Abstract
Background
A disease severity index (SI) for Alzheimer's disease (AD) has been proposed that summarizes MRI-derived structural measures into a single score using multivariate data analysis.
Objectives
To longitudinally evaluate the use of the SI to monitor disease progression and predict future progression to AD in mild cognitive impairment (MCI). Further, to investigate the association between longitudinal change in the SI and cognitive impairment, Apolipoprotein E (APOE) genotype as well as the levels of cerebrospinal fluid amyloid-beta 1–42 (Aβ) peptide.
Methods
The dataset included 195 AD, 145 MCI and 228 control subjects with annual follow-up for three years, where 70 MCI subjects progressed to AD (MCI-p). For each subject the SI was generated at baseline and follow-ups using 55 regional cortical thickness and subcortical volumes measures that extracted by the FreeSurfer longitudinal stream.
Results
MCI-p subjects had a faster increase of the SI over time (p < 0.001). A higher SI at baseline in MCI-p was related to progression to AD at earlier follow-ups (p < 0.001) and worse cognitive impairment (p < 0.001). AD-like MCI patients with the APOE ε4 allele and abnormal Aβ levels had a faster increase of the SI, independently (p = 0.003 and p = 0.004).
Conclusions
Longitudinal changes in the SI reflect structural brain changes and can identify MCI patients at risk of progression to AD. Disease-related brain structural changes are influenced independently by APOE genotype and amyloid pathology. The SI has the potential to be used as a sensitive tool to predict future dementia, monitor disease progression as well as an outcome measure for clinical trials.
Keywords: Alzheimer disease, Cognitive dysfunction, Atrophy, Disease progression, Longitudinal
Highlights
-
•
A disease severity index (SI) summarizes the brain atrophy patterns.
-
•
The SI can be used to monitor disease progression in MCI.
-
•
FreeSurfer longitudinal stream is used for the first time to generate the SI.
-
•
The SI is strongly associated with cognitive status and progression incident.
-
•
APOE genotype and amyloid pathology may independently modulate the rate of atrophy.
1. Introduction
Alzheimer's disease (AD), the most common form of dementia, is a neurodegenerative disorder that is clinically characterized by gradual loss of cognitive functions. While the definitive AD diagnosis requires postmortem autopsy-confirmed existence of amyloid plaques and neurofibrillary tangles in the brain tissue (Khachaturian, 1985, Markesbery, 1997), the diagnosis of possible or probable AD is based on clinical signs and symptoms. However, the new diagnostic criteria recommend that the clinical diagnosis should be supported by the use of biomarkers from magnetic resonance imaging (MRI), positron emission tomography (PET) or cerebrospinal fluid (CSF) (McKhann et al., 2011). The disease onset is estimated to be a decade or more before clinical appearance (Jack et al., 2013, Jack et al., 2010a). Therefore, at the time of diagnosis, irreversible brain damage has already occurred. An early and accurate diagnosis of AD will be extremely important when disease-modifying treatments exist. Unfortunately, at the moment, only symptomatic treatments are available.
Mild cognitive impairment (MCI) is an intermediate condition between normal cognition and dementia that involves noticeable decline in cognitive abilities. MCI often represents a prodromal form of dementia, conferring a significantly higher risk of converting to AD (Gauthier et al., 2006). However, MCI is a heterogeneous condition, i.e. not all MCI subjects develop dementia even after several years of follow-up. Therefore, studying MCI is useful because it offers opportunities for relatively early diagnosis of AD by identifying MCI patients at risk of developing AD.
There is still a need to validate existing biomarkers for early detection of the disease and investigate new diagnostic tools. MRI has been widely studied in clinical trials for early detection of AD and MCI. Cognitive impairment in AD and MCI subjects is associated with neuron loss and progressive cerebral atrophy (Serrano-Pozo et al., 2011). Advanced image processing and data analysis techniques provide tools to detect these structural changes in MRI and extract disease-related information (Falahati et al., 2014). Atrophy of medial temporal structures, particularly in the hippocampus and entorhinal cortex is consistent findings in AD (Morra et al., 2009). However, atrophy of medial temporal structures or ventricular enlargement is not specific to AD. The absence of atrophy does not exclude a diagnosis of AD either. Measures of single structures are probably insufficient for accurate AD diagnosis. It has previously been shown that a combination of different structural measures is more accurate for distinguishing AD from cognitively normal (CN) subjects and predicting future conversion from MCI to AD (Kloppel et al., 2008, Westman et al., 2013, Zhang et al., 2011). Recently, we proposed a methodology using multivariate analysis of automated MRI-derived structural measures (global and regional thickness and volumes) to generate a disease severity index (SI) (Aguilar et al., 2014, Spulber et al., 2013). The SI summarizes the patterns of structural brain changes as a single score for each individual subject, based on the observed patterns in AD and CN subjects. The methodology has been evaluated in the AddNeuroMed (large European multi-center study) and the Alzheimer's Disease Neuroimaging Initiative (ADNI) and the Australian Imaging Biomarkers and Lifestyle flagship study of aging (AIBL) cohorts (Ferreira et al., 2017, Mangialasche et al., 2013, Westman et al., 2011).
The present study builds on our previous work to generate the SI, but we have made several methodological improvements. Longitudinal image processing in FreeSurfer is employed for the first time to generate regional volume and thickness measures. This reduces the inter-individual variability and increases the accuracy of the extracted structural measures (Reuter et al., 2012). By improving the input features for multivariate analysis, we can study changes in the patterns of atrophy, investigate disease progression and evaluate the performance of the SI more accurately. We have added age correction (Falahati et al., 2016) to further improve the accuracy of the SI. This has not previously been applied longitudinally. We have only described longitudinal changes in the SI once before in a small sample of MCI subject with short follow-up time (one year). In the present study we have a much larger sample size and subjects are followed-up for three years (baseline, 12-, 24- and 36-month). To have a sufficient follow-up time is important to evaluate small change as well as study the relationship between the SI to cognition and APOE genotype. Finally, changes in the SI have not previously been investigated in relation to amyloid burden in the brain as well as how sensitive it is to predict and monitor time to conversion to AD.
2. Material and methods
2.1. Participants
A total of 568 subjects (AD = 195, MCI = 145, CN = 228) were included in the current study. For AD, follow-up data were available up to 24 months. All MCI and 129 CN subjects had a 36 months follow-up. Out of 145 MCI subjects, 70 subjects progressed from MCI to AD (MCI-p) and 75 subjects remained stable or returned to normal cognition (MCI-s) within the 36 months follow-up period. Besides, during the 36 months period, 11 CN subjects progressed to MCI or AD. The demographics of participants are given in Table 1.
Table 1.
Baseline demographic and clinical characteristics of subjects.
| CN | MCI-s | MCI-p | AD | |
|---|---|---|---|---|
| Count | 228 | 75 | 70 | 195 |
| Age, years | 75.9 ± 5.0 (59.9–89.6) |
74.5 ± 7.4 (55.1–86.4) |
74.3 ± 6.9 (55.2–87.8) |
75.5 ± 7.5 (55.1–90.9) |
| Education, years | 16.0 ± 2.8 (6–20) |
16.2 ± 2.8 (7–20) |
15.8 ± 3.2 (6–20) |
14.7 ± 3.2 (4–20) |
| MMSE score | 29.1 ± 1.0 (25–30) |
27.5 ± 1.7 (24–30) |
26.6 ± 1.7 (24–30) |
23.3 ± 2.0 (18–27) |
| CDR total score | 0 | 0.5 | 0.5 | 0.74 ± 0.25 (0.5–1) |
| Gender, male/female | 111/117 | 15/60 | 27/43 | 95/100 |
| CSF Aβ levels | 207.1 ± 53.4 (79.0–297.7) |
177.1 ± 62.5 (90.8–298.8) |
142.5 ± 38.8 (84.7–272.8) |
142.7 ± 40.0 (75.0–295.8) |
| CSF Aβ−/Aβ+ | 69/45 | 16/23 | 3/37 | 8/91 |
| APOE ε4−/ε4+ | 168/60 | 42/33 | 24/46 | 67/128 |
Continuous data is represented as mean ± SD (minimum – maximum); CN, cognitively normal subjects; MCI, mild cognitive impairment; MCI-s, stable MCI; MCI-p, progressive MCI; AD, Alzheimer's disease; MMSE, mini mental state examination; CDR, clinical dementia rating; CSF, cerebrospinal fluid; Aβ, amyloid-beta 1–42; Aβ−, Aβ > 192 pg/ml; Aβ+, Aβ ≤ 192 pg/ml; APOE, apolipoprotein E; ε4+, carrier of one or two ε4 alleles; ε4−, without any ε4 allele.
The dataset used was obtained from the Alzheimer's disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations (Mueller et al., 2005). The primary goal of ADNI is to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI subjects were recruited from over 50 sites across the U.S. and Canada. For up-to-date information, see www.adni-info.org.
2.2. Inclusion and diagnostic criteria
Briefly, for AD the dementia diagnosis was based on DSM-IV and they had to meet the NINCDS-ADRDA criteria for probable AD, as well as have a total Clinical Dementia Rating (CDR) score of 0.5 or above. MCI diagnosis required a MMSE score between 24 and 30; memory complaints; normal activities of daily living; total CDR score of 0.5; and Geriatric Depression Scale (GDS) score of ≤ 5. The inclusion criteria for control participants were a MMSE score between 24 and 30; total CDR score of 0; and GDS score ≤ 5. No significant neurological or psychiatric illness, no significant unstable systemic illness or organ failure, and no history of alcohol or substance abuse or dependence were required for all three groups. MRI information was not used for the diagnosis. The complete inclusion and exclusion criteria are described by Petersen et al. (2010).
2.3. MRI data acquisition
1.5T MRI data was collected from a variety of MR-systems with protocols optimized for each type of scanner. The MRI protocol included a high resolution sagittal 3D T1-weighted MPRAGE volume (voxel size 1.1 × 1.1 × 1.2 mm3) acquired using a custom pulse sequence specifically designed for the ADNI study to ensure compatibility across scanners (Jack et al., 2008a).
2.4. Longitudinal MRI processing
MRI scans were automatically processed with the FreeSurfer pipeline (version 5.3.0), http://freesurfer.net/. The Freesurfer longitudinal stream (Reuter et al., 2012) was used to extract regional cortical thickness and subcortical volumetric measures. The longitudinal processing pipeline includes three steps. Initially all baseline and follow-up time points were cross-sectionally processed with the default workflow. Then an unbiased within-subject template volume (Reuter and Fischl, 2011) was created from all time points using inverse consistent registration (Reuter et al., 2010). Afterwards, all time points were longitudinally processed. In the latter step, the unbiased template was used as initial guess of several segmentation and reconstruction steps for processing each time point. Since segmentation and parcellation procedures are highly sensitive to algorithm initialization, using the unbiased template reduces the random variation in the processing procedure and increases reliability and robustness of the overall longitudinal analysis (Reuter et al., 2012).
Finally, for each time point 55 MRI measures including 34 regional cortical thickness measures and 21 regional subcortical volumes were extracted. Both cortical and subcortical regions are involved in AD and the combination of the two has previously been shown to give the best results for discriminating between AD and CN as well as predicting future conversion to AD from MCI (Lerch et al., 2008, Li et al., 2012, Westman et al., 2013). Measures from the left and right sides of the brain were averaged. All subcortical volumetric measures from each subject were normalized by the subject's intracranial volume (Voevodskaya et al., 2014), which was estimated based on an affine transform in FreeSurfer. Cortical thickness measures were not normalized and were used in their raw form (Westman et al., 2013). Data was processed through the hive database system (theHiveDB) (Muehlboeck et al., 2014).
2.5. APOE genotyping and CSF Aβ
APOE genotyping was performed at screening using DNA obtained from subjects peripheral blood samples (Saykin et al., 2010). Based on the APOE ε4 status subjects are divided in two groups, carrier subjects (ε4+) with one or two ε4 alleles and non-carrier subjects (ε4−) without any ε4 allele.
The concentration levels of CSF Aβ at baseline were available for 392 subjects (for 79 out of 145 MCI subjects with 36 moths of follow-up). The details of the measurement method are provided elsewhere (Shaw et al., 2009). A cut-off value equal to 192 pg/ml was used to divide subjects in two groups (Shaw et al., 2009), Aβ negative subjects (Aβ−) with concentration levels > 192 pg/ml, and Aβ positive subjects (Aβ+) with concentration levels ≤ 192 pg/ml.
2.6. Data analysis
An age correction method was applied to MRI measures prior to any statistical analysis to eliminate the confounding effect of age, previously described in detail (Falahati et al., 2016). Briefly, the age-related structural changes are estimated as a linear association between each MRI-derived variable and age at baseline, in the CN group only. The age correction is based only on the CN group to remove age related changes, while keep the changes related to disease. Then, the age-related changes were detrended from all individuals by subtracting the estimated linear trend. Afterwards, pre-processing was performed on the unbiased age corrected MRI data using mean-centering and unit variance scaling in order to transform the data into a suitable form (Eriksson et al., 2013). In addition, all data (including CN, MCI and AD subjects) was tested using a scatter plot of the first and second PCA components and Hotelling's T-squared elliptical range with 95% and 99% confidence intervals, to reveal the homogeneity, check whether data was normally distributed and detect outlier samples.
Orthogonal projection to latent structures (OPLS) (Bylesjö et al., 2006, Trygg and Wold, 2002), a supervised multivariate data analysis method was used to classify AD patients and CN individuals as well as to predict progression in the MCI patients. The OPLS method is an extension to the projection to latent structures (PLS) method (Wold et al., 1984). PLS has been developed for the purpose of modelling complex data based on the assumption that there are latent variables that generate the observed data. PLS extracts these latent variables by maximizing the covariance between two sets of data, descriptor and response variables. In OPLS, the systematic variation in descriptor data is separated into two blocks, predictive variation correlated to response data and non-predictive variation orthogonal to response data. This separation improves the model transparency and reduces the model complexity. For the two-class discriminant problem OPLS has an advantage over PLS that provides only one single predictive component (first component) and the other orthogonal components (if any) are not important for class separation. Accordingly, one single loading vector describes the class discriminating variables. The OPLS model generates an index (the SI) for each subject which is the estimated value of the response variable. The generated index is a scalar value in the range of − 1 to 2, which is close to one for AD subjects and close to zero for CN subjects.
In this work, the extracted data from baseline AD and CN scans is used to train an OPLS model. The performance of the OPLS model was quantified by the goodness of fit (R2) and the goodness of prediction (Q2) parameters (Eriksson et al., 2013). R2 shows how well the model fits the training data and Q2 shows how reliable the model predicts new data. In addition the receiver operating characteristic (ROC) curve (Metz, 1978) and the area under the ROC curve (AUC) were used as a performance metric of the classification model.
Following the training phase, the trained model is used to predict the rest of the scans (i.e. baseline and follow-up MCI scans as well as follow-up AD and CN scans). Correspondingly, a SI is assigned to each predicted subject, where a SI close to 1 displays a pattern similar to AD (AD-like) and a SI close to 0 displays a pattern similar to CN (CN-like). We used the distribution of the baseline SI of CN subjects (in the training model) to calculate the cut-off for labeling subjects as AD-like or CN-like. A Gaussian distribution model fitted to baseline SI of non-progressive CN subject (excluding 11 subjects who progressed to MCI or AD within 36 months follow-up). The cut-off was through this procedure set to 0.372 (mean SI of CN subjects = 0.172 + one standard deviation = 0.2). We based the calculation of the cut-off on a previous study for determining deviation from normality to define cognitive impairment (Busse et al., 2006). The authors reported that the criteria based on a 1 SD cut-off to define MCI lead to the highest relative predictive power for the development of dementia (Busse et al., 2006).
Vertex analysis using the FreeSurfer software was performed to compare cortical thickness between AD-like and CN-like MCI subjects. In this approach a general linear model is fitted at each vertex, where the dependent variable is cortical thickness and diagnostic group is the independent variable. In order to correct for multiple comparisons, Monte Carlo Null-Z simulations were used with a cluster-forming threshold of p < 0.05 (two-sided).
Mixed ANOVA/ANCOVA with Bonferroni adjustment for multiple comparisons were conducted to study time effect, group effect and time × group interaction on longitudinal SI changes. A Greenhouse-Geisser correction was applied when the assumption of sphericity was violated. All mixed ANOVA analyses were adjusted for age and gender as covariates. The results of tests are reported in terms of F-statistic as: F(df-variable, df-error) = F-value, p-value. Linear regression analyses were conducted to study the associations between the SI and MMSE as well as their changes. The MMSE score as the dependent variable and the SI, baseline age and gender as independent variables were included in the regression models. To compare regression models, the interaction of the SI and predicted diagnosis were included in the model. Linear regression results were reported as: standardized coefficient, p-value.
3. Results
Initially, an OPLS model was trained with baseline CN and AD scans. The trained model resulted in cross validated classification accuracy = 87.2%, sensitivity = 91.3% and specificity = 83.8%. The classification model resulted in R2 = 0.887 and Q2 = 0.609. Fig. 1A shows the histograms of the baseline SI for CN and AD subjects. Fig. 1B shows the receiver operating characteristic (ROC) curve for the trained model, with the area under ROC curve of 0.95.
Fig. 1.
A) Histograms and normal curves fitted to the histograms of the baseline SI for CN and AD subjects. B) The receiver operating characteristic (ROC) curve for classification of CN and AD subjects. The area under ROC curve is equal to 0.95. C) The boxplot of the SI in different diagnostic groups at baseline and follow-up. The SI of the CN and AD subjects at baseline were generated by the cross-validated classification model. The SI of the CN and AD subjects at the different time points as well as the SI of MCI subjects were predicted by the training model.
SI, disease severity index; AD, Alzheimer's disease; CN, cognitively normal subjects; MCI, mild cognitive impairment; PDF, probability density function.
The trained model was used to assign a SI to each MCI subject at baseline and follow-up time points as well as follow-up time points of CN and AD subjects. Fig. 1C shows the boxplot of the SI at different time points and diagnoses. As expected, the average SI was the highest in AD subjects followed by MCI and then CN subjects.
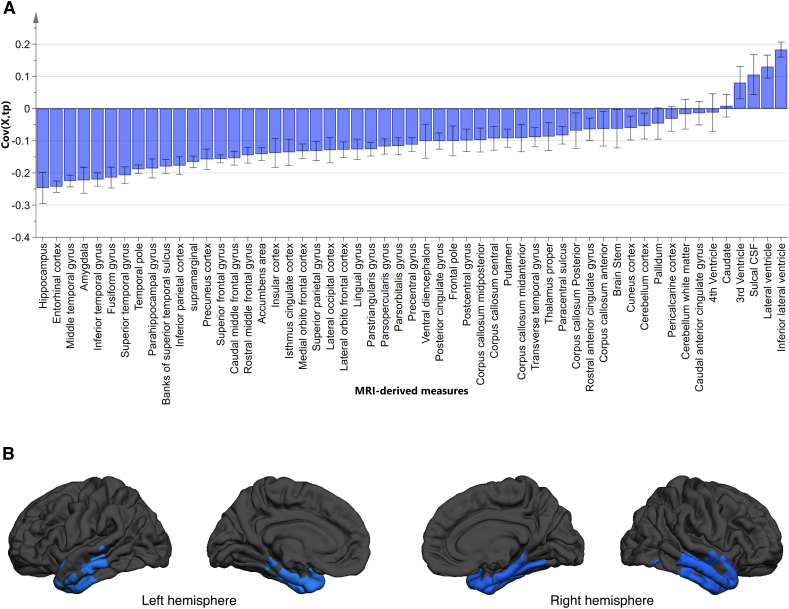
Fig. 2A shows the variables of importance for AD vs. CN classification model. The most influential measures in class separation were structures in medial temporal lobe such as hippocampus, entorhinal cortex and amygdala. The cortical thickness map of AD-like and CN-like MCI subjects from vertex analysis (Fig. 2B) showed that AD-like MCI subjects had reduced thickness in entorhinal cortex, fusiform, temporal pole as well as superior-, middle-, and inferior-temporal gyrus.
Fig. 2.
A) Loadings plot of the variables of importance for AD vs. CN classification model. A variable with a higher magnitude is more influential in group separation than a variable with low magnitude. A variable with a positive covariance has a higher value in AD patients compared to CN subjects and a variable with negative covariance has a lower value in AD patients. B) The cortical thickness map from vertex analysis between AD-like and CN-like MCI subjects. Significant differences in cortical thickness were observed in entorhinal cortex, fusiform, temporal pole as well as superior-, middle-, and inferior-temporal.
Table 2 summarizes the number of correctly predicted MCI-p subjects at different time points. Using the baseline SI, 95.7% of the MCI-p subjects that progressed during the first year of follow-up were correctly identified as AD-like, 78.1% of subjects who progressed during the second year and 60% of the subjects that progressed during the third year (totally 56 out of 70, i.e. 80%). Using the baseline SI, 43 MCI-s subjects (57.3%) were predicted as CN-like and 32 as AD-like (42.7%). Based on 3 years follow-up diagnosis, the baseline SI resulted in an accuracy = 68.3%, sensitivity = 80% and specificity = 57.3% for prediction of progression from MCI to AD.
Table 2.
Number of correctly predicted MCI-p subjects. The SI prior to each follow-up was used to predict MCI subjects as AD-like or CN-like. At each time point, the number of MCI-p subjects that are correctly predicted as AD-like are reported.
| Predicted by | Progressed during |
|||
|---|---|---|---|---|
| Baseline to month-12 (MCI-p = 23) |
Month-12 to month-24 (MCI-p = 32) |
Month-24 to month-36 (MCI-p = 15) |
Baseline to month-36 (MCI-p = 70) |
|
| SI at baseline | 22 (95.7%) | 25 (78.1%) | 9 (60.0%) | 56 (80.0%) |
| SI at month-12 | 22 (95.7%) | 27 (84.4%) | 11 (73.3%) | 60 (85.7%) |
| SI at month-24 | – | 27 (84.4%) | 11 (73.3%) | 61 (87.1%) |
| SI at month-36 | – | – | 13 (86.6%) | 66 (94.2%) |
SI, disease severity index; MCI, mild cognitive impairment; MCI-p, progressive MCI.
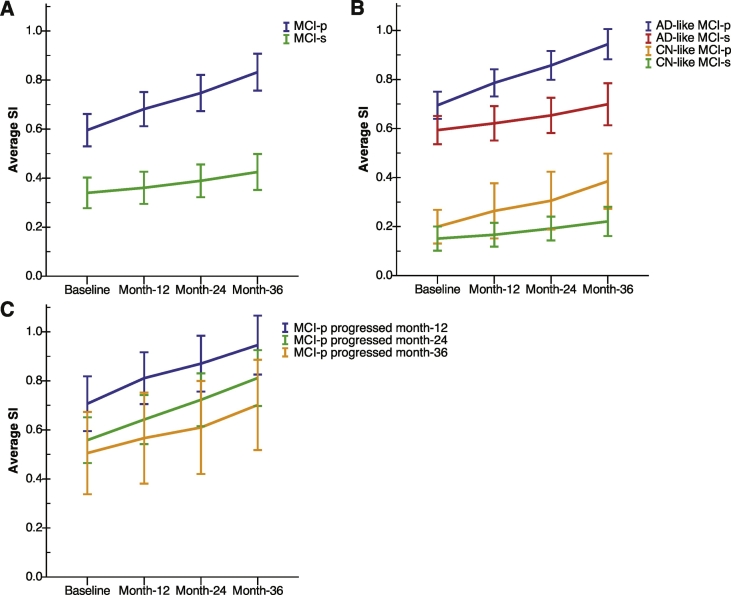
A mixed ANOVA was conducted to compare changes in the SI in MCI-p and MCI-s subjects over 36 months. A significant time × group interaction was observed (F(2,294) = 39.5, p < 0.001) where the SI of MCI-p subjects increased faster compared to MCI-s (Fig. 3A). Further, MCI-p and MCI-s subjects were stratified by the predicted patterns of atrophy (i.e. AD-like and CN-like) using the baseline SI. Within both the AD-like and CN-like group, significant time × group interactions were observed (F(2,168) = 14.9, p < 0.001 and F(2,120) = 4.9, p = 0.006, respectively) where the SI increased faster in MCI-p subjects compared to MCI-s, independently of MCI subjects being AD-like or CN-like (Fig. 3B).
Fig. 3.
A) The average SI in MCI-p and MCI-s subjects over 36 months. The SI was higher and increased faster in MCI-p (p < 0.001). B) The average SI of MCI-p and MCI-s subjects stratified by the predicted patterns (AD-like and CN-like). Within both AD-like and CN-like groups, the SI was higher and increased faster in MCI-p subjects (p < 0.001 and p = 0.006, respectively). C) The average SI of MCI-p stratified by time of conversion. The average SI of subjects who progressed at month-12 was significantly higher than the average of those who progressed at month-36 (p = 0.026). There was no significant difference between MCI-p that progressed at month-24 and those who progressed at month-36.
SI, disease severity index; AD, Alzheimer's disease; CN, cognitively normal subjects; MCI, mild cognitive impairment; MCI-p, progressive MCI; MCI-s, stable MCI; Error bars show 95% confidence intervals.
The longitudinal changes of the SI were compared with regard to the time of progression of MCI-p subjects and the predicted patterns (AD-like or CN-like) of MCI-s subjects. The mixed ANOVA showed a significant time × group interaction (F(8,287) = 10.7, p < 0.001). Bonferroni corrected post hoc tests showed that the SI of subjects who progressed at month-12 were significantly higher than subjects who progressed at month-36 and MCI-s with an AD-like pattern (p = 0.026 and p = 0.032, respectively). Interestingly, no significant differences were observed between subjects who progressed at month-24, subjects who progressed at month-36 and MCI-s with an AD-like pattern. The SI of MCI-s subjects with CN-like pattern was significantly lower than the rest of the subjects (p < 0.001). Fig. 3C shows the average SI of MCI-p subjects that progressed within the first, second and third year of follow-up.
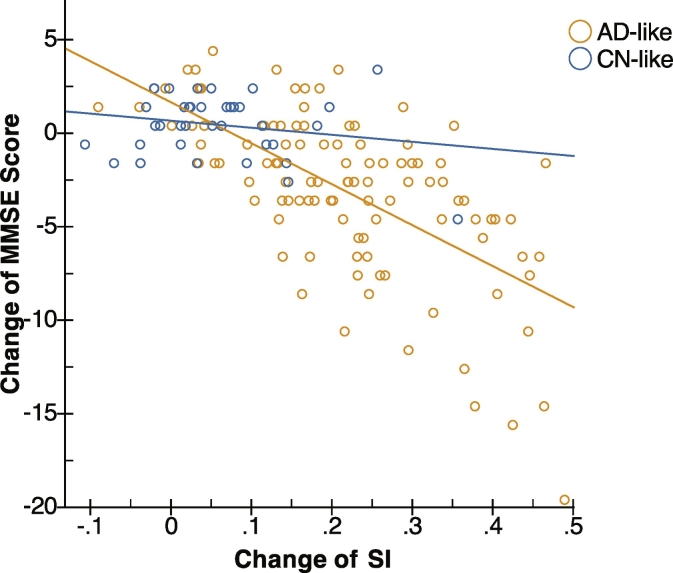
In the next step the association between the SI and cognitive performance (thorough MMSE score) was studied in MCI subjects. At baseline, a significant association (β = − 0.325, p = 0.001) was observed. The associations became stronger at follow-up, where the strongest association was seen at month-36 (β = − 0.605, p < 0.001) followed by month-24 (β = − 0.561, p < 0.001) and month-12 (β = − 0.507, p < 0.001). Moreover, the association between the changes in the SI and in MMSE over 36 months was calculated which resulted in an even stronger association (β = − 0.663, p-value < 0.001). Further, the association between MMSE change and SI change over 36 months was compared between MCI subjects with AD-like and CN-like pattern (Fig. 4), where a significant difference (p-value = 0.001) was observed between regression coefficients of subjects with AD-like patterns (β = − 0.631, p-value < 0.001) and subjects with CN-like patterns (β = − 0.206, p-value = 0.221).
Fig. 4.
Scatter plot showing changes over time (36 month) in MMSE and the SI. MCI subjects with a CN-like and AD-like pattern are shown in blue and green symbols respectively. The blue and green lines show the linear regression lines that fitted to CN-like (β = − 0.206, p-value = 0.221) and AD-like (β = − 0.631, p-value < 0.001) subjects.
SI, disease severity index; AD, Alzheimer's disease; CN, cognitively normal subjects; MMSE, mini mental state examination. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
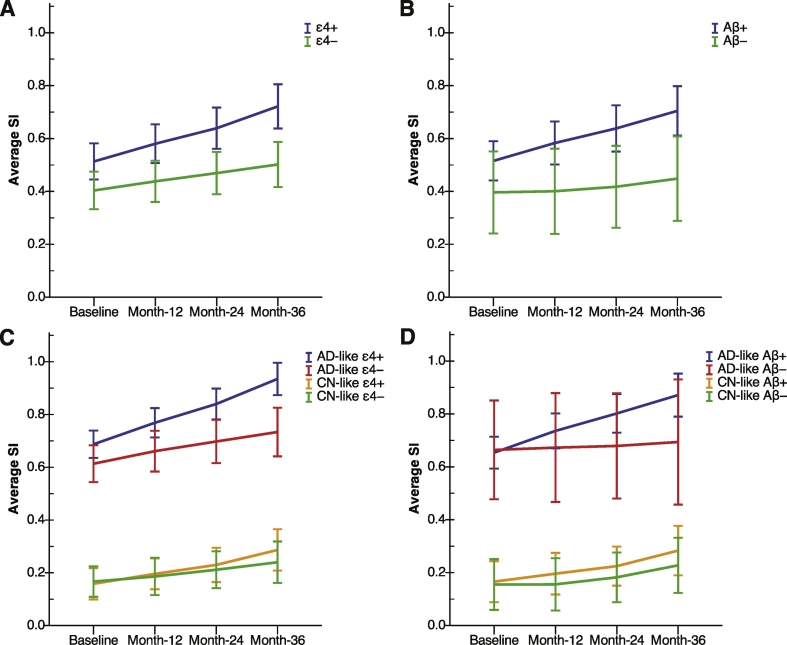
The change of the SI in MCI subjects was studied with regard to the presence of the APOE ε4 allele and levels of CSF Aβ (Fig. 5). This was investigated in two separate mixed ANCOVAs since the reduced size of some of the subgroups did not allow testing for a triple interaction, i.e. time × APOE group × Aβ group. The mixed ANCOVA showed a significant time × APOE group interaction (F(2,156) = 5.1, p = 0.006, adjusted for Aβ group as covariate), where the SI increased faster over time in ε4+ compared to ε4− (Fig. 5A). Likewise, a significant time × Aβ group interaction was found (F(2,156) = 3.9, p = 0.021, adjusted for APOE group) where Aβ+ subjects had a faster increase of the SI compared to Aβ− subjects (Fig. 5B).
Fig. 5.
Average SI in MCI subjects with regard to presence of APOE ε4 allele and levels of CSF Aβ. A) The SI increased faster in ε4+ subjects compared to ε4− subjects (p = 0.006). B) The SI increased faster in Aβ+ subjects compared to Aβ− subjects (p = 0.021). C) Within AD-like subjects, the ε4+ group had a greater increase of the SI compared to the ε4− group (p = 0.003), but there is no significant difference within CN-like subjects. D) Within AD-like subjects, Aβ+ had a greater increase of the SI compared to the Aβ− group (p = 0.004), but there was no significant difference within CN-like subjects.
CSF, cerebrospinal fluid; SI, disease severity index; AD, Alzheimer's disease; CN, cognitively normal subjects; MCI, mild cognitive impairment, Error bars show 95% confidence intervals.
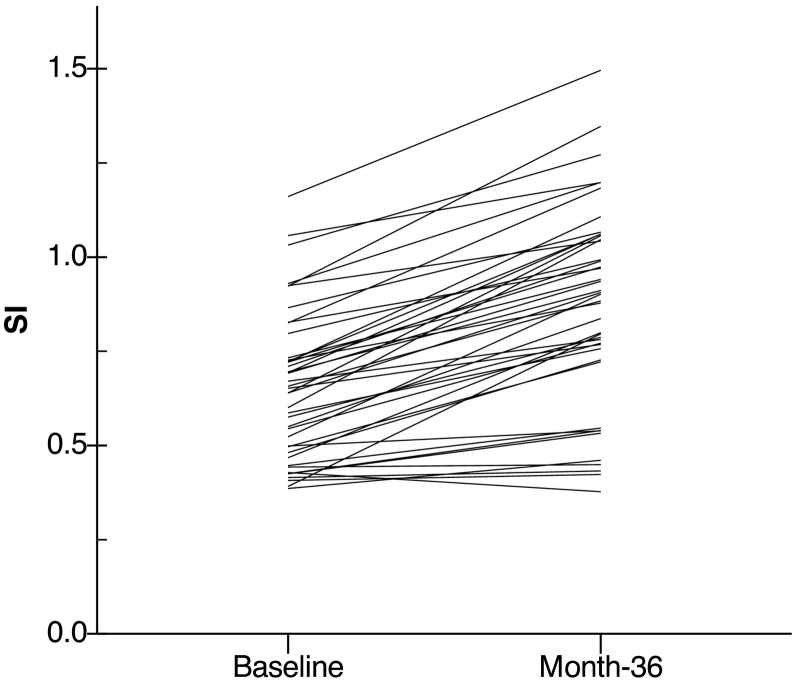
Next, the MCI subjects were divided into AD-like and CN-like and analogous mixed ANOVA were performed to evaluate the effect of APOE and Aβ within each group. The results showed a significant time × APOE group interaction within AD-like subjects (F(2,92) = 6.2, p = 0.003, Fig. 5C), which indicated a faster increase of the SI in ε4+ (n = 53) compared to ε4− (n = 35). Also a significant time × Aβ group interaction was found within AD-like subjects (F(2,92) = 6.0, p = 0.004, Fig. 5D) that indicated a faster increase of the SI in Aβ+ subjects (n = 43) compared to Aβ− subjects (n = 9). No significant effect of APOE or Aβ was observed within CN-like subjects. Fig. 6 shows the SI at baseline and month-36 for each MCI individual with abnormal Aβ and an AD-like structural pattern. This figure illustrates the change of the SI over 36 months follow-up at the individual level. Additional analyses were conducted on CN subjects to compare the effect of APOE genotype and Aβ levels, which resulted in no significant differences (Supplementary Fig. S1).
Fig. 6.
The SI of Aβ+ AD-like MCI subjects at baseline and month-36. Each line represents a MCI individual with abnormal Aβ and an AD-like structural pattern.
SI, disease severity index.
4. Discussion
Structural MRI is an important examination in the clinical assessment of patients with suspected dementia and AD (Falahati et al., 2015, Frisoni et al., 2010). The proposed method for generating the SI summarizes multiple MRI-derived structural measures of the brain in a single score. The multivariate OPLS model that generates the SI is trained using AD and CN subjects. Therefore, the model captures the most discriminative patterns of AD and CN subjects. Based on the SI, the MCI subjects can be divided into AD-like or CN-like. In the CN-like group, the dominant structural patterns are close to the structural patterns of CN subjects i.e. less atrophied structures and smaller ventricles. On the other hand, the AD-like group expresses structural patterns similar to AD, i.e. greater atrophy in medial temporal structures as well as in other regions of the brain.
The potential of the SI as a tool for prediction of progression from MCI to AD has been demonstrated previously (Aguilar et al., 2014, Spulber et al., 2013). In this work the SI was studied longitudinally. The average SI was higher in AD subjects compared to MCI and CN subjects. The results in this study show that in AD and MCI subjects the average SI increased over time, but in CN subjects, it remained stable. This is in line with previous cross-sectional and longitudinal studies that reported increased rates of regional and whole brain atrophy in MCI and AD subjects (Jack et al., 2004, Leung et al., 2013, Schuff et al., 2012, Sluimer et al., 2009, Spulber et al., 2012). A higher average SI was also observed in MCI-p subjects compared to MCI-s subjects, which is in line with previous findings of higher rates of atrophy in progressive MCI subjects (Desikan et al., 2008, Jack et al., 2004, Jack et al., 2008b, Orellana et al., 2016). The most influential variables in the classification model of AD and CN subjects were measures of structures in medial temporal lobe. Results from the cortical thickness map from vertex analysis showed significant cortical thinning of similar regions in AD-like MCI subjects compared to CN-like MCI subjects.
The accuracy of AD versus CN classification in this work was in the level of the reported accuracies in previous studies using ADNI cohort (between 80% and 90% (Falahati et al., 2014)), though higher classification accuracies are also reported (Liu et al., 2013b, Wee et al., 2013, Westman et al., 2013, Wolz et al., 2011). Compared to a recent study (Hu et al., 2016) that used a very similar longitudinal dataset, we obtained a higher classification accuracy (87.2% compared to 84.1%). The baseline SI showed a high sensitivity, 80% over a period of 3 years (95.7% within the first year) for prediction of progression from MCI to AD. The sensitivity of detecting progressive MCI individuals using baseline SI is higher in the present study compared to the sensitivities (65%–75%) which have been reported in previous studies (Liu et al., 2013b, Wee et al., 2013, Wolz et al., 2011). The accuracy for predicting conversion from MCI to AD was at the same level of these studies. However, 3 years is relatively short follow-up time for MCI subjects. A longer follow-up time is needed for reporting more accurate prediction rates, particularly for MCI-s subjects.
Compared to our previous work, we used a lower cut-off which is calculated based on the distribution of the SI in CN subjects (cut-off = 0.372 instead of a strict arbitrary cut-off of 0.5). Since the current cut-off is based on normal variation in a group of healthy controls, the cut-off has better clinical applicability and should facilitate the use of the SI as diagnostic tool. On the contrary, the previously used cut-off of 0.5 is dependent on variation within the AD. Variation within AD is most likely to be larger than in normal healthy individuals. We know from previous work that AD is a very heterogeneous disorder and several subtypes have been defined with different patterns of atrophy (Ferreira et al., 2015, Lam et al., 2013, Noh et al., 2014, Pereira et al., 2014). Naturally, part of the variation in CN subjects is due to normal aging, which is reduced by applying age correction. Further, with the current cut-off we increase the sensitivity to 91.3% for classifying AD subjects, which is in line with a previous study showing that around 10% of diagnosed AD subjects show no atrophy compared to controls (Byun et al., 2015). With the current cut-off we can also classify 80% of the MCI converters at baseline correctly. This number increases the closer it gets to time of conversion to dementia. This supports the notion that structural changes measured by MRI are a good progression marker rather than a staging marker since they are so closely related to cognitive function (Jack et al., 2013). Further, 43% of the stable MCI subjects have an AD-like pattern of atrophy. Although a yearly conversion rate of 10–15% from MCI to dementia is usually mentioned in the literature (Farias et al., 2009), this figure ranges widely between studies (4–36%) (Bennett et al., 2002). Taking this into account, our stable MCI subjects with an AD-like pattern of atrophy will likely convert to dementia in the future. This is further supported by the fact the MCI-s subjects with an AD-like pattern had a very similar baseline SI and SI progression as the MCI-p subjects whom converted at month 36. However, we cannot confirm this with the current data and a longer follow-up is needed. Finally, 57% of the stable MCI subject had a CN-like pattern (30% of the total sample). These subjects could potentially represent subjects that will never develop dementia (Bennett et al., 2002).
The relation between brain structures and cognitive performance in AD has previously been investigated and positive correlations between MMSE scores and cortical and subcortical volumes and negative correlations with the volume of the ventricles have been reported (Ferrarini et al., 2008, Fjell et al., 2009, Orellana et al., 2016). In this study, we observed strong negative correlations between the SI (as a summary of structural measures) and MMSE score in MCI subjects at baseline and follow-up. In addition, a strong correlation was observed between the change of the SI and the change of MMSE scores during 36 months. This is in line with previous reports on the associations of brain atrophy rates and ventricular enlargement with MMSE changes (Evans et al., 2010, Orellana et al., 2016, Yao et al., 2012). The regression coefficient was larger in AD-like MCI subjects compared to CN-like subjects, which might indicate a higher risk of developing AD in AD-like MCI subjects (Sluimer et al., 2008).
APOE genotype is the main genetic risk factor in sporadic AD. APOE ε4 carriers have an increased risk of developing AD compared to ε3 and ε2 carriers (Chartier-Harlin et al., 1994). The effect of APOE ε4 on atrophy rates in AD and MCI subjects has been studied and increased atrophy rates were reported in different brain structures particularly in the medial temporal lobe and hippocampus (Goni et al., 2013, Li et al., 2016, Soldan et al., 2015, Wahlund et al., 1999). In this work, a significant higher average SI and a faster increase of the SI was observed in ε4+ MCI subjects compared to ε4− MCI subjects. Since the measures of medial temporal lobe structures are given greater weights in generating the SI, this result intensify the previous findings that ε4 allele drives atrophy to the medial-temporal lobe (Manning et al., 2014). However, this result might also indicate that the presence of the ε4 allele affects other brain areas. Interestingly, within AD-like MCI subjects a faster increase of SI in ε4+ subjects compared to ε4− subjects was observed, but not in CN-like MCI subjects. This suggests that the presence of the ε4 allele accelerates atrophy in subjects with positive disease biomarkers (Khan et al., 2017).
The longitudinal changes of the SI were also studied in relation to CSF Aβ levels. AD is associated with deposition of Aβ in the brain, which is reflected by low concentration of the Aβ peptide in the CSF (Portelius et al., 2010). Low CSF Aβ levels are also associated with an increased risk of progressing from MCI to AD (Hansson et al., 2006, Herukka et al., 2005, Jack et al., 2010b). Our results show an increased rate of atrophy in Aβ+ subjects compared to Aβ− subjects, which is compatible with a previous study that reported a significant correlation between lower Aβ levels and lower brain volume as well as larger ventricular volume in progressive MCI subjects (Wahlund and Blennow, 2003). Moreover, a faster increase of the SI was observed in AD-like Aβ+ subjects, suggesting that the abnormal Aβ levels can accelerate brain atrophy. This result is analogous to the above findings about AD-like ε4+ subjects. APOE genotype has a strong role in Aβ metabolism where altered APOE alleles affect the Aβ deposition differently (Castellano et al., 2011, Liu et al., 2013a). Because of limited number of subjects with Aβ− and ε4+ we could not test for an interaction between amyloid pathology and APOE genotype. Alternatively, APOE genotype and Aβ group were included as covariates in the mixed ANCOVA. These results suggest that disease-related structural brain changes are influenced independently by APOE genotype and amyloid pathology. However, unlike APOE genotype, the Aβ levels are variable and the baseline levels here may become abnormal in the future. Further investigations on the association between longitudinal changes in CSF biomarkers and the SI are necessary.
Additionally, the effect of APOE and CSF Aβ on SI was examined in 129 CN subjects with 36 months follow-up. Although similar trends were visible, the effects of APOE and Aβ on the SI were not statistically significant. This is compatible with previous studies that reported no significant association of APOE and Aβ with hippocampal loss in CN subjects (Khan et al., 2014, Schuff et al., 2009). Future studies with longer follow-up time and larger sample sizes could be beneficial to assess the effect of APOE and Aβ on the SI in CN subjects.
In this study, the SI is evaluated using a series of MRI scans that are processed longitudinally. An advantage of the longitudinal processing pipeline compared to cross sectional analysis is the increased reliability and robustness of the overall image processing analysis (Reuter et al., 2012). Consequently, the extracted MRI measures, the multivariate model and finally the generated SI are more reliable. Moreover, a longitudinal SI reflects changes in brain structures over a period of time, not only at a single time point. This provides information on disease dynamics rather than static information, which helps to monitor progression in MCI subjects as well as finding diversities in this group. This study is conducted in the ADNI cohort, which is a highly selective cohort. Therefore, these results should be validated in other cohorts, preferably in non-selected clinical data.
5. Conclusion
The SI and its longitudinal changes show great potential for identifying MCI patients at risk of progressing to AD. Progressive MCI patients evidenced a greater SI and a faster increase of the SI. The SI was not only different between progressive and stable MCI, but we could also clearly see differences in the index within the progressive group. The value of the SI was significantly higher and the rate of decline was faster depending on at which time point subjects converted. This is important for prognosis and inclusion in clinical trials. A greater SI in MCI patients was also as expected associated with worse cognitive impairment. Further, MCI patients carrying the APOE ε4 allele and abnormal CSF Aβ levels had greater SI and faster rate of atrophy. Our results suggest that disease-related brain structural changes are modulated independently by APOE genotype and amyloid pathology. Therefore, the SI has a great potential to be employed in clinical trials as an inclusion criteria, monitoring disease progression or a simplified outcome. Ultimately, we believe that the SI has the potential to be implemented in clinical setup to aid early diagnosis and predict MCI progression to AD.
The following is the supplementary data related to this article.
Average SI of 129 CN subjects with 36 months follow-up, stratified by: A) APOE ε4 + and ε4 −; B) CSF Aβ+ and Aβ− groups.
Acknowledgments
Acknowledgements
This project is financially supported by the Swedish Foundation for Strategic Research (SSF, 4-3193/2014), the Swedish Research Council (VR, 2016-02282), the Strategic Research Programme in Neuroscience at Karolinska Institutet (StratNeuro, 1188/2011-223), the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, Hjärnfonden, Alzheimerfonden and Åke Wibergs Stiftelse (H-3550/2016). We also thank Birgitta och Sten Westerberg (2-3079/2015) for additional financial support.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Acknowledgments
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- Aguilar C., Muehlboeck J.S., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Lovestone S., Wahlund L.O., Simmons A., Westman E., AddNeuroMed C. Application of a MRI based index to longitudinal atrophy change in Alzheimer disease, mild cognitive impairment and healthy older individuals in the AddNeuroMed cohort. Front. Aging Neurosci. 2014;6:145. doi: 10.3389/fnagi.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D.A., Wilson R.S., Schneider J.A., Evans D.A., Beckett L.A., Aggarwal N.T., Barnes L.L., Fox J.H., Bach J. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59:198–205. doi: 10.1212/wnl.59.2.198. [DOI] [PubMed] [Google Scholar]
- Busse A., Hensel A., Guhne U., Angermeyer M.C., Riedel-Heller S.G. Mild cognitive impairment: long-term course of four clinical subtypes. Neurology. 2006;67:2176–2185. doi: 10.1212/01.wnl.0000249117.23318.e1. [DOI] [PubMed] [Google Scholar]
- Bylesjö M., Rantalainen M., Cloarec O., Nicholson J.K., Holmes E., Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006;20:341–351. [Google Scholar]
- Byun M.S., Kim S.E., Park J., Yi D., Choe Y.M., Sohn B.K., Choi H.J., Baek H., Han J.Y., Woo J.I., Lee D.Y., Alzheimer's Disease Neuroimaging, I Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer's disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J.M., Kim J., Stewart F.R., Jiang H., DeMattos R.B., Patterson B.W., Fagan A.M., Morris J.C., Mawuenyega K.G., Cruchaga C., Goate A.M., Bales K.R., Paul S.M., Bateman R.J., Holtzman D.M. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin M.C., Parfitt M., Legrain S., Perez-Tur J., Brousseau T., Evans A., Berr C., Vidal O., Roques P., Gourlet V. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum. Mol. Genet. 1994;3:569–574. doi: 10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- Desikan R.S., Fischl B., Cabral H.J., Kemper T.L., Guttmann C.R., Blacker D., Hyman B.T., Albert M.S., Killiany R.J. MRI measures of temporoparietal regions show differential rates of atrophy during prodromal AD. Neurology. 2008;71:819–825. doi: 10.1212/01.wnl.0000320055.57329.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson L., Byrne T., Johansson E., Trygg J., Vikström C. 2013. Multi- and Megavariate Data Analysis Basic Principles and Applications. [Google Scholar]
- Evans M.C., Barnes J., Nielsen C., Kim L.G., Clegg S.L., Blair M., Leung K.K., Douiri A., Boyes R.G., Ourselin S., Fox N.C., Alzheimer's Disease Neuroimaging, I Volume changes in Alzheimer's disease and mild cognitive impairment: cognitive associations. Eur. Radiol. 2010;20:674–682. doi: 10.1007/s00330-009-1581-5. [DOI] [PubMed] [Google Scholar]
- Falahati F., Westman E., Simmons A. Multivariate data analysis and machine learning in Alzheimer's disease with a focus on structural magnetic resonance imaging. J. Alzheimers Dis. 2014;41:685–708. doi: 10.3233/JAD-131928. [DOI] [PubMed] [Google Scholar]
- Falahati F., Fereshtehnejad S.M., Religa D., Wahlund L.O., Westman E., Eriksdotter M. The use of MRI, CT and lumbar puncture in dementia diagnostics: data from the SveDem registry. Dement. Geriatr. Cogn. Disord. 2015;39:81–91. doi: 10.1159/000366194. [DOI] [PubMed] [Google Scholar]
- Falahati F., Ferreira D., Soininen H., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Lovestone S., Eriksdotter M., Wahlund L.O., Simmons A., Westman E., AddNeuroMed, c., the Alzheimer's Disease Neuroimaging, I The effect of age correction on multivariate classification in Alzheimer's disease, with a focus on the characteristics of incorrectly and correctly classified subjects. Brain Topogr. 2016;29:296–307. doi: 10.1007/s10548-015-0455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias S.T., Mungas D., Reed B.R., Harvey D., DeCarli C. Progression of mild cognitive impairment to dementia in clinic- vs community-based cohorts. Arch. Neurol. 2009;66:1151–1157. doi: 10.1001/archneurol.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini L., Palm W.M., Olofsen H., van der Landen R., Jan Blauw G., Westendorp R.G., Bollen E.L., Middelkoop H.A., Reiber J.H., van Buchem M.A., Admiraal-Behloul F. MMSE scores correlate with local ventricular enlargement in the spectrum from cognitively normal to Alzheimer disease. NeuroImage. 2008;39:1832–1838. doi: 10.1016/j.neuroimage.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Ferreira D., Cavallin L., Larsson E.M., Muehlboeck J.S., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Lovestone S., Simmons A., Wahlund L.O., Westman E., AddNeuroMed, c., the Alzheimer's Disease Neuroimaging, I Practical cut-offs for visual rating scales of medial temporal, frontal and posterior atrophy in Alzheimer's disease and mild cognitive impairment. J. Intern. Med. 2015;278:277–290. doi: 10.1111/joim.12358. [DOI] [PubMed] [Google Scholar]
- Ferreira D., Falahati F., Linden C., Buckley R.F., Ellis K.A., Savage G., Villemagne V.L., Rowe C.C., Ames D., Simmons A., Westman E. A 'Disease Severity Index' to identify individuals with Subjective Memory Decline who will progress to mild cognitive impairment or dementia. Sci Rep. 2017;7:44368. doi: 10.1038/srep44368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A.M., Amlien I.K., Westlye L.T., Walhovd K.B. Mini-mental state examination is sensitive to brain atrophy in Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 2009;28:252–258. doi: 10.1159/000241878. [DOI] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Jr., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier S., Reisberg B., Zaudig M., Petersen R.C., Ritchie K., Broich K., Belleville S., Brodaty H., Bennett D., Chertkow H., Cummings J.L., de Leon M., Feldman H., Ganguli M., Hampel H., Scheltens P., Tierney M.C., Whitehouse P., Winblad B., International Psychogeriatric Association Expert Conference on mild cognitive, I Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. [DOI] [PubMed] [Google Scholar]
- Goni J., Cervantes S., Arrondo G., Lamet I., Pastor P., Pastor M.A. Selective brain gray matter atrophy associated with APOE epsilon4 and MAPT H1 in subjects with mild cognitive impairment. J. Alzheimers Dis. 2013;33:1009–1019. doi: 10.3233/JAD-2012-121174. [DOI] [PubMed] [Google Scholar]
- Hansson O., Zetterberg H., Buchhave P., Londos E., Blennow K., Minthon L. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: a follow-up study. Lancet Neurol. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- Herukka S.K., Hallikainen M., Soininen H., Pirttila T. CSF Abeta42 and tau or phosphorylated tau and prediction of progressive mild cognitive impairment. Neurology. 2005;64:1294–1297. doi: 10.1212/01.WNL.0000156914.16988.56. [DOI] [PubMed] [Google Scholar]
- Hu K., Wang Y.J., Chen K.W., Hou L.K., Zhang X.Q. Multi-scale features extraction from baseline structure MRI for MCI patient classification and AD early diagnosis. Neurocomputing. 2016;175:132–145. [Google Scholar]
- Jack C.R., Jr., Shiung M.M., Gunter J.L., O'Brien P.C., Weigand S.D., Knopman D.S., Boeve B.F., Ivnik R.J., Smith G.E., Cha R.H., Tangalos E.G., Petersen R.C. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D., Borowski B., Britson P.J., J L.W., Ward C., Dale A.M., Felmlee J.P., Gunter J.L., Hill D.L., Killiany R., Schuff N., Fox-Bosetti S., Lin C., Studholme C., DeCarli C.S., Krueger G., Ward H.A., Metzger G.J., Scott K.T., Mallozzi R., Blezek D., Levy J., Debbins J.P., Fleisher A.S., Albert M., Green R., Bartzokis G., Glover G., Mugler J., Weiner M.W. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Weigand S.D., Shiung M.M., Przybelski S.A., O'Brien P.C., Gunter J.L., Knopman D.S., Boeve B.F., Smith G.E., Petersen R.C. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008;70:1740–1752. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W., Petersen R.C., Trojanowski J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Wiste H.J., Vemuri P., Weigand S.D., Senjem M.L., Zeng G., Bernstein M.A., Gunter J.L., Pankratz V.S., Aisen P.S., Weiner M.W., Petersen R.C., Shaw L.M., Trojanowski J.Q., Knopman D.S., Alzheimer's Disease Neuroimaging, I Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer's disease. Brain. 2010;133:3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D., Lesnick T.G., Pankratz V.S., Donohue M.C., Trojanowski J.Q. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachaturian Z.S. Diagnosis of Alzheimer's disease. Arch. Neurol. 1985;42:1097–1105. doi: 10.1001/archneur.1985.04060100083029. [DOI] [PubMed] [Google Scholar]
- Khan W., Giampietro V., Ginestet C., Dell'Acqua F., Bouls D., Newhouse S., Dobson R., Banaschewski T., Barker G.J., Bokde A.L., Buchel C., Conrod P., Flor H., Frouin V., Garavan H., Gowland P., Heinz A., Ittermann B., Lemaitre H., Nees F., Paus T., Pausova Z., Rietschel M., Smolka M.N., Strohle A., Gallinat J., Westman E., Schumann G., Lovestone S., Simmons A., Consortium I. No differences in hippocampal volume between carriers and non-carriers of the ApoE epsilon4 and epsilon2 alleles in young healthy adolescents. J. Alzheimers Dis. 2014;40:37–43. doi: 10.3233/JAD-131841. [DOI] [PubMed] [Google Scholar]
- Khan W., Giampietro V., Banaschewski T., Barker G.J., Bokde A.L., Buchel C., Conrod P., Flor H., Frouin V., Garavan H., Gowland P., Heinz A., Ittermann B., Lemaitre H., Nees F., Paus T., Pausova Z., Rietschel M., Smolka M.N., Strohle A., Gallinat J., Vellas B., Soininen H., Kloszewska I., Tsolaki M., Mecocci P., Spenger C., Villemagne V.L., Masters C.L., Muehlboeck J.S., Backman L., Fratiglioni L., Kalpouzos G., Wahlund L.O., Schumann G., Lovestone S., Williams S.C., Westman E., Simmons A., Alzheimer-s Disease Neuroimaging, I, AddNeuroMed Consortium, A.I.B., Lifestyle Study Research, G., consortium, I A multi-cohort study of ApoE varepsilon4 and amyloid-beta effects on the hippocampus in Alzheimer's disease. J. Alzheimers Dis. 2017;56:1159–1174. doi: 10.3233/JAD-161097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel S., Stonnington C.M., Chu C., Draganski B., Scahill R.I., Rohrer J.D., Fox N.C., Jack C.R., Jr., Ashburner J., Frackowiak R.S. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B., Masellis M., Freedman M., Stuss D.T., Black S.E. Clinical, imaging, and pathological heterogeneity of the Alzheimer's disease syndrome. Alzheimers Res. Ther. 2013;5:1. doi: 10.1186/alzrt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch J.P., Pruessner J., Zijdenbos A.P., Collins D.L., Teipel S.J., Hampel H., Evans A.C. Automated cortical thickness measurements from MRI can accurately separate Alzheimer's patients from normal elderly controls. Neurobiol. Aging. 2008;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Leung K.K., Bartlett J.W., Barnes J., Manning E.N., Ourselin S., Fox N.C., Alzheimer's Disease Neuroimaging, I Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology. 2013;80:648–654. doi: 10.1212/WNL.0b013e318281ccd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Wu G., Shi F., Zhou L., Lin W., Shen D., Alzheimer's Disease Neuroimaging, I Discriminant analysis of longitudinal cortical thickness changes in Alzheimer's disease using dynamic and network features. Neurobiol. Aging. 2012;33(427):e415–430. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Shi J., Gutman B.A., Baxter L.C., Thompson P.M., Caselli R.J., Wang Y., Alzheimer's Disease Neuroimaging, I Influence of APOE genotype on hippocampal atrophy over time - an N = 1925 surface-based ADNI study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Tosun D., Weiner M.W., Schuff N., Alzheimer's Disease Neuroimaging, I Locally linear embedding (LLE) for MRI based Alzheimer's disease classification. NeuroImage. 2013;83:148–157. doi: 10.1016/j.neuroimage.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangialasche F., Westman E., Kivipelto M., Muehlboeck J.S., Cecchetti R., Baglioni M., Tarducci R., Gobbi G., Floridi P., Soininen H., Kloszewska I., Tsolaki M., Vellas B., Spenger C., Lovestone S., Wahlund L.O., Simmons A., Mecocci P., AddNeuroMed, c. Classification and prediction of clinical diagnosis of Alzheimer's disease based on MRI and plasma measures of alpha-/gamma-tocotrienols and gamma-tocopherol. J. Intern. Med. 2013;273:602–621. doi: 10.1111/joim.12037. [DOI] [PubMed] [Google Scholar]
- Manning E.N., Barnes J., Cash D.M., Bartlett J.W., Leung K.K., Ourselin S., Fox N.C., Alzheimer's Disease NeuroImaging, I APOE epsilon4 is associated with disproportionate progressive hippocampal atrophy in AD. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery W.R. Neuropathological criteria for the diagnosis of Alzheimer's disease. Neurobiol. Aging. 1997;18:S13–19. doi: 10.1016/s0197-4580(97)00064-x. [DOI] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Avedissian C., Madsen S.K., Parikshak N., Hua X., Toga A.W., Jack C.R., Jr., Schuff N., Weiner M.W., Thompson P.M., Alzheimer's Disease Neuroimaging, I Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum. Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlboeck J.-S., Westman E., Simmons A. TheHiveDB image data management and analysis framework. Front. Neuroinform. 2014;7:49. doi: 10.3389/fninf.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C.R., Jagust W., Trojanowski J.Q., Toga A.W., Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y., Jeon S., Lee J.M., Seo S.W., Kim G.H., Cho H., Ye B.S., Yoon C.W., Kim H.J., Chin J., Park K.H., Heilman K.M., Na D.L. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology. 2014;83:1936–1944. doi: 10.1212/WNL.0000000000001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana C., Ferreira D., Muehlboeck J.S., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Lovestone S., Simmons A., Wahlund L.O., Westman E., AddNeuronMed, c., for the Alzheimer's Disease Neuroimaging, I Measuring global brain atrophy with the brain volume/cerebrospinal fluid index: normative values, cut-offs and clinical associations. Neurodegener. Dis. 2016;16:77–86. doi: 10.1159/000442443. [DOI] [PubMed] [Google Scholar]
- Pereira J.B., Cavallin L., Spulber G., Aguilar C., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Spenger C., Aarsland D., Lovestone S., Simmons A., Wahlund L.O., Westman E., AddNeuroMed, c., for the Alzheimer's Disease Neuroimaging, I Influence of age, disease onset and ApoE4 on visual medial temporal lobe atrophy cut-offs. J. Intern. Med. 2014;275:317–330. doi: 10.1111/joim.12148. [DOI] [PubMed] [Google Scholar]
- Petersen R.C., Aisen P.S., Beckett L.A., Donohue M.C., Gamst A.C., Harvey D.J., Jack C.R., Jr., Jagust W.J., Shaw L.M., Toga A.W., Trojanowski J.Q., Weiner M.W. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E., Andreasson U., Ringman J.M., Buerger K., Daborg J., Buchhave P., Hansson O., Harmsen A., Gustavsson M.K., Hanse E., Galasko D., Hampel H., Blennow K., Zetterberg H. Distinct cerebrospinal fluid amyloid beta peptide signatures in sporadic and PSEN1 A431E-associated familial Alzheimer's disease. Mol. Neurodegener. 2010;5:2. doi: 10.1186/1750-1326-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Fischl B. Avoiding asymmetry-induced bias in longitudinal image processing. NeuroImage. 2011;57:19–21. doi: 10.1016/j.neuroimage.2011.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Rosas H.D., Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage. 2010;53:1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmansky N.J., Rosas H.D., Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. NeuroImage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin A.J., Shen L., Foroud T.M., Potkin S.G., Swaminathan S., Kim S., Risacher S.L., Nho K., Huentelman M.J., Craig D.W., Thompson P.M., Stein J.L., Moore J.H., Farrer L.A., Green R.C., Bertram L., Jack C.R., Jr., Weiner M.W., Alzheimer's Disease Neuroimaging, I Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N., Woerner N., Boreta L., Kornfield T., Shaw L.M., Trojanowski J.Q., Thompson P.M., Jack C.R., Jr., Weiner M.W., Alzheimer's Disease Neuroimaging, I MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N., Tosun D., Insel P.S., Chiang G.C., Truran D., Aisen P.S., Jack C.R., Jr., Weiner M.W., Alzheimer's Disease Neuroimaging, I Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol. Aging. 2012;33:845–855. doi: 10.1016/j.neurobiolaging.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Pozo A., Frosch M.P., Masliah E., Hyman B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011;1:a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M., Trojanowski J.Q., Alzheimer's Disease Neuroimaging, I Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer J.D., van der Flier W.M., Karas G.B., Fox N.C., Scheltens P., Barkhof F., Vrenken H. Whole-brain atrophy rate and cognitive decline: longitudinal MR study of memory clinic patients. Radiology. 2008;248:590–598. doi: 10.1148/radiol.2482070938. [DOI] [PubMed] [Google Scholar]
- Sluimer J.D., van der Flier W.M., Karas G.B., van Schijndel R., Barnes J., Boyes R.G., Cover K.S., Olabarriaga S.D., Fox N.C., Scheltens P., Vrenken H., Barkhof F. Accelerating regional atrophy rates in the progression from normal aging to Alzheimer's disease. Eur. Radiol. 2009;19:2826–2833. doi: 10.1007/s00330-009-1512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldan A., Pettigrew C., Lu Y., Wang M.C., Selnes O., Albert M., Brown T., Ratnanather J.T., Younes L., Miller M.I., Team B.R. Relationship of medial temporal lobe atrophy, APOE genotype, and cognitive reserve in preclinical Alzheimer's disease. Hum. Brain Mapp. 2015;36:2826–2841. doi: 10.1002/hbm.22810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spulber G., Niskanen E., Macdonald S., Kivipelto M., Padilla D.F., Julkunen V., Hallikainen M., Vanninen R., Wahlund L.O., Soininen H. Evolution of global and local grey matter atrophy on serial MRI scans during the progression from MCI to AD. Curr. Alzheimer Res. 2012;9:516–524. doi: 10.2174/156720512800492486. [DOI] [PubMed] [Google Scholar]
- Spulber G., Simmons A., Muehlboeck J.S., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Spenger C., Lovestone S., Wahlund L.O., Westman E., dNeuroMed, c., for the Alzheimer Disease Neuroimaging, I An MRI-based index to measure the severity of Alzheimer's disease-like structural pattern in subjects with mild cognitive impairment. J. Intern. Med. 2013;273:396–409. doi: 10.1111/joim.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J., Wold S. Orthogonal projections to latent structures (O-PLS) J. Chemom. 2002;16:119–128. [Google Scholar]
- Voevodskaya O., Simmons A., Nordenskjold R., Kullberg J., Ahlstrom H., Lind L., Wahlund L.O., Larsson E.M., Westman E., Alzheimer's Disease Neuroimaging, I The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer's disease. Front. Aging Neurosci. 2014;6:264. doi: 10.3389/fnagi.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlund L.O., Blennow K. Cerebrospinal fluid biomarkers for disease stage and intensity in cognitively impaired patients. Neurosci. Lett. 2003;339:99–102. doi: 10.1016/s0304-3940(02)01483-0. [DOI] [PubMed] [Google Scholar]
- Wahlund L.O., Julin P., Lannfelt L., Lindqvist J., Svensson L. Inheritance of the ApoE epsilon4 allele increases the rate of brain atrophy in dementia patients. Dement. Geriatr. Cogn. Disord. 1999;10:262–268. doi: 10.1159/000017130. [DOI] [PubMed] [Google Scholar]
- Wee C.Y., Yap P.T., Shen D., Alzheimer's Disease Neuroimaging, I Prediction of Alzheimer's disease and mild cognitive impairment using cortical morphological patterns. Hum. Brain Mapp. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman E., Simmons A., Muehlboeck J.S., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., Soininen H., Weiner M.W., Lovestone S., Spenger C., Wahlund L.O., AddNeuroMed, c., Alzheimer's Disease Neuroimaging, I AddNeuroMed and ADNI: similar patterns of Alzheimer's atrophy and automated MRI classification accuracy in Europe and North America. NeuroImage. 2011;58:818–828. doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- Westman E., Aguilar C., Muehlboeck J.S., Simmons A. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer's disease and mild cognitive impairment. Brain Topogr. 2013;26:9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S., Ruhe A., Wold H., Dunn W.J. The collinearity problem in linear regression. The Partial Least Squares (PLS) approach to generalized inverses. SIAM J. Sci. Stat. Comput. 1984;5:735–743. [Google Scholar]
- Wolz R., Julkunen V., Koikkalainen J., Niskanen E., Zhang D.P., Rueckert D., Soininen H., Lotjonen J., Alzheimer's Disease Neuroimaging, I Multi-method analysis of MRI images in early diagnostics of Alzheimer's disease. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Hu B., Liang C., Zhao L., Jackson M., Alzheimer's Disease Neuroimaging, I A longitudinal study of atrophy in amnestic mild cognitive impairment and normal aging revealed by cortical thickness. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wang Y., Zhou L., Yuan H., Shen D., Alzheimer's Disease Neuroimaging, I Multimodal classification of Alzheimer's disease and mild cognitive impairment. NeuroImage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average SI of 129 CN subjects with 36 months follow-up, stratified by: A) APOE ε4 + and ε4 −; B) CSF Aβ+ and Aβ− groups.