Abstract
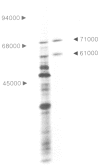
By the use of glucoamylase-specific synthetic oligodeoxyribonucleotides and molecular cloning of cDNA synthesized from Aspergillus niger total poly(A) + RNA, the primary structure of the glucoamylase G1 mRNA was determined. Glucoamylase G1 is synthesized as a precursor of 640 amino acid residues containing a putative signal peptide of 18 residues, a short propeptide of six residues and the 616 residues long mature enzyme. In vitro translations of mRNA and immunoprecipitations with glucoamylase-specific antisera showed that two glucoamylase polypeptides are synthesized. The larger form with an apparent mol. wt. of 71 000 corresponds to the precursor of glucoamylase G1, and the shorter form with an apparent mol. wt. of 61 000 corresponds to the precursor of glucoamylase G2. From the nucleotide sequencing data of several glucoamylase-specific cDNA recombinants it is shown that the G1 mRNA contains a 169 bp long intervening sequence that can be spliced out to generate a G2 mRNA. Only the 3' part of the G1 mRNA is modified by this splicing event. This kind of differential mRNA processing to give different protein products from one primary transcript has previously only been demonstrated in higher eukaryotes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel E., Vuust J., Norris F., Norris K., Wind A., Rehfeld J. F., Marcker K. A. Molecular cloning of human gastrin cDNA: evidence for evolution of gastrin by gene duplication. Proc Natl Acad Sci U S A. 1983 May;80(10):2866–2869. doi: 10.1073/pnas.80.10.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Crabtree G. R., Kant J. A. Organization of the rat gamma-fibrinogen gene: alternative mRNA splice patterns produce the gamma A and gamma B (gamma ') chains of fibrinogen. Cell. 1982 Nov;31(1):159–166. doi: 10.1016/0092-8674(82)90415-9. [DOI] [PubMed] [Google Scholar]
- Cullen S. E., Schwartz B. D. An improved method for isolation of H-2 and Ia alloantigens with immunoprecipitation induced by protein A-bearing staphylococci. J Immunol. 1976 Jul;117(1):136–142. [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz H. J., Belagaje R., Brown E. L., Fritz R. H., Jones R. A., Lees R. G., Khorana H. G. High-pressure liquid chromatography in polynucleotide synthesis. Biochemistry. 1978 Apr 4;17(7):1257–1267. doi: 10.1021/bi00600a020. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi K., Ohnishi M., Tanaka A. Subsite structure and ligand binding mechanism of glucoamylase. Mol Cell Biochem. 1983;51(1):79–95. doi: 10.1007/BF00215589. [DOI] [PubMed] [Google Scholar]
- Inokuchi N., Iwama M., Takahashi T., Irie M. Modification of a glucoamylase from Aspergillus saitoi with 1-cyclohexyl-3-(2-morpholinyl-(4)-ethyl)carbodiimide. J Biochem. 1982 Jan;91(1):125–133. doi: 10.1093/oxfordjournals.jbchem.a133669. [DOI] [PubMed] [Google Scholar]
- Ito H., Ike Y., Ikuta S., Itakura K. Solid phase synthesis of polynucleotides. VI. Further studies on polystyrene copolymers for the solid support. Nucleic Acids Res. 1982 Mar 11;10(5):1755–1769. doi: 10.1093/nar/10.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Vibe-Pedersen K., Baralle F. E. Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J. 1984 Jan;3(1):221–226. doi: 10.1002/j.1460-2075.1984.tb01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lineback D. R., Russell I. J., Rasmussen C. Two forms of the glucoamylase of Aspergillus niger. Arch Biochem Biophys. 1969 Nov;134(2):539–553. doi: 10.1016/0003-9861(69)90316-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCleary B. V., Anderson M. A. Hydrolysis of alpha-D-glucans and alpha-D-gluco-oligosaccharides by Cladosporium resinae glucoamylases. Carbohydr Res. 1980 Nov 1;86(1):77–96. doi: 10.1016/s0008-6215(00)84583-8. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazur J. H., Knull H. R., Cepure A. Glycoenzymes: structure and properties of the two forms of glucoamylase from Aspergillus niger. Carbohydr Res. 1971 Nov;20(1):83–96. doi: 10.1016/s0008-6215(00)84951-4. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983 Jan 10;258(1):143–149. [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983 Dec;35(2 Pt 1):421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Inokuchi N., Irie M. Purification and characterization of a glucoamylase from Aspergillus saitoi. J Biochem. 1981 Jan;89(1):125–134. doi: 10.1093/oxfordjournals.jbchem.a133172. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Tsuchida Y., Irie M. Isolation of two inactive fragments of a Rhizopus sp. glucoamylase: relationship among three forms of the enzyme and the isolated fragments. J Biochem. 1982 Nov;92(5):1623–1633. doi: 10.1093/oxfordjournals.jbchem.a134088. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C. Yeast gene TRP5: structure, function, regulation. J Biol Chem. 1982 Feb 10;257(3):1491–1500. [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]