Abstract
Cysteine proteases are among the most abundant hydrolytic enzymes produced by bacteria, and this diverse family of proteins have significant biological roles in bacterial viability and environmental interactions. Members of the clostripain-like (C11) family of cysteine proteases from distal gut commensal strains have recently been shown to mediate immune responses by inducing neutrophil phagocytosis and activating bacterial pathogenic toxins. Development of substrates, inhibitors, and probes that target C11 proteases from enteric bacteria will help to establish the role of these proteins at the interface of the host and microbiome in health and disease. We employed mass spectrometry-based substrate profiling method to identify an optimal peptide substrate of PmC11, a C11 protease secreted by the commensal bacterium Parabacteroides merdae. Using this substrate sequence information, a panel of fluorogenic substrates were synthesized to calculate kcat and KM and to evaluate the importance of the P2 amino acid for substrate turnover. A potent and irreversible tetrapeptide inhibitor with an C-terminal acyloxymethyl ketone warhead, Ac-VLTK-AOMK, was then synthesized. We determined the crystal structure of PmC11 in complex with this inhibitor and uncovered key active-site interactions that govern PmC11 substrate recognition and specificity. This is the first C11 protease structure in complex with a substrate mimetic and is also the highest resolution crystal structure of a C11 protease to date at 1.12 Å resolution. Importantly, subjecting human epithelial cell lysates to PmC11 hydrolysis in combination with subtiligase-based N-terminal labeling and tandem mass spectrometry proteomics complemented the stringent substrate specificity observed in the in vitro substrate profiling experiment. The combination of chemical biological, biophysical, and biochemical techniques presented here to elucidate and characterize PmC11 substrate selectivity can be expanded to other proteases and the development of chemical tools to study these essential proteins in biologically relevant samples, such as the highly complex distal gut microbiome.
Secreted bacterial proteases are essential to microbial fitness, as they are critical to several fundamental processes related to metabolism, development, and nutrient acquisition.1,2 Members of this broad class of bacterial enzymes, including Streptococcus pyogenes streptopain3,4 and Staphylococcus spp. staphopain,5,6 have historically been linked to human diseases and help propagate bacterial pathogenesis by promoting adherence to epithelial cells, hydrolysis of extracellular host proteins, permeabilization of the epithelial barrier, and escape from the innate and adaptive immune systems.1,2,7–9 Critically, elevated proteolytic activity and increased levels of free amino acids correlate with dysbiosis of the human distal gut microbiome in patients suffering from inflammatory bowel disease7,10,11 and likely promote the degradation of the protective mucus layer or epithelial cell barrier that, in turn, can lead to inflammation. The gut microbiome is densely populated (approximately 1013 bacterial cells) with hundreds of bacterial species.12,13 Chemical tools such as substrates, inhibitors, and probes are imperative to begin elucidating the origin of the proteases (i.e., host and/or microbial), as well as quantitating activity in health and disease. Here, our goal was to determine the canonical substrate sequence specificity of PmC11, a C11 clostripain-like protease from Parabacteroides merdae that is highly conserved across both Gram+ and Gram− bacteria (Figure S1), and characterize the protease with a tailored peptide-based fluorogenic substrate and an irreversible inhibitor.
Since the first isolation of clostripain from supernatants of Clostridium histolyticum almost 80 years ago,14 sequencing technology in combination with structural genomics initiatives have identified over 2,000 C11 clostripain homologues.15–19 Several members of this protease class have been linked to biological and pathogenic processes of the human gut microbiome. For example, the secreted protease fragipain from the commensal microbe Bacteroides fragilis activates the B. fragilis toxin (BFT, fragilysin) and promotes anaerobic sepsis in mice.20 Likewise, a secreted clostripain-like protease (Clp) from the commensal Clostridium perfringens targets a unique degradation signal on the surface of neutrophils that stimulates phagocytosis by macrophages.21 Of the 2,000 currently known C11 homologues, over 60% are derived from members of the Bacteroidetes and Firmicutes phyla that account for the majority of commensal bacterial species in intestinal microbiomes.22,23 The ability to quantitate protease activity in microbiome samples from healthy vs. diseased individuals, and identify possible host targets will shed light on the role of these enzymes in microbiome-related diseases.
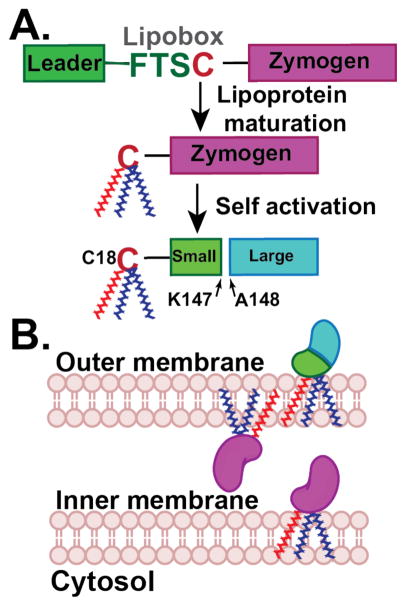
In order to uncover the functional role of an extracellular protease, it is important to know where the protein localizes, how it becomes active and what substrates can be cleaved. Most extracellular bacterial proteases are synthesized and secreted as inactive zymogens as a mechanism to protect the host cell from proteolytic activity.6,24 Analysis of PmC11 protein sequence reveals that it is synthesized as a zymogen and consists of a lipobox motif (-FTSC18-) in the N-terminal signal sequence typical of a membrane-associated protein. This 4-amino acid sequence is found among all lipoproteins and results in the transfer of a phosphatidylglycerol to the lipobox Cys residue25,26 (Cys18 in PmC11), thereby attaching a saturated lipid tail for insertion of the protease into the bacterial membrane as a mature lipoprotein (Figure 1A).27,28 For Gram− species, such as P. merdae, lipoproteins are localized to either the inner or outer membranes, and the presence of an aspartic acid directly C-terminal to the lipidated cysteine (e.g., +2 rule) confers retention to the inner membrane.29,30 PmC11 does not contain this Asp residue and is therefore likely shuttled to the outer membrane and can target hydrolysis of extracellular targets or other secreted P. merdae proteins depending on whether the protease faces outward and/or towards the interstitial peptidoglycan layer, respectively (Figure 1B). Addressing these fundamental questions with respect to PmC11 localization and activation kinetics will be possible with specific probes, inhibitors, and substrates that target PmC11.
Figure 1.
Schematic of PmC11 activation and localization. (A) PmC11 is synthesized as an inactive zymogen. The leader sequence contains a lipobox sequence that results in the posttranslational addition of a lipid tail onto the lipobox Cys18 residue. The protease likely self-activates in cis once it becomes a mature lipoprotein and inserted into the membrane. (B) PmC11 will be inserted into the outer membrane facing either the extracellular or interstitial space. Chemical probes will assist in understanding the sequence of events and final location of PmC11, including self-activation vs. maturation by an orthogonal protease, activation before or after secretion, lipid addition, and orientation in the outer membrane.
In this study, we show that the PmC11 self-activates by cleaving at Lys147 and identify a single synthetic peptide that is rapidly cleaved by this enzyme under in vitro conditions. We exploited the peptide sequence to synthesize a fluorogenic substrate and irreversible inhibitor. Michaelis-Menten kinetics with several PmC11 substrates demonstrate preference for specific substrates, and the x-ray co-crystal structure with a peptide-based inhibitor clearly shows how the protease active site can only accommodate certain substrates. We then show PmC11 proteolysis of human proteins in cell culture with N-terminal labeling and LC-MS/MS proteomics, confirming the in vitro cleavage profile. Our ultimate aim is to understand the specific roles of bacterially secreted proteases within the microbiome. Through this work, we demonstrate a process by which we can discover, synthesize, and characterize chemical tools and methodologies toward this goal.
RESULTS AND DISCUSSION
PmC11 activates in cis
We focused our efforts on the characterization of PmC11 due to its high conservation across the Bacterial Kingdom18, with the intent to establish protease characterization methods that could be expanded to include other C11 proteases of interest. PmC11 self-activates through an internal cleavage event after residue Lys14716, and overexpression of recombinant PmC11 lacking its N-terminal signal sequence (residues 1-22) in E. coli verified that it purifies as a soluble heterodimer (Figure S2). Autoprocessing during maturation is conserved across the clostripain family of proteases; however, the location and number of cleavage events that occur varies by protease. For example, C. histolyticum clostripain removes a 9-amino acid linker peptide during self-activation,15 while B. fragilis fragipain relies on a single cleavage event after Arg147.31 Gel electrophoresis SDS-PAGE of WT PmC11 suggests that the estimated molecular weights of the N-terminal light chain (including the N-terminal affinity tag MGSDKI-H6-ENLYFQG-) and C-terminal heavy chain are approximately 16.2 and 26.4 kDa, respectively, and correlate to cleavage after Lys147 (Figure 1A, Fig S3). Addition of PmC11 to an inactive PmC11 mutant (PmC11C179A) failed to generate the 16.2 and 26.4 kDa protein fragments. Therefore, the zymogen of PmC11 is unable to activate in trans, indicating that Lys147 is inaccessible to cleavage by neighboring protease molecules. In E. coli and in the natural environment of P. merdae, cis activation of PmC11 is predicted to occur. Interestingly, PmC11 can hydrolyze the inactive mutant into two fragments corresponding to 28 and 15 kDa that are clearly different from the heavy and light chains of the mature enzyme. The recombinant WT and mutant proteins differ by only 0.032 kDa (Cys to Ala) and therefore this molecular weight difference is due to cleavage at a region distinct from Lys147 (Figure S2).
Optimizing PmC11 assay conditions
We used the ‘alternative’ processing of PmC11C179A as a macromolecular substrate to assess activity of PmC11 under a variety of assay buffer conditions. Hydrolysis of PmC11C179A by the WT enzyme was initially investigated across a pH spectrum ranging from pH 4.0 to 8.0, and in the presence and absence of a reducing environment. PmC11 is optimally active at pH 8.0 with considerably lower activity evident at pH 7.2 (D-PBS) and below (Figure S2). These conditions correlate with the related B. fragilis fragipain, which also undergoes in trans self-activation during maturation, as evidenced by the presence of the cleavage residue Arg147 in the active site of crystal contact.20 Addition of dithiothreitol (DTT) had no effect on cleavage efficiency (Figure S2). Using pH 8.0 phosphate buffer, we further assessed the effect of Ca2+ ions and detergent on activity. Ca2+ ions have been shown to improve catalysis by clostripain32,33 and trace amounts of detergent is a common additive. PmC11 cleavage of the inactive PmC11C179A mutant was increased in the presence of 0.1% CHAPS and 5 mM CaCl2. A combination of CaCl2 and CHAPS increased activity further (Figure S3). With optimized PmC11 proteolysis conditions, we proceeded to assess proteolytic activity on a library of highly diverse synthetic peptides.
In vitro PmC11 substrate specificity
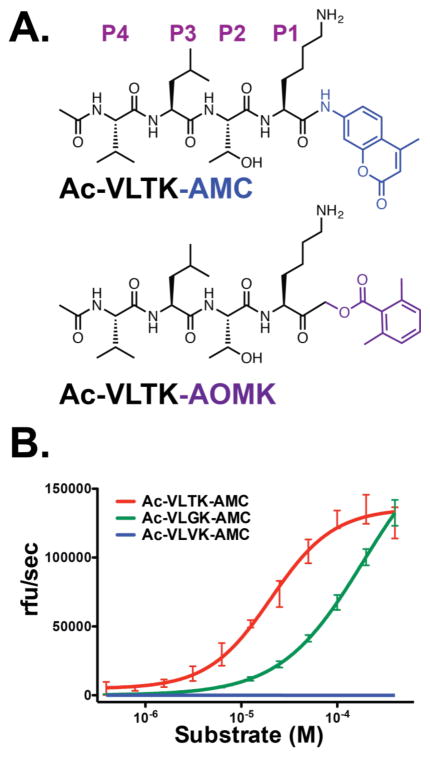
PmC11 auto-activates by cleaving between Lys147-Ala148 and has been shown to hydrolyze Z-Gly-Gly-Arg-AMC, Bz-Arg-AMC, and BOC-Val-Leu-Lys-AMC.16 These data are consistent with C11 proteases that have been shown to hydrolyze on the C-terminal side of Arg and Lys residues.6,19 An in-depth multiplex substrate profiling by mass spectrometry (MSP-MS) study was performed using a library of 228 diverse tetradecapeptides to qualitatively search for specific canonical substrates. These peptides have been used previously to identify key substrate specificity differences between the human and malarial proteasome for the design of an anti-malarial compound.34 Peptide cleavage was evaluated using LC-MS/MS after 15, 60, 240 and 1200 min incubation with a final concentration of 5 nM WT PmC11 and 0.5 μM of each peptide. A control assay containing PmC11C179A was employed to identify attributable hydrolytic activity to any low abundance E. coli proteases that may have co-purified with the recombinant proteins. No detectable activity was observed in the control. After a 15-min incubation, a single 10-mer cleavage product, RnENYnVLTK, was detected, where lowercase ‘n’ corresponds to norleucine. When this cleavage product was mapped to the parent 14-mer peptide, we identified the P4 to P4′ residues as VLTK-AAPV. No other cleavage products were detected until PmC11 had incubated with the peptide library for 4 hr (Figure 2). Although these other cleavage events are considerably slower we were able to determine that PmC11 can cleave peptides with Lys or Arg at P1, Thr, Ser, Val and Ala at P2 and hydrophobic residues at P3. On the carboxyl prime side of the cleavage site, Ala, Val, Gly, His and Ser were found at P1′ (Figure 2). We synthesized a fluorogenic tetrapeptide substrate, Ac-VLTK-AMC, based on the best substrate where AMC corresponds to 7-amino-4-methylcourmarin (Figure 3A). PmC11 cleaves this substrate with high affinity (KM of 20.4 μM ± 2.0). The substrate turnover (kcat) and catalytic efficiency (kcat/KM) were calculated to be 3.3 s−1 ± 0.1 and 1.6 x 105 s−1M−1, respectively (Figure 3B).
Figure 2.
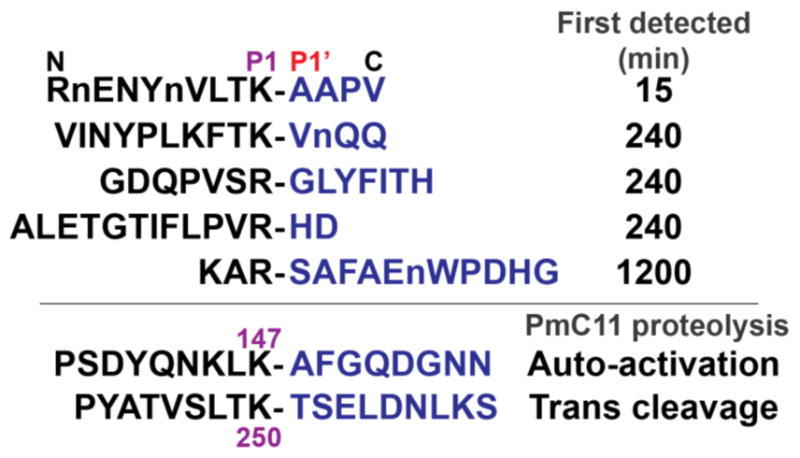
In vitro substrate specificity of PmC11. MSP-MS substrate profiling with 5 nM PmC11 showed stringent specificity of the protease for target peptides. Only 5 peptide sequences (out of 228) were hydrolyzed by PmC11 after 20-hr incubation with the cleavage location denoted by a hyphen and time when first detected by LC-MS/MS (n = norleucine). The PmC11 self-cleavage sites are Lys147 for self-activation and Lys250 for trans cleavage, as observed for WT incubation with PmC11C179A.
Figure 3.
Design and application of PmC11 substrate and irreversible inhibitor. (A) Structure of PmC11 AMC-based substrate and AOMK-based inhibitor synthesized by standard SPPS. AOMK acts as a leaving group that permits nucleophilic attack and irreversible covalent attachment of Ac-VLTK- to the active-site Cys179. (B) Michaelis-Menten kinetics with a range of substrate concentrations against 25 nM PmC11 shows that substitution of the P2 Thr for Val is not tolerated and the kcat and KM for Ac-VLTK-AMC are 3.3 s−1 ± 0.1 and 20.4 μM ± 2.0, respectively.
Our substrate profiling data suggested a preference for Thr, Ser, Val and Ala in the P2 position. To assess the importance of a polar P2 residue for substrate recognition by PmC11, two additional fluorogenic tetrapeptide substrates were synthesized with either Gly or Val at P2 (Ac-VLGK-AMC and Ac-VLVK-AMC). While PmC11 cleaved both Ac-VLTK-AMC and Ac-VLGK-AMC at similar rates (kcat of 3.3 s−1 and ~3.6 s−1, respectively), affinity for the Ac-VLGK-AMC substrate was an order of magnitude lower (KM ~185.8 μM). However, substitution of Thr with a hydrophobic Val side chain ablated substrate turnover, as PmC11 exhibited no activity against the Ac-VLVK-AMC substrate (Figure 3B). Taken together, these results suggest that PmC11 strongly favors a small and polar residue to bind the S2 subsite (Ser or Thr) but will permit sequences with Gly.
Mapping PmC11 specificity to PmC11-C179A processing at Lys250
Discovery of a substrate sequence with high catalytic efficiency allowed us to identify the ‘alternative’ cleavage site in PmC11C179A. Analysis of protein sequence identified a motif near residue Lys250, SLTK250-T that closely matched the preferred cleavage site in the peptide library VLTK-A (Figure 2). Mass spectrometry analysis of the digested heavy and light chain confirmed that the cleavage site was located between Lys250 and Thr251. The SLTK250-T sequence is surface-exposed and lies at the end of the core mixed β-sheet on strand β8, approximately 25 Å away from the active site Cys179 (Figure S4). This site may represent a biologically relevant PmC11 inactivation site to reduce the local level of activity. Directed mutagenesis of these residues and application of specific probes to isolate the protease from P. merdae cultures will help shed light on the role of this additional cleavage site and its effects on PmC11 maturation, activity, and termination.
Design of a selective irreversible inhibitor
We synthesized a specific irreversible peptide inhibitor containing a C-terminal acyloxymethyl ketone warhead (AOMK) that is widely used for cysteine proteases (Figure 3A).35,36 The inhibitor Ac-VLTK-AOMK is susceptible to nucleophilic attack by the active site thiol group, resulting in irreversible alkylation of Cys179 and loss of the inhibitor acyloxy group.35,37 Michaelis-Menten kinetics showed that Ac-VLTK-AOMK inhibited PmC11 with an IC50 <25 nM, which is equivalent to the PmC11 concentration used in the assay (Figure S5A). Interestingly, incubation of PmC11 with a 4-fold excess of the broad-spectrum cysteine protease inhibitor E64 resulted in only a 30% loss of proteolytic activity (Figure S5B). These results are consistent with previous reports of E64 inhibiting clostripain in a reversible competitive manner rather than covalently modifying the catalytic cysteine residue,38,39 and no crystal structures of C11 proteases have been determined in complex with E64. This places PmC11 and similar clostripain-like proteases in a unique group with other select members of the CD cysteine protease clan,59 suggesting they have unique active site interactions that bind E64 in such a way to prevent the epoxide ring of E64 from contacting the active site thiol. We performed x-ray structural analysis on PmC11 to characterize interactions with the VLTK sequence motif and identify the unique active site architecture for substrate recognition.
Co-crystal structure of PmC11 and Ac-VLTK-AOMK
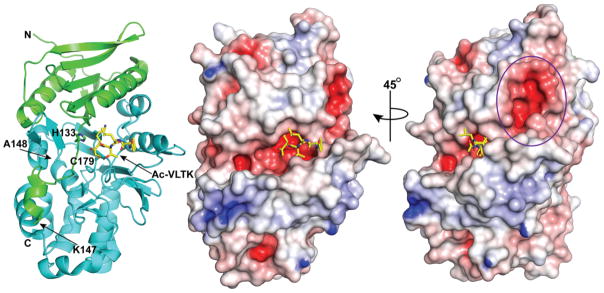
We determined the x-ray crystal structure of PmC11 in complex with the peptide inhibitor Ac-VLTK-AOMK to 1.12 Å resolution (PDB ID: 4YEC) to better understand the stringent substrate specificity of the protease (Table S1). Mature PmC11 (residues 23-375) was incubated with a 2-fold excess of Ac-VLTK-AOMK for 2 hours at room temperature prior to crystallization with conditions similar to those used for the apo structure.16 The PmC11/Ac-VLTK-AOMK complex structure was determined using the apo PmC11 structure (PDB ID: 3UWS) as the search model for molecular replacement (MR). The naïve electron density maps clearly outlined the entire Ac-VLTK- peptide within the PmC11 active site covalently attached to Cys179 (Figure S6). After several iterations of refinement and manual building, the final Rcryst and Rfree values were 15.8% and 19.1%, respectively, with 93% of the residues residing in the most favored region of the Ramachandran plot (Table S1). Despite having clear density for both the main chain and side chain, Asn238 did not adhere to standard phi/psi rules and the odd main chain positioning is likely due to crystal contacts. All of the residues were visible within the electron density and no unexplained regions of positive density were present in the final Fo-Fc maps. Superposition of the Ac-VLTK-AOMK complex and 3UWS apo structure showed that very few conformational changes occur upon binding of the peptide to the PmC11 active site, as evidenced by an average RMSD = 0.4 Å with a maximum displacement of 6.3 Å.
The overall structure of active PmC11 contains 15 β-strands and 14 α-helices, with a core architecture consisting of an extended mixed β-sheet through the center of the protease and has been previously described in detail (Figure 4).16 The surface electrostatic potential suggests that the active site pockets are generally electronegative with some hydrophobic patches for substrate side chain recognition (Figure 4). Approximately 15 Å away from the active site is a highly negatively charged cavity comprised of side chains from residues Glu52, Ser53, Glu56, Glu212, and Glu213. The B. fragilis fragipain sequence and structure is conserved with PmC11 and has a similar electronegative pocket. However, fragipain contains an extended loop region (fragipain residues 240-257) that covers this cavity, where PmC11 has a much shorter loop (residues 237-245) and thus exposes the pocket. The PmC11 cavity is bridged by an electronegative belt to the active site pocket and may represent an exosite recognition site for PmC11 protein substrates (Figure 4).
Figure 4.
Structure of PmC11 in complex with Ac-VLTK-AOMK. Overall structure of the protease bound with the irreversible peptide inhibitor Ac-VLTK shown in stick representation (yellow carbon, blue nitrogen, and red oxygen). The N-terminal small domain and C-terminal large domain are colored green and aqua, respectively. Surface representation and electrostatic potential show that the active site pocket of PmC11 is negatively charged and a large electronegative cavity connected to the active site is approximately 15 Å away and may represent an exosite for biologically relevant substrates (purple circle). Blue, positive potential (≥10 mV); white, neutral potential (0 mV); red, negative potential (≤−10 mV).
Previous structural comparisons to the apo PmC11 3UWS structure identified legumain, gingipain, and caspase-7 with similar structural characteristics.16 Dali structure similarity searches40 with our co-complex PmC11 4YEC model identified additional related structures, including B. fragilis fragipain (PDB 5DN, Z-score = 17.5, rmsd 2.1 Å, 114/118 residues aligned),20 human αXβ2 integrin (PDB 1N3Y: Z-score = 6.2, rmsd 3.2 Å, 93/189 residues),41 and human caspases, such as caspase-3 (PDB 4JJE, Z-score = 3.9, rmsd 5.3 Å, 79/146 residues aligned)42 (Figure S7). Most surprising is the high structural conservation with the human integrin, as the putative binding site of this protein superimposes with the active site of PmC11. αXβ2 integrin is expressed on leukocytes and is key to host innate immune defense, as it acts as a receptor for fibrinogen, the complement factor fragment iC3b, and denatured proteins.41 As secreted proteases are involved in evasion of host immunity and cleavage of host matrix proteins, structural mimicry between PmC11 (and other C11 proteases) with integrins may provide the bacteria with access to integrin ligands.
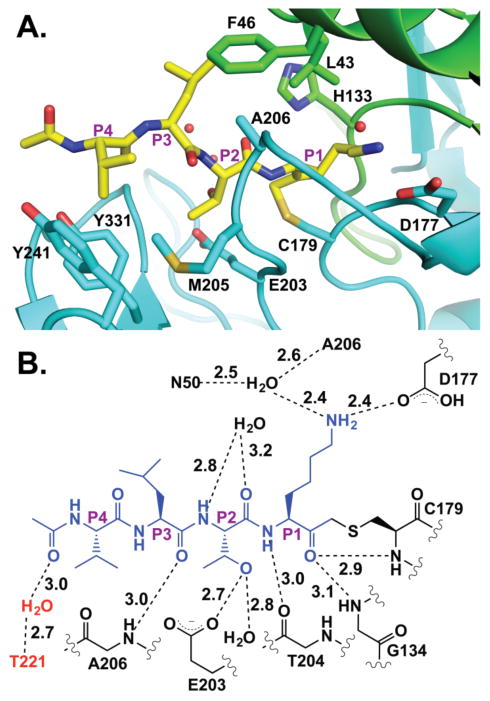
The PmC11 active site is composed of a cysteine-histidine dyad for peptide bond cleavage17,43 and strong electron density was observed for the Ac-VLTK- portion of the inhibitor covalently attached to C179 (Figure S6). The co-crystal structure illustrates the selectivity of PmC11 for the VLTK-based substrates identified from the multiplex substrate screen (Figure 5A). The P1 Lys main-chain carbonyl is positioned toward the standard oxyanion hole configuration of main-chain amides provided by Gly134 and Cys179 with the Lys side chain nestled into a deep pocket that directly hydrogen bonds with the aspartate side chain of Asp177 (Figure 5B). Along with the P1 Lys side chain, an ordered water molecule is positioned within hydrogen bonding distance to the peptide Lys e-amine and residues Asn50 and Ala206. The size and shape of the PmC11 S1 pocket can readily accommodate substrates with a P1 Arg residue (Figure 5A). An additional main-chain potential hydrogen bond occurs between the P1 amide and the main-chain carbonyl of Thr204 (Figure 5B). The P2 Thr side chain interacts directly with a side-chain carboxylate from Glu203 and an ordered water molecule and explains the lack of Ac-VLVK-AMC cleavage (Figure 3B). Replacement of Thr with Val would result in van der Waal’s repulsion against the Glu203 side chain. Of note, the Thr side chain rotamer is perfectly positioned, as the side-chain methyl orients itself in a relatively hydrophobic portion of the protease active site composed of side chains from Met205 and Tyr241. A water molecule is located within hydrogen-bonding distance to both the main-chain carbonyl and amide of the P2 position. The P3 Leu main-chain carbonyl is located within hydrogen bonding distance to the main-chain amide of Ala206 with the side chain pointed into a hydrophobic pocket formed by side chains from Leu43, Phe46, and Ala206 (Figure 5A). The only interactions provided to the peptide P4 Val are contributed by a hydrophobic pocket made up of side chains from Met205, Tyr241, and Tyr331 (Figure 5A). The PmC11 active site provides no interactions with the inhibitor N-terminal acetyl group; however, the acetyl is locked into position via a water-mediated hydrogen bond to residue Thr221 of a crystal contact (Figure 5B). This interaction was likely key to helping position the C-terminal region of the peptide to obtain strong electron density (Figure S6). While His133 does not directly interact with the peptide, it is clearly well-positioned to assist in the pKa perturbation of Cys179 for nucleophilicity. The number of hydrogen bonds provided by the PmC11 active site, primarily focused to the first 2 residues of the peptide inhibitor, combined with the shallow electrophilic S2 binding pocket that would allow only Ser/Thr/Gly side chains suggests PmC11 would likely have a narrow range of canonical substrates and supports the limited spectrum of substrates found within the synthetic peptide library used in our study.
Figure 5.
Ac-VLTK-AOMK inhibitor complex. (A) Side chains involved in binding the peptide Ac-VLTK are shown, including those that provide hydrogen bonds and residues that make hydrophobic pocket (Ac-VLTK and PmC11 are colored as in Figure 4). (B) Schematic of the potential hydrogen bonding network between Ac-VLTK (blue) and PmC11 (black). Interactions provided by a crystal contact are shown in red.
Human cell lysates treated with PmC11 exhibit similar substrate profile
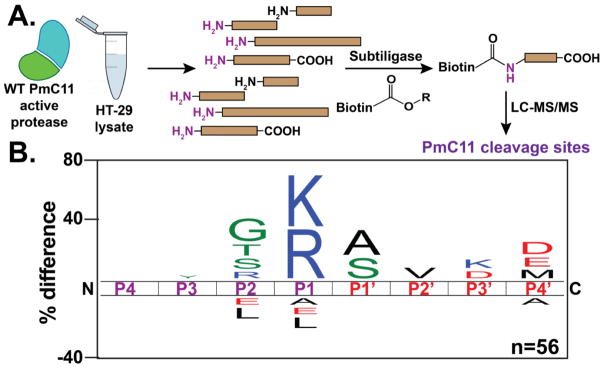
To determine if human proteins susceptible to PmC11 cleavage generated a similar substrate profile as the MSP-MS screen (Figure 2B), we subjected the active protease to the well-established N-terminal labeling technique (Figure 6A).44–47 Briefly, human intestinal epithelial HT-29 cell lysates were treated with 500 nM PmC11, allowing for the generation of new N-termini on host target proteins. Cell extracts were subsequently treated with subtiligase, which specifically ligates peptide esters onto naked N-termini. Therefore, the freshly generated N-termini created by PmC11 will be covalently labeled with a biotin-containing TEV-ester tag via subtiligase treatment (Figure 6A). Once labeled, the biotinylated proteins were enriched and purified from the HT-29 proteome by streptavidin bead affinity. Samples were subjected to tryptic digestion, and subsequent TEV protease digestion of the remaining streptavidin-bound peptide releases an unnatural amino acid (aminobutyric acid, abu) bound to the most N-terminal peptide of the enriched host protein.45 By searching the data collected on high-resolution LC-MS/MS for the presence of an N-terminal Abu modification, we can identify the cleavage site sequences of PmC11 in these host proteins. After subtraction of targets identified in our PBS-treated negative control samples, we identified 56 Abu-modified peptide sequences cleaved by PmC11 across 5 experiments (Table S1). The cleavage sequence logo generated from this data shows a preference for lysine and arginine at P1, and glycine, serine, threonine at P2 (Figure 6B). This sequence preference is strikingly similar to the canonical sequence generated by the MSP-MS screen (Figure 2B), thus providing further validation of the preferred cleavage sequence. Additionally, these experiments demonstrate that there are limited number of host (human) proteins that can be cleaved by PmC11. Future experiments using live immortalized or primary intestinal epithelial cell monolayers, as opposed to cell lysates, will help address questions about biologically relevant host intestinal targets of PmC11.
Figure 6.
PmC11 Cleavage specificity of human proteins. (A) Addition of active PmC11 to HT-29 lysate generates new N-termini that are labeled with a biotin tag via subtiligase treatment.44–46 Streptavidin enrichment and subsequent LC-MS/MS peptide identification provide the proteins and cleavage sequences that are susceptible to PmC11 proteolysis (magenta N-termini generated from PmC11 activity). (B) Sequence logo of the 56 peptides identified to be targeted by PmC11 in HT-29 lysates correlates with the sequence specificity determined by MSP-MS (Figure 2B) and the crystal structure (Figure 5B).
CONCLUSIONS
A combination of in vitro techniques, including MSP-MS, kinetics, and x-ray structure have allowed us to determine the substrate specificity of PmC11, a highly conserved secreted bacterial protease from the clostripain-like C11 family. PmC11 demonstrates precise sequence specificity for substrates, and the x-ray co-complex structure provides evidence that subsite S2 requires a small hydrophilic residue, such as Ser or Thr, and the S1 pocket can accommodate both Lys and Arg. Our N-terminal labeling experiments on human HT-29 cell lysates confirms both the MSP-MS and structure and shows that PmC11 can cleave human proteins. With the canonical substrate sequence in hand, we can now develop probes and additional substrates to elucidate the spatial and temporal information of PmC11 on P. merdae (i.e., location on membrane and activity). Moreover, application of these molecules to highly complex microbial mixtures, such as the human gut microbiome will allow us to find, characterize, and quantitate secreted proteases with similar substrate specificities generated by other bacteria.
METHODS
PmC11 expression and purification
The PmC11 WT clone consisted of residues 23-375 without the N-terminal secretion leader sequence (residues 1-22) and was kindly provided by the Joint Center for Structural Genomics (JCSG). PmC11 was over-expressed as an N-terminal His6-tag fusion with a TEV cleavage site (additional amino acids for the affinity tag include MGSDKI-H6-ENLYFQG-) from E. coli BL21DE3pLysS (Stratagene) in a pBAD vector. Cells were grown in 2xYT media supplemented with 50 μg/ml kanamycin at 37 °C to an OD600nm of 0.6–0.8. Flasks were then transferred to 25 °C and protein expression was induced with 0.2% L-Arabinose for 16 hr. PmC11 self-maturates during expression and results in the fully active protease with small (residues 23-146) and large (residues 147-375) domains. Cells were immediately harvested and resuspended in ice cold 100 mM Tris, pH 8.0, 100 mM NaCl and 10 mM Imidazole (buffer A) and subjected to 3 cycles of lysis by microfluidization (Microfluidics). The cell lysate was clarified by centrifugation at 14,000xg for 8 minutes at 4 °C and soluble fractions were loaded onto a 5 mL HisTrap FF crude Ni-NTA affinity column (GE Amersham) pre-equilibrated with buffer A and eluted with buffer A containing 250 mM Imidazole. The eluted protein was immediately diluted 5-fold with buffer B (20 mM Tris, pH 8.0) and purified by anion-exchange chromatography (HiTrap Q HP, GE Amersham) with a 20-column volume gradient to 50% of buffer B containing 1 M NaCl. Fractions corresponding to PmC11 were pooled and immediately stored at −80 °C. Presence of the His6-tag had no effect on PmC11 activity. Protein used for crystallography was treated with TEV protease (1 mg TEV per 15 mg of PmC11) and dialyzed overnight at 4 °C in 20 mM Tris, pH 8.0, 150 mM NaCl and 0.5 mM TCEP. PmC11 was reloaded over a 5 ml Ni-NTA column pre-equilibrated with 20 mM Tris, pH 8.0, 150 mM NaCl and 0.5 mM TCEP. The FT was collected and pooled and immediately stored at −80 °C.
Cleavage of inactive C179A zymogen SP511E
PmC11C179A was expressed and purified from E. coli as outlined above with the notable exception that autoprocessing into a heavy and light chain did not occur. This protein was incubated at 5 μM with 50 nM active WT PmC11 in a reaction buffer consisting of 10 mM buffer (i.e., citrate, pH 4.0; citrate, 5.0; phosphate, pH 6.0; phosphate, pH 7.0; or phosphate pH 8.0), 50 mM NaCl, and 0.1% CHAPS with and without 10 mM DTT (Figure S2) or 5 mM CaCl2 (Figure S3). The mixtures were agitated at 37 °C and aliquots were quenched after 3 hr with addition of 1% SDS reducing gel loading buffer and boiled. Samples were subjected to SDS-PAGE gel electrophoresis to determine the extent of substrate cleavage in the inactive PmC11C179A.
Multiplex substrate profiling
PmC11, PmC11C179A (control), or buffer (control) were incubated at a final concentration of 5 nM in 10 mM phosphate, pH 8.0, 50 mM NaCl and 2 mM DTT with two pools of 114 synthetic peptides. Each pool contained 0.5 μM of each peptide and digestion assays were performed at 25 °C. Aliquots of 30 μL each were removed after 15, 60, 240, 1200 min incubation and quenched with a final concentration of 4% formic acid. The enzyme reactions containing the peptides were desalted with C18 tips (Rainin) and rehydrated in 0.1% formic acid. Samples were subjected to LC-MS/MS analysis on an LTQ-FT mass spectrometer (Thermo Scientific) equipped with 10,000 psi system nanoACUITY (Waters) UPLC instrument. Reversed phase chromatography was performed with a C18 column (BEH130, 1.7 μm bead size, 100 μm x 100 mm). The LC was operated at 600 nL/min low rate, and peptides were separated using a linear gradient over 42 min from 2% B to 30% B, with solvent A: 0.1% formic acid in water and solvent B: 0.1% formic acid in 70% acetonitrile. Survey scans were recorded over 350–1800 m/z range, and MS/MS was performed with CID fragmentation on the six most intense precursor ions. Data analysis and peptide cleavage assay analysis were performed as described previously.48
PmC11 fluorogenic substrate synthesis
The PmC11 Ac-VLTK-AMC peptide substrate was designed based on the multiplex substrate profiling hit peptide and synthesized using standard Fmoc solid phase synthesis chemistry starting with Fmoc-Lys(carbamate)-AMC Wang resin (EMD Biosciences). After completion of the peptide synthesis and its N-terminal acetylation with acetic anhydride and diisopropylethylamine (DIEA) in dichloromethane (DCM), the substrate was released from the resin with a cocktail of trifluoroacetic acid (TFA), triisopropylsilane (TIPS), and water (95%:2.5%:2.5%). Crude Ac-VLTK-AMC was purified by reverse-phase HPLC using a C18 Xterra column (Waters), and all fractions containing the desired product were lyophilized. The final purity of Ac-VLTK-AMC exceeded 95% and was verified by mass spectrometry: expected m/z 658.3, LC/MS (ESI) m/z 659.3 (MH+). Ac-VLGK-AMC (m/z 614.3, LC/MS (ESI) m/z 615.3 (MH+)) and Ac-VLVK-AMC (m/z 656.4, LC/MS (ESI) m/z 657.4 (MH+)) were synthesized identically with similar yields.
Kinetic assays
Unless otherwise stated, PmC11 was incubated at 25 nM in 50 μL of PBS (pH 7.4), 5 mM DTT, 0.1 mM EDTA, 1 mM CaCl2, 0.1% CHAPS at 25 °C with the activity measurement initiated by introduction of increasing amounts of Ac-VLTK-AMC from 400 pM to 400 μM. Increase in fluorescence due to substrate hydrolysis was measured every 20 seconds for a 25-minute duration in 96-well plates on a PerkinElmer EnVision plate reader (excitation 355 nm, emission 460 nm). All components of the assay are stored as frozen aliquots and thawed immediately prior to the assay. Michaelis-Menten (KM and kcat) values were determined using GraphPad Prism software (GraphPad, Inc.) and the rate of hydrolysis for Ac-VLTK-AMC by 25 nM of the WT PmC11 yielded a kcat = 3.3 s−1 ± 0.1, KM = 20.4 μM ± 2.0, and a kcat/KM = 1.6 x 105 s−1M−1 (Figure 2D).
Synthesis Fmoc-Lys-CMK
Fmoc-Lys(Boc)-OH was (1 g, 2.13 mmol) was added to DCM at 0 °C within a 200 mL flame polished round bottom, N-methylmorpholine (227 mg, 2.24 mmol, 1.05 equiv) and isobutyl chloroformate (306 mg, 2.24 mmol, 1.05 equiv) were added dropwise to yield the mixed anhydride. After 15 min, ethereal diazomethane was generated and distilled into the stirred solution (prepared according to Aldrich Technical Bulletin, AL-180) from Diazald® (2.3 g, 10.7 mmol, 5 equiv) over the course of 45 min. After distillation, the reaction was allowed to proceed at 25 °C for 2 hr. The resulting Fmoc-Lys(Boc)-diazoketone was dried under vacuum and purified by flash chromatography in 1:1 mix of ethylacetate and hexanes. Fractions containing the product were dried under vacuum, dissolved in 4.5 mL 1,4-dioxane and 1.5 mL of conc. HCl was added dropwise to the reaction mixture and simultaneously create the CMK and release the Boc protecting group. After 10 min at 0 °C, diethyl ether and water was added to the reaction. The organic layer was washed with sat. aq. NaHCO3 twice, sat. NaCl, and dried over MgSO4. Concentration in vacuo yielded Fmoc-Lys-CMK (85% yield), which was used without further purification.
Synthesis Ac-VLTK-AOMK inhibitor
The Ac-VLTK-AOMK peptide inhibitor was synthesized using standard Fmoc solid phase synthesis chemistry starting with Fmoc-Lys-CMK added to 2-chlorotrityl resin (EMD Biosciences). After N-terminal acetylation with acetic anhydride and DIEA in DCM, 2,6-dimethylbenzoic acid (5 equiv) and potassium fluoride (10 equiv) were incubated overnight in DMF at 25 °C as previously described to create the C-terminal AOMK leaving group.35,36 Crude Ac-VLTK-AOMK was purified by reverse-phase HPLC using a C18 Xterra column (Waters) and the desired product fractions were lyophilized to produce the final product. The purity of Ac-VLTK-AOMK exceeded 90% and was verified by mass spectrometry: expected m/z 647.8, LC/MS (ESI) m/z 488.2 (MH2+). All intermediates and reactions were monitored by LC/MS.
Ac-VLTK-AOMK IC50 calculations
PmC11 was incubated at 25 nM in the presence of decreasing amounts of Ac-VLTK-AOMK (5 μM to 10 nM) in a reaction buffer consisting of PBS (pH 7.4), 5 mM DTT, 0.1 mM EDTA, 1 mM CaCl2, 0.1% CHAPS and incubated for 30 min at 37 °C. 50 μM Ac-VLTK-AMC was subsequently added and the rate of substrate hydrolysis was measured for 25 min.
Crystallization and x-ray data collection
Inhibitor Ac-VLTK-AOMK was added in a 2-fold molar excess to PmC11 (10 mg/ml), incubated for 2 hr at 25 °C and immediately used for co-crystallization experiments. Crystals were grown by sitting drop-vapor diffusion by mixing equal volumes (2 μl) of the PmC11:VLTK complex and reservoir solution consisting of 0.05 M ammonium chloride and 20% PEG 3500 at 25 °C.16 Data was collected on single, flash-cooled crystals at 100 K in a cryoprotectant consisting of mother liquor and 20% PEG 400 and were processed with HKL200049 in orthorhombic space group P21 (Table 1). The calculated Matthews’ coefficient (VM = 1.82 Å3 Da−1) suggested one monomer per asymmetric unit with a solvent content of 32%. X-ray data was collected to 1.12 Å resolution on beamline 12.2 at the Stanford Synchrotron Radiation Lightsource (SSRL) (Menlo Park, CA). Data collection and processing statistics are summarized in Table S1.
Structure solution and refinement
The PmC11:VLTK structure was determined by molecular replacement (MR) with Phaser50 using the previously published apo structure (PDB ID: 3UWS) determined by the JCSG as the initial search model. The structure was manually built with Coot51 and iteratively refined using Phenix52 with cycles of conventional positional refinement with anisotropic B-factor refinement (excluding water molecules). The electron density maps clearly identified that Ac-VLTK-AOMK was covalently attached to Cys179 within the active site (Figure S6). Water molecules were automatically positioned by Phenix using a 2.5σ cutoff in Fo-Fc maps and manually inspected. The final Rcryst and Rfree are 15.8% and 19.1%, respectively (Table 1). The model was analyzed and validated with PROCHECK,53 WHATCHECK,54 and Molprobity55 on the JCSG webserver. Analysis of backbone dihedral angles with the program PROCHECK indicated that all residues are located in the most favorable and additionally allowed regions in the Ramachandran plot except Asn238, located in a loop region near the active site. Coordinates and structure factors have been deposited in the PDB56 with accession entry 4YEC. Structure refinement statistics are shown in Table S1.
Cell Lines
HT-29 cells were purchased from ATCC (HTB-38), and grown in McCoy’s 5a Medium Modified, supplemented with 10% (v/v) FBS and antibiotics (100 units of penicillin, 0.1 mg/ml streptomycin). Cells were incubated at 37 °C, in the presence of 5% CO2. Cells were harvested or sub-cultured every 3 days at a ratio of 1:5 into 150 mm x 25 mm cell culture dishes. The harvested cells were washed with PBS and the pellets (1 plate per aliquot) were stored in −80 °C until use.
N-Terminal Isolation
HT-29 cell pellet aliquots were thawed and lysed by sonication (Qsonica Q700 sonicator with cup horn attachment, temperature: 4 °C; amplitude: 40; time: 10 min; pulse: 10 sec on, 20 sec off) in 200 mM Bicine pH 8.0, 0.1% Triton X-100, 2 mM AEBSF, 10 mM EDTA, and 10 mM chloracetamide. After sonication, DTT was added at a final concentration of 20 mM to quench the chloracetamide reaction, and centrifuged at 1000xg for 5 min to remove cell debris. The lysates were then treated with 500 nM PmC11 (or an equivalent volume of PBS) for 2 hr at 37 °C with gentle rocking. After this incubation, further protease inhibition was performed for 30 min at 37 °C with gentle rocking using 200 μM E64, 2 mM AEBSF, 10 mM EDTA, 0.1% Triton X-100, and 10 mM chloracetamide. 20 mM DTT was used to quench the chloracetamide reaction before the N-terminal labeling reaction with 1 mM biotin-Abu-TEV-ester45 and 1 μM subtiligase47,57, performed for 2 hr at 25 °C. Proteins were reduced in the presence of 6 M guanidine HCl by 2 mM TCEP at 95 °C for 15 min, and alkylated with 6 mM chloracetamide in dark for 1 hr at 25 °C. The chloracetamide reaction was then quenched with 10 mM DTT. Samples were desalted using PD MidiTrap G-25 columns (GE Healthcare). The biotinylated proteins were captured on immobilized high capacity streptavidin agarose beads (Thermo Scientific Pierce) in a 100 mM Tris, pH 8.0 buffer. The beads were washed and resuspended in 50 mM Tris pH 7.6 containing 1 mM CaCl2, and on-bead digestion was performed overnight using sequence-grade trypsin (Promega V5111) at 37 °C. The beads were washed and N-terminal peptides were isolated by 1.4 μM TEV protease in 50 mM Tris pH 8.0, 0.5 mM EDTA, and 5 mM DTT at 30 °C overnight. A total of 5 independent experiments was performed for analysis by LC-MS/MS.
Preparation of MudPIT LC Column
Tev-digested peptides were loaded onto a biphasic MudPIT column (250 μm fused silica (Agilent), packed with 3 cm of 5 μm Aqua C18 resin followed by 3 cm of Partisphere strong cation exchange resin (SCX)). An analytical column was prepared from 100 μm fused silica pulled to a 5 μm tip by a micropipette puller (Sutter Instrument Company, Model P-2000). This column was then pressure loaded with 10 cm of 3 μm Aqua C18 resin.
LC-MS/MS
Standard MudPIT tandem mass spectrometry was performed using a Thermo Scientific LTQ XL mass spectrometer. The sample and analytical columns were joined by a zero-dead volume union (Waters). Peptides were eluted at 0.2 mL/min using a 6-step MudPIT program. The first step began with 100% buffer A (95% H2O, 5% acetonitrile, 0.1% formic acid) with a 2.5 min gradient of 0 to 15% buffer B (20% H2O, 80% acetonitrile, 0.1% formic acid). This was followed by a 30-min gradient of 15 to 45% buffer B, 10 min gradient of 45 to 75% buffer B, 5-min gradient of 75% to 100% buffer B, 2.5 min of 100% buffer B, and finally 20 min of 100% buffer A. Each subsequent step began with 1 min of 100% buffer A, a 4-min salt pulse with x% buffer C (500 mM ammonium acetate, 95% H2O, 5% acetonitrile, 0.1% formic acid), then 5 min 100% buffer A, followed by a 105-min gradient from 5 to 65% buffer B, and finally 5 min of 100% buffer A. The 4-min buffer C salt pulses (x) were as follows: 20%, 40%, 60%, 80%, 100%. Precursor ions were recorded by scanning in the range of m/z 400.00–1800.00 with the FTMS analyzer. The top 5 peaks were selected for fragmentation using CID with normalized collision energy set to 35.0. Dynamic exclusion was enabled with exclusion duration set to 60.0 s.
Peptide Identification
Precursor and fragmentation ion data were extracted from the Xcalibur RAW files via rawXtract 1.9.9.2 (http://fields.scripps.edu/yates/wp/?page_id=17) to the MS1 and MS2 file formats. The files were searched using Prolucid58 (protein database search algorithm; http://fields.scripps.edu/yates/wp/?page_id=17) within IP2 (commercially available Integrated Proteomic Analysis environment; http://integratedproteomics.com/) using the June 2013 release of the UniProt Human protein database which contained a decoy database of reversed proteins. Precursor/peptide and fragment mass tolerances were 3 ppm and 600 ppm, respectively. The digest protease specified was trypsin, allowing for unlimited missed cleavages. A cysteine carbamidomethylation (+57.02146 Da) was specified as a static modification, methionine oxidation (+15.9949 Da) was specified as a variable modification, and Abu modification of (+86.06059 Da) was specified as a N-terminal differential modification. Filtering was performed using DTASelect 2.1.3 (http://fields.scripps.edu/yates/wp/?page_id=17), requiring one peptide per protein and a false discovery rate (FDR) of 1% with respect to proteins (-pfp). Abu-modified peptides identified in both the control and PmC11-treated samples were eliminated from analysis, and the remaining Abu-modified peptides were used to identify the specific cleavage sequence for PmC11 (Table S2). Weblogo (http://weblogo.berkeley.edu/logo.cgi) was used to create a sequence logo of the Abu-labeled cleavage sites.
Supplementary Material
Acknowledgments
Funding Sources
The authors gratefully acknowledge financial support from The Scripps Research Institute, the National Institutes of Health U54 GM094586-03S1 and R21 CA181027 (to D.W.W.), P41 CA196276 and R21 CA186077 (to C.S.C.), UCSD and the Skaggs School of Pharmacy and Pharmaceutical Sciences (A.J.O), the National Science Foundation (to E.J.R.), NIH Training Grant No. T32AI007244 (to C.M.).
We thank I. Wilson and the Joint Center for Structural Genomics for the PmC11 clone, J. Wells for subtiligase, R. Stanfield, M. Elsliger, and X. Dai for computational assistance, J. Moresco, J. Diedrich, and J. Yates for technical assistance with sample preparation and mass spectrometry instrumentation, H. Rosen for access to instrumentation, G. Stupp for suggestions, and the staff of the Stanford Synchrotron Radiation Lightsource.
ABBREVIATIONS
- Abu
aminobutyric acid
- AMC
7-amino-4-methylcourmarin
- AOMK
acyloxymethyl ketone warhead
- BFT
B. fragilis toxin
- Clp
clostripain-like protease
- DCM
dichloromethane
- DIEA
diisopropylethylamine
- DTT
dithiothreitol
- JCSG
Joint Center for Structural Genomics
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MR
molecular replacement
- MSP-MS
multiplex substrate profiling by mass spectrometry
- SCX
strong cation exchange resin
- RMSD
root-mean-square deviation
- SSRL
Stanford Synchrotron Radiation Lightsource
- TFA
tri-fluoroacetic acid
- TIPS
triisopropylsilane
Footnotes
Author Contributions
D.W.W. and S.C. conceived of the project. S.C. and A.J.O. performed in vitro substrate profiling with assistance from C.S.C.. D.W.W. and E.J.R. synthesized PmC11 substrates and inhibitor and E.J.R. performed all of the in vitro kinetics studies and analysis. G.E.G.P. purified all the PmC11 proteins and optimized PmC11 crystallization. D.W.W. and E.J.R. performed the crystallography and structure analysis. C.M. performed the N-terminal labeling experiments and collected and analyzed tandem LC-MS/MS data. All authors contributed to the preparation and editing of the manuscript and have given approval to the final version of the manuscript.
Notes
No competing financial interests have been declared.
Additional figures, 4YEC x-ray data collection and refinement table, and N-terminal labeling proteomics data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll IM, Maharshak N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J Gastroenterol. 2013;19:7531–7543. doi: 10.3748/wjg.v19.i43.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connolly KL, Roberts AL, Holder RC, Reid SD. Dispersal of Group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS ONE. 2011;6:e18984. doi: 10.1371/journal.pone.0018984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumitomo T, Nakata M, Higashino M, Terao Y, Kawabata S. Group A streptococcal cysteine protease cleaves epithelial junctions and contributes to bacterial translocation. J Biol Chem. 2013;288:13317–13324. doi: 10.1074/jbc.M113.459875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imamura T, Tanase S, Szmyd G, Kozik A, Travis J, Potempa J. Induction of vascular leakage through release of bradykinin and a novel kinin by cysteine proteinases from Staphylococcus aureus. J Exp Med. 2005;201:1669–1676. doi: 10.1084/jem.20042041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Supuran CT, Scozzafava A, Clare BW. Bacterial protease inhibitors. Med Res Rev. 2002;22:329–372. doi: 10.1002/med.10007. [DOI] [PubMed] [Google Scholar]
- 7.Steck N, Mueller K, Schemann M, Haller D. Bacterial proteases in IBD and IBS. Gut. 2012;61:1610–1618. doi: 10.1136/gutjnl-2011-300775. [DOI] [PubMed] [Google Scholar]
- 8.Róka R, Rosztóczy A, Leveque M, Izbéki F, Nagy F, Molnár T, Lonovics J, Garcia-Villar R, Fioramonti J, Wittmann T, Bueno L. A pilot study of fecal serine-protease activity: a pathophysiologic factor in diarrhea-predominant irritable bowel syndrome. Clin Gastroenterol Hepatol. 2007;5:550–555. doi: 10.1016/j.cgh.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Collin M, Olsén A. Extracellular enzymes with immunomodulating activities: variations on a theme in Streptococcus pyogenes. Infect Immun. 2003;71:2983–2992. doi: 10.1128/IAI.71.6.2983-2992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Midtvedt T, Zabarovsky E, Norin E, Bark J, Gizatullin R, Kashuba V, Ljungqvist O, Zabarovska V, Möllby R, Ernberg I. Increase of faecal tryptic activity relates to changes in the intestinal microbiome: analysis of Crohn’s disease with a multidisciplinary platform. PLoS ONE. 2013;8:e66074. doi: 10.1371/journal.pone.0066074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustos D, Negri G, De Paula JA, Di Carlo M, Yapur V, Facente A, De Paula A. Colonic proteinases: increased activity in patients with ulcerative colitis. Medicina. 1998;58:262–264. [PubMed] [Google Scholar]
- 12.Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. Amer J Gastroenterol. 2012;1:15–21. [Google Scholar]
- 13.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Kocholaty W, Weil L, Smith L. Proteinase secretion and growth of Clostridium histolyticum. Biochemical J. 1938;32:1685. doi: 10.1042/bj0321685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manabe S, Nariya H, Miyata S, Tanaka H, Minami J, Suzuki M, Taniguchi Y, Okabe A. Purification and characterization of a clostripain-like protease from a recombinant Clostridium perfringens culture. Microbiology. 2010;156:561–569. doi: 10.1099/mic.0.031609-0. [DOI] [PubMed] [Google Scholar]
- 16.McLuskey K, Grewal JS, Das D, Godzik A, Lesley SA, Deacon AM, Coombs GH, Elsliger M-A, Wilson IA, Mottram JC. Crystal structure and activity studies of the C11 cysteine peptidase from Parabacteroides merdae in the human gut microbiome. J Biol Chem. 2016;291:9482–9491. doi: 10.1074/jbc.M115.706143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labrou NE, Rigden DJ. The structure–function relationship in the clostripain family of peptidases. Eur J Biochem. 2004;271:983–992. doi: 10.1111/j.1432-1033.2004.04000.x. [DOI] [PubMed] [Google Scholar]
- 18.McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem J. 2015;466:219–232. doi: 10.1042/BJ20141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44:D343–50. doi: 10.1093/nar/gkv1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi VM, Herrou J, Hecht AL, Teoh WP, Turner JR, Crosson S, Bubeck Wardenburg J. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat Med. 2016;22:563–567. doi: 10.1038/nm.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzik K, Bzowska M, Smagur J, Krupa O, Sieprawska M, Travis J, Potempa J. A new insight into phagocytosis of apoptotic cells: proteolytic enzymes divert the recognition and clearance of polymorphonuclear leukocytes by macrophages. Cell Death Differ. 2007;14:171–182. doi: 10.1038/sj.cdd.4401927. [DOI] [PubMed] [Google Scholar]
- 22.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Antolin M, Artiguenave F, Blottiere H, Borruel N, Bruls T, Casellas F, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Forte M, Friss C, van de Guchte M, Guedon E, Haimet F, Jamet A, Juste C, Kaci G, Kleerebezem M, Knol J, Kristensen M, Layec S, Le Roux K, Leclerc M, Maguin E, Melo Minardi R, Oozeer R, Rescigno M, Sanchez N, Tims S, Torrejon T, Varela E, de Vos W, Winogradsky Y, Zoetendal E, Bork P, Ehrlich SD, Wang J MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wandersman C. Secretion, processing and activation of bacterial extracellular proteases. Mol Microbiol. 1989;3:1825–1831. doi: 10.1111/j.1365-2958.1989.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama H, Kurokawa K, Lee BL. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 2012;279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 26.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol. 2006;188:2761–2773. doi: 10.1128/JB.188.8.2761-2773.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold ‘em, knowing when to fold ‘em. Trends Microbiol. 2009;17:13–21. doi: 10.1016/j.tim.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zückert WR. Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim Biophys Acta. 2014;1843:1509–1516. doi: 10.1016/j.bbamcr.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juncker AS, Willenbrock H, Heijne von G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrou J, Choi VM, Bubeck Wardenburg J, Crosson S. Activation mechanism of the Bacteroides fragilis cysteine peptidase, fragipain. Biochemistry. 2016;55:4077–4084. doi: 10.1021/acs.biochem.6b00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell WM, Harrington WF. Purification and properties of clostridiopeptidase B (Clostripain) J Biol Chem. 1968;243:4683–4692. [PubMed] [Google Scholar]
- 33.Kembhavi AA, Buttle DJ, Rauber P, Barrett AJ. Clostripain: characterization of the active site. FEBS Lett. 1991;283:277–280. doi: 10.1016/0014-5793(91)80607-5. [DOI] [PubMed] [Google Scholar]
- 34.Li H, O’Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PCA, Bogyo M. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530:233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KAH, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 36.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 38.Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sreedharan SK, Shah HN, Brocklehurst K. Trans-epoxysuccinyl-L-leucylamido (4-guanidino)-butane (E-64) inhibits trypsin-catalysed hydrolysis possibly by binding in the S1-subsite. Biochem Soc Trans. 1994;22:212S. doi: 10.1042/bst022212s. [DOI] [PubMed] [Google Scholar]
- 40.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorup-Jensen T, Ostermeier C, Shimaoka M, Hommel U, Springer TA. Structure and allosteric regulation of the αXβ2 integrin I domain. Proc Nat Acad Sci USA. 2003;100:1873–1878. doi: 10.1073/pnas.0237387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vickers CJ, González-Páez GE, Wolan DW. Selective detection of caspase-3 versus caspase-7 using activity-based probes with key unnatural amino acids. ACS Chem Biol. 2013;8:1558–1566. doi: 10.1021/cb400209w. [DOI] [PubMed] [Google Scholar]
- 43.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–9. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Nat Acad Sci USA. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wildes D, Wells JA. Sampling the N-terminal proteome of human blood. Proc Nat Acad Sci USA. 2010;107:4561–4566. doi: 10.1073/pnas.0914495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braisted AC, Judice JK, Wells JA. Synthesis of proteins by subtiligase. Meth Enzymol. 1997;289:298–313. doi: 10.1016/s0076-6879(97)89053-2. [DOI] [PubMed] [Google Scholar]
- 48.O’Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, Anderson MO, Derisi JL, McKerrow JH, Burlingame AL, Craik CS. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9:1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallog D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 54.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang TK, Jackson DY, Burnier JP, Wells JA. Subtiligase: a tool for semisynthesis of proteins. Proc Nat Acad Sci USA. 1994;91:12544–12548. doi: 10.1073/pnas.91.26.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CCL, Fonslow B, Delahunty C, Gao Y, Shah H, Yates JR. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rangarajan M, Smith SJUS, Curtis MA. Biochemical characterization of the arginine-specific proteases of Porphyromonas gingivalis W50 suggests a common precursor. Biochem J. 1997;323:701–709. doi: 10.1042/bj3230701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.