Summary
The pathogenicity of the clinically important yeast, Candida albicans, is dependent on robust responses to host‐imposed stresses. These stress responses have generally been dissected in vitro at 30°C on artificial growth media that do not mimic host niches. Yet host inputs, such as changes in carbon source or temperature, are known to affect C. albicans stress adaptation. Therefore, we performed screens to identify novel regulators that promote stress resistance during growth on a physiologically relevant carboxylic acid and at elevated temperatures. These screens revealed that, under these ‘non‐standard’ growth conditions, numerous uncharacterised regulators are required for stress resistance in addition to the classical Hog1, Cap1 and Cta4 stress pathways. In particular, two transcription factors (Sfp1 and Rtg3) promote stress resistance in a reciprocal, carbon source‐conditional manner. SFP1 is induced in stressed glucose‐grown cells, whereas RTG3 is upregulated in stressed lactate‐grown cells. Rtg3 and Sfp1 regulate the expression of key stress genes such as CTA4, CAP1 and HOG1 in a carbon source‐dependent manner. These mechanisms underlie the stress sensitivity of C. albicans sfp1 cells during growth on glucose, and rtg3 cells on lactate. The data suggest that C. albicans exploits environmentally contingent regulatory mechanisms to retain stress resistance during host colonisation.
Introduction
Of the circa 1.5 million fungal species thought to inhabit our planet, only around 600 have been reported to be pathogenic for humans. The yeast Candida albicans is a common cause of mucosal infection (oral and vaginal thrush), and is the most frequent cause of nosocomial fungal infections (Brown et al., 2007; Brock, 2009; Calderone and Clancy, 2012). The fate of this opportunistic fungal pathogen is intertwined with its mammalian host, in which it is normally found as a relatively harmless commensal in the oral, urogenital and gastrointestinal microbiota (Bouza and Muñoz, 2008; Calderone and Clancy, 2012). However, infections can arise when our immunological defenses become compromised, allowing C. albicans to thrive in niches where it would normally be subject to phagocytic clearance (Gow et al., 2012; Brown et al., 2014a,).
The ability of C. albicans to colonise diverse host niches is dependent on its rapid adaptation to the local conditions in these microenvironments, including changes in the availability of key nutrients such as the carbon source (Staib et al., 1999; Barelle et al., 2006; Ene et al., 2013; Brown et al., 2014b). For example, glucose levels are minimal in the colon, between 0.06 and 0.1% in the bloodstream, and are reported to be about 0.5% in vaginal secretions (Brown et al., 2014b). Transcript profiling studies that have examined the in vivo gene expression patterns of C. albicans cell populations from the blood or internal organs suggest that both glycolytic and gluconeogenic pathways are active in these fungal populations (Andes et al., 2005; Fradin et al., 2005; Barelle et al., 2006; Walker et al., 2008). This counterintuitive finding could be explained either by an ability of individual C. albicans cells to express both pathways simultaneously (Sandai et al., 2012; Childers et al., 2016) or by the complexity of host niches, in which individual C. albicans cells can be exposed to glucose‐containing or glucose‐lacking microenvironments depending on their location (Hube, 2004; Barelle et al., 2006; Miramón et al., 2012). In the gut, most dietary sugars are absorbed in the small intestine before the remaining nutrients enter the large intestine. This view is supported by bacterial expression profiling studies, which suggest that sugar concentrations are minimal in colon microenvironments (Kröger et al., 2013; Avican et al., 2015). It has been reported that glycolytic genes are upregulated in C. albicans cells colonizing the mouse caecum (Rosenbach et al., 2010), but these experiments involved the pretreatment of mice with antibiotics to deplete the gut microbiota. Indeed, the view that sugars are limiting in gut microenvironments is reinforced by the observation that C. glabrata mutants that cannot utilize the organic acid lactate are unable to colonize the gut (Ueno et al., 2011).
The pathogenicity of C. albicans is further enhanced by its ability to counteract local environmental stresses. This yeast is relatively resistant to certain stresses compared with other fungi (Jamieson et al., 1996; Nikolaou et al., 2009). Host‐imposed stresses include oxidative, nitrosative and cationic stresses, as well as thermal fluctuations in febrile hosts (Enjalbert et al., 2003; Hube, 2004; Enjalbert et al., 2006; Enjalbert et al., 2007; Leach et al., 2012a,b,c; Miramón et al., 2012). C. albicans mounts robust responses to these stresses via specific signaling pathways. The Hog1‐dependent MAP kinase pathway promotes resistance to cationic, osmotic and oxidative stresses (San José et al., 1996; Alonso‐Monge et al., 2003; Smith et al., 2004). Additional MAP kinase signaling pathways, characterized by the Mkc1 and Cek1 MAP kinases, promote the resistance of C. albicans to cell wall stresses (Navarro‐García et al., 1995; Alonso‐Monge et al., 2006; Eisman et al., 2006). The AP‐1‐like transcription factor, Cap1, plays a major role in driving the transcriptional response to oxidative stress (Alarco and Raymond, 1999; Znaidi et al., 2009; Kos et al., 2016), and the response‐regulator Skn7 contributes to this response (Singh et al., 2004). Meanwhile, the transcription factors Cta4 and Hsf1 play key roles in the transcriptional responses to nitrosative stress and heat shock, respectively (Hromatka et al., 2005; Chiranand et al., 2008; Nicholls et al., 2009). These signaling pathways protect C. albicans against many of the stresses imposed by the host (Alonso‐Monge et al., 2006; Herrero‐de‐Dios et al., 2010). Consequently, the inactivation of key stress responses attenuates the virulence of this fungus (Wysong et al., 1998; Alonso‐Monge et al., 1999; Hwang et al., 2002; Fradin et al., 2005; Nicholls et al., 2011).
The in vitro dissection of these signaling pathways and their contributions to stress adaptation has generally been performed using C. albicans cells grown at 30°C on rich media containing 2% glucose. However, as described above, many host niches colonized by this pathogenic yeast contain low levels of glucose or lack this sugar. Also, C. albicans is subjected to changes in ambient temperature, in the febrile host for example. Therefore, during host colonization, C. albicans must respond to local environmental stresses while adapting to alternative carbon sources or thermal fluctuations. Changes in temperature or carbon source have been shown to affect the stress resistance of C. albicans cells. For example, ambient temperature influences their resistance to osmotic and cell wall stresses (Leach et al., 2012a). Exposure to glucose enhances the resistance of C. albicans to oxidative stress (Rodaki et al., 2009). Also, growth on lactate rather than glucose confers elevated resistance to osmotic stress (Ene et al., 2012a, 2012b; Ene et al., 2015).
These observations suggest crosstalk between carbon assimilation and stress adaptation in C. albicans, but the mechanisms that underlie this crosstalk remain to be defined. Therefore, we performed high‐throughput robotic screens to identify C. albicans mutants that display carbon source‐ or temperature‐conditional resistance to oxidative, osmotic or nitrosative stresses. Our screens, which have revealed extensive environmentally conditional stress sensitivities, have highlighted two transcription factors that play complementary roles in the carbon‐conditional modulation of stress responses. Rtg3 promotes stress adaptation in lactate‐grown cells, whereas Sfp1 enhances stress adaptation in glucose‐grown cells. Mechanisms such as these presumably allow C. albicans to maintain robust stress responses as it colonizes host microenvironments with different nutrient profiles.
Results
Carbon‐conditional stress sensitivity in C. Albicans
High‐throughput robotic screens of previously constructed mutant collections (Supporting Information Table S1) were used to identify genes that promote environmentally contingent stress adaptation in C. albicans. These collections included the set of regulatory transposon insertion mutants generated by the Mitchell laboratory (Norice et al., 2007; Nobile and Mitchell, 2009), the transcription factor deletion mutants constructed by Sanglard's group (Vandeputte et al., 2011), and the library of null mutants created by Noble and co‐workers (Noble et al., 2010). These mutants represent approximately 16% of C. albicans protein coding genes. The stress resistance of each mutant was compared to its congenic control strain using plate assays under a range of growth conditions. Moderate doses of oxidative (0.4 mM H2O2) and nitrosative stress (5 mM NaNO2) were used (Chiranand et al., 2008). We also used a relatively moderate dose of salt (1 M NaCl) (Kaloriti et al., 2012), which imposes osmotic and cationic stress (Hohmann, 2002). In control experiments these doses were shown to significantly attenuate the growth of hallmark deletion mutants, such as hog1, cap1 or cta4, but not the growth of the corresponding wild type control strains under standard growth conditions (YPD at 30°C: Experimental Procedures). The stress sensitivities of all mutants were then compared in 96 array format on rich glucose‐containing medium and minimal media containing glucose or lactate as sole carbon source at 30°C, 37°C and 42°C (36 conditions in total, including the unstressed controls) (Supporting Information Fig. S1). The plates were then imaged, and the growth of each strain, relative to its isogenic wild type control, was recorded by computational analysis of these raw images. A strain was defined as displaying stress sensitivity if it consistently displayed more than an 80% reduction in growth in the presence of that stress relative to the control plate without the stress but with matching carbon and temperature conditions (Experimental Methods). The data were filtered to exclude mutants that were unable to grow under the control unstressed condition. The output was then used to construct carbon‐ and temperature‐conditional stress networks.
Mutants with defects in the HOG signaling module did not display carbon conditional sensitivity to cationic stress: pbs2 and hog1 cells were sensitive to NaCl whether cells were growth on rich glucose‐containing medium (YPD) or on minimal medium containing glucose or lactate as sole carbon source (Fig. 1). Cells lacking Pbs2 seemed more sensitive to cationic stress than hog1 cells when grown on glucose as sole carbon source (GYNB; Fig. 1), but in the context of our screen, both mutants displayed greater stress sensitivity than wild type cells under this growth condition. Therefore, this key MAP kinase module contributes to cationic stress adaptation under conditions outwith the standardized conditions that have generally been used to dissect C. albicans stress adaptation in vitro. However, our screens revealed many other C. albicans mutants that did display carbon‐conditional sensitivity to cationic stress. In comparison with glucose‐grown cells, cells growing on lactate required many additional functions for adaptation to cationic stress. This did not simply reflect the additional biochemical functions required for growth on lactate, because in our screen, the stress sensitivity of a mutant grown on lactate was defined in comparison to its growth on lactate in the absence of stress. The carbon‐conditional cationic stress sensitive mutants included cdc10 (lacking a septin required for virulence and tissue invasion), frp1 (ferric reductase) and sfp1 (C2H2 transcription factor involved in regulation of biofilm formation: Chen and Lan, 2015) (Fig. 1). Interestingly, both cap1 and cta4 cells were sensitive to cationic stress during growth on lactate, but not on glucose (Fig. 2A). Under standard growth conditions (YPD), Cap1 and Cta4 play central roles in the transcriptional responses to oxidative and nitrosative stresses, respectively (Alarco and Raymond, 1999; Chiranand et al., 2008; Znaidi et al., 2009). Our data suggest that these transcription factors also contribute to cationic stress adaptation under other growth conditions. Mutants with defects in cell wall biosynthesis (crz1, kre62, mnn4, phr3), cellular morphogenesis and biofilm formation (efg1/cph1, ace2, bcr1, cbk1, cdc10, pde2, swe1, wsc1) also displayed carbon conditional cationic stress sensitivity. These genes are required for normal levels of NaCl resistance during growth on lactate (Fig. 2A).
Figure 1.
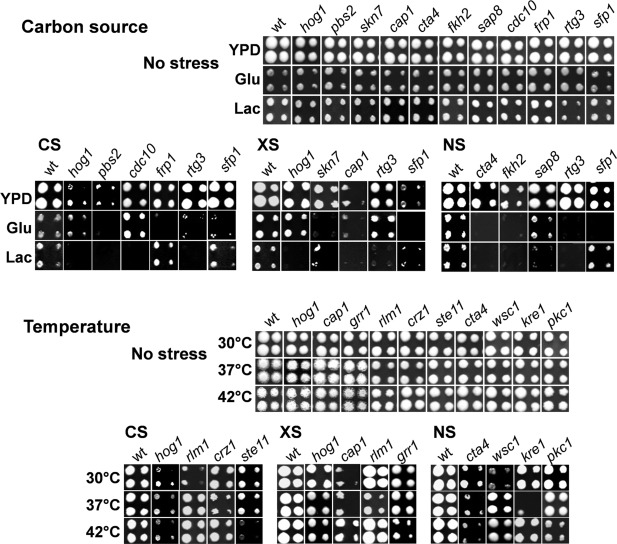
C. albicans mutants that display carbon‐ and temperature‐conditional stress sensitivity. To examine the impact of carbon source, C. albicans mutants were robotically plated onto YPD, GYNB (Glu) or LacYNB (Lac) containing cationic stress (CS; 1 M NaCl), oxidative stress (XS; 0.4 mM H2O2), nitrosative stress (NS; 5 mM NaNO2) or no stress, and grown at 30°C (Experimental Procedures). The robot generated four spots per strain: two at higher, and two at lower cell densities. To examine the impact of ambient temperature, C. albicans strains were plated on YPD and grown at the temperature indicated. Examplar strains are shown in this figure, whilst the full sets of environmentally contingent stress sensitive mutants are shown in Figs. 2 and 3.
Figure 2.
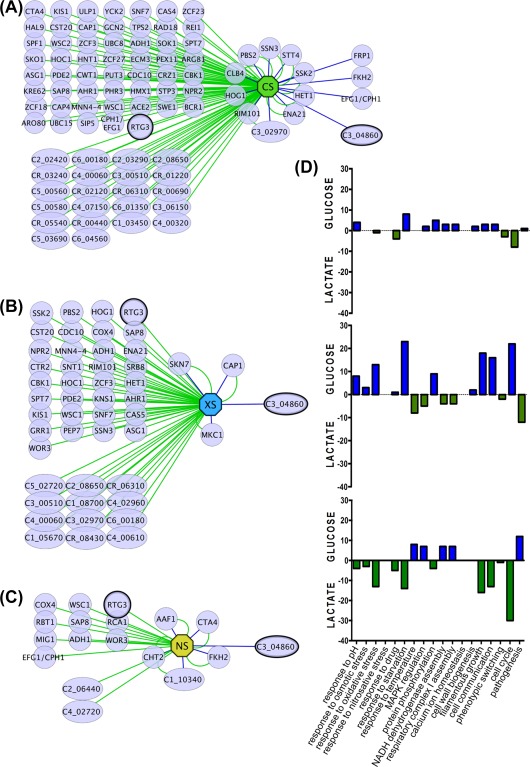
Networks of C. albicans mutants revealed by the screens for carbon‐conditional stress sensitivity. The networks of carbon‐conditional mutants (at 30°C) are displayed for:
A. Cationic stress (CS, 1 M NaCl, green octagon).
B. Oxidative stress (XS, 0.4 mM H2O2, blue octagon).
C. nitrosative stress (NS, 5 mM NaNO2, yellow octagon) using edges between these hubs and the gene nodes that are either green (sensitivity on lactate) or blue (sensitivity on glucose). Nodes with double edges represent mutants with sensitivity on both carbon sources. Standard name genes are represented with circles and systematic name genes with diamonds.
D. Significant enrichment of specific GO terms (biological processes) for lactate‐conditional (green) and glucose‐conditional mutants (blue).
Other C. albicans mutants showed carbon‐conditional sensitivity to oxidative stress. These included rtg3 (C1_10990C), which encodes a putative transcription factor orthologous to Saccharomyces cerevisiae Rtg3 and recently shown to be involved in galactose metabolism in C. albicans (Dalal et al., 2016), and also sfp1 (C3_04860W), which as mentioned above, encodes a partially characterized transcription factor involved in biofilm formation (Fig. 1). Interestingly, hog1 cells were sensitive to oxidative stress during growth on lactate, but not on minimal medium containing glucose (Fig. 1). This was also the case for ssk2 and pbs2 cells, suggesting that the HOG MAPK module makes environmentally contingent contributions to oxidative stress resistance (Fig. 2B). In contrast, cap1 and skn7 cells, which lack key players in the oxidative stress response under standard growth conditions, were sensitive to oxidative stress during growth on lactate as well as glucose. Therefore, Cap1 and Skn7 would appear to promote oxidative stress adaptation under conditions additional to the standardized in vitro growth conditions that have generally been used to examine these regulators (Alarco and Raymond, 1999; Zhang et al., 2000; Alonso‐Monge et al., 2003; Singh et al., 2004; Wang et al., 2006; Enjalbert et al., 2007).
Carbon‐conditional sensitivity to nitrosative stress was also displayed by some specific C. albicans mutants (Fig. 1). These included morphogenetic mutants such as cph1/efg1 and rbt1 (Fig. 2C). They also included the rtg3 and sfp1 mutants, which also displayed carbon‐conditional sensitivities to cationic and oxidative stresses (Fig. 1). Once again the significance of carbon‐conditional nitrosative stress phenotypes was emphasized by the observation that cta4 cells, which lack the key regulator that drives transcriptional responses to nitrosative stress, displayed sensitivity to this stress during growth on glucose and lactate (Fig. 2C). Interestingly, mutants lacking Fkh2 (forkhead transcription factor involved in morphogenetic regulation), Cht2 (chitinase) or Aaf1 (adhesin‐like protein) were sensitive to nitrosative stress, irrespective of the carbon source. These findings reinforce the previously reported links between metabolic adaptation, cell wall biogenesis and morphogenesis (Ene et al., 2012a, 2012b; Brown et al., 2014a).
The carbon‐conditional stress mutants identified in our screens displayed significant enrichment of specific functional categories (GO terms), relative to the functional categories represented in the entire mutant set used in the screens (Fig. 2D). For example, genes related to ‘Pathogenesis’ were enriched in lactate‐dependent oxidative stress genes, and genes related to ‘Filamentous Growth’ were enriched in lactate‐dependent nitrosative stress genes (Fig. 2D). A significant proportion of the carbon‐conditional stress mutants we identified carry defects in uncharacterized genes (33% of the carbon‐conditional cationic stress mutants) (Fig. 2A). This reflects our ignorance about the ways in which environmental changes within host niches impact upon stress adaptation mechanisms in this major pathogen.
Temperature‐conditional stress sensitivity in C. Albicans
Our screens also revealed numerous C. albicans mutants that display temperature‐conditional stress phenotypes. For example, the sensitivity of rlm1 cells to cationic and oxidative stress was higher at 30°C than at 37°C and 42°C. In contrast, crz1 cells were more sensitive to cationic stress at 37°C (Fig. 3). The Rlm1 and Crz1 transcription factors promote cell wall remodelling in C. albicans, and the cell wall provides some protection against environmental insults. Therefore, the temperature‐conditional stress phenotypes of rlm1 and crz1 cells might suggest that Rlm1 and Crz1 make differential thermally contingent contributions to cell wall remodelling in C. albicans (Leach et al., 2012c), for example during caspofungin exposure (Walker et al., 2008). Both Rlm1 and Crz1 promote resistance to this antifungal drug in vitro (Hahn and Thiele, 2002; Selvaggini et al., 2004; Lesage and Bussey, 2006).
Figure 3.
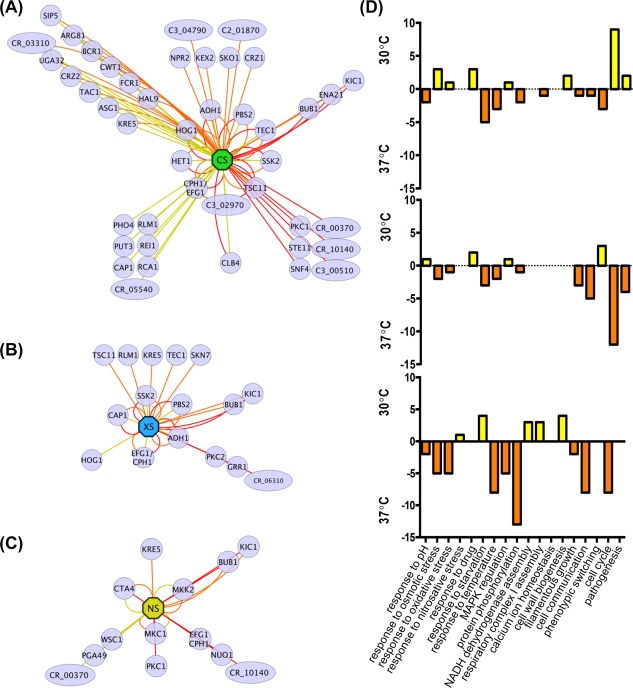
C. albicans gene networks revealed by screens for temperature‐conditional stress sensitivity. The networks of temperature‐conditional mutants (on YPD) are displayed for:
A. cationic stress (CS, 1 M NaCl, green octagon)
B. oxidative stress (XS, 0.4 mM H2O2, blue octagon)
C. nitrosative stress (NS, 5 mM NaNO2, yellow octagon) using edges between these hubs and gene nodes that are either yellow (sensitivity at 30°C), orange (sensitivity at 37°C) or red (sensitivity at 42°C). Nodes with double or triple edges indicate stress sensitivity at two or more temperatures. Standard name genes are represented with circles and systematic name genes with diamonds.
D. Significant enrichment of specific GO terms (biological processes) at 30°C (yellow) and 37°C (orange).
Mutants with defects in the HOG pathway (ssk2, pbs2, hog1) did not display temperature conditional cationic stress sensitivity (Fig. 3A). Furthermore, cap1 cells showed no temperature conditionality in their oxidative stress sensitivity (Fig. 3B), and the cta4 mutant did not display temperature conditional nitrosative stress sensitivity (Fig. 3C). These observations are consistent with the view that the Hog1, Cap1 and Cta4 signalling pathways play critical roles in cationic, oxidative and nitrosative stress adaptation, respectively, at ambient temperatures associated with host colonisation and invasion. However, hog1 cells did not display significant oxidative stress sensitivity at 37°C or 42°C (Fig. 3C), which was consistent with the observation that Hog1 activation is reduced at higher temperatures in C. albicans (Smith et al., 2004), and that Hog1 is an Hsp90 client protein (Hawle et al., 2007; Diezmann et al., 2012). This might suggest that Hog1 plays a minor role in oxidative stress adaptation in vivo. In contrast, skn7 cells were more sensitive to oxidative stress at 37°C, suggesting that Skn7 could play a greater role during host colonisation and invasion than might be predicted based on in vitro analyses performed at 30°C (Singh et al., 2004).
Temperature‐conditional stress mutants displayed significant enrichment in specific functional categories (Fig. 3D). As might be expected, ‘Temperature Stimulus’ genes were enriched in those mutants that displayed elevated stress sensitivity at 37°C, irrespective of the type of stress examined. In contrast, ‘Pathogenesis’‐related genes were enriched in the subsets of mutants that were sensitive to oxidative and nitrosative stress at 37°C, whereas ‘Pathogenesis’ genes were enriched in mutants that were cationic stress sensitive at 30°C. This observation highlights the significance of oxidative and nitrosative stresses to C. albicans cells in vivo.
Environmental contingency of classical stress modules
The carbon‐ and temperature‐conditional stress networks (Figs. 2 and 3) suggested that key stress signalling pathways might display differential contributions to stress resistance under certain growth conditions. To better illustrate this we mapped condition‐dependent stress sensitivities against four key pathways: the Cap1, Hog1, cell integrity (Mkc1) and hyphal MAP kinase (Cek1) pathways (Fig. 4). This exercise clearly showed that the Hog1 MAP kinase module (Ssk2, Pbs2, Hog1) is essential for cationic stress adaptation under all of the conditions analyzed. However, the Hog1 module contributes more to oxidative stress resistance during growth on lactate and at 30°C, than during growth on glucose or at 37°C or 42°C. Meanwhile, Cap1 contributes to oxidative stress resistance under all of the growth conditions tested, but contributes to cationic stress resistance in a carbon‐ and temperature‐conditional manner. Few components of the cell integrity pathway displayed significant stress sensitivity under the conditions tested, although mkc1 cells displayed oxidative stress sensitivity under most of these conditions. Surprisingly, cells lacking the morphogenetic regulator Efg1 (Stoldt et al., 1997) displayed cationic stress sensitivity and oxidative stress sensitivity on rich, but not minimal growth media. With respect to the hyphal MAP kinase (Cek1) pathway, Cph1 and Tec1 displayed cationic stress sensitivity under all conditions tested, and interestingly, temperature‐conditional oxidative stress sensitivity. Under some conditions, certain components on each pathway were required for stress resistance whilst others were not (Fig. 4). This might reflect differences in the relative contribution of a signalling module or transcription factor to stress resistance under a particular growth condition. Nevertheless, taken together as a whole, the data indicate that these well‐studied signalling pathways mediate environmentally contingent outputs that are likely to have relevance in vivo during infection, but that have not been subjected to detailed examination in vitro so far.
Figure 4.
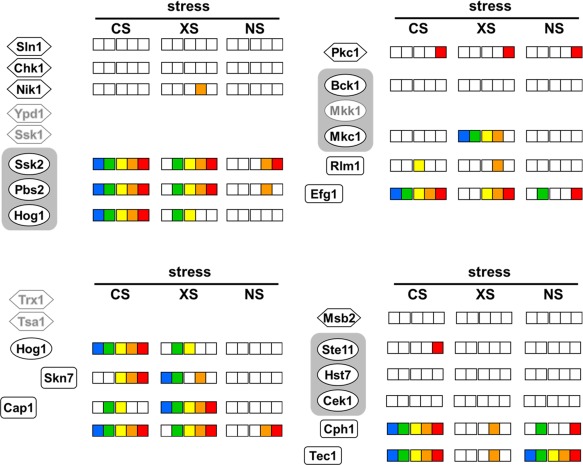
Temperature‐ and carbon‐conditional stress sensitivities for components of major stress pathways in C. albicans. Components of the Hog1 (top left quarter of the figure), Mkc1 (top right), Cap1 (bottom left) and Cek1 pathways (bottom right) are examined. On each pathway, those components for which cationic (CS), oxidative (XS) and nitrosative stress (NS) screening data are available are each named in black on the left, and have panels of boxes on the right: from left to right, sensitivity on glucose (blue), on lactate (green), at 30°C (yellow), 37°C (orange) or at 42°C (red). If no stress sensitivity was observed under a particular growth condition, then the corresponding box in the panel is white. MAP kinase modules are highlighted by rounded grey rectangles. Those components for which stress screening data were not available are named in grey and have no panels of boxes.
Complementary Rtg3 and Sfp1 regulons in C. Albicans
Two mutants, namely rtg3 and sfp1, appeared to display complimentary carbon‐conditional stress sensitivities. The rtg3 mutant appeared more sensitive to cationic, oxidative and nitrosative stresses when grown on lactate, whereas sfp1 cells were more sensitive to these stresses during growth on glucose (Fig. 1). Rtg3 is a bZIP transcription factor that is thought to be involved in galactose metabolism, cationic stress resistance, antifungal drug resistance and filamentous growth (Inglis et al., 2012; Yan et al., 2014; Dalal et al., 2016). Its orthologue in S. cerevisiae is a downstream effector of the TOR (Target of Rapamycin) pathway, which regulates growth in response to nutrients. Sfp1 is predicted to be a C2H2 transcription factor, the expression of which is induced in the rat catheter biofilm model (Nett et al., 2009; Inglis et al., 2012; Chen and Lan, 2015). In S. cerevisiae, Sfp1 regulates ribosomal protein gene transcription as well as responses to nutrients and stress (Inglis et al., 2012). On this basis we reasoned that C. albicans Rtg3 and Sfp1 might act to modulate stress responses in a complimentary carbon‐conditional manner (Fig. 1). Before pursuing this idea further we confirmed the carbon‐conditional stress sensitivities of rtg3 and sfp1 cells by comparing them against control reintegrant strains. The cationic, oxidative and nitrosative stress sensitivities of lactate‐grown rtg3 cells were suppressed by reintroduction of the wild type RTG3 gene. Also, the stress sensitivities of glucose‐grown sfp1 cells were suppressed by transformation with wild type SFP1 (Fig. 5). This confirmed that rtg3 and sfp1 cells display complimentary carbon‐conditional stress sensitivities.
Figure 5.
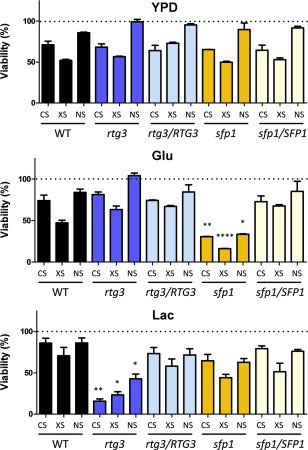
Carbon‐conditional contributions of RTG3 and SFP1 to stress resistance. The susceptibility of rtg3 and sfp1 mutants to cationic (CS), oxidative (XS) and nitrosative stress (NS) during growth on YPD, or on glucose (GLC) or lactate (LAC) as sole carbon source at 30°C. Percentage viability relative to the unstressed controls is presented. Statistically significant differences between the rtg3 (blue) and sfp1 (yellow) mutants and their corresponding reintegrant control strains (pale blue and pale yellow, respectively) are highlighted: *, P < 0.05; **, P < 0.01; ***, P < 0.001***; ****, P < 0.0001. Both the mutant and reintegrant strains were transformed with CIp30 to repair their remaining auxotrophies and to prevent URA3 position effects (see Experimental Procedures).
We then performed a bioinformatic analysis of putative Rtg3 and Sfp1 transcriptional target genes in C. albicans based on the presence of consensus Rtg3 (5′‐GTCACGT‐3′) or Sfp1 binding sites (5′‐AAA(A/T)TTT‐3′) in their promoter regions (Zhu et al., 2009; Perez et al., 2013). Genes encoding other transcription factors, translation and ribosomal proteins, kinases, oxidoreductases and transporters were amongst those identified using this approach (Fig. 6). Interestingly, this analysis suggested that Rtg3 and Sfp1 regulate complementary sets of genes in these functional categories. The small number of genes that may be regulated by both transcription factors include HOG1, PBS2 and CAT1 (catalase) (Fig. 6). Therefore, Rtg3 and Sfp1 are predicted to control complementary regulons involved in C. albicans growth and stress adaptation.
Figure 6.
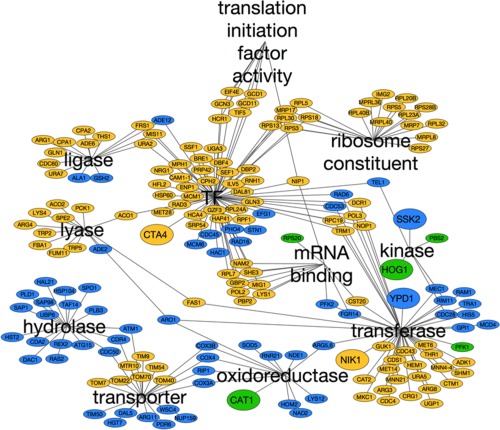
Rtg3 and Sfp1 regulons in C. albicans. Networks of putative gene targets of Rtg3 (blue) and Sfp1 (yellow) based on the presence of their consensus binding sites in the promoters of these genes. Those genes that might be targets for both transcription factors are highlighted in green. The networks are organized into significantly enriched GO categories (TF, transcription factors).
RTG3 and SFP1 display carbon‐conditional stress induction
We reasoned that differential RTG3 and SFP1 expression patterns might contribute to the complementary carbon‐conditional stress sensitivities of rtg3 and sfp1 cells. Therefore, using qRT‐PCR, we tested whether RTG3 and SFP1 transcript levels are induced in response to stress in wild type cells grown on lactate or glucose. Interestingly, the RTG3 mRNA was induced in response to cationic, oxidative and nitrosative stress, but only in cells grown on lactate (Fig. 7A). In contrast, the SFP1 mRNA was up‐regulated in response to cationic, oxidative and nitrosative stress in glucose‐grown cells. SFP1 expression was induced in lactate‐grown cells following exposure to cationic stress, but not oxidative and nitrosative stress (Fig. 7B). Therefore, with only one exception (the cationic stress‐mediated induction of SFP1 in lactate‐grown cells), RTG3 and SFP1 display complementary carbon‐conditional stress induction patterns that match the complementary carbon‐conditional stress sensitivities of rtg3 and sfp1 cells (Fig. 5).
Figure 7.
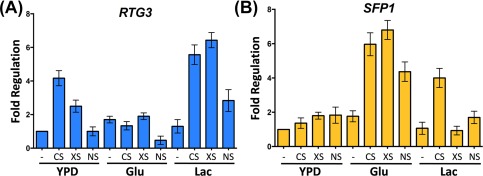
Carbon‐conditional induction of RTG3 or SFP1 genes in response to stresses. Induction of the RTG3 (A) and SFP1 (B) transcripts (relative to the unstressed YPD control) after 10 min of exposure to cationic, oxidative or nitrosative stress.
Rtg3 and Sfp1 control key stress genes in carbon‐conditional manner
We then tested the impact of Rtg3 and Sfp1 on the expression of genes encoding key stress regulators in C. albicans that carry Rtg3 and Sfp1 consensus sequences in their promoter regions: CAT1, CTA4, NIK1, YPD1, SSK2, PBS2 and HOG1. The levels of these transcripts were measured by qRT‐PCR in untreated and stressed wild type, rtg3 and sfp1 cells growing in YPD or in minimal medium containing glucose or lactate as sole carbon source. Both Rtg3 and Sfp1 were found to play major roles in the regulation of these genes in response to stress (Figs. 8 and 9).
Figure 8.
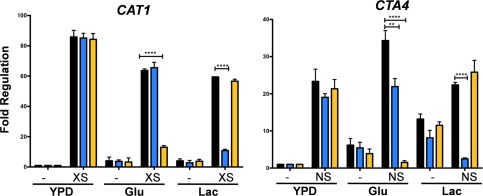
Impact of RTG3 or SFP1 inactivation upon the regulation of key oxidative and nitrosative stress genes in C. albicans. The levels of the CAT1 transcripts were measured by qRT‐PCR, relative to the internal ACT1 mRNA control, after 10 min of exposure to oxidative stress (XS) during growth on YPD, glucose or lactate: wild type, black; rtg3, blue; sfp1, yellow. Fold regulation was then calculated by normalizing CAT1 transcript levels to those on YPD in the absence of stress. Using analogous procedures, CTA4 transcript levels were measured in wild type, rtg3 and sfp1 cells following nitrosative stress (NS). Data represent the means and standard deviations from three independent experiments: *, P < 0.05; **, P < 0.01; ***, P < 0.001***; ****, P < 0.0001.
Figure 9.
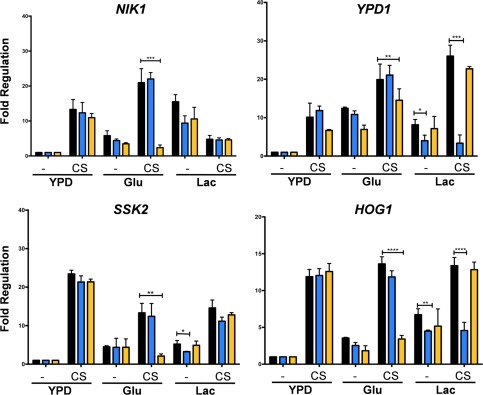
Impact of RTG3 or SFP1 inactivation upon the regulation of key cationic stress genes in C. albicans. The levels of the NIK1, YPD1, SSK2 and HOG1 transcripts were measured by qRT‐PCR, relative to the internal ACT1 mRNA control, after 10 min of exposure to cationic stress (CS), during growth on YPD, glucose or lactate: wild type, black; rtg3, blue; sfp1, yellow. Fold regulation was then calculated by normalizing transcript levels to those on YPD in the absence of stress. Data represent the means and standard deviations from three independent experiments: *, P < 0.05; **, P < 0.01; ***, P < 0.001***; ****, P < 0.0001.
In wild type cells, CAT1 expression was strongly induced in response to oxidative stress under all three growth conditions (Fig. 8). The inactivation of Rtg3 or Sfp1 did not affect the up‐regulation of CAT1 in cells grown on rich media (YPD). However, CAT1 induction was significantly reduced in lactate‐grown rtg3 cells and in glucose‐grown sfp1 cells (Fig. 8). Therefore, the transcription factors Rtg3 and Sfp1 play complementary roles, up‐regulating CAT1 expression in response to oxidative stress in cells growing on different carbon sources.
Similar observations were made for CTA4 in the context of nitrosative stress (Fig. 8). In the absence of stress, basal CTA4 transcript levels were higher in cells grown on minimal medium than in rich medium. Also, modest CTA4 up‐regulation (about two‐fold) was observed following nitrosative stress treatment in lactate‐grown cells. Nevertheless, CTA4 was upregulated by nitrosative stress under all growth conditions analysed. Interestingly, CTA4 induction was significantly attenuated in lactate‐grown rtg3 cells and in glucose‐grown sfp1 cells (Fig. 8), reflecting the responses of CAT1 to oxidative stress (Fig. 8). Therefore, Rtg3 and Sfp1 also play complementary roles in regulating CTA4 expression in response to nitrosative stress.
We then analysed the regulation of NIK1, YPD1, SSK2 and HOG1 in response to cationic stress. The basal levels of these transcripts varied between cells grown on rich or minimal medium, but almost without exception, these mRNAs were induced in response to cationic stress in wild type cells grown in YPD, glucose or lactate (Fig. 9). The only exception was NIK1, which was not induced in NaCl‐treated lactate‐grown cells (Fig. 9). Significantly, the NIK1, SSK1 and HOG1 transcripts were not induced in NaCl‐treated sfp1 cells growing on glucose. Also, the YPD1 and HOG1 mRNAs were not up‐regulated in NaCl‐treated rtg3 cells grown on lactate (Fig. 9). Therefore, Rtg3 and Sfp1 play complementary carbon‐conditional roles in regulating the expression of key regulators of the cationic stress response in C. albicans.
Discussion
The success of C. albicans as a pathogen is dependent on its ability to adapt to multifarious environmental challenges and cues in host niches. Our data support the view that the responses of C. albicans to some key environmental cues – stresses and nutrients – are tightly coordinated (Rodaki et al., 2009; Ene et al., 2012a,b). In addition to those regulators that have been shown to contribute to stress adaptation under standardized growth conditions in vitro (Fig. 4), we have demonstrated that many additional factors contribute to stress adaptation under alternative growth conditions (Figs. 1, 2, 3). This is particularly important because the standardized growth conditions that have generally been used to dissect stress responses in this pathogenic yeast (YPD at 30°C) do not accurately reflect host niches, where glucose is often limiting and the ambient temperature often approximates to 37°C. Our robotic screens of approximately 16% of C. albicans genes revealed novel regulators that promote C. albicans stress resistance in a carbon source‐ and temperature‐conditional manner (Figs. 1, 2, 3, 4). Many of these novel regulators are uncharacterised transcription factors (Fig. 2). This suggests that much remains to be discovered about the mechanisms that underlie environmentally contingent stress adaptation in this pathogen. This is significant because these mechanisms are likely to promote the physiological robustness of C. albicans in host niches, and hence could conceivably present novel targets for therapeutic intervention.
We then focussed on two regulators, Rtg3 (C1_10990c) and Sfp1 (C3_04860w), the inactivation of which caused complementary carbon‐conditional stress sensitivities in C. albicans. Rtg3 cells were sensitive to cationic, oxidative and nitrosative stresses during growth on lactate, whereas sfp1 cells were stress sensitive during growth on glucose (Figs. 1 and 5). The expression of RTG3 and SFP1 was induced in lactate‐ and glucose‐grown cells, respectively (Fig. 7), which was consistent with the carbon‐conditional stress sensitivities of rtg3 and sfp1 cells. Many C. albicans transcription factors bind similar DNA sequences to their S. cerevisiae orthologues (e.g. Tripathi et al. 2002; Nicholls et al., 2009, 2004; Ihmels et al., 2005; Tsong et al., 2006). This has been confirmed for Rtg3 (Perez et al., 2013), but not for Sfp1. Nevertheless, on this basis, Rtg3 or Sfp1 appear to regulate complementary sets of genes with related functions, such as transcription factors, kinases, transporters, transferases and ligases (Fig. 6). This does not simply reflect the nature of the mutants in the libraries that were screened because these represent many other functional categories (e.g. Figs. 2 and 3). We then showed that Rtg3 and Sfp1 regulate the expression of genes encoding key regulators of the cationic, oxidative and nitrosative stress responses, and that they do so in a carbon conditional manner that again reflects the carbon‐conditional sensitivities of rtg3 and sfp1 cells to these stresses (Figs. 8 and 9). Therefore, Rtg3 and Sfp1 appear to maintain key stress pathways and promote stress resistance under different growth conditions – Rtg3 during growth on lactate, and Sfp1 during growth on glucose (Fig. 10).
Figure 10.
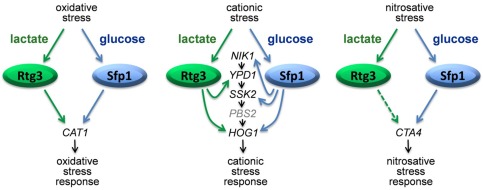
Model illustrating the carbon‐dependent modulation of stress adaptation in C. albicans by Rtg3 and Sfp1. Continuous arrows suggest direct effects of Rtg3 and Sfp1 upon gene expression, while dashed arrows suggest indirect effects (because a perfect match to the consensus binding site was not observed in the promoter of the target gene).
How does Rtg3 contribute to stress resistance during growth on lactate? In addition to promoting the induction of key stress regulators in lactate‐growing C. albicans cells (Figs. 8 and 9), Rtg3 appears to regulate the expression of a range of transporters, hydrolases and transcription factors, some of which may contribute to stress adaptation (Fig. 6). These include Hal21 (a phosphatase that is predicted to be involved in the hyperosmotic response) and Hac1 (a transcription factor that regulates the endoplasmic reticulum (ER) stress response in C. albicans (Wimalasena et al., 2008). They also include numerous mitochondrial functions (COX3A, COX3B, COX4, TIM50). In S. cerevisiae, Rtg3 is regulated by TOR signalling (Crespo et al., 2002) and contributes to mitochondrion‐to‐nucleus signalling via the retrograde response pathway (Rothermel et al., 1997; Jia et al., 1997; Jazwinski, 2014). While this type of intra‐organellar communication has not been studied extensively in C. albicans, mitochondrial functionality is known to influence stress resistance in this pathogenic yeast. For example, the inactivation of Goa1 (which is required for respiratory function and localizes to the mitochondrion under stress conditions) or Sam37 (a component of the mitochondrial outer membrane Sorting and Assembly Machinery complex) renders C. albicans cells sensitive to stresses, affects cell wall integrity and attenuates their virulence (Jia et al., 1997 ; Rothermel et al., 1997; Bambach et al., 2009; Leach et al., 2012c; Qu et al., 2012; Yan et al., 2014). Also the attenuation of mitochondrial functionality by Rtg3 inactivation might reduce the ability of lactate‐growing cells to generate the metabolic energy required for stress adaptation.
How does Sfp1 enhance stress resistance during growth on glucose? In S. cerevisiae, Sfp1 is thought to be a downstream effector of the TORC kinase (Crespo et al., 2002; Jorgensen et al., 2004; Lempiäinen et al., 2009). Following activation via TORC signaling, Sfp1 activates a large number of S. cerevisiae genes (10%), mainly driving ribosomal protein synthesis and ribosome biogenesis under favorable nutrient conditions (Xu and Norris, 1998; Crespo et al., 2002; Jorgensen et al., 2004; Marion et al., 2004; Cipollina et al., 2008). Interestingly, in C. albicans Sfp1 appears to regulate a large number of ribosomal protein genes (RPL5/8/20B/23A/28B/30/32/40B, RPS3/5/13/18/27) (Fig. 6). In S. cerevisiae, the activity of the TORC kinase is low on lactate, compared to glucose. Also, TORC signaling has been shown to inhibit the retrograde response especially in the presence of glutamine (Dilova et al., 2002). Thus, the differential regulation of Sfp1 and Rtg3 by TORC signaling in response to carbon source might underlie the differential contributions of these transcription factors to stress resistance in C. albicans.
To summarize, this study has revealed that numerous regulators, in addition to the classical stress regulators, contribute to stress resistance of C. albicans cells under growth conditions that better reflect some of the key environmental inputs encountered in host niches. These genes were not previously identified using the standardized in vitro growth conditions that have generally been used in the past. Our analyses of two transcription factors, Rtg3 and Sfp1, have revealed mechanisms by which C. albicans retains stress resistance under different growth conditions and have highlighted the complexity of crosstalk between nutrient and stress signaling in this pathogen.
Experimental procedures
Strains and growth conditions
The mutant libraries used in the screens (Supporting Information Table S1), representing a total of 1158 strains, were generously provided by Suzanne Noble, Aaron Mitchell and Dominique Sanglard. C. albicans rtg3 (ura3Δ/ura3Δ, his1Δ/his1Δ, arg4Δ/arg4Δ, rtg3::ARG4/rtg3::URA3) and sfp1 mutants (ura3Δ/ura3Δ, his1Δ/his1Δ, arg4Δ/arg4Δ, orf19.5953::ARG4/orf19.5953::URA3) (Vandeputte et al., 2011) were subjected to more detailed analysis. The genotypes of these strains were confirmed by diagnostic PCR using the primers described in Supporting Information Table S2. To confirm the phenotypes of the rtg3 and sfp1 mutants, the corresponding wild type gene was cloned into the vector CIp30 and integrated into the RPS1 locus (Murad et al. 2000; Dennison et al., 2005). These reintegrant strains were compared with mutant strains transformed with the empty CIp30 vector to ensure that: (i) the histidine and arginine auxotrophies was repaired in both mutant and control and (ii) both mutant and control strains carried URA3 at the RPS1 locus to avoid URA3‐related position effects (Brand et al., 2004).
C. albicans were grown on YPD (2% glucose, 2% Mycopeptone, 1% yeast extract, 2% BactoAgar), GYNB (2% glucose, 0.67% yeast nitrogen base without amino acids, 2% BactoAgar), or LacYNB (2% sodium lactate, pH 7, 0.67% yeast nitrogen base without amino acids, 2% BactoAgar) containing uridine, histidine, arginine and leucine (400 μ/ml) at the specified temperatures (Sherman, 2002; Tillmann et al., 2011; You et al., 2012). To impose stress, 0.4 mM H2O2, 5 mM NaNO2 or 1 M NaCl were used (Chiranand et al., 2008 ; Kaloriti et al., 2012). These concentrations were selected because in control experiments they were found to attenuate the growth of hallmark cap1, cta4 and hog1 strains, respectively, but not the wild type control strain.
To test cell viability, strains were grown on YPD at 30°C. Exponential phase cells were harvested, washed with YPD, GYNB or LacYNB, and then exposed to osmotic (1 M NaCl), oxidative (0.4 mM H2O2) or nitrosative stress (5 mM NaNO2 plus 25 mM succinic acid) or to no stress (control cells) for one hour. Cells were then washed using the same medium and left to recover for 120 min in the absence of stress. Percentage viability is presented, relative to the unstressed control. Mutants are compared with the corresponding re‐integrant strain using a 1‐way ANOVA ‐Dunnett's Multiple Comparison Test.
Genetic screen
The 1158 C. albicans strains subjected to screening (Supporting Information Table S1) were organized into twelve 96‐well plates. Using a Singer RoToR robot (Singer Instruments, Watchet, UK), each strain was pinned (four spots per strain) onto plates prepared using RoToR Singer plates using the above media. Replicate plates were incubated for 24 h at 30°C, 37°C and 42°C and photographed using a GeneFlash (Syngene UK, Cambridge, UK) gel imager. Image analysis was conducted using Proteus pilot software, and growth assessed electronically via pixel quantification (0 = 0–20%; 1= 21–71%; 2 = 72–100% of normal growth under the same conditions except in the absence of stress). Each screen was performed in duplicate. A strain was defined as sensitive to a stress if it reproducibly displayed a > 80% decrease in growth with the stress, relative to no‐stress control plate with matching carbon source and temperature.
The screening output was validated by retesting the stress sensitivities of a selection of mutants using drop tests. To achieve this, strains were grown overnight in YPD at 30°C, subcultured into fresh YPD, and then grown at 30°C up to an OD600 of 1. Cells were then serially diluted, plated on to GYNB or LacYNB media containing the relevant stress, and then grown at the appropriate temperature.
Data analysis and network visualization
The screening data generated by Proteus in Excel format were used to generate Venn Diagrams using Venny open source software (http://omictools.com/venny-s6319.html), and the output from these analyses was used to create filtered excel files that were subsequently analyzed through Cytoscape V3 (www.cytoscape.org/cy3.html) to construct biological networks. The analyses of GO terms was performed through the Term Finder tool of the Candida Genome Database (www.candidagenome.org). Significant differences of the input cluster in comparison to the background gene database were considered if the e‐value was < 0.001. Promoter analyses were performed via PatMatch in the Candida Genome Database. Positive hits contained at least one identical match to the consensus sequence of interest within the 5′‐region of annotated C. albicans genes. Graphs were constructed and statistical analyses performed using GraphPad Prism 6 software.
Gene expression
Total RNA was isolated from exponentially growing C. albicans cells using the YeaStarTM RNA Kit (Zymo Research, Irvine, U.S.A) according to the manufacturer's instructions. The RNA was treated with DNase I (Invitrogen, Paisley, UK) in the presence of RNase inhibitor (Rnase OUT: Invitrogen), and then the levels of specific transcripts subjected to qRT‐PCR using published procedures (Leach et al., 2012a). The primers are described in Supporting Information Table S2.
Supporting information
Supporting Figure S1
Supporting Table S1
Supporting Table S2
Supporting Table S3
Acknowledgements
We thank Aaron Mitchell, Dominique Sanglard and Suzanne Noble for their generosity in providing mutant collections, and Linghuo Jiang for generously providing strains. We also thank Susan Budge for her support and excellent technical assistance. We also thank the qPCR Facility in the Institute of Medical Sciences, and particularly Fiona Saunders for her great advice and help. SLK was supported by a PhD scholarship from the University of Aberdeen. AJPB was supported by the UK Biotechnology and Biological Research Council (BB/F00513X/1; BB/K017365/1), by the European Research Council (STRIFE Advanced Grant; ERC‐2009‐AdG‐249793), and by the UK Medical Research Council (MR/M026663/1). AJPB and CAM were also supported by the Wellcome Trust (088858; 097377), and by the MRC Centre for Medical Mycology and the University of Aberdeen (MR/N006364/1).
References
- Alarco, A.M. , and Raymond, M. (1999) The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans . J Bacteriol 181: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Monge, R. , Navarro‐García, F. , Molero, G. , Diez‐Orejas, R. , Gustin, M. , Pla, J. , Sánchez, M. , et al (1999) Role of the mitogen‐activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans . J Bacteriol 181: 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Monge, R. , Navarro‐García, F. , Román, E. , Negredo, A.I. , Eisman, B. , Nombela, C. , and Pla, J. (2003) The Hog1 mitogen‐activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans . Eukaryot Cell 2: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Monge, R. , Román, E. , Nombela, C. , and Pla, J. (2006) The MAP kinase signal transduction network in Candida albicans . Microbiology 152: 905–912. [DOI] [PubMed] [Google Scholar]
- Andes, D. , Lepak, A. , Pitula, A. , Marchillo, K. , and Clark, J. (2005) A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J Infect Dis 192: 893–900. [DOI] [PubMed] [Google Scholar]
- Avican, K. , Fahlgren, A. , Huss, M. , Heroven, A.K. , Beckstette, M. , Dersch, P. , and Fällman, M. (2015) Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA‐seq analysis. PLoS Pathog 11: e1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambach, A. , Fernandes, M.P. , Ghosh, A. , Kruppa, M. , Alex, D. , Li, D. , et al (2009) Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell 8: 1706–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle, C.J. , Priest, C.L. , Maccallum, D.M. , Gow, N.A. , Odds, F.C. , and Brown, A.J.P. (2006) Niche‐specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol 8: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. , MacCallum, D.M. , Brown, A.J.P. , Gow, N.A.R. , and Odds, F.C. (2004) Ectopic expression of URA3 can influence the virulence phenotype and proteome of C. albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryotic Cell 3: 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. , Vacharaksa, A. , Bendel, C. , Norton, J. , Haynes, P. , Henry‐Stanley, M. , et al (2008) An internal polarity landmark is important for externally induced hyphal behaviors in Candida albicans . Eukaryotic Cell 7: 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouza, E. , and Muñoz, P. (2008) Epidemiology of candidemia in intensive care units. Int J Antimicrob Agents 32: S87–S91. [DOI] [PubMed] [Google Scholar]
- Brock, M. (2009) Fungal metabolism in host niches. Curr Opin Microbiol 12: 371–376. [DOI] [PubMed] [Google Scholar]
- Brown, A.J.P. , Brown, G.D. , Netea, M.G. , and Gow, N.A.R. (2014a) Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol 22: 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J.P. , Budge, S. , Kaloriti, D. , Tillmann, A. , Jacobsen, M.D. , Yin, Z. , et al (2014b) Stress adaptation in a pathogenic fungus. J Exp Biol 217: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J.P. , Odds, F.C. , and Gow, N.A.R. (2007) Infection‐related gene expression in Candida albicans . Curr Opin Microbiol 10: 307–313. [DOI] [PubMed] [Google Scholar]
- Calderone, R.A. , and. Clancy, C.J. (2012) Candida and Candidiasis. ASM Press, Washington, DC. [Google Scholar]
- Chen, H.F. , and Lan, C.Y. (2015) Role of SFP1 in the regulation of Candida albicans biofilm formation. PLoS One 10: e0129903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers, D.S. , Raziunaite, I. , Avelar, G.M. , Potrykus, J. , Budge, S. , Stead, D. , et al (2016) The rewiring of metabolic ubiquitination targets in a pathogenic yeast promotes metabolic flexibility, host colonization and virulence. PLoS Pathogens 12: e1005566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiranand, W. , McLeod, I. , Zhou, H. , Lynn, J.J. , Vega, L.A. , Myers, H. , et al (2008) CTA4 transcription factor mediates induction of nitrosative stress response in Candida albicans . Eukaryot Cell 7: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollina, C. , van den Brink, J. , Daran‐Lapujade, P. , Pronk, J.T. , Porro, D. , and de Winde, J.H. (2008) Saccharomyces cerevisiae SFP1: at the crossroads of central metabolism and ribosome biogenesis. Microbiology 154: 1686–1699. [DOI] [PubMed] [Google Scholar]
- Crespo, J.L. , Powers, T. , Fowler, B. , and Hall, M.N. (2002) The TOR‐controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA 99: 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal, C.K. , Zuleta, I.A. , Mitchell, K.F. , Andes, D.R. , El‐Samad, H. , and Johnson, A.D. (2016) Transcriptional rewiring over evolutionary timescales changes quantitative and qualitative properties of gene expression. Elife 5: e18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison, P.M.J. , Ramsdale, M. , Manson, C.L. , and Brown, A.J.P. (2005) Gene Disruption in Candida albicans using a synthetic, codon‐optimised Cre‐loxP system. Fungal Genet Biol 42: 737–748. [DOI] [PubMed] [Google Scholar]
- Diezmann, S. , Michaut, M. , Shapiro, R.S. , Bader, G.D. , and Cowen, L.E. (2012) Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet 8: e1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilova, I. , Chen, C.Y. , and Powers, T. (2002) Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae . Curr Biol 12: 389–395. [DOI] [PubMed] [Google Scholar]
- Dunkel, N. , Liu, T.T. , Barker, K.S. , Homayouni, R. , Morschhäuser, J. , and Rogers, P.D. (2008) A gain‐of‐function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryotic Cell 7: 1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman, B. , Alonso‐Monge, R. , Román, E. , Arana, D. , Nombela, C. , and Pla, J. (2006) The Cek1 and Hog1 mitogen‐activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans . Eukaryot Cell 5: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene, I.V. , Adya, A.K. , Wehmeier, S. , Brand, A.C. , MacCallum, D.M. , Gow, N.A. , and Brown, A.J.P. (2012a) Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14: 1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene, I.V. , Cheng, S.C. , Netea, M.G. , and Brown, A.J.P. (2013) Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun 81: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene, I.V. , Heilmann, C.J. , Sorgo, A.G. , Walker, L.A. , de Koster, C.G. , Munro, C.A. , Klis, F.M. , and Brown, A.J.P. (2012b) Carbon source‐induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans . Proteomics 12: 3164–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene, I.V. , Walker, L.A. , Schiavone, M. , Lee, K.K. , Martin‐Yken, H. , Dague, E. , et al (2015) Cell wall remodelling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6: e00986–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert, B. , MacCallum, D.M. , Odds, F.C. , and Brown, A.J.P. (2007) Niche‐specific activation of the oxidative stress response by the pathogenic fungus Candida albicans . Infect Immun 75: 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert, B. , Nantel, A. , and Whiteway, M. (2003) Stress‐induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell 14: 1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert, B. , Smith, D.A. , Cornell, M.J. , Alam, I. , Nicholls, S. , Brown, A.J.P. , and Quinn, J. (2006) Role of the Hog1 stress‐activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans . Mol Biol Cell 17: 1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin, C. , De Groot, P. , MacCallum, D. , Schaller, M. , Klis, F. , Odds, F.C. , and Hube, B. (2005) Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56: 397–415. [DOI] [PubMed] [Google Scholar]
- Gow, N.A. , van de Veerdonk, F.L. , Brown, A.J.P. , and Netea, M.G. (2012) Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10: 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, J.S. , and Thiele, D.J. (2002) Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress‐inducible Sdp1 dual specificity phosphatase. J Biol Chem 277: 21278–21284. [DOI] [PubMed] [Google Scholar]
- Hawle, P. , Horst, D. , Bebelman, J.P. , Yang, X.X. , Siderius, M. , and van der Vies, S.M. (2007) Cdc37p is required for stress‐induced high‐osmolarity glycerol and protein kinase C mitogen‐activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p). Eukaryot Cell 6: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero‐de‐Dios, C. , Román, E. , Monge, R.A. , and Pla, J. (2010) The role of MAPK signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: implications in virulence. Curr Protein Pept Sci 11: 693–703. [DOI] [PubMed] [Google Scholar]
- Hohmann, S. (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromatka, B.S. , Noble, S.M. , and Johnson, A.D. (2005) Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell 16: 4814–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube, B. (2004) From commensal to pathogen: stage‐ and tissue‐specific gene expression of Candida albicans . Curr Opin Microbiol 7: 336–341. [DOI] [PubMed] [Google Scholar]
- Hwang, C.S. , Rhie, G.E. , Oh, J.H. , Huh, W.K. , Yim, H.S. , and Kang, S.O. (2002) Copper‐ and zinc‐containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148: 3705–3713. [DOI] [PubMed] [Google Scholar]
- Ihmels, J. , Bergmann, S. , Gerami‐Nejad, M. , Yanai, I. , McClellan, M. , Berman, J. , and Barkai, N. (2005) Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309: 938–940. [DOI] [PubMed] [Google Scholar]
- Inglis, D.O. , Arnaud, M.B. , Binkley, J. , Shah, P. , Skrzypek, M.S. , Wymore, F. , et al (2012) The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata . Nucleic Acids Res 40: D667–D674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson, D.J. , Stephen, D.W. , and Terrière, E.C. (1996) Analysis of the adaptive oxidative stress response of Candida albicans . FEMS Microbiol Lett 138: 83–88. [DOI] [PubMed] [Google Scholar]
- Jazwinski, S.M. (2014) The retrograde response: a conserved compensatory reaction to damage from within and from without. Prog Mol Biol Transl Sci 127: 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Rothermel, B. , Thornton, J. , and Butow, R.A. (1997) A basic helix‐loop‐helix‐leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 17: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P. , Rupes, I. , Sharom, J.R. , Schneper, L. , Broach, J.R. , and Tyers, M. (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloriti, D. , Tillmann, A. , Cook, E. , Jacobsen, M. , You, T. , Lenardon, M. , et al (2012) Combinatorial stresses kill pathogenic Candida species . Med Mycol 50: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos, I. , Patterson, M.J. , Znaidi, S. , Kaloriti, D. , da Silva Dantas, A. , Herrero‐de‐Dios, C.M. , et al (2016) Mechanisms underlying the delayed activation of the Cap1 transcription factor in Candida albicans following combinatorial oxidative and cationic stress are important for phagocytic potency. mBio 7: e00331–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger, C. , Colgan, A. , Srikumar, S. , Händler, K. , Sivasankaran, S.K. , Hammarlöf, D.L. , et al (2013) An infection‐relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium . Cell Host Microbe 14: 683–695. [DOI] [PubMed] [Google Scholar]
- Leach, M.D. , Budge, Walker, S. , Munro, L.C. , Cowen, L.E. , and Brown, A.J.P. (2012a) Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog 8: e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, M.D. , Klipp, E. , Cowen, L.E. , and Brown, A.J.P. (2012b) Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat Rev Microbiol 10: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, M.D. , Tyc, K.M. , Brown, A.J.P. , and Klipp, E. (2012c) Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS One 7: e32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen, H. , Uotila, A. , Urban, J. , Dohnal, I. , Ammerer, G. , Loewith, R. , and Shore, D. (2009) Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell 33: 704–716. [DOI] [PubMed] [Google Scholar]
- Lesage, G. , and Bussey, H. (2006) Cell wall assembly in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 70: 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, R.M. , Regev, A. , Segal, E. , Barash, Y. , Koller, D. , Friedman, N. , and O'Shea, E.K. (2004) Sfp1 is a stress‐ and nutrient‐sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA 101: 14315–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón, P. , Dunker, C. , Windecker, H. , Bohovych, I.M. , Brown, A.J.P. , Kurzai, O. , and Hube, B. (2012) Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7: e52850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad, A.M.A. , Lee, P.R. , Broadbent, I.D. , Barelle, C.J. , and Brown, A.J.P. (2000) CIp10, an efficient and convenient integrating vector for Candida albicans . Yeast 16: 325–327. [DOI] [PubMed] [Google Scholar]
- Navarro‐García, F. , Sánchez, M. , Pla, J. , and Nombela, C. (1995) Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen‐activated protein kinase homolog related to cell integrity. Mol Cell Biol 15: 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett, J.E. , Lepak, A.J. , Marchillo, K. , and Andes, D.R. (2009) Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, S. , Leach, M.D. , Priest, C.L. , and Brown, A.J.P. (2009) Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm‐blooded animals. Mol Microbiol 74: 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, S. , MacCallum, D.M. , Kaffarnik, F.A. , Selway, L. , Peck, S.C. , and Brown, A.J.P. (2011) Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans . Fungal Genet Biol 48: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, S. , Straffon, M. , Enjalbert, B. , Nantel, A. , Macaskill, S. , Whiteway, M. , and Brown, A.J.P. (2004) Msn2/4‐like transcription factors play no obvious roles in the stress responses of the fungal pathogen, Candida albicans . Eukaryotic Cell 3: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou, E. , Agrafioti, I. , Stumpf, M. , Quinn, J. , Stansfield, I. , and Brown, A.J.P. (2009) Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile, C.J. , and Mitchell, A.P. (2009) Large‐scale gene disruption using the UAU1 cassette. Methods Mol Biol 499: 175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, S.M. , French, S. , Kohn, L.A. , Chen, V. , and Johnson, A.D. (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norice, C.T. , Smith, F.J. , Solis, N. , Filler, S.G. , and Mitchell, A.P. (2007) Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot Cell 6: 2046–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, J.C. , Kumamoto, C.A. , and Johnson, A.D. (2013) Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11: e1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Y. , Jelicic, B. , Pettolino, F. , Perry, A. , Lo, T.L. , Hewitt, V.L. , et al (2012) Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryot Cell 11: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaki, A. , Bohovych, I.M. , Enjalbert, B. , Young, T. , Odds, F.C. , Gow, N.A. , and Brown, A.J.P. (2009) Glucose promotes stress resistance in the fungal pathogen Candida albicans . Mol Biol Cell 20: 4845–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbach, A. , Dignard, D. , Pierce, J.V. , Whiteway, M. , and Kumamoto, C.A. (2010) Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9: 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel, B.A. , Thornton, J.L. , and Butow, R.A. (1997) Rtg3p, a basic helix‐loop‐helix/leucine zipper protein that functions in mitochondrial‐induced changes in gene expression, contains independent activation domains. J Biol Chem 272: 19801–19807. [DOI] [PubMed] [Google Scholar]
- San José, C. , Monge, R.A. , Pérez‐Díaz, R. , Pla, J. , and Nombela, C. (1996) The mitogen‐activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans . J Bacteriol 178: 5850–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandai, D. , Yin, Z. , Selway, L. , Stead, D. , Walker, J. , Leach, M.D. , et al (2012) The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast, Candida albicans . mBio 3: e00495–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaggini, S. , Munro, C.A. , Paschoud, S. , Sanglard, D. , and Gow, N.A. (2004) Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae . Microbiology 150: 921–928. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (2002) Getting started with yeast. Methods Enzymol 350: 3–41. [DOI] [PubMed] [Google Scholar]
- Singh, P. , Chauhan, N. , Ghosh, A. , Dixon, F. , and Calderone, R. (2004) SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect Immun 72: 2390–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.A. , Nicholls, S. , Morgan, B.A. , Brown, A.J.P. , and Quinn, J. (2004) A conserved stress‐activated protein kinase regulates a core stress response in the human pathogen Candida albicans . Mol Biol Cell 15: 4179–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib, P. , Kretschmar, M. , Nichterlein, T. , Köhler, G. , Michel, S. , Hof, H. , Hacker, J. , and Morschhäuser, J. (1999) Host‐induced, stage‐specific virulence gene activation in Candida albicans during infection. Mol Microbiol 32: 533–546. [DOI] [PubMed] [Google Scholar]
- Stoldt, V.R. , Sonneborn, A. , Leuker, C.E. , and Ernst, J.F. (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 16: 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann, A. , Gow, N.A. , and Brown, A.J.P. (2011) Nitric oxide and nitrosative stress tolerance in yeast. Biochem Soc Trans 39: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, G. , Wiltshire, C. , Macaskill, S. , Tournu, H. , Budge, S. , and Brown, A.J.P. (2002) CaGcn4 co‐ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans . EMBO J 21: 5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong, A.E. , Tuch, B.B. , Li, H. , and Johnson, A.D. (2006) Evolution of alternative transcriptional circuits with identical logic. Nature 443: 415–420. [DOI] [PubMed] [Google Scholar]
- Ueno, K. , Matsumoto, Y. , Uno, J. , Sasamoto, K. , Sekimizu, K. , Kinjo, Y. , and Chibana, H. (2011) Intestinal resident yeast Candida glabrata requires Cyb2p‐mediated lactate assimilation to adapt in mouse intestine. PLoS One 6: e24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte, P. , Ischer, F. , Sanglard, D. , and Coste, A.T. (2011) In vivo systematic analysis of Candida albicans Zn2‐Cys6 transcription factors mutants for mice organ colonization. PLoS One 6: e26962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, L.A. , Munro, C.A. , de Bruijn, I. , Lenardon, M.D. , McKinnon, A. , and Gow, N.A. (2008) Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog 4: e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cao, Y.Y. , Jia, X.M. , Cao, Y.B. , Gao, P.H. , Fu, X.P. , et al (2006) Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans . Free Radic Biol Med 40: 1201–1209. [DOI] [PubMed] [Google Scholar]
- Wimalasena, T.T. , Enjalbert, B. , Guillemette, T. , Plumridge, A. , Budge, S. , Yin, Z. , et al (2008) Impact of the unfolded protein response upon genome‐wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans . Fungal Genet Biol 45: 1235–1247. [DOI] [PubMed] [Google Scholar]
- Wysong, D.R. , Christin, L. , Sugar, A.M. , Robbins, P.W. , and Diamond, R.D. (1998) Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun 66: 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , and Norris, D. (1998) The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA‐damage response. Genetics 150: 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Zhao, Y. , and Jiang, L. (2014) The putative transcription factor CaRtg3 is involved in tolerance to cations and antifungal drugs as well as serum‐induced filamentation in Candida albicans . FEMS Yeast Res 14: 614–623. [DOI] [PubMed] [Google Scholar]
- You, T. , Ingram, P. , Jacobsen, M. , Cook, D. , McDonagh, E. , Thorne, A.T. , et al (2012) A systems biology analysis of long and short‐term memories of osmotic stress adaptation in fungi. BMC Res Notes 5: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , De Micheli, M. , Coleman, S.T. , Sanglard, D. , and Moye‐Rowley, W.S. (2000) Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 36: 618–629. [DOI] [PubMed] [Google Scholar]
- Zhu, C. , Byers, K.J. , McCord, R.P. , Shi, Z. , Berger, M.F. , Newburger, D.E. , et al (2009) High‐resolution DNA‐binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znaidi, S. , Barker, K.S. , Weber, S. , Alarco, A.M. , Liu, T.T. , Boucher, G. , et al (2009) Identification of the Candida albicans Cap1p regulon. Eukaryot Cell 8: 806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1
Supporting Table S1
Supporting Table S2
Supporting Table S3


