Abstract
Measuring regulatory T cell suppression provides important insight into T cell dysfunction in autoimmune disease. However, to date, suppression assays are limited by the requirement for freshly isolated cells, and significant cell numbers. Here, we present a novel and rapid in vitro assay using effector T cell surface expression of both CD25 and CD134 as a surrogate marker of regulatory T cell-mediated suppression. This surface marker-based suppression assay works for frozen samples and for samples with limited cell numbers. It is also shorter taking two days to complete compared to the four days required for proliferation-based assays. Furthermore, this assay works with both in vitro expanded and natural Tregs, as well as anti-CD3/anti-CD28 bead-based and APC stimulation conditions. In conclusion, we have developed and validated a new suppression assay for cryopreserved samples with limited cell numbers that may be helpful to investigate T cell regulation in the context of infection or autoimmune diseases.
Keywords: Suppression Assay, regulatory T cells, CD25, CD134, proliferation
1. Introduction
Quantification of immune cell function is paramount to our understanding of the pathogenesis of immunological diseases and to identifying successful therapies. In patients with autoimmune disease, the regulation of the immune system is of particular interest and CD4+ regulatory T cells (Tregs) have been an important research area. CD4+ CD25+FOXP3+ Tregs suppress CD4 T cells via multiple mechanisms, leading to decreased cell proliferation, changes in expression of cell surface proteins, and diminished production of cytokines. In the study of human disease, measurement of suppression in vitro is vital to research and has been achieved through co-culture of Tregs and T effector cells (Teff). Methods include measuring cell proliferation (thymidine incorporation or CFSE dye dilution); cytokine production (typically IFNγ and TNFα) (McMurchy and Levings, 2012); and activation markers (CD69 and CD154) on the cell surface (Canavan et al., 2012). Although in vitro assays may not fully reflect in vivo suppression, co-culture assays do identify defective Treg function (Bacchetta et al., 2006) and demonstrate resistance of effector cells (CD4+CD25−) to suppression by Treg in the setting of several autoimmune diseases (Lawson et al., 2008; Schneider et al., 2008; Goodman et al., 2009; Schneider and Buckner, 2011; Schneider et al., 2013).
T cell proliferation is dependent on signals via the TCR and co-stimulatory molecule CD28. Among the proteins that are expressed in response to T cell activation is IL2Rα (CD25), which is very tightly controlled through transcriptional regulation. CD25 is minimally expressed in resting CD4 T cells, but is rapidly upregulated upon stimulation of the TCR (Kim et al., 2006). A number of other cell surface molecules are rapidly upregulated following TCR signaling including OX40 (CD134), ICAM-1 (CD54), CD69, and CD40L (CD154), which have a variety of functions such as aiding proliferation, survival, cytokine production, and cell adhesion (Tohma et al., 1992; Mardiney et al., 1996; Chatzigeorgiou et al., 2009; Croft et al., 2009).
Overall, the assessment of Treg mediated suppression of effector T cells has been limited due to the number of cells required, the need for long culture times (4–6 days), and the complexity of experimental set up of Treg assays. CSFE based co-culture assays have become the gold standard of in vitro assays, but consume larger numbers of cells, due to cell loss during the CSFE staining process (approx. 50% cells lost), and require 4 days of culture. Measurement of proliferation by uptake of tritiated thymidine can be achieved with a limited number of cells, but does not allow concurrent assessment of number of cell divisions and the characteristics of dividing cells available with flow cytometry techniques. Early activation markers CD69 and CD154 have been used to assess suppression (Canavan et al., 2012), (Ruitenberg et al., 2011), an approach that decreases the time of co-culture to as few as 7 hours and has been used successfully to assess the function of Tregs, which have been enriched or expanded in vitro or the target of IL-2 immunotherapy (Berglund et al., 2013; Hannon et al., 2014; Ito et al., 2014; Landwehr-Kenzel et al., 2014). However, this assay has barriers due to the need to perform the assay within a single extended workday and high reagent costs. In other studies, CD25 and CD134 have been used to identify antigen specific responses and their regulation in the context of viral infection (Endl et al., 2006; Zaunders et al., 2009; Shaw et al., 2011; Keoshkerian et al., 2012).
Our goal was to develop a method to measure events close to the start of cell division, but prior to extensive proliferation. To achieve this, we examined the cell surface molecules known to be upregulated in activated/proliferating cells at 48 hours (the time in which initial T cell division occurs after activation) in order to identify an alternative marker that could be used to measure suppression. This study showed that the combined expression of CD25 and CD134 on effector cells is a good surrogate for, or additional measure to proliferation that is suppressed by Tregs to a similar degree.
2. Materials and methods
2.1. Subjects
This study was approved by the Benaroya Research Institute’s Institutional Review Board, and all subjects signed written informed consent prior to inclusion in the study. Frozen PBMCs and fresh peripheral blood samples were obtained from the Benaroya Research Institute Immune Mediated Disease Registry and Repository. In total, cryopreserved samples from 50 healthy adult volunteers were used for suppression assays. In addition, six volunteers provided fresh blood samples for expanded Tregs, CD4+CD25+ nTregs, or large quantities of Teff from a single source as assay controls.
2.2. Autoantibodies and reagents
Flow cytometry data were acquired on a FACS Canto (BD Biosciences). Teff (CD4+CD25−) were stained with CFSE (Invitrogen) and Tregs with eFluor670 (EF670) (eBiosciences) as previously published (Schneider and Buckner, 2011). In addition, the following were used: FITC anti-CD69; PE CD25, CD54, CD134, CD137, CD152, CD154, CD183 or CD278; PE-Cy5 CD4 or CD25; PE-Cy7 CD25; BV421 CD279 or CD134; APC-Cy7 CD4; and Live/Dead Aqua (Invitrogen). Analysis of cell cycle progression was carried out according to manufacturer’s instructions using a FITC BrdU kit (BD Biosciences). Cells were cultured in RPMI1640 HEPES media containing 10% pooled human serum, additional L-glutamine, penicillin/streptomycin, and sodium pyruvate (Sigma).
2.3. Cell sources
Unless otherwise specified, experiments used an expanded source of regulatory cells (eTregs) from a single healthy control, and effector cells (Teff) were isolated from frozen PBMC. The eTregs were generated by isolation of CD4+CD25HICD127− using a FACS Aria (BD) and expansion over 14 days with IL-2 (Chiron) and anti-CD3/anti-CD28 (aCD3/aCD28)–coated Dynabeads (Invitrogen) (Putnam et al., 2009). The percentage of FOXP3+ cells was verified to be greater than 90%. Effector cells (CD4+CD25−) were isolated using MACs kits through negative selection for CD4 and positive selection for CD25 (Miltenyi Biotec) (Schneider and Buckner, 2011). In experiments using ex vivo Tregs (nTregs), the highest 2% of CD25 stained CD4+ cells were isolated from PBMC using a FACs Aria. In these experiments, freshly sorted Teffs were the lowest 50% of CD25 stained CD4+ cells. The marker CD127 was not included in this isolation strategy, for simplicity, based on previous data showing high (90%) FOXP3 expression in the cells with the highest 3–5% of CD25 expression (Mikacenic et al., 2014).
2.4. Suppression Assays
Analysis was carried out at 7 hours, 1 day (20–24 hours, d1), 2 days (44–48 hours, d2), and/or 4 days (90–96 hours, d4) as indicated. For all time points Teffs (number of cells varied between assays) were co-cultured with several ratios of Tregs cells in 96-well plates with anti-CD3/anti-CD28 Dynabeads (Invitrogen). Tregs and Teff were cultured at ratios between 0:1, 1:2 to 1: 64 in doubling dilutions. Where insufficient cells were available ratios of 1:8>1:4>1:2>1:16 were prioritized. For analysis, Teff cultured in media alone were used to set gates for the various activation markers used or proliferation. EF670 was used to identify Treg; Teff cells were in the EF670− gate. Representative gating for 7 hours, 2 days, and 4 days is shown in Figs. S1–S3. Flow cytometry was standardized between experiments using eight peak beads (Spherotech).
For the 7-hour assay, five wells of 50,000 Teff were prepared per condition. These were stimulated with Dynabeads (Invitrogen) at a bead:Teff ratio of 1:5 (high) or 1:28 (low). Anti-CD154 PE was added at start of assay all other markers were cell surface stained at the end of assay. Total cell requirement for this method was approximately 1.8 million Teff and 250,000 eTregs per person.
For the 1 day, 2 day, and 4 day experiments 100,000 Teff were plated three times for each condition, and Dynabeads were added at a ratio of 1:28 (beads:Teff). Adaptations of this method included the use of a 1:35 (beads:Teff) ratio for nTreg experiments, and a 1:4 ratio (Teff:irradiated CD4− APC) with additional aCD3 (UCHT1, 5μg/mL) and aCD28 (CD28.2, 1μg/mL) for APC experiments. When added, Pam3CSK4 (1μg/ml; Invivogen) was included at the start of co-culture. Analysis of proliferation by CFSE and activation by cell surface marker were carried out on the same cells. For these experiments approximately 2.5 million CD4+CD25− and 300,000 Tregs were used for each day of analysis. An adaptation of this method used 5,000 Teff plated in triplicate, this required 90,000 Teff and 15,000 nTregs per ‘miniature’ assay; In these experiments flow cytometry based sorting of 60 million frozen PBMC yielded between 20–70,000 nTregs after staining with EF670.
Percentage suppression (S) was calculated as follows:
where a is the percentage proliferation or marker+ (e.g. CD25+CD134+) in the absence of Tregs and b is the percentage proliferation or marker+ in the presence of Tregs.
2.5. Statistical analysis
Spearman’s rank correlation was used to analyze the relationship between different measures of activation/proliferation and suppression. Bland-Altman analysis was used to interrogate bias between different measures for activation/proliferation and suppression. Wilcoxon matched pairs test was used to compare suppression in the presence/absence of Pam3CSK4. Kruskal-Wallis test was used to compare suppression based on source of Teff and Treg in ‘miniature’ assays. FlowJo software (Tree Star) was used for all analysis of flow cytometry and Prism (GraphPad Software) was used for all statistical analysis.
3. Results
3.1. CD25 cell surface expression correlates with CFSE-based proliferation at day 4
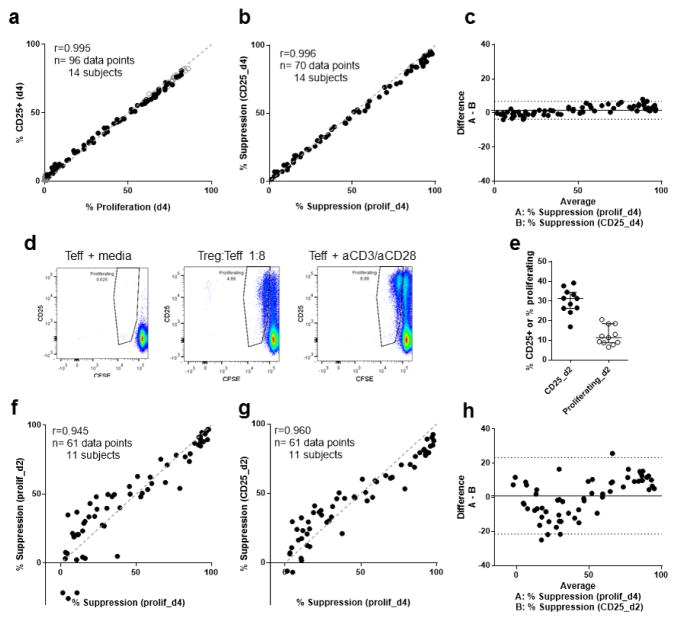
CD25 is upregulated in response to T cell stimulation and by IL-2 within 48 hours of initial activation, and persists for at least 4 days (Wang et al., 2008). To determine whether CD25 expression on Teff correlated with their number of cell divisions based on CFSE dilution, co-culture experiments were performed with Teffs (CD4+CD25−) from 14 healthy volunteers. Teffs were labelled with CFSE, and then placed in culture with in vitro expanded Tregs (eTregs) (Putnam et al., 2009) and anti-CD3/anti-CD28 coated Dynabeads. These experiments showed that the percentage of proliferating cells determined by dye dilution compared to the percentage of CFSE labeled cells that were CD25+ on day 4 of co-culture were highly correlated (r=0.995, Fig. 1a). The suppression of both of these measures by eTregs was also highly correlated (r=0.996, Fig. 1b). Bland-Altman analysis of suppression demonstrated a low overall bias of 1.6% (equivalent to 1.6% difference in suppression between the two methods), but this was not uniform (Fig. 1c). The 95% limits of agreement were −3.6 to 6.9%. The level of expression of CD25 (measured by mean fluorescent intensity) on the surface of the cells also correlated with proliferation but was less well correlated than percentage of CD25+ cells (Fig. S4). These data demonstrate a strong agreement between CD25 expression and proliferation particularly when percentage suppression is in the mid-range (40–60% suppression), which our group has found to be most sensitive to differences between individuals (Schneider and Buckner, 2011).
Figure 1. Suppression of CD25 Teff surface expression correlates with suppression of Teff proliferation.
(a) Correlation of CD25 surface expression on Teff and CFSE-based Teff proliferation at day 4. (b) Correlation of percentage suppression of CD25+ Teff and percentage suppression of CFSE-based Teff proliferation at day 4. (c) Bland-Altman plot of percentage suppression of CD25 surface expression on Teff and percentage suppression of CFSE-based Teff proliferation at day 4. (d) For one representative individual, the dilution of CFSE through cell division and expression of CD25 on Teff are shown for day 2. (e) CD25 surface expression and CFSE-based proliferation on day 2 from Teff cultured with anti-CD3/anti-CD28 beads alone. (f) Correlation of percentage suppression of CFSE-based Teff proliferation at days 2 and 4. (g) Correlation of percentage suppression of CD25+ Teff at day 2 and percentage suppression of CFSE-based Teff proliferation at day 4. (h) Bland-Altman plot of percentage suppression of CD25+ Teff at day 2 and percentage suppression of CFSE-based Teff proliferation at day 4. For day 4 (a–c), 96 data points representing different Treg:Teff ratios for 14 healthy controls are shown (7 experiments with no replicated samples). From the same experiments on day 2 (e–h), 70 data points from 11 of 14 healthy controls due to low cell numbers from three participants. Spearman’s rank correlation coefficient: a (r=0.995, P<0.0001), b (r=0.996, P<0.0001), f (r=0.945, P<0.0001), and g (r=0.960, P<0.0001); dashed line shows y=x, solid circles show Teff+Treg, and open circles in a show Teff alone. Bland-Altman analysis was used to compare methods in c (bias=1.6%) and h (bias= 0.9%). The solid line shows average bias, dotted lines 95% CI for agreement. No statistical comparison was made for (e), median and interquartile range are shown.
3.2. The suppression of surface CD25 at day 2 predicts suppression of proliferation at day 4
To shorten the duration of this assay, cell surface markers were assessed at days 1 and 2, and compared to day 4 proliferation. A single cell division is typically demonstrable by CFSE dilution on day 2 of co-culture. At this time-point only 6–21% of Teff cultured alone have divided (Fig. 1d) while 16–40% of Teff expressed CD25 (Fig. 1e). In co-culture assays, cell division at day 2 correlated with proliferation on day 4 (r=0.945, Fig. 1f) and suppression of CD25 expression on day 2 correlated with suppression of proliferation on day 4 (r=0.960, Fig. 1g). The overall bias for both proliferation and CD25 as measures was low (0.8% and −1.0% respectively). However, limits of agreement were higher than 20% for both these comparisons (Fig. 1h) and were particularly variable when suppression was low. Thus, neither cell division as measured by CFSE or expression of CD25 alone were adequate markers for suppression on day 2.
In experiments concurrent with the above assays, the 7-hour suppression assay developed by Canavan et al were performed (Canavan et al., 2012). We performed the assay using two different levels of anti-CD3/anti-CD28 beads: a “high” 1:5 ratio of Teff:beads, as per the published method, and a “low” 1:28 ratio of Teff:beads as per our standard 4 day assay. Suppression of CD69+CD154+ at 7 hours was correlated with suppression of proliferation on day 4 of culture when either high (r=0.721, p<0.001) or low (p=0.926, p<0.001) concentrations of beads were used. However, despite the high correlation, the level of suppression observed utilizing this method, with either high or low beads, was lower than suppression measured on day 4 (bias was 53% and 24%, respectively, Fig. S5). Thus, we went on to perform further experiments with a focus on development of a 2-day assay.
3.3. Additional surface markers improve assessment of suppression at earlier time points
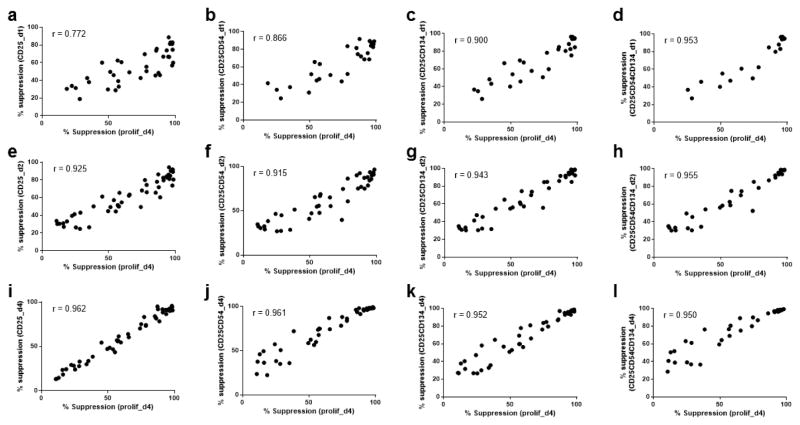
Complementary cell surface markers were investigate in an attempt to remove the non-uniform bias described above (Fig. 1c, h, i). In preliminary experiments (data not shown), the expression and suppression of CD54, CD134, CD137, CD152, CD183, CD278, and CD279 on days 1, 2, and 4 were compared to CD25 and proliferation. From these data, CD54 and CD134 were chosen for further investigation. In experiments with Teff from 10 additional healthy subjects, suppression was calculated based on the percentage of cells on days 1, 2 or 4 that were CD25+, CD25+CD54+, CD25+CD134+, or CD25+CD54+CD134+. On day 4, correlation between CD25 alone and proliferation remained the highest (Fig. 2i–l). However, on day 1 and day 2, combinations of CD25+CD134+CD54+/− had higher correlation values than CD25 alone (r=0.772 vs. r>0.9 Fig. 2a–d, and r=0.925 vs r>0.94 Fig. 2e–h, respectively). Thus, a combination of cell surface markers CD25, CD134 and CD54 could be used to assess suppression at day 2. Although the addition of CD54 on day 1 or day 2 improved correlation coefficients, in future experiments, analysis was restricted to CD25+CD134+ on day 1 and day 2, and analysis of CD25 alone on day 4, in order to simplify the flow cytometry fluorophore scheme.
Figure 2. Suppression of both CD54 and CD134 Teff surface expression also correlate with suppression of Teff proliferation.
Correlation of percentage suppression of CD25+ Teff (a, day 1; e, day 2; i, day 4) and percentage suppression of CFSE-based Teff proliferation at day 4. Correlation of percentage suppression of CD25+CD54+ Teff (b, day 1; f, day 2; j, day 4) and percentage suppression of CFSE-based Teff proliferation at day 4. Correlation of percentage suppression of CD25+CD134+ Teff (c, day 1; g, day 2; k, day 4) and percentage suppression of CFSE-based Teff proliferation at day 4. The median frequency of CD25+CD134+ on day 2 in Teff cultured without Treg suppression was 25% (range 10–41%). Correlation of percentage suppression of CD25+CD54+CD134+ Teff (d, day 1; h, day 2; l, day 4) and percentage suppression of CFSE-based Teff proliferation at day 4. Spearman’s rank correlation coefficients: a (r=0.772; P<0.0001), b (r=0.866; P<0.0001), c (r=0.900; P<0.0001), d (r=0.953; P<0.0001), e (r=0.925; P<0.0001), f (r=0.915; P<0.0001), g (r=0.943; P<0.0001), h (r=0.955; P<0.0001), i (r=0.962; P<0.0001), j (r=0.961; P<0.0001), k (r=0.952; P<0.0001), and l (r=0.950; P<0.0001). Depending on the number of cells available ratios of 1:2–1:16 Treg:Teff were included for each sample and compared to Teff with anti-CD3/anti-CD28 beads for between 5 to 14 healthy controls from 3 experiments with no replicates. Not all combinations of markers were stained in all assays.
3.4 Surface marker-based suppression assay correlates with proliferation-based suppression assay when using nTregs or APC activation
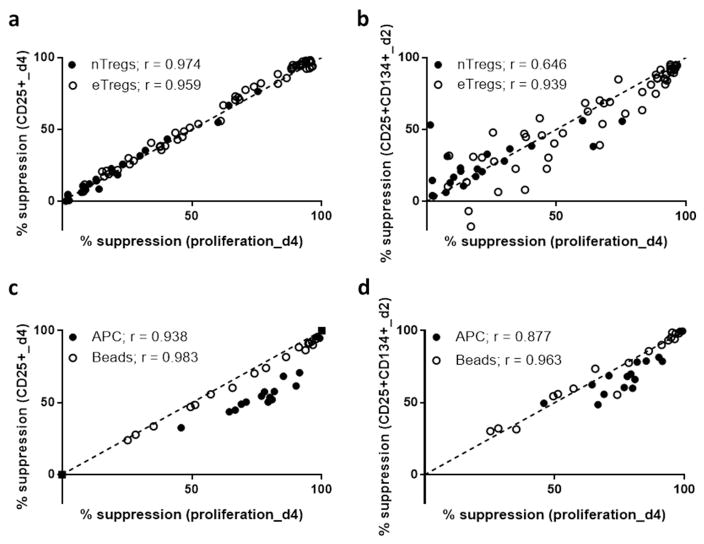
To investigate the applicability of this method to alternative in vitro systems, the expression of CD25 or the combination of CD25+ CD134 was compared to measurement of proliferation in co-culture with different sources of regulation (nTregs) and stimulation (APC + soluble anti-CD3 + soluble anti-CD28).
We found that despite a lower maximum suppression with nTregs (suppression of 76%, 67%, 60% at a dilution of 1:2, for 3 subjects) compared to eTregs (suppression of 97%, 95%, 92%, for the same subjects), the correlation between suppression of proliferation and suppression of CD25 on day 4 was strong for both either nTregs and eTregs (r=0.974 and r=0.959, Fig. 3a). Suppression by nTregs of CD25+CD134+ on day 2, and proliferation on day 4 were significantly correlated (r=0.647, p=0.002) but to a lesser degree than these measures by eTregs (r=0.939). This may reflect differences in the function of these two types of Tregs with respect to early activation. Alternatively, this result may be due to the increased variance of the assay when low level of suppression are present, as was the case in this specific assay containing nTreg.
Figure 3. Surface marker-based suppression assay correlates with proliferation-based suppression assay when using nTregs or APC activation.
(a) Correlation of percentage suppression of CD25+ Teff at day 4 and percentage suppression of CFSE-based Teff proliferation at day 4; solid circles = nTregs, and open circles = eTregs. (b) Correlation of percentage suppression of CD25+CD134+ Teff at day 2 and percentage suppression of CFSE-based Teff proliferation at day 4; solid circles = nTregs, and open circles = eTregs. The median frequency of CD25+CD134+ on day 2 in Teff cultured without Treg suppression was 20% (range 7–22%). (c) Correlation of percentage suppression of CD25+ Teff at day 2 and percentage suppression of CFSE-based Teff proliferation at day 4; solid circles = APC activation, and open circles = Bead activation. (d) Correlation of percentage suppression of CD25+CD134+ Teff at day 2 and percentage suppression of CFSE-based Teff proliferation at day 4; solid circles = APC activation, and open circles = Bead activation. Spearman’s rank correlation coefficients: a (nTregs: r= 0.974, p<0.0001 eTregs: r=0.959, p<0.0001), b (nTregs: r= 0.646, p=0.0016 eTregs: r=0.939, p<0.0001), c (APC: r= 0.938, p<0.0001 Beads: r=0.983, p<0.0001), d (APC: r= 0.877, p<0.0001 Beads: r=0.963, p<0.001). nTregs were sorted from two subjects, eTregs were expanded from three subjects, and grouped for analysis, and Teff were from six subjects.
An alternative form of activation was also studied by using a co-culture assay in which soluble anti-CD3 and autologous antigen presenting cells (APC) were used for stimulation. Suppression of cell surface markers was lower than suppression of proliferation using APC compared to bead based activation. However, suppression of proliferation was correlated with suppression of cell surface markers on both days 2 and 4 for APC (r=0.8765 and r=0.9382, respectively) and bead based conditions (r=0.963 and r=0.983, respectively; Fig. 3c, d). This indicates that our assay could be used to assess Treg mediated suppression with a variety of Tregs and activation conditions.
3.5. CD25 cell surface expression correlates with BrdU-based proliferation
An alternative measure of proliferation is the incorporation of BrdU into DNA during S-phase of the mitosis. We performed co-culture assays with BrdU and found low incorporation of BrdU into cells during 0–24 hours, but between 24 hours and 48 hours cells as cells entered S-phase; BrdU staining correlated well with CD25+CD134+ expression (r=0.9823, p<0.001) and with the number of cells proliferating in concurrent CFSE based assays (r=0.978, p<0.001, Fig. S6). These findings indicate that staining for cell surface CD25 and CD134 mirrors proliferation using two distinct measures.
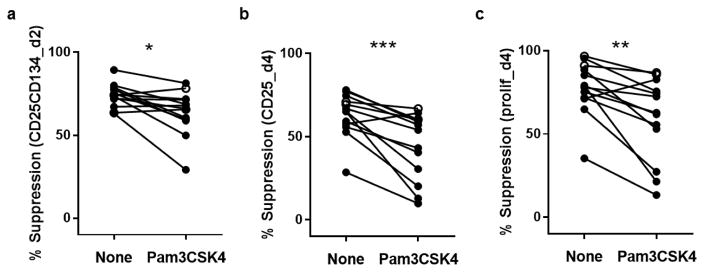
3.6. Suppression of surface markers mirrors the reduction in suppression of proliferation caused by addition of the TLR 1/2 agonist Pam3CSK4
To address the ability of this assay to detect differences in the response of effector T cells to Treg mediated suppression in vitro, we performed suppression assays in the presence or absence of the TLR1/2 agonist Pam3CSK4. The toll-like receptor 1/2 agonist Pam3CSK4 reduces suppression measured by proliferation (CFSE) in a 4-day suppression assay (Mikacenic et al., 2014). We tested the suppression of proliferation and CD25/CD134 after 2–4 days of co-culture of Tregs and Teff from 13 healthy individuals with and without addition of Pam3CSK4. The relationship between suppression of proliferation and suppression of CD25 on day 4 or CD25+CD134+ on day 2 was not different with Pam3CSK4 compared to without Pam3CSK4. As expected, addition of Pam3CSK4 reduced suppression of proliferation on day 4 of the assay (p=0.024, 1:4 ratio), this was mirrored by a reduction in suppression of CD25+ on day 4 (p=0.007), and CD25+CD134+ on day 2 (p=0.0105, Fig. 4a–c). These results indicate an ability to demonstrate TLR mediated impaired suppression with the 2-day assay.
Figure 4. Comparison of surface marker-based suppression assay and CFSE-based suppression assay in the presence of Pam3CSK4.
(a) Percentage suppression of CD25+CD134+ Teff at day 2 in the absence and presence of Pam3CSK4. The median frequency of CD25+CD134+ on day 2 in Teff cultured without Pam3CSK4 was 57% (range 30–82%). (b) Percentage suppression of CD25+ Teff at day 4 in the absence and presence of Pam3CSK4. (c) Percentage suppression of CFSE-based proliferation at day 4 in the absence and presence of Pam3CSK4. Wilcoxon test of paired data: (a) *P=0.011, (b) ***P=0.001, and (c) **P=0.002. Data from 13 healthy controls are shown with cells at Tregs:Teff ratios of 1:4.
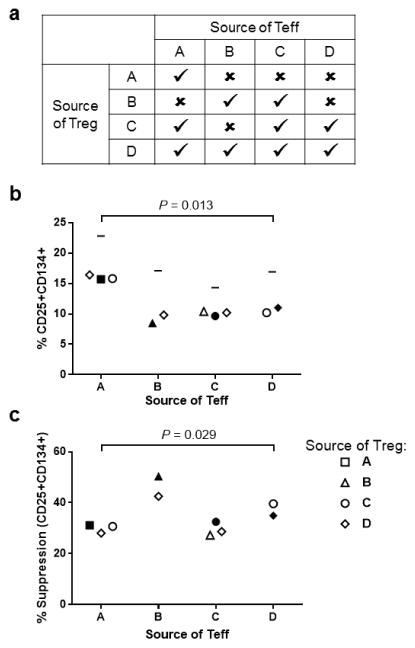
3.7 Surface marker-based suppression assay is effective for frozen PBMC and low volume blood samples
A goal of developing this assay was to not only shorten and simplify the Treg assay, but also to miniaturize the assay for use with frozen PBMC or very low volume blood samples. Additionally, our goal was to be able to use this assay to demonstrate differences in both the function of Treg and the ability of Teff to be suppressed between individual subjects. To test this frozen cells from four donors (referred to as A–D; Fig. 5a) were thawed and sorted using a FACS Aria. Teffs (CD4+ T cells in the lowest 50% of CD25 expression) and nTregs (highest 2% of CD25 expression CD4+ T cells) were co-cultured. Assays were performed with 5000 Teffs per well (plated in triplicate and combined for analysis) and Teff:Treg ratios of 1:2 to 1:16, Tregs were stained with EF670. Combinations of Tregs and Teffs from different samples tested reproducibility and demonstrated differences between subjects. Of 16 possible combinations of nTregs and effectors, there were sufficient cells for 10 different pairs (Fig. 5a). On day 2 the number of effectors expressing CD25 in response to bead stimulation, without suppression by Tregs, were for donor A 23%, donor B 17%, donor C 14%, and donor D 17% (horizontal line in Fig. 5b). In this set of assays, there were differences in activation measured at a 1:4 Treg:Teff ratio between donors (p=0.013, Fig. 5b). There were also significant differences in suppression dependent on the donor of the Teff (p=0.029, Fig. 5c) but not driven by the source of Treg (p=0.987, data not plotted) nor by the level of activation of the Teff. These data demonstrate the consistency of this miniaturized assay and its ability to detect differences in the responsiveness of Teff to Treg mediated suppression between individuals.
Figure 5. Measurement of suppression of CD25+CD134+ expression on day 2 using 5,000 Teff per well.
(a) Outline of the source of Tregs and Teff from four donors (A, B, C and D). Cells were combined in four autologous and six allogenic pairings depending on number of cells recovered from sorting. (b) Percentage CD25+CD134+ Teff at a 1:4 ratio of Tregs:Teff. Horizontal line indicates the maximum proliferation for Teff incubated with anti-CD3/anti-CD28 beads without Tregs (not included in Kruskal-Wallis Test). (c) Percentage CD25+CD134+ suppression calculated from the data in Figure B. The median frequency of CD25+CD134+ on day 2 in Teff cultured without Treg suppression was 17% (range 14–23%). For b and c, the x-axis indicates the source of the Teff for the assay. The shape of the symbol indicates the source of Treg: subject A = square, subject B = triangle, subject C = circle, subject D = diamond. Filled symbols indicate cells from the same subject. Kruskal-Wallis Test: (b) P=0.013, and (c) P=0.029.
4. Discussion
In this study, we demonstrate the use of cell surface activation markers to determine suppression in in vitro co-culture suppression assays. Notably we demonstrate a very strong correlation between CD25 expression of CD4 effectors at day 4 and CFSE based cell division. Application of an assay that utilizes percentage CD25 as a measure at day 4, would limit manipulation, and the resulting loss of viable CD4 effector T cells prior to the co-culture assay. Further, we have shown that there is a good correlation between the percentage CD25+ CD134+ T cells at day 2 with CFSE-based proliferation on day 4. Application of this assay has the benefit of a shortened assay and manageable time constraints on the performance of the assay. This assay yields similar results under different stimulation conditions and multiple regulatory T cell types, and captures the influence of known factors that impair suppression. Finally, an adaption of this method to a miniaturized assay, using 5,000 Teffs per well, showed good intra assay reproducibility and demonstrated differences between subjects in suppression. Application of this assay could dramatically reduce cell requirements in addition to the other benefits of the 2-day assay.
The strengths of this study are the examination of a progression of assay adaptations in a variety of subjects and testing in different experimental settings. Although agreement between ‘gold standard’ CFSE-based assays and the alternative assays examined was effected by nonuniform bias the middle range of the assay (30–60% suppression) is probably the most biologically important and is similar in both the day 4 CD25 and day 2 CD25/CD134 assays. In this range both of these assays should be able to detect differences of 20% in suppression, which is of a similar magnitude to the differences between patients with remitting-relapsing multiple sclerosis or diabetes and healthy controls previously published by our group (Schneider et al., 2008; Schneider et al., 2013). Our group has also shown that a TLR 1/2 agonist, Pam3CSK4, reduces suppression when added to co-culture assays (Mikacenic et al., 2014). In this study, we were able to replicate this observation using the day 2 CD25/CD134 assay.
The role of CD25 and CD134 in the immune system could mean that in some circumstances they may provide additional information to analysis of proliferation but their expression could also be altered by some immune defects. In the current study, CD25 staining was consistent between individuals suggesting common polymorphisms in the IL-2RA gene did not significantly alter binding of the CD25 antibody or the upregulation of CD25, as it correlates with proliferation in response to activation. In addition, in vitro CD134 antagonizes Treg mediated suppression (Croft et al., 2009), therefore if OX40 signaling is of interest these assays would not be appropriate.
Despite these caveats, the measurement of CD25 and CD134 surface expression at 2–5 days of culture has been successfully employed by others to detect antigen specific cells from fresh and frozen samples (Endl et al., 2006; Zaunders et al., 2009; Keoshkerian et al., 2012). Following this example Shaw and colleagues investigated both suppression of proliferation and suppression of CD25+CD134+ cells by peripheral and mucosal (rectal) responders and Tregs from seronegative individuals and individuals with HIV able to maintain their viral RNA load (Shaw et al., 2011). They showed no difference in suppression of proliferation, at 60–84 hours of culture, between these three groups of individuals; however, suppression calculated from CD25+CD134+ was different between these three groups. The reasons for this discrepancy are unknown but support our observation (Fig. 3c) that suppression of CD25+CD134+ and suppression of proliferation may be disparate under APC based stimulation, used by Shaw et al., compared to bead-based stimulation. This also highlights the importance of the level of stimulation used in these assays on the outcome. It is vital that the source of stimulation is optimized and suggest that the CD25+CD134+ subset of effector T cells alone at 48 hours be above 10% in order to attain measurable and reliable levels of suppression.
In this study, measurement of CD154 and CD69 at 7 hours was also assessed. Our data showed good correlation of this method with suppression of proliferation at 4 days supporting the initial description of the assay by Canavan et al. (Canavan et al., 2012). Despite the good correlation, the 7-hour assay underestimated suppression of proliferation at 4 days by at least 25% and therefore a 2-day assay was pursued. We chose to quantify CD69+CD154+ cells because in preliminary experiments a combination of markers gave the highest suppression. Whereas, Canavan et al. analyzed CD69+ and CD154+ cells separately, finding that CD154+ had better agreement with suppression of proliferation than CD69+. Upon reanalysis, re-gating to measure CD69+, CD154+, and proliferation precursors instead of all proliferating cells (Wells et al., 1997), our data still showed that activation at 7 hours underestimates proliferation at 4 days.
Here, we have outline three suppression assay methods, all applicable to cryopreserved cells. Although all these assays can measure defects in suppression, they will be most useful in different contexts. Analysis of CD25+ on day 4 of culture, as opposed to proliferation, requires less manipulation of cells, less cell death through CFSE staining, and would simplify experimental procedures. In particular, in studies analyzing large numbers of participants removing the need to CFSE stain should save a substantial amount of time and assay variation. However, Tregs or Teff must be labeled in some way to allow separation of these cells during flow cytometry analysis; in the assays described here, Tregs were stained with EF670. Therefore, this assay would be most useful to measure suppression of multiple Teff by a single source of Treg in research with a large numbers of participants, where there are no stringent constraints on time or cell number. Alternatively, where a shorter assay is required or early events in cell activation are of interest, expression of CD25 and CD134 on day 2 of co-culture would be a good method. In studies where cell number is a constraint, our data suggest that a miniature (5,000 Teff per well) suppression assay measuring CD25+CD134+ at day 2 could be employed. Although, the choice of staining Teff or Treg should be dependent on which cell type is most limiting. Even if CFSE-based analysis of proliferation is employed measurement of CD25 and CD134 is recommended because it is likely to give additional biological information in some experimental, tissue, or disease settings.
In conclusion, we report a number of adaptations of suppression assays that should suit a range of different research questions. The ability to measure suppression more simply should allow researchers to make full use of arduously collected biobank samples. This in turn should provide a more comprehensive understanding of the immunology of a variety of diseases.
Supplementary Material
Highlights.
CD25 is highly correlated with cell division as measured by CFSE.
A CD25/CD134-based assay simplifies and shortens assays of Treg suppression
The CD25/CD134-based assay works for frozen samples and limited cell numbers
The CD25/CD134-based assay can be utilized with bead based and APC based stimuli.
Acknowledgments
The authors would like to thank all the volunteers who provided samples for this research through the Benaroya Research Institute Immune Mediate Disease Registry and Repository. Funding for this work came from JDRF (Collaborative Center for Cell Therapy; 2-SRA-2014-150), and NIDDK of the National Institutes of Health under award number DP3DK104466. In addition, A. E. L was supported by a Diabetes UK-Fulbright scholarship and other fellowship funding from Diabetes UK. C. M. was supported by NHLBI of the National Institutes of Health under award number K23HL120896.
Abbreviations
- eTreg
expanded regulatory CD4+ T cell
- nTreg
ex vivo ‘natural’ CD4+ regulatory T cell
- Teff
effector CD4+ T cell
- d1
day 1
- d2
day 2
- d4
day 4
- CFSE
Carboxyfluorescein succinimidyl ester
- aCD3/aCD28
anti-CD3/anti-CD28 Dynabeads
- PBMC
peripheral blood mononuclear cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, Azzari C, Ziegler SF, Levings MK, Roncarolo MG. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest. 2006;116:1713–22. doi: 10.1172/JCI25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund D, Karlsson M, Biglarnia AR, Lorant T, Tufveson G, Korsgren O, Carlsson B. Obtaining regulatory T cells from uraemic patients awaiting kidney transplantation for use in clinical trials. Clin Exp Immunol. 2013;173:310–22. doi: 10.1111/cei.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavan JB, Afzali B, Scotta C, Fazekasova H, Edozie FC, Macdonald TT, Hernandez-Fuentes MP, Lombardi G, Lord GM. A rapid diagnostic test for human regulatory T-cell function to enable regulatory T-cell therapy. Blood. 2012;119:e57–66. doi: 10.1182/blood-2011-09-380048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou A, Lyberi M, Chatzilymperis G, Nezos A, Kamper E. CD40/CD40L signaling and its implication in health and disease. Biofactors. 2009;35:474–83. doi: 10.1002/biof.62. [DOI] [PubMed] [Google Scholar]
- Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev. 2009;229:173–91. doi: 10.1111/j.1600-065X.2009.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endl J, Rosinger S, Schwarz B, Friedrich SO, Rothe G, Karges W, Schlosser M, Eiermann T, Schendel DJ, Boehm BO. Coexpression of CD25 and OX40 (CD134) receptors delineates autoreactive T-cells in type 1 diabetes. Diabetes. 2006;55:50–60. [PubMed] [Google Scholar]
- Goodman WA, Levine AD, Massari JV, Sugiyama H, McCormick TS, Cooper KD. IL-6 signaling in psoriasis prevents immune suppression by regulatory T cells. J Immunol. 2009;183:3170–6. doi: 10.4049/jimmunol.0803721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon M, Lechanteur C, Lucas S, Somja J, Seidel L, Belle L, Bruck F, Baudoux E, Giet O, Chantillon AM, Delvenne P, Drion P, Beguin Y, Humblet-Baron S, Baron F. Infusion of clinical-grade enriched regulatory T cells delays experimental xenogeneic graft-versus-host disease. Transfusion. 2014;54:353–63. doi: 10.1111/trf.12279. [DOI] [PubMed] [Google Scholar]
- Ito S, Bollard CM, Carlsten M, Melenhorst JJ, Biancotto A, Wang E, Chen J, Kotliarov Y, Cheung F, Xie Z, Marincola F, Tanimoto K, Battiwalla M, Olnes MJ, Perl S, Schum P, Hughes TE, Keyvanfar K, Hensel N, Muranski P, Young NS, Barrett AJ. Ultra-low dose interleukin-2 promotes immune-modulating function of regulatory T cells and natural killer cells in healthy volunteers. Mol Ther. 2014;22:1388–95. doi: 10.1038/mt.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keoshkerian E, Helbig K, Beard M, Zaunders J, Seddiki N, Kelleher A, Hampartzoumian T, Zekry A, Lloyd AR. A novel assay for detection of hepatitis C virus-specific effector CD4(+) T cells via co-expression of CD25 and CD134. J Immunol Methods. 2012;375:148–58. doi: 10.1016/j.jim.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–66. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Landwehr-Kenzel S, Issa F, Luu SH, Schmuck M, Lei H, Zobel A, Thiel A, Babel N, Wood K, Volk HD, Reinke P. Novel GMP-compatible protocol employing an allogeneic B cell bank for clonal expansion of allospecific natural regulatory T cells. Am J Transplant. 2014;14:594–606. doi: 10.1111/ajt.12629. [DOI] [PubMed] [Google Scholar]
- Lawson JM, Tremble J, Dayan C, Beyan H, Leslie RD, Peakman M, Tree TI. Increased resistance to CD4+CD25hi regulatory T cell-mediated suppression in patients with type 1 diabetes. Clin Exp Immunol. 2008;154:353–9. doi: 10.1111/j.1365-2249.2008.03810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardiney M, 3rd, Brown MR, Fleisher TA. Measurement of T-cell CD69 expression: a rapid and efficient means to assess mitogen- or antigen-induced proliferative capacity in normals. Cytometry. 1996;26:305–10. doi: 10.1002/(SICI)1097-0320(19961215)26:4<305::AID-CYTO11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- McMurchy AN, Levings MK. Suppression assays with human T regulatory cells: a technical guide. Eur J Immunol. 2012;42:27–34. doi: 10.1002/eji.201141651. [DOI] [PubMed] [Google Scholar]
- Mikacenic C, Schneider A, Radella F, Buckner JH, Wurfel MM. Cutting edge: Genetic variation in TLR1 is associated with Pam3CSK4-induced effector T cell resistance to regulatory T cell suppression. J Immunol. 2014;193:5786–90. doi: 10.4049/jimmunol.1401185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, Atkinson MA, Bluestone JA. Expansion of human regulatory T-cells from patients with type 1 diabetes. Diabetes. 2009;58:652–62. doi: 10.2337/db08-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruitenberg JJ, Boyce C, Hingorani R, Putnam A, Ghanekar SA. Rapid assessment of in vitro expanded human regulatory T cell function. J Immunol Methods. 2011;372:95–106. doi: 10.1016/j.jim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Schneider A, Buckner JH. Assessment of suppressive capacity by human regulatory T cells using a reproducible, bi-directional CFSE-based in vitro assay. Methods Mol Biol. 2011;707:233–41. doi: 10.1007/978-1-61737-979-6_15. [DOI] [PubMed] [Google Scholar]
- Schneider A, Long SA, Cerosaletti K, Ni CT, Samuels P, Kita M, Buckner JH. In active relapsing-remitting multiple sclerosis, effector T cell resistance to adaptive T(regs) involves IL-6-mediated signaling. Sci Transl Med. 2013;5:170ra15. doi: 10.1126/scitranslmed.3004970. [DOI] [PubMed] [Google Scholar]
- Schneider A, Rieck M, Sanda S, Pihoker C, Greenbaum C, Buckner JH. The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J Immunol. 2008;181:7350–5. doi: 10.4049/jimmunol.181.10.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JM, Hunt PW, Critchfield JW, McConnell DH, Garcia JC, Pollard RB, Somsouk M, Deeks SG, Shacklett BL. Increased frequency of regulatory T cells accompanies increased immune activation in rectal mucosae of HIV-positive noncontrollers. J Virol. 2011;85:11422–34. doi: 10.1128/JVI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohma S, Ramberg JE, Lipsky PE. Expression and distribution of CD11a/CD18 and CD54 during human T cell-B cell interactions. J Leukoc Biol. 1992;52:97–103. doi: 10.1002/jlb.52.1.97. [DOI] [PubMed] [Google Scholar]
- Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics. 2008;9:225. doi: 10.1186/1471-2164-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells AD, Gudmundsdottir H, Turka LA. Following the fate of individual T cells throughout activation and clonal expansion. Signals from T cell receptor and CD28 differentially regulate the induction and duration of a proliferative response. J Clin Invest. 1997;100:3173–83. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M, Xu Y, Brown K, Dyer WB, Kim M, de Rose R, Kent SJ, Jiang L, Breit SN, Emery S, Cunningham AL, Cooper DA, Kelleher AD. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) J Immunol. 2009;183:2827–36. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.