonstrated that the Rev3/Rev7 heterodimer is the minimal assembly sufficient for the in vitro activity of pol ζ, but this complex lacks essential components required for proper functioning of the polymerase in vivo. According to the current paradigm, pol ζ, similar to other B-family polymerases, is a tetrameric holoenzyme (ζ4), which shares two essential accessary subunits (POLD2/p50 and POLD3/p66 in humans and Pol31 and Pol32 in S.cerevisiae) with the replicative pol δ (Baranovskiy et al., 2012; Johnson et al., 2012; Makarova et al., 2012).
The catalytic activity of yeast Rev3 on undamaged DNA is stimulated 20–30 fold by the Rev7 subunit and increases another 5–10-fold with the addition of Pol31 and Pol32 subunits (Makarova et al., 2012). Similar results were obtained using damaged templates where primer extensions were 2–5 fold more efficient when pol ζ holoenzyme was used (Johnson et al., 2012). An increase in the efficiency of nucleotide incorporation by the purified human ζ4 compared to ζ2 was even higher (~30-fold) (Lee et al., 2014). The presence of pol δ accessory subunits also enhanced the processivity of both human and yeast pol ζ4 (Lee et al., 2014; Makarova et al., 2012). The increase in the catalytic activity and processivity are likely due to enhanced DNA binding by pol ζ holoenzyme. In contrast, it is not significantly modified by the presence of the accessory proteins RPA (replication protein A), RFC (replication factor C), or PCNA (Zhong et al., 2006).
It has been long known that even though pol ζ belongs to the B-family of DNA polymerases that are composed of accurate replicative polymerases, its 3′-5′ exonuclease domain is inactive and it is responsible for more than half of spontaneous and virtually all DNA damage-induced mutagenesis in yeast. In humans, the low fidelity of pol ζ is manifested by chromosome instability and carcinogenesis caused by alterations in its expression (Diaz et al., 2003; Gan et al., 2008; Lange et al., 2011; Wittschieben et al., 2006). It was originally believed that pol ζ-dependent mutagenesis resulted primarily from the exceptional ability of the enzyme to extend mispairs made by other polymerases. Indeed, steady-state kinetic studies of yeast pol ζ using primed single-stranded oligonucleotide templates revealed about 100-fold higher discrimination against nucleotide misincorporation (103–104 fold) compared to mismatched primer extension (10–100 fold) (Johnson et al., 2000b). However, subsequent studies using undamaged gapped plasmid DNA substrates have shown that yeast pol ζ is also able to directly generate its own mismatches. The mutational spectra determined in these studies correlate well with the in vivo findings. They indicate that pol ζ-dependent errors are confined to specific sequence contexts and are often clustered in short patches, at a rate that is unprecedented in comparison with other polymerases. The average base substitution fidelity of yeast pol ζ in gap-filling reactions was significantly lower than that of related pols α, δ and ε (Zhong et al., 2006). Thus, pol ζ2 was roughly 100-times more error-prone than pols δ and ε with intact 3′ – 5′ proofreading capacities and ~10-times more inaccurate than naturally proofreading-deficient pols α and exo-deficient δ and ε (Zhong et al., 2006). However, the overall error rate of pol ζ was still lower than that of Y-family polymerases. Thus in a gap-filling assay, yeast pol ζ made 1 mistake per 770 nucleotides copied, which makes it almost 5-times more accurate than the most faithful Y-family human pol κ (Zhong et al., 2006). Despite the fact that the base substitution fidelity of pol ζ is substantially lower than that of replicative polymerases, discrimination against incorporation of nucleotides with the wrong sugar by yeast pol ζ is only slightly lower than the sugar selectivity of high fidelity enzymes (Makarova et al., 2014).
In vitro studies using damaged DNA templates suggested that among the large variety of DNA lesions that pol ζ is able to bypass, some are replicated accurately (Johnson et al., 2003; Lin et al., 2014b), while TLS past others is highly error-prone (Lin et al., 2014a). In agreement with in vitro data, genetic studies have demonstrated that yeast pol ζ is not only necessary for damage-induced nuclear and mitochondrial genome mutagenesis, it also promotes accurate bypass of some DNA lesions (Baruffini et al., 2012; Baynton et al., 1999; Bresson and Fuchs, 2002; Kalifa and Sia, 2007; Zhang et al., 2006). On all damaged templates tested, pol ζ was a great deal less efficient at incorporating a nucleotide opposite the damaged bases than extending the resulting primer terminus, consistent with the idea that its primary role in lesion bypass is confined to the elongation step, while the incorporation step depends on other polymerases. However, pol ζ has the capacity to catalyze all steps of TLS by itself acting as both the inserter and an extender (Stone et al., 2011). It should be noted that studies related to the fidelity and TLS activity of yeast and human pol ζ4 are very limiting at the present time, but the available data do suggest that in the presence of all accessory subunits, pol ζ4 catalyzes TLS much more efficiently compared to the pol ζ2 complex (Lee et al., 2014; Makarova et al., 2012).
Y-family pols η, ι, κ and Rev1
In contrast to pol ζ, all eukaryotic Y-family polymerases are currently believed to be encoded by a single polypeptide enzyme consisting of a conserved N-terminal domain of 350–450 amino acids containing the catalytic active site, and a variable-length C-terminal region important for regulatory protein–protein interactions (Figure 2B). Despite being characterized by generally common features, each member of the Y-family of DNA polymerases has a unique personality. For example, pol η is exceptionally efficient and accurate while incorporating nucleotides opposite the CPDs; pol κ specializes in the extension step of lesion bypass; Rev1 abstains from incorporating nucleotides other than dCTP and functions in mitochondria, while pol ι in some cases favors the formation of non-Watson-Crick base pairs and, like X-family pols β and λ, has a dRP lyase activity important for Base Excision Repair (BER) and prefers to use Mn2+ as an activator for catalysis. The characteristics of each polymerase have been extensively studied using an in vitro approach. Although these studies have sometimes produced conflicting results and still require in vivo verification, they provide an important clue to understanding how these polymerases function in a living cell.
Figure 2. Structural organization of catalytic domains of TLS polymerases.

A. Comparison of yeast (S. cerevisiae) and human (H. sapiens) REV1 and REV3 gene structures. The domain arrangement in yeast and human Rev proteins are very similar (the percent identity for the related regions are indicated within the gray areas between the human and yeast proteins). The human enzymes are larger than the yeast enzymes due to long inserts in the respective genes (one in Rev3 and two in Rev1 [I1 and I2], see the text for details). The conserved sequence motifs in Rev3 characteristic for the B-family DNA polymerases are indicated by roman numerals (I-VI). B. Arrangement of the functional domains in human Y-family polymerases. All polymerases (panels A & B) are aligned relative to the N-terminus of the catalytic core and all diagrams are drawn to the scale. The name of each polymerase and its length (number of amino acids) are indicated on the right-hand side of the figure and the gene designation is indicated on the left side of each protein. The catalytic domains of the polymerases are color-coded as follows: red – palm; green – thumb; light blue – fingers; purple – little finger; aquamarine –catalytic core of pol ζ with polymerase (light pink) and inactive 3′-5′ exonuclease (dark pink) domains. C. Interaction map of the proteins important for TLS. In all panels (A-C) the domains involved in protein-protein interactions are color-coded: NTD – N-terminal domain; MLS – mitochondrial localization signal; BRCT – breast cancer-associated protein-1 carboxyl-terminal domain; N-Clasp – structural feature of pol κ; NLS – nuclear localization signal in pols ζ, η, ι, and κ (pol ι contains non-classical NLS motif; in pol ζ the signal is shown for the yeast protein, but is not shown for the human enzyme since it is located within the omitted ~1550 amino acid region); UBM – ubiquitin binding motif; UBZ – ubiquitin binding zinc-finger motif; CTD – C-terminal domain of Rev3 which in addition to the N-terminal zinc finger [ZF] motif contains C-terminal iron–sulfur [FS] cluster, a binding site for other polymerase subunits; dRP – dRP lyase domain (the ~40 kDa region in pol ι to which the dRP lyase activity has been mapped (Prasad et al., 2003) is indicated below the schematic representation of the primary protein structure); PIP – PCNA-interacting protein motif; RIR – Rev1-interacting region motif; PIR–protein interaction regions (two PIR sites [PIR1 & 2]) in human Rev3 and yeast Rev1, as well as one PIR site in yeast Rev3 and human Rev1 are involved in the interaction with Rev7; in addition the PIR site in the CTD of human Rev1 is required for the binding to pols η, κ, and ι, while in yeast the PIR site in the catalytic domain of Rev1 [PIR1] is involved in the interaction with pol η, Rev3 and Rev7. The PIR in the CTD of human Rev1 [PIR2] has two interfaces: the C-terminal part (C) is required for the interaction with Rev7, while the N-terminal part (N) is involved in an interaction with PolD3 and pols η, ι and κ. The PIP and RIR motifs have similar consensus sequences, but have traditionally been viewed as separate entities. In several recent studies, it has been proposed that these motifs be considered as parts of a single “PIP-like” motif capable of binding multiple target proteins (Boehm and Washington, 2016). The position of the star (*) above the pol η sequence corresponds to the F1 motif necessary for the interaction with the POLD2 subunit of pol δ (Baldeck et al., 2015).
Pols η and κ, similar to the high-fidelity replicative polymerases, form all four possible correct base pairs with comparable catalytic efficiencies, while the catalytic activity of pol ι and Rev1 are highly template base dependent: Rev1 specializes in the incorporation of dCTP opposite template G, whereas pol ι is most efficient while incorporating T opposite template A. In vitro replication reactions have clearly established that Y-family polymerases are significantly less processive than the high fidelity replicative polymerases. However, the absolute number of nucleotides incorporated per DNA binding event varies among individual members, from very low, such as 2–3 nucleotides per DNA binding event in the case of pol ι in the presence of physiological activator Mg+2 and Rev1 on a poly(dG) template, to moderate, ~ 20–30 nucleotides for pol κ. Y family polymerases are also characterized by significantly different abilities to discriminate against incorrect nucleotide substrates which include those that do not comply with Watson-Crick base complementarity, as well as those that have the wrong sugar moiety. Ribonucleotide incorporation by DNA polymerases has only recently attracted long overdue attention and very little information about the discrimination between dNTPs and rNTPs by the eukaryotic members of Y-family is currently available. But even the limited data that is available suggest that these enzymes tolerate nucleotides with an incorrect sugar differently. Thus, on undamaged templates, human pol κ hesitates to incorporate ribonucleotides at any detectible levels, while Rev1 discriminates between deoxy- and ribo-cytosine with moderate efficiency, incorporating rCTP 280 times less frequently than dCTP (Brown et al., 2010). The sugar selectivity of pol η and pol ι during TLS indicates that ribonucleotide incorporation is only observed only for pol ι (Donigan et al., 2014; Donigan et al., 2015). However, even then, the efficiency of pol ι-catalyzed rNTP insertion was more than 1000 times lower compared to incorporation of dNTPs and extension of ribonucleotide-terminated primers was efficient only in the presence of dNTPs.
In contrast to sugar selectivity, the base substitution fidelity of Y-family polymerases has been investigated extensively. These studies revealed that in the presence of Mg+2, base substitution error rates range from 6×10−3 for the most faithful pol κ, to 3.5×10−2 for pol η, and 3 ×100 for pol ι (Matsuda et al., 2001; Ohashi et al., 2000a; Tissier et al., 2000b; Vaisman et al., 2004). Furthermore, the nucleotide misincorporation specificity and the mode of introducing mutations are different for each polymerase. Thus, while pol η and pol ι readily misincorporate nucleotides, pol κ is most efficient in the extension of mismatched, or misaligned, primer termini and readily generates frameshift errors especially single-nucleotide deletions. Pol ι is exceptional in its nucleotide misincorporation specificity: it is most accurate opposite template A (with a misincorporation frequency of 1–2 × 10−4) and most unfaithful opposite template T/U, where it favors misincorporation of dG over the correct dA (by 3 to 10-fold), and it also misinserts dT opposite template T almost as frequently as the correct base, dA. Such unusual substrate specificity might play an incredibly important role in maintaining genome integrity by restoring the coding properties of cytosines that have undergone spontaneous or damage-induced deamination. Cytosine deamination occurs at a significant rate in cells, making pol ι uniquely equipped to single-handedly combat CG to TA transition mutations that would be generated if the modified base was copied accurately (Vaisman and Woodgate, 2001; Vaisman et al., 2006). Consistent with this idea are studies with mice lacking functional pol ι that indicate that despite its extreme replication infidelity, pol ι contributes to maintaining genomic stability and preventing mutagenesis and tumorigenesis (Iguchi et al., 2014; McDonald et al., 2003). However, on the “flip side of the coin”, the exceptional ability of pol ι to insert the “correct” dG opposite deaminated cytosines could hinder somatic hypermutation in immunoglobulin genes. By preventing generation of CG to TA transitions, pol ι would interfere with an adequate immune response to infection where enhanced mutagenesis induced by the targeted deamination of Cs and accurate copying of the resulting Us plays a pivotal role. This potentially helps explain why cells generally avoid recruiting highly error-prone pol ι for somatic hypermutation despite its potential usefulness for the alternative mechanisms of generating mutations (McDonald et al., 2003). As with all DNA polymerases, replacing Mg2+ with Mn2+ results in a general reduction of pol ι’s fidelity at template A, but interestingly, the preference for the formation of a wobble base pair between the incoming dG and template T disappears (Frank and Woodgate, 2007). The latter phenomena could be considered as a peculiar increase in fidelity or, alternatively, as a typical Mn2+−dependent decrease in fidelity, if incorporation opposite template T by pol ι represents a case of mistaken identity.
The Y-family polymerases also exhibit a remarkable diversity in their substrate specificity, efficiency and fidelity during replication of damaged DNA. A great body of literature with detailed characterization of the in vitro replication of DNA containing various lesions by different TLS polymerases has accumulated in recent years. We do not have the intention to list all these studies, but rather will devote our discussion to several which emphasize the notion that Y-family polymerases are specialized for the roles they play in lesion bypass. For example, pol η is best characterized for very efficient and quite accurate bypass of UV-induced CPDs, but also platinum interstrand crosslinks formed at adjacent guanines by anticancer drugs (Masutani et al., 1999a; Masutani et al., 2000; McCulloch et al., 2004; Vaisman et al., 2000; Washington et al., 2000). These properties on one hand put pol η in a unique position as a protector of mammals from UV-induced carcinogenesis that is manifested by the XPV syndrome in humans lacking POLH gene (Masutani et al., 1999b), but on the other hand, they paint pol η as a facilitator of tumor progression that reduces the efficacy of Platinum-based chemotherapy. While pol κ is unable to incorporate nucleotides opposite the 3′T of a CPD, pol ι replicates the CPD-containing DNA templates with moderate efficiency. Pol ι’s ability to bypass CPDs is dependent on the surrounding sequence context and even more on the identity of the divalent metal ions coordinated within the active site of the polymerase (Frank and Woodgate, 2007; Tissier et al., 2000a; Vaisman et al., 2003). Thus, although pol ι -catalyzed bypass of CPDs is still substantially less efficient than that of pol η, it is stimulated dramatically by Mn2+ and can reach up to 60% bypass, which is more than that observed for any other human polymerase (except pol η). In fact, the reduced efficiency in the presence of Mg2+ could be important in suppressing the translesion capacity of pol ι in normal cells. In the absence of pol η, pol ι can be activated by Mn2+ which although present at much lower concentration than Mg2+, can accumulate intracellularly through specific import pathways (Madejczyk and Ballatori, 2012; Bouron et al., 2015). The ability of pol ι to replicate past CPDs makes it a likely candidate to collaborate with other TLS polymerases in substituting for pol η in XPV cells (Frank and Woodgate, 2007; Tissier et al., 2000a; Vaisman et al., 2003; Vaisman et al., 2006). Several subsequent in vivo studies provided multiple lines of evidence supporting this hypothesis (Gueranger et al., 2008; Jansen et al., 2014a; Livneh et al., 2010; Wang et al., 2007; Ziv et al., 2009).
Both pol η and pol ι but not pol κ, are able to incorporate nucleotides opposite the 3′ base of a thymine-thymine 6–4 photoproduct (Yamamoto et al., 2008; Zhang et al., 2000a; Zhang et al., 2000b). On the other hand, whereas human pol η and pol ι bypass benzo[a]pyrene diol epoxide deoxyguanosines adducts (a naturally occurring metabolite of carcinogens commonly found in cigarette smoke) inefficiently and inaccurately (Chiapperino et al., 2002; Frank et al., 2002; Rechkoblit et al., 2002; Zhang et al., 2002a), pol κ bypasses the lesion with high efficiency and predominantly incorporates the correct base, dC, opposite the adducted G (Rechkoblit et al., 2002; Suzuki et al., 2002; Zhang et al., 2000b) (Zhang et al., 2002a) (Huang et al., 2003) (Zhang et al., 2002b). In fact, it has been proposed that pol κ may have evolved specifically to catalyze accurate bypass of various related lesions generated by polycyclic aromatic hydrocarbons, abundant and ubiquitous environmental pollutants, and studies with transgenic mice devoid of pol κ support such hypotheses (Ogi et al., 2002). All four Y family polymerases can incorporate nucleotides opposite an abasic site although with different efficiencies and base and sugar specificities and therefore could be involved in translesion replication past this common non-instructional lesion.
Finally, recent studies suggest that Y-family polymerases participate in metabolic pathways unrelated to TLS. For example, pol η appears to be involved in somatic hypermutation of immunoglobulin genes (Delbos et al., 2005; Martomo et al., 2005; Pavlov et al., 2002; Rogozin et al., 2001; Seki et al., 2005; Wilson et al., 2005), replication of common fragile sites (Bergoglio et al., 2013; Despras et al., 2016; Rey et al., 2009), telomeres (Betous et al., 2009; Pope-Varsalona et al., 2014) and homologous recombination repair (Kawamoto et al., 2005; McIlwraith et al., 2005; Nicolay et al., 2012a); pol κ has been implicated in nucleotide excision repair (Ogi and Lehmann, 2006); and pol ι may play a role in specialized forms of BER (Bebenek et al., 2001).
Structural features of TLS polymerases
Despite the lack of primary amino acid sequence homology between DNA polymerases from different families, they all share a common structural topology of a catalytic core often described as a right hand with finger, palm, and thumb subdomains holding DNA and positioning the incoming dNTP for incorporation (Figure 2). The active center of the polymerases resides in the palm domain where the essential metal ions required for the nucleotidyl transferase reaction are coordinated. The thumb subdomain plays an important role in binding to DNA and determining the enzyme’s processivity and translocation. The fingers domain is largely responsible for the positioning of DNA template required for optimal nucleotide pairing. Besides the conserved polymerase domains, DNA polymerases in general often have additional domains that have evolved to fulfill specific functions. Thus, most replicative polymerases contain a 3′ – 5′ exonuclease domain that proofreads newly synthesized DNA and corrects mismatched base pairs. In contrast, the majority of specialized polymerases lack a 3′ – 5′ exonuclease domain. The exception is pol ζ, the catalytic core of which is homologous to the B-family replicative polymerases, and thus contains sequences corresponding to the 3′–5′ exonuclease domain, even though it is non-functional (Figure 2A). All Rev proteins (Rev1, Rev3, and Rev7) have a mitochondrial localization domain located at their N-terminus (Figure 2A), which allows them to assist pol γ in the replication of a damaged mitochondrial genome (Zhang et al., 2006). The domain that separates Y-family enzymes from other polymerases is located in the C-terminus and is connected to the catalytic core by a flexible linker. This domain mediates the contact of the polymerase with DNA and has been referred to as the “little finger” (LF) (Ling et al., 2001), “wrist” (Silvian et al., 2001), or “polymerase-associated domain” (PAD) (Trincao et al., 2001) (Figure 2B). We think that the “LF” acronym is more appropriate, because it keeps in line with the general structural topology of a hand and we will use this term throughout the review. While the palm, thumb and finger domains of the Y-family polymerases are highly conserved, the LF domain is unique for each family member and plays an important role in determining the biochemical properties and biological function of each individual enzyme (Boudsocq et al., 2004; Wilson et al., 2013).
Primary structure and domain organization of pol ζ
Pol ζ is the only error-prone TLS polymerase which shares structural complexity and sequence homology with high-fidelity B-family replicative polymerases. Like other B-family polymerases, pol ζ is a multi-subunit complex. The catalytic domain of pol ζ (Rev3) is homologous to that of other family members (pols α, ε, and δ) in both amino-terminal and carboxyl-terminal regions. A cysteine-rich C-terminal domain (CTD) of Rev3 contains two conserved metal binding sites– a zinc finger [ZF] motif towards the N-terminal side of the CTD and an iron–sulfur [4Fe-4S] cluster in the C-terminal portion of the CTD (Figure 2A). The ZF motif has been proposed to mediate the DNA-dependent interactions of pol ζ with PCNA similar to that shown for pol δ (Netz et al., 2012). The [4Fe-4S] cluster serves as a docking site for additional polymerase subunits, such as POLD2 in mammalian cells and Pol31 in yeast, which pol ζ shares with pols δ (Baranovskiy et al., 2012; Netz et al., 2012). Rev3 differs from other B-family polymerases by the presence of a large insert containing short motifs required for binding to Rev7, a regulatory subunit specific for pol ζ (Figure 2). The size of the insert in Rev3 varies between species. For example, the human Rev3L protein is about twice the size of its S.cerevisiae counterpart (Figure 2). The amino acid sequence of the inserts in the human and S.cerevisiae Rev3 proteins show little similarity, except for the 55-residue region containing the Rev7 binding site (gray area between yeast and human enzymes on the Figure 2). The internal sequence of REV3L contains a second site required for the interaction with Rev7 and is located ~100 amino acids away from the first Rev7 binding site (Tomida et al., 2015). This site is conserved in vertebrates, but it has not been found in Rev3 homologs from fungi (Figure 2).
A 3D structural model of the four-subunit yeast pol ζ generated using electron microscopy suggests an overall elongated structure with separate catalytic and regulatory lobes that resembles the bilobal architecture of replicative polymerases (Gómez-Llorente et al., 2013). Based on sequence conservation, pol ζ is predicted to have the same overall topology and mechanism of nucleotide incorporation as replicative polymerases.
Structural insight into Y-family polymerases functions
Common structural features
In contrast to pol ζ, various structural studies involving the eukaryotic Y-family enzymes have been determined that have helped us to understand their unique biochemical activities. As expected, these studies revealed the general structural organization separating this group from polymerases belonging to other families, i.e. an unusually flexible and solvent accessible active site pocket that is able to tolerate a variety of damaged template bases. The finger and thumb domains which play an important role in choosing and positioning of the correctly paired nucleotide in the active site of the replicative polymerases are generally shorter and make fewer contacts with a DNA substrate and an incoming nucleotide and the unique LF domain helps to constrain the target nucleotide of the DNA backbone within the polymerase active site.
One might predict that the relaxed constraints of the active sites of Y-family polymerases should not only reduce their base-substitution fidelity, but also make them less discriminatory with regard to sugar selection. However, quite unexpectedly, no such correlation has been found. Instead, discrimination factors vary greatly and are not directly related to the base-fidelity of the polymerase [reviewed in (Vaisman and Woodgate, 2014)]. This apparent discrepancy is explained by the fact that most DNA polymerases, including those from the Y-family, utilize the same major mechanism of sugar discrimination. In order to prevent incorporation of nucleotides with the wrong sugar, DNA polymerases rely on a specific amino acid within the nucleotide-binding pocket in the polymerase active site. This highly conserved residue (in the Y-family it is Phe or Tyr) serves as a “steric gate” clashing with the 2′-OH of the incoming rNTP. Therefore, it is this region of the polymerase cleft, rather than general active site architecture, which controls sugar selection of a particular polymerase.
Conformational changes during catalytic cycle
While the general structural features of the active site of high fidelity DNA polymerases can be characterized (as above), it is not static, but rather goes through conformational changes during substrate binding and catalysis. Upon correct nucleotide binding, these polymerases undergo “an induced fit” structural change from the open conformation of the binary complex (polymerase-DNA template), to the closed conformation of the catalysis-ready ternary complex (polymerase-DNA template-dNTP), due to the large finger subdomain motions (Freudenthal et al., 2013; Johnson, 2008; Moscato et al., 2016). Subsequently, the correct nucleotide incorporation occurs from a closed conformation that ensures the tight fit of the nascent base pair within the binding pocket. This conserved mechanism safeguards the high-fidelity of replicative polymerases, but is not shared by the error-prone Y-family polymerases, because their spacious and solvent-accessible active sites are preformed with the fingers already in a closed position, even in the absence of DNA or dNTP. However, a significant conformational change does take place at least in some Y-family members, although it occurs during the transition from the apo state to DNA-bound binary state, by a rotation of the little finger domain relative to the polymerase catalytic core. In eukaryotes such movement has been shown for pol κ (Lone et al., 2007; Uljon et al., 2004). Although crystallographic studies did not detect a large finger subdomain motion upon the nucleotide substrate binding, several fluorescence studies have provided some insight into the conformational dynamics in the archaeal enzymes [reviewed in (Maxwell and Suo, 2014)]. These studies indicated conformational rearrangements in the active sites of Y-family polymerases taking place during nucleotide incorporation and provided the first evidence that TLS polymerases may adjust their conformational changes, so as to accommodate the proper positioning of lesion-containing DNA substrates.
Unique characteristics
All TLS polymerases are characterized by a similar general structural organization, they share multiple primary sequence motifs, and have the same ultimate goal – to ensure that replication of DNA is completed without leaving behind any unfilled gaps or discontinuances. However, even within one family, each individual member chooses its own strategy to tackle this task and each TLS polymerase is characterized by its own distinctive features, which define its specific catalytic activities.
DNA Polymerase η
The signature feature of the active site of pol η is its particularly large size, allowing it to accommodate the linked thymine bases of a cyclobutane pyrimidine dimer and to stabilize them for the correct pairing with an incoming dA (Biertümpfel et al., 2010; Yang, 2014). The fascinating feature of pol η is that it is not only able to efficiently and accurately interpret information encoded by the damaged Ts, but also ensures that the reading frame is properly maintained. This is possible because unlike pol κ, there is no gap between the catalytic core and the LF domain. As a result, extensive interactions between these domains stabilize the entire polymerase region and act as a “molecular splint” to keep the newly synthesized duplex in a stable B-form (Biertümpfel et al., 2010). Furthermore, Pol η processively inserts three more bases past the lesion, at which point it switches back to a distributive mode of DNA synthesis. Further extension is prevented by steric clashes displacing DNA from the enzyme and thus ensuring re-engagement of a high fidelity DNA polymerase. Interestingly, for the accurate bypass of CPDs pol η relies on two uniquely conserved residues (Arg61 and Gln38 in humans) located near the active site and synergistically contributing to the efficiency and fidelity of the enzyme (Biertümpfel et al., 2010; Su et al., 2015). However, these two residues also play a key role in the efficient misincorporation of dG opposite template T, especially when the template T is immediately preceded by an AT primer/template base pair, known as the WA motif (Zhao et al., 2013). Indeed, such a misincorporation pattern on undamaged DNA correlates with the well-known mutation signature of pol η generated during somatic hypermutation of immunoglobulin genes.
DNA Polymerase ι
The catalytic core of pol ι has two partially overlapping catalytic domains; one responsible for DNA polymerase activity and one that harbors dRP lyase activity (Figure 2B). Significant insight into the unusual base selectivity of pol ι that is characterized by a 105-fold difference in fidelity depending upon the template base has been gained from the crystal structure of its ternary complex with DNA substrates and an incoming nucleotide (Nair et al., 2004; Nair et al., 2006a; Nair et al., 2006b). Several large aliphatic residues in the active cleft of pol ι prevent the DNA template and dNTP from binding in its normal position and from forming a canonical Watson–Crick base pair. Instead, the unusually perturbed active site holds the template A fixed in the syn conformation limiting hydrogen-bonding opportunities with any incoming nucleotide other than dT in an anti conformation and thus promoting Hoogsteen base pairing that results in the accurate replication of template A (Nair et al., 2004). However, the same active site configuration is also responsible for the extreme infidelity of pol ι. The unique base selectivity opposite template T is explained by the fact that it is always held by pol ι in an anti conformation, irrespective of the identity of the incoming dNTP. Since an incoming dA adopts a syn conformation and exhibits reduced base stacking, pol ι gives preference to dG remaining in an anti conformation that is stabilized via hydrogen bonding with glutamine (Q59) in the finger subdomain (Nair et al., 2006a). At the same time, the enzyme’s narrow groove width imposition on the nascent base pair makes pol ι more accurate than most polymerases while copying DNA containing the oxidative lesion, 8-oxoG (Kirouac and Ling, 2011). In this case, pol ι firmly holds the damaged base in the syn conformation that would normally cause pairing with the incorrect dA. However, the restrictive active site prevents this from occurring and instead promotes stable pairing with the smaller and correct dC (Kirouac and Ling, 2011).
DNA Polymerase κ
Pol κ differs from other family members by an N-terminal extension (N-clasp) involved in DNA binding (Fig. 2B). A large structural gap divides the catalytic core and the LF in pol κ because of limited interactions between the domains, even in ternary complexes with DNA and dNTPs. The N-clasp stabilizes the polymerase by holding the catalytic core and LF together to encircle DNA (Lone et al., 2007; Uljon et al., 2004; Yang, 2014). It has been hypothesized that these features of pol κ make it specifically adept for the extension of a diverse range of primer termini distorted by base mismatches, or lesions in the template, even though in vivo findings suggest that the main “extender” in cells is pol ζ. Structural studies also suggested that pol κ has a more restrictive active-site cleft compared to other Y-family polymerases and is only able to accommodate only a single Watson–Crick base pair (Lone et al., 2007). Such a configuration explains the relatively higher fidelity of pol κ on undamaged DNA, its limited selection of lesions it can tolerate, and inability to bypass dinucleotide lesions (Bavoux et al., 2005). On the other hand, the structural gap between the catalytic core and LF appears to allow pol κ to accommodate bulky polycyclic aromatic hydrocarbons during lesion bypass (Liu et al., 2014).
Rev 1
The unique features of Rev1 include an extended N-terminal region containing BRCT (breast cancer-associated protein-1 carboxy-terminal) and MTS (mitochondrial targeting signal) domains and a C-terminal protein-binding domain. Similar to pol κ, the three-dimensional structure of Rev1 has a large gap that divides the LF domain and the ring-shaped catalytic core encircling DNA (Yang, 2014). This gap is filled by the N-terminal extension from the catalytic core (Nair et al., 2005; Swan et al., 2009). Two large inserts in the finger and palm domains specific for the catalytic core of human Rev1 (Figure 2, I1 and I2) have been suggested to play an important role in supporting the peculiar properties and unique functions of the protein in TLS (Swan et al., 2009). One insert (I1), has been proposed to serve as an additional platform for the protein-protein interactions needed for a specific non-catalytic role of human Rev1 in TLS, while the second insert (I2), is apparently important for the extremely limiting selectivity of Rev1 in the choice of nucleotide and template substrates in human cells.
Unlike other polymerases that depend on DNA to identify the nucleotide best suited for incorporation, Rev1 itself dictates the identity of both the templating base it uses as a substrate (mostly dG and a limited number of N2-adducted dG) and the incoming nucleotide insertion of dC. In order to do so, Rev 1 utilizes an unusual strategy. Instead of the direct pairing of the DNA template with the incoming dC, it swings the template base (dG) out of the helix replacing it with an arginine residue (R324), which forms a hydrogen bond with the incoming dC. The displaced template dG is temporarily accommodated within the hydrophobic pocket in the little finger domain (Nair et al., 2005; Nair et al., 2008), which in the human enzyme is supported by the large insert (Fig 2A, I2) in the fingers domain (Swan et al., 2009).
TLS protein interaction network
Although the overall TLS pathway appears to be conserved throughout evolution (Woodgate, 1999), there is some level of variability in its components between species. This includes the distribution of DNA polymerases themselves. Pol η and Rev1 have been found in all eukaryotic organisms checked thus far, while pols ι and κ have not been detected in some species (Ohmori et al., 2001). It is likely that this variability reflects environmental specificity and variations in DNA damage to which the different organisms are exposed.
Soon after the general mechanism of TLS had been uncovered, the focus in the field began shifting from the biochemical characterization of polymerases in vitro towards finding and characterization of their protein partners in vivo. While substantial progress has been made in this field, there are still a lot of “gray areas” and the proposed mechanisms governing protein interactions are being constantly reexamined. What is evident is that the access of error-prone TLS polymerases to a replication fork is strictly regulated through multiple levels of an elaborate protein interaction network.
Dynamic network of protein-protein interactions involving Rev1 and Rev7
The hREV7 protein is an integral subunit of pol ζ and its interaction with REV1 plays an important role in the recruitment of pol ζ for the bypass of many DNA lesions. Multiple studies have demonstrated that Rev7 not only interacts with Rev3 and REV1 (Murakumo et al., 2001), but also with various proteins that function in different pol ζ-dependent pathways unrelated to TLS (Iwai et al., 2007; Nelson et al., 1999; Weterman et al., 2001; Zhang et al., 2007). While Rev7 has its own protein interaction network independent of TLS, the ability of Rev1 to interact with various proteins is instrumental to TLS regulation. In fact, Rev1 works as a binding platform targeting different TLS polymerases to the stalled replication fork and coordinating their rearrangement in order to ensure the best possible outcome (Boehm et al., 2016; Guo et al., 2003; Ohashi et al., 2004; Waters et al., 2009).
Several studies indicated that Rev1 and Rev7 utilize two modes of protein binding, where one is driven by the recognition of a short motif sequence, while the other involves recognition of the protein tertiary structure. For example, it has been reported that the large region of hREV7 (~130 amino acids) is required for the interaction with the C-terminal domain of hREV1 (Murakumo et al., 2001) and an approximately 100 amino acid region in C-terminal domain of human REV1 (Figure 2A, Protein Interaction Region, PIR) is required for the interaction with all other polymerases from the Y-family, as well as with Rev7 and PolD3 subunits of pol ζ (Figure 2C) (Guo et al., 2003; Masuda et al., 2003; Murakumo et al., 2001; Ohashi et al., 2004; Pustovalova et al., 2016). On the other hand, sequences as short as 16 amino acids in pols η, ι and κ (Figure 2B, one Rev1-interacting region (RIR) in pols ι and κ and two such motifs in pol η) are sufficient for recognition by hRev1 (Figure 2C) (Ohashi et al., 2009). Similarly, two short 9 amino acid-motifs mapped to a region in the N-terminus of human Rev3 (Figure 2A, PIR 1 & 2 also called MCS, Minimum Core Sequence) must be intact to ensure binding to hRev7 (Hanafusa et al., 2010; Tomida et al., 2015). It has been proposed that presence of two binding sites in Rev3 can stabilize a homodimer of REV7 thereby expanding the opportunities for simultaneous interactions with its other protein partners and facilitating multi-protein complex formation, at least in vertebrates.
In a recent study, a second Rev7-interacting site has also been detected in human Rev1 (not shown on the Figure 2A because the exact size and position of this site within the region between amino acids 1–884 has yet to be determined) (Tomida et al., 2015). However, unlike hRev3, in which both Rev7 binding motifs (Fig 2A, PIR 1 & 2) cooperate in order to ensure proper functioning of pol ζ in the DNA damage tolerance pathway (Tomida et al., 2015), Rev7 binding to two sites in Rev1 has opposite biological consequences (Chun et al., 2013). Thus, binding of Rev7 near the C-terminus of Rev1 (Figure 2A, PIR) is important for the recruitment of pol ζ for TLS, while its binding to the N-terminal end promotes polyubiquitination and degradation of REV1(Chun et al., 2013).
Extensive structural studies revealed that an assembly of a multi-protein complexes is not only stimulated by the multiple independent binding sites, but it is also determined by the presence of more than one distinct interface in a specific binding site. For example, it has been shown that there are two different interfaces within a 100 amino acid region in the C-terminal domain of human Rev1 (Figure 2A, PIR) (Kikuchi et al., 2012; Pustovalova et al., 2012). The C-terminal part of this domain (C) is required for the interaction with the Rev7 subunit of pol ζ, while the opposite surface (N-terminal part) of the CTD of Rev1 (N) is involved in an interaction with the short RIR motifs of pols η, ι and κ (Figure 2B, RIR) (Kikuchi et al., 2012; Pustovalova et al., 2012; Wojtaszek et al., 2012b), as well as of PolD3 subunit of pol ζ (Pustovalova et al., 2016). These studies provide a molecular basis for a deeper insight into the highly complex mechanism mediating regulation and assembly of the TLS machinery. Thus, according to the current understanding, instead of competing for Rev1 binding, implied by the original model (Guo et al., 2003; Ohashi et al., 2004), pol ζ and one of the Y-family TLS polymerases cooperate in lesion bypass by simultaneous binding to the Rev1 scaffold (Kikuchi et al., 2012; Liu et al., 2013; Pozhidaeva et al., 2012; Pustovalova et al., 2012; Pustovalova et al., 2016; Wojtaszek et al., 2012a; Wojtaszek et al., 2012b; Xie et al., 2012; Xing et al., 2009) (Figure 3). This model suggests a mechanism by which Rev1 facilitates polymerase switching that is required to catalyze the two consecutive steps of TLS (Figure 3B & C) without the need for polymerase dissociation: 1) nucleotide insertion opposite the damaged site and 2) extension of the nascent strand past the lesion [for the origins of the two-step TLS model see (Bridges and Woodgate, 1985; Goodman and Woodgate, 2013; Livneh et al., 2010; Prakash and Prakash, 2002)]. Conformational changes in REV7 induced by binding to PolD3 mediates the interaction with Rev1 (Hanafusa et al., 2010; Hara et al., 2010) thereby ensuring efficient assembly of the “extender” complex (multi-subunit pol ζ) and facilitating displacement of the “inserter” polymerase (one of the members of the Y-family) (Kikuchi et al., 2012; Pustovalova et al., 2016) (Figure 3C). Furthermore, Rev1 seems to be instrumental in the selection of the best-suited polymerase for each TLS step via coordination of the replacement of a suboptimal DNA polymerase with a more appropriate enzyme, i.e., the one which is able to complete the task by catalyzing TLS of a specific lesion most efficiently and accurately (Ohashi et al., 2009).
Figure 3. Basic mechanism of TLS in eukaryotic cells.

TLS is a multi-step process involving (A) stalling of the replicative DNA polymerase (here shown for pol δ consisting of PolD1, PolD2, PolD3, and PolD4 subunits) at damaged sites (red); (B) PCNA ubiquitination, recruitment of a TLS polymerase to the primer terminus (here shown for pol η) and nucleotide (mis)insertion opposite the lesion; (C) extension (catalyzed by pol ζ) of the resulting primer termini by several nucleotides; (D) de-ubiquitination of PCNA and release of a TLS polymerase after bypass completion allowing the normal replication restart. TLS is regulated through an elaborate network of protein-protein interactions and posttranslational modifications (shown for different polymerases in A-E; see text for the details). TLS polymerases are recruited to the replication factories via interactions with PCNA (E), which due to the trimeric structure and various binding sites can accommodate multiple partners (A-E). The initial interactions through the PIP boxes (1) are too unstable, which makes them hard to be detected in replication foci. DNA damage reinforces the PCNA/TLS polymerases interactions due to mono-ubiquitination of PCNA (2) that is catalyzed by the RAD6/RAD18 complex (3). In normal cells, Rad18 forms a dimer consisting of a mono-ubiquitinated and non-ubiquitinated proteins (4). Non-modified Rad18 (5) that is required for PCNA ubiquitination is released from the dimer via an interaction with Rev1 (6) and is stabilized by an interaction with polη (7). Damage-induced phosphorylation of pol η and Rad18 facilitates their interaction and localization to replication foci (8). PCNA ubiquitination is further promoted by the binding of the polymerase molecules via their UBD domains (2). Pol η has a higher affinity for ubiquitinated PCNA and this interaction is sufficiently stable for pol η to form detectable foci, while the accumulation of pol ι requires its interaction with pol η (9 in the panel A). Mono-ubiquitination of either polη or polι enhances the interaction between these two polymerases (9). Cells use another protein binding platform, Rev1, which serves as a “polymerase bridge” for the assembly of two TLS polymerases [shown in panel B for pol ζ (through Rev7 as shown here, and through the PolD3 subunit) and pol κ (10)] and might facilitate the switch from the “inserter” (here shown for pol η) to the “extender” (pol ζ). Interactions between Rev3 (through Rev7 and PolD3) and Rev1 are possibly facilitated by Rev1 phosphorylation (11). Such interactions stabilize binding of pol ζ (through the PIP motif of PolD3) to ubiquitinated PCNA and thereby enhances pol ζ-dependent TLS.
Rev1 has also been shown to play an important structural role in yeast. It appears to be required for TLS catalyzed by pol ζ, but not by η, which lacks a PIR motif. Rev1 can nevertheless bind to pol η in vitro at a site within the LF domain, rather than the C-terminal domain, as in humans (Acharya et al., 2007) (Figure 2A). Furthermore, the C-terminal domain of Rev1, which in human is indispensible for TLS regulation, plays only a minor role in mediating structural function of Rev1 in yeast. Interestingly, the physical interaction of Rev1 with both Rev3 and pol η is inhibited by Rev7, whose binding to Rev1 involves the CTD, LF, and BRCT domains (Acharya et al., 2006; Acharya et al., 2007; D’Souza and Walker, 2006) (Figure 2A, BRCT & PIR 1 & 2). In contrast, S. cerevisiae Rev3 contains only a single Rev7-binding site (Gómez-Llorente et al., 2013; Makarova and Burgers, 2015), which is closer to the N-terminal end and larger (residues 532–588) than the two Rev7-binding sites in the human Rev3 (Figure 2A, compare PIR in Hs Rev3 to the PIRs 1 & 2 in Sc Rev3).
Proliferating cell nuclear antigen as an essential partner of TLS polymerases
Rev 1 and PCNA, cooperation or coordination?
Cells utilize different means to regulate TLS polymerases. Multiple lines of evidence indicate that the recruitment of these enzymes to replication factories is mainly controlled through their interactions with two scaffolding proteins: the TLS polymerase Rev1, and the sliding clamp, PCNA. Interaction between REV1 and PCNA themselves (Figure 3B) has been shown, suggesting a mechanism whereby Rev1 serves as an adaptor between the clamp and the other polymerases (de Groote et al., 2011; Guo et al., 2006a; Ross et al., 2005; Wood et al., 2007).
More recent studies using single-molecule fluorescence microscopy implied that REV1 and PCNA function as a hub for the formation of two multi-protein complexes having different architectures, the so-called PCNA “tool belt” and Rev1 “polymerase bridge” (Boehm et al., 2016). Analysis of the ternary complexes formed by PCNA, Rev1 and pol η suggests that these structures are dynamic and can interconvert without dissociation providing additional means for the facilitation of polymerase sampling and switching that ensures selection of the best candidate for TLS (Boehm et al., 2016). Nevertheless, none of the proposed recruitment mechanisms clarifies a hierarchy for targeting the TLS polymerases to one of the protein scaffolds.
Interaction between PCNA and TLS polymerases
It is well known that PCNA, a sliding platform that encircles duplex DNA and mediates protein-DNA interactions, is essential for the function of high-fidelity replicative polymerases and it is also crucial for the biological and biochemical modes of damage-induced mutagenesis performed by low fidelity polymerases. The physical interaction with PCNA loaded onto DNA by the clamp loader, RFC, with the help of the single-stranded DNA binding protein RPA, stimulates the localization of TLS polymerase in replication foci at sites of arrested replication (Figure 3A). Even though several lines of evidence suggest that the accumulation of Y-family polymerases in replication foci is not required for TLS [reviewed in (Masuda et al., 2015)], PCNA significantly stimulates the catalytic efficiency of all TLS polymerases on both non-damaged and damaged templates (Acharya et al., 2008; Bienko et al., 2005; Garg and Burgers, 2005; Haracska et al., 2001a; Haracska et al., 2001b; Haracska et al., 2002; Haracska et al., 2005a; Masuda et al., 2015; Vidal et al., 2004; Wit et al., 2015), with pol η stimulation being the most efficient. Known as a processivity factor of high fidelity polymerases, PCNA also improves the processivity of TLS enzymes allowing them to continue copying DNA past the lesion in a single binding event until the synthesized stretch is long enough to prevent degradation by the exonuclease activity of the replicative polymerase. Therefore, PCNA has at least two key roles in TLS; one as a modulator of polymerase activity; and the second, as a core structural component of a large multi-protein complex to which the polymerases can attach. Furthermore, the homotrimeric structure of PCNA suggests that it could, in principal, bind multiple proteins simultaneously through dynamic protein–protein interactions, thereby facilitating polymerase switching (De Biasio and Blanco, 2013).
Like most of the PCNA-interacting replication and repair proteins, TLS polymerases contain a short, conserved (PCNA-interacting protein) motif (Figure 2, A-C) which binds a hydrophobic pocket on the front face of the PCNA ring near the inter-domain connector loop (Jonsson et al., 1998; Maga and Hubscher, 2003; Moldovan et al., 2007; Warbrick, 2000). According to crystal structures of PCNA bound to the PIP motifs of pols η, ι and κ (Hishiki et al., 2009), there is some variability in the mode of the interaction of Y-family polymerases with PCNA. Thus, whereas conformation of the PIP motif of pol η is similar to that of other PCNA-binding proteins, the PIP motif of pol ι forms a different structure (Hishiki et al., 2009). Furthermore, pol ι has a single PIP box, while pol κ and pol η have two and three functionally distinct canonical sequence motifs, respectively (Figure 2B) (Masuda et al., 2015), and Rev1 has no readily identifiable canonical PIP box. Instead, a PCNA-binding signal is located in the C -terminal and possibly the N -terminal BRCT domain of Rev1 (Figure 2A) (Murakumo et al., 2006; Tissier et al., 2004).
The ubiquitin cycle and other means to regulate recruitment of TLS polymerases
The interaction between PCNA and Y-family polymerases through their PIP domains is weak, but can be significantly strengthened by mono-ubiquitination of PCNA (Figure 2C, 3B & E), which creates additional interacting surfaces. All four Y-family polymerases possess small ubiquitin-binding domains (UBDs) in their C-terminal regions known as UBZ (ubiquitin binding zinc domain of ~20 amino acids) and UBM (ubiquitin-binding motif of ~30 amino acids) which have a capacity to interact with mono-ubiquitinated PCNA (Ub-PCNA) (Figure 2A-C, 3E) (Yang and Woodgate, 2007). Pol ι and Rev1 have two UBMs while pols η and κ possess UBZs (one in pol η and two in κ) (Figure 2) (Bomar et al., 2007; Bomar et al., 2010; Burschowsky et al., 2011). Upon DNA damage, the Rad6 (the E2 ubiquitin-conjugating enzyme) in a complex with single-stranded binding protein Rad18 (E3 ubiquitin ligase) catalyzes binding of ubiquitin to Lysine-164 of PCNA (Figure 3E). Although such posttranslational modification does not significantly disrupt the interaction of replicative polymerases with PCNA, it nevertheless promotes binding of translesion polymerases (Figure 3B) (Bi et al., 2006; Watanabe et al., 2004).
Interestingly, binding to PCNA can also be regulated by the mono-ubiquitination of TLS polymerases themselves [this and other types of posttranslational modifications of proteins involved in TLS is reviewed in (McIntyre and Woodgate, 2015)]. It has been unambiguously shown that all Y-family polymerases are ubiquitinated in vivo and the effect of this posttranslational modification varies for different family members. The biological function of pol κ and Rev1 modification remains unclear. In contrast, significant information about the modes and consequences of ubiquitination of pol η and pol ι has been documented. In the case of pol η, ubiquitination occurs at one of four lysine residues within the nuclear localization signal (NLS) at its C-terminus. In contrast to pol η, pol ι has over 27 potential sites for ubiquitination that are located within different functional domains (McIntyre et al., 2015). Why pol ι is ubiquitinated at so many sites is puzzling, especially since the monoubiquitination status does not change after cells are exposed to a wide array of DNA-damaging agents (McIntyre et al., 2015). In contrast, UBM or UBZ-mediated covalent self-binding of ubiquitin to pol η is down regulated by DNA damage (Bienko et al., 2010). It has been shown that ubiquitination of pol η in undamaged cells causes a conformational change that blocks its PCNA-binding sites and thus inhibits the interaction with PCNA (Bienko et al., 2005; Bienko et al., 2010; Guo et al., 2006b; Jung et al., 2011; Plosky et al., 2006). Therefore, pol η de-ubiquitination in cells exposed to DNA damaging agents is required for the efficient interaction with the Ub-PCNA and subsequent recruitment to the site of replication (Bienko et al., 2005; Bienko et al., 2010; Plosky et al., 2006). On the other hand, mono-ubiquitination of either pol η or pol ι enhances the interaction between the two polymerases (Figure 3A), which it thought to occur through their respective UBZ and UBM ubiquitin-binding domains (McIntyre et al., 2013). Since the interaction with pol η facilitates the localization of pol ι into replication factories (Kannouche and Stary, 2003), it is possible that the ubiquitination of polι is dictated by the need to retain it in the vicinity of pol η.
DNA damage not only inhibits ubiquitination of pol η and induces PCNA mono-ubiquitination, but also stimulates deubiquitination of Rad18 (Figure 3E, 5–6), which in normal cells exists as a dimer consisting of a mono-ubiquitinated and non-ubiquitinated proteins (Figure 3E, 4) (Zeman et al., 2014). Ubiquitination prevents Rad18 from localizing to sites of DNA damage and inducing PCNA mono-ubiquitination. Furthermore, TLS polymerases themselves can influence the ubiquitination status of Rad18 and, as a consequence, ubiquitination of PCNA (Durando et al., 2013; Masuda et al., 2015). Very recently, Rev1 has been found to bind ubiquitinated Rad18 (Figure 3E, 4–5), leading to the release of the non-modified protomer from the dimer (Figure 3E, 5–6) and allowing it to ubiquitinate PCNA (Figure 3E, 2) (Wang et al., 2016). Furthermore, it has been proposed that pol η has the capacity to enhance and stabilize Rad18 (Figure 3E, 7). According to this hypothesis, DNA damage-induced phosphorylation of Rad18 (Figure 3E, 8–7) increases its affinity for pol η and promotes their mutual recruitment to the replication fork and ubiquitination of PCNA (Figure 3E, 3-2) (Barkley et al., 2012; Day et al., 2010).
On the other hand, the physical interaction with Rev18 mediates DNA damage–induced phosphorylation of pol η, which also requires an intact UBZ and facilitates localization of pol η to replication foci (Figure 3E, 7-3-2) (Gohler et al., 2011). An interaction with the Rad18 protein has also been reported for pol κ, suggesting that Rad18 can help to chaperone different TLS polymerases to a stalled replication fork (Bi et al., 2006; Watanabe et al., 2004).
Damage-dependent phosphorylation has also been reported for Rev1 (Pages et al., 2009; Sabbioneda et al., 2007) (Figure 3C. 11). Phosphorylation does not appear to regulate Rad18 de-ubiquitination, or affect Rev1 trans-localization to PCNA, but instead appears to contribute to increasing the efficiency of pol ζ-catalyzed TLS (Figure 3C), as seen in nucleotide excision repair-deficient cells. Therefore, it seems likely that phosphorylation does not affect the PCNA-dependent polymerase activity of REV1, but rather it regulates the function of Rev1 in its structural role by increasing the effectiveness of complex formation with pol ζ and possibly other polymerases.
Taken together, these findings point to ubiquitin (and ubiquitin-like proteins as discussed in McIntyre and Woodgate, 2015) as a key player in TLS regulation. Virtually no step within the extremely complex network of interconnecting processes takes place without the participation of this small molecule. This includes orchestrating access of different polymerases with partially overlapping functions to the replication fork, modulating the residence time of individual enzymes in the fork vicinity (Sabbioneda et al., 2008), and determining the probability of each TLS polymerase being used in replication past the lesion.
According to the current model, modification of PCNA by ubiquitin plays a major role in determining of the affinity of a particular polymerase for the sliding clamp and in governing polymerase switching (Kannouche et al., 2004) (Figure 3A-B). Structural and computational modeling studies suggest that although mobile, the ubiquitin moiety attached to Lysine-164 of PCNA has a preferred position on the side and back face of the ring-shaped PCNA (Figure 3B) that is a significant distance from the PIP-box-binding sites, such that it potentially increases the number of proteins that PCNA can simultaneously bind (Figure 3) (Tsutakawa et al., 2011; Zhang et al., 2012). Based on these data, it has been proposed that Y-family polymerases non-covalently bound to these sites can be held in an inactive orientation without interfering with the activity of other DNA replication and repair proteins which bind via their PIP motifs to the front surface of PCNA (Figure 2B & C, 3A). When required, the flexible TLS complex can readily move to the front face of PCNA (Figure 3B) and begin to take on its responsibilities. If a “first responder” TLS polymerase fails to replicate across the lesion, its ubiquitination will prevent the enzyme from rebinding. This will allow engagement of an alternative polymerase optimal for a particular lesion. Using such a mechanism in combination with the Rev1-based regulatory module, cells can probe different TLS polymerases until the appropriate one is selected.
After the lesion is successfully bypassed, the switch from TLS to replicative polymerases (Figure 3D) is also controlled by PCNA modification. In addition to various de-ubiquitination mechanisms which reduce the affinity of TLS polymerases for PCNA, it has been shown that PCNA ISGylation (binding of the ubiquitin-like protein, interferon-stimulated gene 15) promotes release of the TLS polymerase (pol η) from PCNA thus “turning off” the error-prone polymerase and preventing excessive mutagenesis (Park et al., 2014). Subsequent de-ISGylation of PCNA allows replicative DNA polymerases to reload and resume normal DNA replication.
It is also possible that cells might utilize alternative means to control the access of TLS polymerases to damaged DNA. For example, it has been reported that the Rev7 subunit of the pol ζ physically interacts with Mec3 and Ddc1 proteins in S.cerevisiae (Sabbioneda et al., 2005). These proteins, together with Rad17 form a heterotrimeric “9-1-1” checkpoint complex (consisting of Rad9, Hus1, and Rad1 in humans) that is structurally and functionally similar to the homotrimeric PCNA clamp (Venclovas and Thelen, 2000; Xu et al., 2009). Whether an interaction of pol ζ with the 9-1-1 complex is important for translesion synthesis similar to its interaction with PCNA, remains to be determined. However, the fact that a great deal of spontaneous pol ζ-dependent frameshift mutagenesis requires an intact 9-1-1 complex suggests that different polymerases and/or cellular pathways might utilize alternative clamps for the optimal control of access to damaged DNA.
Conclusions
The importance of understanding the regulation of inherently error-prone TLS is highlighted by its implications for human health, particularly with respect to cancer prevention and treatment. A surge of interest in this topic is illustrated by the fact that it has been the focus of numerous recent review articles (Jansen et al., 2015; Knobel and Marti, 2011; Korzhnev and Hadden, 2016; Lange et al., 2011; Maga and Hubscher, 2008; Makridakis and Reichardt, 2012; Tomasso et al., 2014). Uncontrolled error-prone nucleotide incorporation during TLS results in excessive accumulation of mutations that can lead to the transformation of normal cells to a malignant state. Among the many reasons for TLS deregulation are changes in the expression of the polymerase catalyzing lesion bypass; mutations in the TLS polymerase itself reducing its fidelity or improving its competitiveness; and modulation of the interactions of a TLS polymerase with its protein partners. An increase in genomic instability, a hallmark of cancer, has been linked to the (i) hindering of the normal replicase operation, (ii) imbalances in expression of DNA polymerases, (iii) lack of a relatively accurate TLS “polymerase of choice” responsible for bypassing a particular lesion in normal cells, or (iv) outcompeting the polymerase by another, less accurate enzyme [reviewed in (Beagan and McVey, 2016; Ghosal and Chen, 2013; Guo et al., 2013; Jansen et al., 2015; Knobel and Marti, 2011; Lange et al., 2011; Loeb and Monnat, 2008; Luo et al., 2012; Makridakis and Reichardt, 2012; Nicolay et al., 2012b; Parsons et al., 2013; Pillaire et al., 2014; Sharma et al., 2013; Shcherbakova and Fijalkowska, 2006; Sweasy et al., 2006; Yousefzadeh et al., 2014)].
All TLS polymerases have been shown to act as both contributors and suppressors of carcinogenesis. The most prominent example is the defect in DNA polymerase η that is directly connected with the development of skin cancers. No gross deregulation (either up- or down) of any other TLS polymerase has been linked to human disease. This can probably be explained by the fact that large changes in the cellular amounts of these enzymes are incompatible with organism survival. However, polymorphisms and moderate alterations in their expression levels in cancer cells have been reported [reviewed in (Jansen et al., 2014b; Stallons and McGregor, 2010)].
TLS polymerases not only modulate tumor development, but also influence the outcome of cancer chemotherapy [reviewed in (Maga and Hubscher, 2008; Salehan and Morse, 2013)]. Indeed, efficient replication past DNA lesions induced by DNA-damaging antineoplastic agents can reduce the susceptibility of cancer cells to these agents. Furthermore, mutations introduced by error-prone enzymes during bypass synthesis might promote secondary tumor formation and contribute to the acquisition of drug resistance. Both these mechanisms have been demonstrated in cell lines and in mice (Korzhnev and Hadden, 2016; Lange et al., 2011; Maga and Hubscher, 2008; Salehan and Morse, 2013; Xie et al., 2010). Therefore, suppression of the TLS pathway during chemotherapy can be used to enhance drug efficacy, prevent development of secondary neoplasia, and sensitize otherwise drug resistant tumor cells to anticancer treatment (Korzhnev and Hadden, 2016). However development of new potentially fruitful treatment strategies should be approached with caution, so that the beneficial effects of TLS incapacitation would not be overpowered by the life-threatening unbalancing of the overall genomic stability, which may actually cause an acceleration of carcinogenesis.
TLS polymerases have also been implicated in other pathways connected with human health. For example, an important role of pol ζ in the preservation of the genetic integrity of the mitochondrial genome (Singh et al., 2015) suggests that deregulation of Rev3 can lead to cellular dysfunctions and a wide variety of mitochondrial disease.
Despite the tremendous progress in the field of TLS polymerases that have been made this century, many questions remain to be answered. Just to highlight a few: detailed biochemical and structural characterization of human pol ζ still awaits to be accomplished; further advances should be made in uncovering of biological pathways involving other error-prone polymerases; deeper understanding of the precise mechanisms of TLS regulation and targeting that ensures DNA synthesis with the highest accuracy and efficiency should be acquired; and, most importantly, greater attention should be paid to finding ways to use all this information to prevent and fight human disease.
Acknowledgments
Funding
This work was supported by funds from the National Institutes of Health/National Institute of Child Health and Human Development Intramural Research Program to RW.
Biographies
Dr. Alexandra Vaisman is an Interdisciplinary Scientist in the Laboratory of Genomic Integrity at the National Institute of Child Health and Human Development. She has worked at the NIH since 2000, studying various aspects of the damage tolerance pathways. She received her Ph.D., from the Moscow Institute of Fine Chemical Technology.
Dr. Roger Woodgate is Chief of the Laboratory of Genomic Integrity at the National Institute of Child Health and Human Development. He has devoted his scientific career to understanding the molecular mechanisms of damage-induced mutagenesis. He received his Ph.D., from the University of Sussex, England, and joined the NIH in 1989.
Footnotes
Disclosure statement
The authors declare no conflict of interest
References
- Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–63. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase η. Mol Cell Biol. 2007;27:8401–2408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Gali H, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc Natl Acad Sci U S A. 2008;105:17724–9. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types : Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeck N, Janel-Bintz R, Wagner J, et al. FF483-484 motif of human Polη mediates its interaction with the POLD2 subunit of Polδ and contributes to DNA damage tolerance. Nucleic Acids Res. 2015;43:2116–25. doi: 10.1093/nar/gkv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, et al. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem. 2012;287:17281–7. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril EF, Brown OE, Jenkins MD, Laszlo J. Deoxyribonucleic acid polymerase with rat liver ribosomes and smooth membranes. Purification and properties of the enzymes. Biochemistry. 1971;10:1981–92. doi: 10.1021/bi00787a004. [DOI] [PubMed] [Google Scholar]
- Barkley LR, Palle K, Durando M, et al. c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Polη recruitment to stalled replication forks. Mol Biol Cell. 2012;23:1943–54. doi: 10.1091/mbc.E11-10-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruffini E, Serafini F, Ferrero I, Lodi T. Overexpression of DNA polymerase ζ reduces the mitochondrial mutability caused by pathological mutations in DNA polymerase γ in yeast. PLoS One. 2012;7:e34322. doi: 10.1371/journal.pone.0034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoux C, Hoffmann JS, Cazaux C. Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase κ. Biochimie. 2005;87:637–46. doi: 10.1016/j.biochi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Baynton K, Bresson-Roy A, Fuchs RP. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol Microbiol. 1999;34:124–33. doi: 10.1046/j.1365-2958.1999.01583.x. [DOI] [PubMed] [Google Scholar]
- Beagan K, McVey M. Linking DNA polymerase θ structure and function in health and disease. Cell Mol Life Sci. 2016;73:603–15. doi: 10.1007/s00018-015-2078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Tissier A, Frank EG, et al. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science. 2001;291:2156–59. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- Berger H, Jr, Huang RC, Irvin JL. Purification and characterization of a deoxyribonucleic acid polymerase from rat liver. J Biol Chem. 1971;246:7275–83. [PubMed] [Google Scholar]
- Bergoglio V, Boyer AS, Walsh E, et al. DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Rey L, Wang G, et al. Role of TLS DNA polymerases η and κ in processing naturally occurring structured DNA in human cells. Mol Carcinog. 2009;48:369–78. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Barkley LR, Slater DM, et al. Rad18 regulates DNA polymerase κ and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–40. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi J, Rudd SG, Jozwiakowski SK, et al. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell. 2013;52:566–73. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–24. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, et al. Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- Biertümpfel C, Zhao Y, Kondo Y, et al. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–8. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L. DNA polymerase lambda [Mus musculus] GenBank; 1998. https://www.ncbi.nlm.nih.gov/protein/CAB65241 (November 23, 2016) [Google Scholar]
- Boehm EM, Spies M, Washington MT. PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 2016;44:8250–60. doi: 10.1093/nar/gkw563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EM, Washington MT. R.I.P. to the PIP: PCNA-binding motif no longer considered specific : PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. Bioessays. 2016;38:1117–22. doi: 10.1002/bies.201600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–64. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden A, Noy GP, Weissbach A. DNA polymerase of mitochondria is a γ-polymerase. J Biol Chem. 1977;252:3351–6. [PubMed] [Google Scholar]
- Bollum FJ, Potter VR. Thymidine incorporation of into deoxyribonucleic acid of rat liver homogenates. J Am Chem Soc. 1957;79:3603–04. [Google Scholar]
- Bomar MG, Pai MT, Tzeng SR, et al. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep. 2007;8:247–51. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar MG, D’Souza S, Bienko M, et al. Unconventional ubiquitin recognition by the ubiquitin-binding motif within the Y family DNA polymerases ι and Rev1. Mol Cell. 2010;37:408–17. doi: 10.1016/j.molcel.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, et al. Investigating the role of the little finger domain of Y-family DNA polymerases in low-fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–40. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- Boulet A, Simon M, Faye G, et al. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989;8:1849–54. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouron A, Kiselyov K, Oberwinkler J. Permeation, regulation and control of expression of TRP channels by trace metal ions. Pflugers Arch. 2015;467:1143–64. doi: 10.1007/s00424-014-1590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson A, Fuchs RP. Lesion bypass in yeast cells: Pol η participates in a multi-DNA polymerase process. EMBO J. 2002;21:3881–87. doi: 10.1093/emboj/cdf363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA, Woodgate R. The two-step model of bacterial UV mutagenesis. Mutation Research. 1985;150:133–39. doi: 10.1016/0027-5107(85)90110-1. [DOI] [PubMed] [Google Scholar]
- Brown JA, Fowler JD, Suo Z. Kinetic basis of nucleotide selection employed by a protein template-dependent DNA polymerase. Biochemistry. 2010;49:5504–10. doi: 10.1021/bi100433x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM, Koonin EV, Bruford E, et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–90. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- Burschowsky D, Rudolf F, Rabut G, et al. Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase ι in complex with ubiquitin. J Biol Chem. 2011;286:1364–73. doi: 10.1074/jbc.M110.135038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis KC, Harris PV. A possible functional role for a new class of eukaryotic DNA polymerases. Curr Biol. 1997;7:R743–4. doi: 10.1016/s0960-9822(06)00391-5. [DOI] [PubMed] [Google Scholar]
- Byrnes JJ, Downey KM, Black VL, So AG. A new mammalian DNA polymerase with 3′ to 5′ exonuclease activity: DNA polymerase δ. Biochemistry. 1976;15:2817–23. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]
- Cann IK, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–67. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LM, Bollum FJ. Low molecular weight deoxyribonucleic acid polymerase in mammalian cells. J Biol Chem. 1971;246:5835–7. [PubMed] [Google Scholar]
- Chargaff E. Structure and function of nucleic acids as cell constituents. Fed Proc. 1951;10:654–9. [PubMed] [Google Scholar]
- Chen H, Sun Y, Stark T, et al. Nucleotide sequence and deletion analysis of the polB gene of Escherichia coli. DNA Cell Biol. 1990;9:631–5. doi: 10.1089/dna.1990.9.631. [DOI] [PubMed] [Google Scholar]
- Chiapperino D, Kroth H, Kramarczuk IH, et al. Preferential misincorporation of purine nucleotides by human DNA polymerase η opposite benzo[a]pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts. J Biol Chem. 2002;277:11765–71. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- Choi JY, Lim S, Kim EJ, et al. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun AC, Kok KH, Jin DY. REV7 is required for anaphase-promoting complex-dependent ubiquitination and degradation of translesion DNA polymerase REV1. Cell Cycle. 2013;12:365–78. doi: 10.4161/cc.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2016;44:D67–72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol Cell Biol. 2006;26:8173–82. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daube SS, Tomer G, Livneh Z. Translesion replication by DNA polymerase δ depends on processivity accessory proteins and differs in specificity from DNA polymerase β. Biochemistry. 2000;39:348–55. doi: 10.1021/bi9917784. [DOI] [PubMed] [Google Scholar]
- Day TA, Palle K, Barkley LR, et al. Phosphorylated Rad18 directs DNA polymerase η to sites of stalled replication. J Cell Biol. 2010;191:953–66. doi: 10.1083/jcb.201006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasio A, Blanco FJ. Proliferating cell nuclear antigen structure and interactions: too many partners for one dancer? Adv Protein Chem Struct Biol. 2013;91:1–36. doi: 10.1016/B978-0-12-411637-5.00001-9. [DOI] [PubMed] [Google Scholar]
- de Groote FH, Jansen JG, Masuda Y, et al. The Rev1 translesion synthesis polymerase has multiple distinct DNA binding modes. DNA Repair. 2011;10:915–25. doi: 10.1016/j.dnarep.2011.04.033. [DOI] [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, et al. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–6. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despras E, Sittewelle M, Pouvelle C, et al. Rad18-dependent SUMOylation of human specialized DNA polymerase η is required to prevent under-replicated DNA. Nat Commun. 2016;7:13326. doi: 10.1038/ncomms13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Watson NB, Turkington G, et al. Decreased frequency and highly aberrant spectrum of ultraviolet-induced mutations in the hprt gene of mouse fibroblasts expressing antisense RNA to DNA polymerase ζ. Mol Cancer Res. 2003;1:836–47. [PubMed] [Google Scholar]
- Dominguez O, Ruiz JF, Lain De Lera T, et al. DNA polymerase mu (Pol m), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–42. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, McLenigan MP, Yang W, et al. The steric gate of human DNA polymerase ι regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J Biol Chem. 2014;289:9136–45. doi: 10.1074/jbc.M113.545442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, Cerritelli SM, McDonald JP, et al. Unlocking the steric gate of DNA polymerase η leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair. 2015;35:1–12. doi: 10.1016/j.dnarep.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durando M, Tateishi S, Vaziri C. A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013;41:3079–93. doi: 10.1093/nar/gkt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Godindagger I, Klein U, et al. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–24. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- Focher F, Gassmann M, Hafkemeyer P, et al. Calf thymus DNA polymerase δ independent of proliferating cell nuclear antigen (PCNA) Nucleic Acids Res. 1989;17:1805–21. doi: 10.1093/nar/17.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank EG, Sayer JM, Kroth H, et al. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diolexpoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase ι. Nucleic Acids Res. 2002;30:5284–92. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank EG, Woodgate R. Increased catalytic activity and altered fidelity of DNA polymerase ι in the presence of manganese. J Biol Chem. 2007;282:24689–96. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–68. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RP, Fujii S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012682. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res. 2008;18:174–83. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, et al. DNA polymerase lambda (Pol l), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301:851–67. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- García-Gómez S, Reyes A, Martínez-Jimenez MI, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–53. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Burgers PM. Ubiquitinated PCNA activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci U S A. 2005;102:18361–66. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Aravind L, Gotway G, et al. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci U S A. 1999;96:11922–27. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–29. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler T, Sabbioneda S, Green CM, Lehmann AR. ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J Cell Biol. 2011;192:219–27. doi: 10.1083/jcb.201008076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Llorente Y, Malik R, Jain R, et al. The architecture of yeast DNA polymerase ζ. Cell Reports. 2013;5:79–86. doi: 10.1016/j.celrep.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Woodgate R. Translesion DNA Polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Zhao L, Pence MG, Egli M. Structure and function of the translesion DNA polymerases and interactions with damaged DNA. Perspectives in Science. 2015;4:24–31. [Google Scholar]
- Gueranger Q, Stary A, Aoufouchi S, et al. Role of DNA polymerases η, ι and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008;7:1551–62. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Guilliam TA, Keen BA, Brissett NC, Doherty AJ. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 2015;43:6651–64. doi: 10.1093/nar/gkv625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–30. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sonoda E, Tang TS, et al. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006a;23:265–71. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006b;26:8892–900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66:2363–81. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhou G, Zhang W, et al. A novel POLH mutation causes XP-V disease and XP-V tumor proneness may involve imbalance of numerous DNA polymerases. Oncol Lett. 2013;6:1583–90. doi: 10.3892/ol.2013.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines ME, Holmes AM, Johnston IR. Distinct cytoplasmic and nuclear DNA polymerases from rat liver. FEBS Lett. 1971;17:63–67. doi: 10.1016/0014-5793(71)80564-1. [DOI] [PubMed] [Google Scholar]
- Hanafusa T, Habu T, Tomida J, et al. Overlapping in short motif sequences for binding to human REV7 and MAD2 proteins. Genes Cells. 2010;15:281–96. doi: 10.1111/j.1365-2443.2009.01380.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Hashimoto H, Murakumo Y, et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J Biol Chem. 2010;285:12299–307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, et al. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol. 2001a;21:7199–206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001b;98:14256–61. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, et al. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol Cell Biol. 2002;22:784–91. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Acharya N, Unk I, et al. A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005a;25:1183–90. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Prakash L, Prakash S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol Cell Biol. 2005b;25:10183–9. doi: 10.1128/MCB.25.22.10183-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, et al. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–71. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M, Benkovic SJ. Regulation of Rad6/Rad18 Activity During DNA Damage Tolerance. Annu Rev Biophys. 2015;44:207–28. doi: 10.1146/annurev-biophys-060414-033841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. PrimPol breaks replication barriers. Nat Struct Mol Biol. 2013;20:1348–50. doi: 10.1038/nsmb.2727. [DOI] [PubMed] [Google Scholar]
- Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Tsuda M, Mohiuddin, et al. In vivo evidence for translesion synthesis by the replicative DNA polymerase δ. Nucleic Acids Res. 2016;44:7242–50. doi: 10.1093/nar/gkw439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A, Hashimoto H, Hanafusa T, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284:10552–60. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kolbanovskiy A, Wu X, et al. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol κ. Biochemistry. 2003;42:2456–66. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- Hübscher U, Spadari S, Villani G, Maga G. History and discovery of DNA polymerases DNA Polymerases: Discovery, Characterization and Functions in Cellular DNA Transactions. London: World Scientific Publishing Co. Pte. Ltd.; 2010. pp. 1–58. [Google Scholar]
- Iguchi M, Osanai M, Hayashi Y, et al. The error-prone DNA polymerase ι provides quantitative resistance to lung tumorigenesis and mutagenesis in mice. Oncogene. 2014;33:3612–7. doi: 10.1038/onc.2013.331. [DOI] [PubMed] [Google Scholar]
- Ishino S, Ishino Y. DNA polymerases as useful reagents for biotechnology – the history of developmental research in the field. Front Microbiol. 2014;5:465. doi: 10.3389/fmicb.2014.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–57. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H, Kim M, Yoshikawa Y, et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell. 2007;130:611–23. doi: 10.1016/j.cell.2007.06.043. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–96. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, et al. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–23. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Temviriyanukul P, Wit N, et al. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 2014a;42:11071–82. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Jacobs H, de Wind N. Mutatis mutandis: Error-prone translesion synthesis, fitness and disease. In: Maiorano D, Hoffmann J-S, editors. Function of Translesion DNA polymerases in Genome Stability. Kerala: India Research Signpost; 2014b. pp. 201–32. [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, de Wind N. Roles of mutagenic translesion synthesis in mammalian genome stability, health and disease. DNA Repair. 2015;29:56–64. doi: 10.1016/j.dnarep.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Xu M, Lai Y, et al. Bypass of a 5′,8-cyclopurine-2′-deoxynucleoside by DNA polymerase β during DNA replication and base excision repair leads to nucleotide misinsertions and DNA strand breaks. DNA Repair. 2015;33:24–34. doi: 10.1016/j.dnarep.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J Biol Chem. 2008;283:26297–301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of Xeroderma Pigmentosum. Science. 1999a;285:263–65. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, polη. Science. 1999b;283:1001–04. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Polθ. Proc Natl Acad Sci U S A. 2000a;97:3838–43. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, et al. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000b;406:1015–19. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000c;275:7447–50. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase ζ (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci U S A. 2012;109:12455–60. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–25. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Kelley WS, Grindley ND. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982;257:1958–64. [PubMed] [Google Scholar]
- Jung GH, Leavitt MC, Hsieh JC, Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987;84:8287–91. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Hakem A, Hakem R, Chen X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase η to suppress Translesion DNA Synthesis. Mol Cell Biol. 2011;19:3997–4006. doi: 10.1128/MCB.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalf GF, Ch’ih JJ. Purification and properties of deoxyribonucleic acid polymerase from rat liver mitochondria. J Biol Chem. 1968;243:4904–16. [PubMed] [Google Scholar]
- Kalifa L, Sia EA. Analysis of Rev1p and Pol ζ in mitochondrial mutagenesis suggests an alternative pathway of damage tolerance. DNA Repair. 2007;6:1732–9. doi: 10.1016/j.dnarep.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Stary A. Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie. 2003;85:1123–32. doi: 10.1016/j.biochi.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA; a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Katafuchi A, Nohmi T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: their efficiency and template base preference. Mutat Res. 2010;703:24–31. doi: 10.1016/j.mrgentox.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–31. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, et al. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–9. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Khare V, Eckert KA. The proofreading 3′→5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat Res. 2002;510:45–54. doi: 10.1016/s0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Hara K, Shimizu T, et al. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J Biol Chem. 2012;287:33847–52. doi: 10.1074/jbc.M112.396838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Ling H. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota. Proc Natl Acad Sci U S A. 2011;108:3210–5. doi: 10.1073/pnas.1013909108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y, Akaboshi E, Shinagawa H, et al. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:4336–40. doi: 10.1073/pnas.82.13.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippers R. DNA polymerase II. Nature. 1970;228:1050–53. doi: 10.1038/2281050a0. [DOI] [PubMed] [Google Scholar]
- Knobel PA, Marti TM. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011;11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Lehman IR, Bessman MJ, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–8. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML. DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant. Biochem Biophys Res Commun. 1970;40:1348–55. doi: 10.1016/0006-291x(70)90014-8. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML. Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II. Proc Natl Acad Sci U S A. 1971;68:761–64. doi: 10.1073/pnas.68.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Hadden MK. Targeting the translesion synthesis pathway for the development of anti-cancer chemotherapeutics. J Med Chem. 2016;59:9321–36. doi: 10.1021/acs.jmedchem.6b00596. [DOI] [PubMed] [Google Scholar]
- Krijger PH, Tsaalbi-Shtylik A, Wit N, et al. Rev1 is essential in generating G to C transversions downstream of the Ung2 pathway but not the Msh2+ Ung2 hybrid pathway. Eur J Immunol. 2013;43:2765–70. doi: 10.1002/eji.201243191. [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Koonin EV, McDonald JP, et al. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res. 1996;357:245–53. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- Lambert WC, Lambert MW. Development of effective skin cancer treatment and prevention in xeroderma pigmentosum. Photochem Photobiol. 2015;91:475–83. doi: 10.1111/php.12385. [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nature Reviews Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Bedford E, Reh S, et al. Dual role for mammalian DNA polymerase ζ in maintaining genome stability and proliferative responses. Proc Natl Acad Sci U S A. 2013;110:E687–96. doi: 10.1073/pnas.1217425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer FW, Perry JR, Hardigree AA. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence and functional analysis. J Bacteriol. 1989;171:230–37. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–59. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Arlett CF, et al. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci U S A. 1975;72:219–23. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 2005;579:873–6. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–9. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Lin Q, Clark AB, McCulloch SD, et al. Increased susceptibility to UV-induced skin carcinogenesis in polymerase η-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- Lin YC, Li L, Makarova AV, et al. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis. 2014a;35:1461–8. doi: 10.1093/carcin/bgu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Li L, Makarova AV, et al. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J Biol Chem. 2014b;289:18497–506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–33. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Liu D, Ryu KS, Ko J, et al. Insights into the regulation of human Rev1 for translesion synthesis polymerases revealed by the structural studies on its polymerase-interacting domain. J Mol Cell Biol. 2013;5:204–6. doi: 10.1093/jmcb/mjs061. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang Y, Tang TS, et al. Variants of mouse DNA polymerase κ reveal a mechanism of efficient and accurate translesion synthesis past a benzo[a]pyrene dG adduct. Proc Natl Acad Sci U S A. 2014;111:1789–94. doi: 10.1073/pnas.1324168111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–35. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljon SN, et al. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–14. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Luo Q, Lai Y, Liu S, et al. Deregulated expression of DNA polymerase β is involved in the progression of genomic instability. Environ Mol Mutagen. 2012;53:325–33. doi: 10.1002/em.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madejczyk MS, Ballatori N. The iron transporter ferroportin can also function as a manganese exporter. Biochim Biophys Acta. 2012;1818:651–7. doi: 10.1016/j.bbamem.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–60. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Maga G, Hubscher U. Repair and translesion DNA polymerases as anticancer drug targets. Anticancer Agents Med Chem. 2008;8:431–47. doi: 10.2174/187152008784220348. [DOI] [PubMed] [Google Scholar]
- Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–5. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–26. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, McElhinny SA, Watts BE, et al. Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Repair. 2014;18:63–7. doi: 10.1016/j.dnarep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Burgers PM. Eukaryotic DNA polymerase ζ. DNA Repair. 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makridakis NM, Reichardt JK. Translesion DNA polymerases and cancer. Front Genet. 2012;3:174. doi: 10.3389/fgene.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD. Polν, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–9. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Wersto RP, et al. Different mutation signatures in DNA polymerase η- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci U S A. 2005;102:8656–61. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–60. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Kanao R, Kaji K, et al. Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases. Nucleic Acids Res. 2015;43:7898–910. doi: 10.1093/nar/gkv712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999a;18:3491–501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999b;399:700–04. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–09. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, et al. Error rate and specificity of human and murine DNA polymerase η. J Mol Biol. 2001;312:335–46. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- Maxwell BA, Suo Z. Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases. Biochemistry. 2014;53:2804–14. doi: 10.1021/bi5000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Masutani C, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–68. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Rapic-Otrin V, Epstein JA, et al. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Frank EG, Plosky BS, et al. Identification of a nonsense mutation in DNA polymerase ι from 129-derived strains of mice and its effect on somatic hypermutation. J Exp Med. 2003;198:635–43. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, et al. Human DNA polymerase η promotes DNA synthesis from strand invasion intermediates (D-loops) of homologous recombination. Mol Cell. 2005;20:783–92. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- McIntyre J, Vidal AE, McLenigan MP, et al. Ubiquitin mediates the physical and functional interaction between human DNA polymerases η and ι. Nucleic Acids Res. 2013;41:1649–60. doi: 10.1093/nar/gks1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre J, McLenigan MP, Frank EG, et al. Posttranslational regulation of human DNA polymerase ι. J Biol Chem. 2015;290:27332–44. doi: 10.1074/jbc.M115.675769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre J, Woodgate R. Regulation of translesion DNA synthesis: Posttranslational modification of lysine residues in key proteins. DNA Repair. 2015;29:166–79. doi: 10.1016/j.dnarep.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhou Y, Zhang S, et al. DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–57. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RR, Simpson MV. DNA biosynthesis in mitochondria: partial purification of a distinct DNA polymerase from isolated rat liver mitochondria. Proc Natl Acad Sci U S A. 1968;61:130–7. doi: 10.1073/pnas.61.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J, et al. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–67. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, et al. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–51. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Moscato B, Swain M, Loria JP. Induced fit in the selection of correct versus incorrect nucleotides by DNA polymerase β. Biochemistry. 2016;55:382–95. doi: 10.1021/acs.biochem.5b01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses RE, Richardson CC. A new DNA polymerase activity of Escherichia coli. I. Purification and properties of the activity present in E. coli polA1. Biochem Biophys Res Commun. 1970;41:1557–64. doi: 10.1016/0006-291x(70)90565-6. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Ogura Y, Ishii H, et al. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Mizutani S, Yamaguchi M, et al. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes Cells. 2006;11:193–205. doi: 10.1111/j.1365-2443.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, et al. Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–80. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, et al. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–22. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, et al. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure. 2006a;14:749–55. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, et al. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat Struct Mol Biol. 2006b;13:619–25. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, et al. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure. 2008;16:239–45. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Narita T, Tsurimoto T, Yamamoto J, et al. Human replicative DNA polymerase δ can bypass T-T (6-4) ultraviolet photoproducts on template strands. Genes Cells. 2010;15:1228–39. doi: 10.1111/j.1365-2443.2010.01457.x. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996a;382:729–31. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996b;272:1646–49. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Nelson KK, Schlondorff J, Blobel CP. Evidence for an interaction of the metalloprotease-disintegrin tumour necrosis factor α convertase (TACE) with mitotic arrest deficient 2 (MAD2), and of the metalloprotease-disintegrin MDC9 with a novel MAD2-related protein, MAD2β. Biochem J. 1999;343(Pt 3):673–80. [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8:125–32. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay NH, Carter R, Hatch SB, et al. Homologous recombination mediates S-phase-dependent radioresistance in cells deficient in DNA polymerase η. Carcinogenesis. 2012a;33:2026–34. doi: 10.1093/carcin/bgs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay NH, Helleday T, Sharma RA. Biological relevance of DNA polymerase β and translesion synthesis polymerases to cancer and its treatment. Curr Mol Pharmacol. 2012b;5:54–67. [PubMed] [Google Scholar]
- O’Day CL, Burgers PMJ, Taylor J-S. PCNA-induced DNA synthesis past cis-syn and trans-syn-I thymine dimers by calf thymus DNA polymerase d in vitro. Nucleic Acids Res. 1992;20:5403–06. doi: 10.1093/nar/20.20.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polk protects mammalian cells against the lethal and mutagenic effects of polycyclic hydrocarbons. Proc Natl Acad Sci U S A. 2002;99:15548–53. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR. The Y-family DNA polymerase k functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–2. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, et al. Fidelity and processivity of DNA synthesis by DNA polymerase k, the product of the human DINB1 gene. J Biol Chem. 2000a;275:39786–684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Ogi T, Kusumoto R, et al. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes & Dev. 2000b;14:1589–94. [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, et al. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–31. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Hanafusa T, Kamei K, et al. Identification of a novel REV1-interacting motif necessary for DNA polymerase κ function. Genes Cells. 2009;14:101–11. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkumo T, Kondo Y, Yokoi M, et al. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol Cell Biol. 2006;26:7696–706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Hatada E, Qiao Y, et al. dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutation Research. 1995;347:1–7. doi: 10.1016/0165-7992(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Pages V, Santa Maria SR, Prakash L, Prakash S. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 2009;23:1438–49. doi: 10.1101/gad.1793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Yang SW, Yu KR, et al. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol Cell. 2014;54:626–38. doi: 10.1016/j.molcel.2014.03.031. [DOI] [PubMed] [Google Scholar]
- Parsons JL, Nicolay NH, Sharma RA. Biological and therapeutic relevance of nonreplicative DNA polymerases to cancer. Antioxid Redox Signal. 2013;18:851–73. doi: 10.1089/ars.2011.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Rogozin IB, Galkin AP, et al. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase η during copying of a mouse immunoglobulin κ light chain transgene. Proc Natl Acad Sci U S A. 2002;99:9954–59. doi: 10.1073/pnas.152126799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KL, Elledge SJ, Mitchell B, et al. umuDC and mucAB operons whose products are required for UV light and chemical-induced mutagenesis: UmuD, MucA, and LexA products share homology. Proc Natl Acad Sci U S A. 1985;82:4331–35. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaire MJ, Betous R, Hoffmann JS. Role of DNA polymerase κ in the maintenance of genomic stability. Mol Cell Oncol. 2014;1:e29902. doi: 10.4161/mco.29902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaire MJ, Cazaux C, Hoffmann JS. Hidden behind translesion synthesis: The secret roles of specialized DNA polymerases in unstressed cells. In: Maiorano D, Hoffmann J-S, editors. Function of Translesion DNA polymerases in Genome Stability. Kerala: India Research Signpost; 2015. pp. 233–60. [Google Scholar]
- Plosky BS, Vidal A, Fernández de Henestrosa AR, et al. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006;25:2847–55. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope-Varsalona H, Liu FJ, Guzik L, Opresko PL. Polymerase η suppresses telomere defects induced by DNA damaging agents. Nucleic Acids Res. 2014;42:13096–109. doi: 10.1093/nar/gku1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozhidaeva A, Pustovalova Y, D’Souza S, et al. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochemistry. 2012;51:5506–20. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Dev. 2002;16:1872–83. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Prasad R, Bebenek K, Hou E, et al. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase ι by controlled proteolysis. J Biol Chem. 2003;278:29649–54. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- Pustovalova Y, Bezsonova I, Korzhnev DM. The C-terminal domain of human Rev1 contains independent binding sites for DNA polymerase η and Rev7 subunit of polymerase ζ. FEBS letters. 2012;586:3051–6. doi: 10.1016/j.febslet.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustovalova Y, Magalhaes MT, D’Souza S, et al. Interaction between the Rev1 C-terminal domain and the PolD3 subunit of polζ suggests a mechanism of polymerase exchange upon Rev1/Polζ-dependent translesion synthesis. Biochemistry. 2016;55:2043–53. doi: 10.1021/acs.biochem.5b01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechkoblit O, Zhang Y, Guo D, et al. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–94. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and Is specialized for translesion replication. J Biol Chem. 1999;274:31763–66. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Rey L, Sidorova JM, Puget N, et al. Human DNA polymerase η is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–54. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, et al. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat Immunol. 2001;2:530–6. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- Ropp PA, Copeland WC. Characterization of a new DNA polymerase from Schizosaccharomyces pombe: a probable homologue of the Saccharomyces cerevisiae DNA polymerase λ. Gene. 1995;165:103–7. doi: 10.1016/0378-1119(95)00412-y. [DOI] [PubMed] [Google Scholar]
- Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–9. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda S, Minesinger BK, Giannattasio M, et al. The 9-1-1 checkpoint clamp physically interacts with polζ and is partially required for spontaneous polζ-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280:38657–65. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Bortolomai I, Giannattasio M, et al. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair. 2007;6:121–7. doi: 10.1016/j.dnarep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Gourdin AM, Green CM, et al. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol Biol Cell. 2008;19:5193–202. doi: 10.1091/mbc.E08-07-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff BU, Heath-Pagliuso S, Castano IB, et al. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics. 1995;141:465–79. doi: 10.1093/genetics/141.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–52. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehan MR, Morse HR. DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br J Biomed Sci. 2013;70:31–40. doi: 10.1080/09674845.2013.11669927. [DOI] [PubMed] [Google Scholar]
- Schenten D, Gerlach VL, Guo C, et al. DNA polymerase k deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–60. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase δ. Biochimie. 2009;91:1163–72. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Marini F, Wood RD. Polθ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–26. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–8. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase θ (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- Sharma S, Canman CE. REV1 and DNA polymerase ζ in DNA interstrand crosslink repair. Environ Mol Mutagen. 2012;53:725–40. doi: 10.1002/em.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Frontiers in Bioscience. 2006;11:2496–517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- Silvian LF, Toth EA, Pham P, et al. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struc Biol. 2001;8:984–89. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- Singh B, Li X, Owens KM, et al. Human REV3 DNA polymerase ζ localizes to mitochondria and protects the mitochondrial genome. PLoS One. 2015;10:e0140409. doi: 10.1371/journal.pone.0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Wootton JC. Widespread eukaryotic sequences, highly similar to bacterial DNA polymerase I, looking for functions. Curr Biol. 1997;7:R463–5. doi: 10.1016/s0960-9822(06)00236-3. [DOI] [PubMed] [Google Scholar]
- Stallons LJ, McGregor WG. Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/643857. pii:643857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JE, Kumar D, Binz SK, et al. Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair. 2011;10:826–34. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Patra A, Harp JM, et al. Roles of residues Arg-61 and Gln-38 of human DNA polymerase η in bypass of deoxyguanosine and 7,8-Dihydro-8-oxo-2′-deoxyguanosine. J Biol Chem. 2015;290:15921–33. doi: 10.1074/jbc.M115.653691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Ohashi E, Kolbanovskiy A, et al. Translesion synthesis by human DNA polymerase k on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–06. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, et al. Structure of the human Rev1-DNA-dNTP ternary complex. J Mol Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- Tang M, Bruck I, Eritja R, et al. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD’2C mutagenic complex and RecA. Proc Natl Acad Sci U S A. 1998;95:9755–60. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, et al. UmuD’2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc Natl Acad Sci U S A. 1999;96:8919–24. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Frank EG, McDonald JP, et al. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase ι. EMBO J. 2000a;19:5259–66. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. polι, a remarkably error-prone human DNA polymerase. Genes & Dev. 2000b;14:1642–50. [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, et al. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase polη and REVl protein. DNA Repair. 2004;3:1503–14. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Tomasiewicz HG, McHenry CS. Sequence analysis of the Escherichia coli dnaE gene. J Bacteriol. 1987;169:5735–44. doi: 10.1128/jb.169.12.5735-5744.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasso A, Casari G, Maga G. What makes Y family pols potential candidates for molecular targeted therapies and novel biotechnological applications. Curr Mol Med. 2014;14:96–114. doi: 10.2174/15665240113136660080. [DOI] [PubMed] [Google Scholar]
- Tomida J, Takata K, Lange SS, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 2015;43:1000–11. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η. Implications for translesion DNA synthesis. Mol Cell. 2001;8:417–26. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, et al. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently bound ubiquitin on PCNA. Proc Natl Acad Sci U S A. 2011;108:17672–7. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uljon SN, Johnson RE, Edwards TA, et al. Crystal structure of the catalytic core of human DNA polymerase κ. Structure. 2004;12:1395–404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past Oxaliplatin and Cisplatin GpG adducts by human DNA polymerase η. Biochemistry. 2000;39:4575–80. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. Unique misinsertion specificity of polι may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–29. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Frank EG, McDonald JP, et al. Polι-dependent lesion bypass in vitro. Mutat Res. 2002;510:9–22. doi: 10.1016/s0027-5107(02)00248-8. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Frank EG, Iwai S, et al. Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase ι. DNA Repair. 2003;2:991–1006. doi: 10.1016/s1568-7864(03)00094-6. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Lehmann AR, Woodgate R. DNA polymerases η and ι. In: Yang W, editor. DNA Repair and Replication. San Diego: Elsevier/Academic Press; 2004. pp. 205–28. [Google Scholar]
- Vaisman A, Woodgate R. Translesion DNA polymerases, eukaryotic. In: Lennarz WJ, Lane MD, editors. Encyclopedia of Biological Chemistry. Oxford: Academic Press, Elsevier Science; 2004. pp. 247–50. [Google Scholar]
- Vaisman A, Takasawa K, Iwai S, Woodgate R. DNA polymerase ι-dependent translesion replication of uracil containing cyclobutane pyrimidine dimers. DNA Repair. 2006;5:210–18. doi: 10.1016/j.dnarep.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. Ribonucleotide selectivity of translesion DNA synthesis polymerases. In: Maiorano D, Hoffmann J-S, editors. Function of translesion DNA polymerases in genome stability. Kerala: India Research Signpost; 2014. pp. 1–23. [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–93. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, et al. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J Biol Chem. 2004;279:48360–68. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- Villani G, Hubscher U, Gironis N, et al. In vitro gap-directed translesion DNA synthesis of an abasic site involving human DNA polymerases ε, λ, and β. J Biol Chem. 2011;286:32094–104. doi: 10.1074/jbc.M111.246611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani G, Shevelev I, Orlando E, et al. Gap-directed translesion DNA synthesis of an abasic site on circular DNA templates by a human replication complex. PLoS One. 2014;9:e93908. doi: 10.1371/journal.pone.0093908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, et al. The dinB gene encodes an novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol Cell. 1999;4:281–86. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Woodgate R, McManus TP, et al. Evidence that in Xeroderma Pigmentosum variant cells, which lack DNA polymerase η, DNA polymrase ι causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–26. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- Wang Z, Castano IB, De Las Penas A, et al. Pol κ: a DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–79. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- Wang Z, Huang M, Ma X, et al. REV1 promotes PCNA monoubiquitylation through interacting with ubiquitylated RAD18. J Cell Sci. 2016;129:1223–33. doi: 10.1242/jcs.179408. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci U S A. 2000;97:3094–99. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, et al. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–96. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–54. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Weissbach A, Schlabach A, Fridlender B, Bolden A. DNA polymerases from human cells. Nat New Biol. 1971;231:167–70. doi: 10.1038/newbio231167a0. [DOI] [PubMed] [Google Scholar]
- Weissbach A, Baltimore D, Bollum F, et al. Nomenclature of eukaryotic DNA polymerases. Science. 1975;190:401–2. doi: 10.1126/science.1179222. [DOI] [PubMed] [Google Scholar]
- Weterman MA, van Groningen JJ, Tertoolen L, van Kessel AG. Impairment of MAD2B-PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc Natl Acad Sci U S A. 2001;98:13808–13. doi: 10.1073/pnas.241304198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Jackson MA, Pata JD. Y-family polymerase conformation is a major determinant of fidelity and translesion specificity. Structure. 2013;21:20–31. doi: 10.1016/j.str.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TM, Vaisman A, Martomo SA, et al. MSH2-MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–45. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wit N, Buoninfante OA, van den Berk PC, et al. Roles of PCNA ubiquitination and TLS polymerases κ and η in the bypass of methyl methanesulfonate-induced DNA damage. Nucleic Acids Res. 2015;43:282–94. doi: 10.1093/nar/gku1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Research. 2006;66:134–42. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- Wojtaszek J, Lee CJ, D’Souza S, et al. Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ. J Biol Chem. 2012a;287:33836–46. doi: 10.1074/jbc.M112.394841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek J, Liu J, D’Souza S, et al. Multifaceted recognition of vertebrate Rev1 by translesion polymerases ζ and κ. J Biol Chem. 2012b;287:26400–8. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SW, Wahl AF, Yuan PM, et al. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Garg P, Burgers PM. A Ubiquitin-binding motif in the translesion DNA polymerase rev1 mediates its essential functional interaction with ubiquitinated PCNA in response to DNA damage. J Biol Chem. 2007;282:20256–63. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes & Dev. 1999;13:2191–95. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20792–7. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Yang X, Xu M, Jiang T. Structural insights into the assembly of human translesion polymerase complexes. Protein Cell. 2012;3:864–74. doi: 10.1007/s13238-012-2102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Kirouac K, Shin YJ, et al. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. MolMicrobiol. 2009;71:678–91. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Bai L, Gong Y, et al. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–61. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lai Y, Jiang Z, et al. A 5′, 8-cyclo-2′-deoxypurine lesion induces trinucleotide repeat deletion via a unique lesion bypass by DNA polymerase β. Nucleic Acids Res. 2014;42:13749–63. doi: 10.1093/nar/gku1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto J, Loakes D, Masutani C, et al. Translesion synthesis across the (6–4) photoproduct and its Dewar valence isomer by the Y-family and engineered DNA polymerases. Nucleic Acids Symp Ser (Oxf) 2008:339–40. doi: 10.1093/nass/nrn171. [DOI] [PubMed] [Google Scholar]
- Yang K, Weinacht CP, Zhuang Z. Regulatory role of ubiquitin in eukaryotic DNA translesion synthesis. Biochemistry. 2013;52:3217–28. doi: 10.1021/bi400194r. [DOI] [PubMed] [Google Scholar]
- Yang W. Portraits of a Y-family DNA polymerase. FEBS Lett. 2005;579:868–72. doi: 10.1016/j.febslet.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–98. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase η. Biochemistry. 2014;53:2793–803. doi: 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S, Yavuz AS, Kraemer KH, Lipsky PE. The role of polymerase h in somatic hypermutation determined by analysis of mutations in a patient with xeroderma pigmentosum variant. J Immunol. 2002;169:3825–30. doi: 10.4049/jimmunol.169.7.3825. [DOI] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Wyatt DW, Takata K, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Lin JR, Freire R, Cimprich KA. DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis. J Cell Biol. 2014;206:183–97. doi: 10.1083/jcb.201311063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chatterjee A, Singh KK. Saccharomyces cerevisiae polymerase ζ functions in mitochondria. Genetics. 2006;172:2683–8. doi: 10.1534/genetics.105.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang SH, Sharrocks AD. Rev7/MAD2B links c-Jun N-terminal protein kinase pathway signaling to activation of the transcription factor Elk-1. Mol Cell Biol. 2007;27:2861–9. doi: 10.1128/MCB.02276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, et al. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000a;28:4717–24. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, et al. Error-free and error-prone lesion bypass by human DNA polymerase κ In vitro. Nucleic Acids Res. 2000b;28:4138–46. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol Cell Biol. 2000c;20:7099–108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D, et al. Two-step error-prone bypass of the (+)- and (-)-trans-anti-BPDE-N2-dG adducts by human DNA polymerases η and κ. Mutat Res. 2002a;510:23–35. doi: 10.1016/s0027-5107(02)00249-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D, et al. Activities of human DNA polymerase κ in response to the major benzo[a]pyrene adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair. 2002b;1:559–69. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang S, Lin SH, et al. Structure of monoubiquitinated PCNA: implications for DNA polymerase switching and Okazaki fragment maturation. Cell cycle. 2012;11:2128–36. doi: 10.4161/cc.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Gregory MT, Biertumpfel C, et al. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase η. Proc Natl Acad Sci U S A. 2013;110:8146–51. doi: 10.1073/pnas.1303126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM, et al. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Research. 2006;34:4731–42. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, et al. DNA polymerase ζ cooperates with polymerases κ and ι in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci U S A. 2009;106:11552–7. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmudzka BZ, SenGupta D, Matsukage A, et al. Structure of rat DNA polymerase β revealed by partial amino acid sequencing and cDNA cloning. Proc Natl Acad Sci U S A. 1986;83:5106–10. doi: 10.1073/pnas.83.14.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]


