Abstract
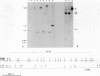
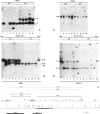
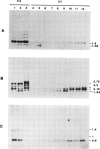
During detailed restriction enzyme mapping of the human X-linked gene Gd, specifying the enzyme glucose 6-phosphate dehydrogenase (G6PD), we have observed the presence, over a 14-kb DNA region spanning across the 3' end of the G6PD transcript, of a large number of methylatable sites. These include 60 HpaII sites, 13 SmaI sites, 22 AvaI sites and 46 HhaI sites. In male leukocyte DNA the majority of HpaII sites are resistant to digestion, indicating that they are in the Cm5CGG form. However, a few sites are found reproducibly unmethylated in 24 samples analyzed. By double and triple digestions we have mapped five unmethylated sites, four of which are within the gene transcript and one distal to the end of transcription. We have also identified a number of sites which are fully methylated, whereas for others the methylation status could not be positively assessed. Thus, in a housekeeping gene expressed in leukocytes, the 3' end is extensively methylated, but some specific sites are unmethylated. In female leukocyte DNA, we found that all sites methylated in males were also methylated. However, of the five sites that are unmethylated in males two are partly methylated in females. This additional site-specific methylation involves approximately 50% of the female leukocyte DNA, and we show evidence that it is associated with the inactive X-chromosome.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Cooper D. N. Eukaryotic DNA methylation. Hum Genet. 1983;64(4):315–333. doi: 10.1007/BF00292363. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Hieter P. A., Max E. E., Seidman J. G., Maizel J. V., Jr, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell. 1980 Nov;22(1 Pt 1):197–207. doi: 10.1016/0092-8674(80)90168-3. [DOI] [PubMed] [Google Scholar]
- Kuo M. T., Mandel J. L., Chambon P. DNA methylation: correlation with DNase I sensitivity of chicken ovalbumin and conalbumin chromatin. Nucleic Acids Res. 1979 Dec 20;7(8):2105–2113. doi: 10.1093/nar/7.8.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Migeon B. R., Wolf S. F., Mareni C., Axelman J. Derepression with decreased expression of the G6PD locus on the inactive X chromosome in normal human cells. Cell. 1982 Jun;29(2):595–600. doi: 10.1016/0092-8674(82)90175-1. [DOI] [PubMed] [Google Scholar]
- Persico M. G., Toniolo D., Nobile C., D'Urso M., Luzzatto L. cDNA sequences of human glucose 6-phosphate dehydrogenase cloned in pBR322. Nature. 1981 Dec 24;294(5843):778–780. doi: 10.1038/294778a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Gartler S. M., Wassman E. R., Yen P., Mohandas T., Shapiro L. J. Transformation with DNA from 5-azacytidine-reactivated X chromosomes. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2352–2354. doi: 10.1073/pnas.79.7.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S. F., Migeon B. R. Studies of X chromosome DNA methylation in normal human cells. Nature. 1982 Feb 25;295(5851):667–671. doi: 10.1038/295667a0. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]