Abstract
The CD4+-lineage specific transcription factor Thpok is required for intrathymic CD4+ T cell differentiation and, together with its homolog LRF, supports CD4+ T cell helper effector responses. However, it is not known if these factors are needed for the T regulatory (Treg) arm of MHC-II responses. Here, by inactivating in mice the genes encoding both factors in differentiated Tregs, we show that Thpok and LRF are redundantly required to maintain the size and functions of the post-thymic Treg pool. They support IL 2-mediated gene expression and the functions of the Treg-specific factor Foxp3. Accordingly, Treg-specific disruption of Thpok and Lrf causes a lethal inflammatory syndrome similar to that resulting from Treg deficiency. Unlike in conventional T cells, Thpok and LRF functions in Tregs are not mediated by their repression of the transcription factor Runx3. Additionally, we found Thpok needed for the differentiation of thymic Treg precursors, an observation in line with the fact that Foxp3+ Tregs are CD4+ cells. Thus, a common Thpok-LRF node supports both helper and regulatory arms of MHC-II responses.
Introduction
MHC II-restricted CD4+ T cells are essential for both immune responses and immune tolerance: in addition to a profound immunodeficiency, patients with CD4+ T cell lymphopenia or defective MHC-II expression experience auto-immune manifestations (1). Although seemingly paradoxical, auto-immunity in this context reflects the critical role of MHC II-restricted CD4+ regulatory T cells (Treg)(2–4) for immune homeostasis. Most of these cells require the transcription factor Foxp3 for their differentiation and function (5–8); indeed, both in humans and mice, Foxp3 mutations cause a severe, early onset auto-immune syndrome affecting the endocrine system, skin and gut (9, 10). Tregs develop in the thymus from CD4+ single positive (SP) thymocytes, and from naïve post-thymic CD4+ T cells upon TGFβ signaling (11, 12). They constitutively express the IL-2 receptor α chain (CD25), which associates with the β and common γ chains to form the high affinity IL-2 receptor required for Treg cell differentiation and function (13). Among other functions, Foxp3 serves to sustain expression of CD25 ensuring Treg cell responsiveness to IL-2.
The zinc finger transcription factor Thpok (14, 15) is expressed in CD4+ but not CD8+ T cells and couples CD4+ T cell differentiation to MHC-II restriction in the thymus (14, 16). It is also important for the differentiation of post-thymic CD4+ T cells into cytokine-producing effectors, including type 1 and type 2 helper cells (Th1 and Th2 cells, defined by the production of IFNγ and IL-4, respectively) (17, 18). In contrast, although Thpok is expressed in Foxp3+ Tregs (19), there is little evidence that it is needed for their function. While it has been reported that Thpok maintains Treg stability in the gut mucosa (20), it is not known how immune homeostasis depends on Treg expression of Thpok. Thpok disruption, whether specifically induced in Tregs (20) or enforced in all T cells, does not cause detectable auto-immune or inflammatory disease. However, these previous studies did not address the functional overlap between Thpok and the related transcription factor LRF, which serves redundantly with Thpok to promote helper T cell differentiation and functions (17, 18).
To study the impact of these factors on Treg functions, we inactivated the genes encoding Thpok and LRF (Zbtb7b and Zbtb7a, called Thpok and Lrf here, respectively) in Tregs. We demonstrate that Thpok and LRF promote Treg survival and homeostasis and are essential for Foxp3-directed gene expression. Accordingly, Treg-specific disruption of Thpok and Lrf causes a lethal inflammatory syndrome similar to that of Scurfy mice. Furthermore, gene expression and genetic analyses indicate that Thpok and LRF serve distinct functions in Treg and conventional T cells. Thus, Thpok and LRF are needed for regulatory MHC II-restricted T cell responses.
Materials and Methods
Mice
Thpokfl and ThpokGFP mice were described previously (19). B2m−/−(21), Cd4-Cre (22), Scurfy (10) animals were from Taconic, Taconic, and JAX, respectively, and CD45.1, CD45.2, and C57BL/6 mice from the NCI Animal Production Facility. Lrff and Foxp3YFP-Cre mice were gifts from Drs. P.P. Pandolfi and A. Rudensky, respectively, and Runx3f and Cd4Silf mice from Dr. D. Littman (23–26). In experiments with Foxp3YFP-Cre mediated deletion, control mice were of the same gender as experimental mice and carried a single Foxp3YFP-Cre allele on a Thpok+/+ Lrf+/+ background unless specified otherwise. Transgenic mice were heterozygous for the transgene they carry. Mice were housed in specific pathogen-free facilities and analyzed between 8–16 weeks of age unless stated otherwise. All animal procedures were approved by NCI and NIAID Animal Care and Use Committees.
Bone marrow chimeras
Bone marrow was isolated from CD45 disparate animals, T cell-depleted with Mouse Pan T (Thy1.2) Dynabeads (ThermoFisher Scientific), mixed at a 1:1 ratio and injected into lethally irradiated (900rad) recipients heterozygous for CD45.1 and CD45.2 or in a second experiment for CD45.1. Mice were analyzed at least eight weeks post transplant.
Antibodies
The following antibodies were from either BD Pharmingen or Affymetrix eBioscience: TCR (H57-597), CD4 (RM4.4 or GK1.5), CD8α (53–6.7), CD24 (M1/69), CD44 (IM7), CD62L (MEL-14), CD25 (PC61.5), CD45.1 (A20); CD45.2 (104), CD8β (53–5.8), MHC Class II (M5/114.15.2), IFNγ (XMG1.2), IL-4 (11B11), IL17A (eBio17B7), Foxp3 (FJK-16S), Gata3 (L50-823), Tbet (4B10), RORγt (AFKJS9), LRF (13E9), Ox40 (OX-86), GITR (DTA-1), pStat5 (47/Stat5), ICOS (7E.17G9) and Thy1.1 (HIS51). Nrp1 (3E12), Granzyme B (GB12), Runx (EPR3099), and β-Actin (AC-15) were from BioLegend, ThermoFisher Scientific, Abcam Epitomics and Sigma-Aldrich respectively. Immunoblotting analyses were performed as described (27), using SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific).
Histology and histological scores
Tissue was removed from euthanized animals, fixed in buffered formalin, embedded in paraffin, sliced and stained with hematoxylin and eosin (H&E). The resulting slides were scored from 0–2 according to methodology previously used for scoring Scurfy mice (28).
Cell preparation and flow cytometric staining
Lymph node, thymus, spleen, siLP, and ear cells were prepared and stained as previously described (15, 19, 27). Flow cytometry data was acquired on LSRII, LSR Fortessa, LSR Fortessa X-20, and Canto II cytometers (BD Biosciences) and analyzed with FlowJo (TreeStar) software. Purification of lymphocytes by cell sorting was performed on a FACSAriaII (BD Biosciences). Dead cells and doublets were excluded by DAPI and a combination of forward light scatter height and width gating for live cell analyses.
Analyses of intracellular cytokine expression were performed as described (19). Transcription factor expression was detected using the Foxp3/Transcription Factor Staining Buffer Set (Affymetrix eBioscience) according to the manufacturer’s instructions, except for RORγt and LRF staining for which the cell fixation-permeabilization step was extended to four-16 hours at 4°C and a 5-minute incubation with 2% paraformaldehyde for combined staining with YFP. Dead cells were excluded using the LIVE/DEAD Fixable Blue Dead Cell Stain Kit (ThermoFisher Scientific).
Stat5 phosphorylation analysis
For in vitro Stat5 phosphorylation analyses, splenocytes were incubated with IL-2 and LiveDead Blue for 30 minutes at 37°C, fixed with 4% PFA in PBS for 10 minutes, permeabilized in 90% methanol for 30 minutes on ice, stained with anti-pStat5 and processed for flow cytometry.
Ex vivo Stat5 phosphorylation was conducted essentially as previously described (29). Briefly, axillary, brachial, and inguinal lymph nodes were removed two at a time, and immediately processed through a 70 µm filter into a 4% PFA PBS solution. After a 10 min incubation, cells were washed once in PBS, resuspended in ice-cold 90% methanol, and incubated overnight at −20 degrees. The following day, cells were stained with anti-pSTAT5 or isotype control for 1 hour at room temperature, washed, and processed for flow cytometry.
In vitro cell analyses
Retroviral transductions were performed as previously described (19), using either MIGR-mFoxP3 (a gift from Dr. Dan Littman [Addgene plasmid # 24067]) or PMRX-Thy1.1-mFoxP3, except activation was performed with only anti-CD3ɛ, and without IL-2 supplementation. Annexin V staining was performed with the PE-Annexin V staining kit (BD Biosciences) on cells incubated for 3 hours at 37°C or 4°C in medium. Suppression assays were performed as previously described (18).
Infections
L. major infections were performed as described (30) with 5 × 104 L. major clone V1 metacyclic promastigotes by intradermal ear injection. Skin lesions were measured at indicated time points, and animals were subsequently euthanized. Cells were harvested from the infected ear and draining lymph node as described (31).
RNAseq
Total lymph node and spleen cells from dKO, animal control and genetic control female mice were stained for MHC Class II and CD25, and double-sorted to prepare YFP+ MHC Class II− (dKO and genetic control) or YFP− MHC Class II− CD25+ (animal control) cells. Mutant samples yielded an average total of 10,000 cells and purity was >97%. RNA was prepared from pelleted cells using the RNeasy Plus Micro kit (Qiagen). From an average total of 2ng RNA per sample, 100pg RNA (RIN values of >9.6) was put through the SMARTer Ultra Low Input RNA Kit for Illumina Sequencing (Clontech Laboratories, Inc.) according to the manufacturer’s instructions. The resulting libraries were then sequenced on Illumina’s HiSeq 2500 Sequencing System. For each genotype, data is derived from four biological replicates (mice) processed separately from cell sorting to sequencing; two of which (per genotype) were merged together into a single sample prior to bioinformatics analyses to achieve consistency in read numbers, whereas the other two were processed each as a single sample. This resulted in three separate samples for each genotype, as shown in Tables S1 and S2. Raw RNAseq fastq reads were aligned to mouse genome (mm10) using STAR (v. 2.4.0h) on 2-pass mode with mouse gencode (release 4) gtf (32). Genes were subsequently counted using Rsubread and analyzed for gene expression changes using limma-voom with quantile normalization (33, 34). The gene and sample-specific normalization factors were then used to correct counts to generate bedgraph files used for further visualization. RNAseq data has been deposited in the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE85532.
Statistical analyses
Statistical analyses were performed using Prism (GraphPad Software, Inc.). Bars in graphs indicate average ± s.e.m. Significance on bar plots was determined using an unpaired t-test with Welch’s correction except where indicated otherwise; when significant, levels (P-values) are indicated on figures. Significance on survival curves was assessed using Log-rank (Mantel-Cox) test and Gehan-Breslow-Wilcoxon test.
Results
Thpok promotes thymic but not induced Treg differentiation
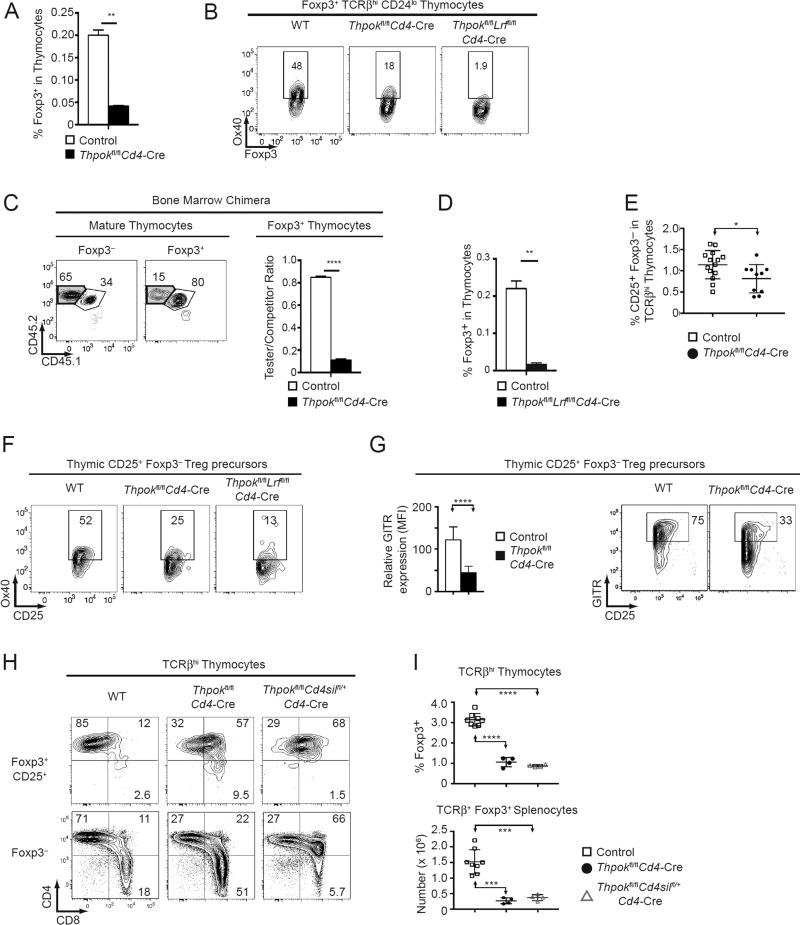
We previously found that germline or T cell-targeted disruption of Thpok, which causes MHC II-restricted thymocytes to develop into CD8+ T cells (14, 16), reduces peripheral Treg numbers (19), and that disruption of both Thpok and the Thpok-related factor LRF resulted in an almost complete absence of Tregs in peripheral lymphoid organs (18). To examine if this resulted from impaired thymic Treg development, we analyzed Thpokfl/fl mice carrying a Cd4-Cre transgene (22), which deletes in CD4+CD8+ thymocytes. Thpok disruption reduced the numbers of Treg thymocytes, and their expression of the Treg marker Ox40 (Fig. 1AB). Thpok-deficient precursors minimally contributed to Treg thymocyte populations in competitive mixed (wild-type: Thpokfl/fl Cd4-Cre, 1:1) bone marrow chimeras (Fig. 1C). Thus, Thpok is needed for the fitness of Treg precursors in the competitive environment that characterizes their intrathymic differentiation (35). While deletion of LRF per se had no effect on Treg thymocyte development (data not shown), deletion of both Thpok and LRF, in Thpokfl/fl Lrffl/fl Cd4-Cre mice, had a greater impact than deletion of Thpok only on Treg thymocyte numbers and their expression of Ox40 (Fig. 1B, D). We conclude from these experiments that Thpok is needed for thymic Treg differentiation, with modest functional overlap with LRF.
Figure 1. Thpok is important for Treg development in the thymus.
(A, D) Percentage of Foxp3+ cells among total thymocytes from mice of the indicated genotype. Thpokfl/fl Lrffl/fl Cd4-Cre mice and corresponding controls were B2m−/− to exclude MHC I-restricted cells from analyses (D). Data is from three mice of each genotype analyzed in three independent experiments (**: P<0.01).
(B) Expression of intra-cellular Foxp3 vs. surface Ox40 in Foxp3+ TCRβhi CD24lo thymocytes from mice of the indicated genotype. Data is representative of four mice of each genotype analyzed in four independent experiments.
(C) Contour plots (left) show CD45.1 vs. CD45.2 expression on Foxp3− and Foxp3+ TCRβhi CD24lo thymocytes from CD45.1+B2m−/− host mice transplanted with a 1:1 mix of CD45.2+ Thpokfl/flCd4-Cre (tester [Test]) and CD45.1+ CD45.2+ wild-type (WT) (competitor[Comp]) bone marrow. Boxes define populations of Test (purple shaded) and Comp (plain line) origin. Numbers near boxes indicate cell percentages and were used to compute Test/Comp ratios for Foxp3+ and Foxp3− subsets. The bar graph shows [(Test/Comp)Foxp3+/ (Test/Comp)Foxp3−] ratios, respectively on gated Foxp3+ and Foxp3− subsets, in chimeras made with Thpokfl/flCd4-Cre Test bone marrow (filled bar) and in control chimeras made from Thpokfl/fl Test bone marrow (open bar). Data is representative of three recipients per genotype, obtained from two separate bone marrow transplants, each with one donor from each genotype (****: P<0.0001).
(E) Percentage of CD25+ Foxp3− cells among TCRβhi thymocytes from mice of the indicated genotype. Each symbol represents one mouse; data is from eight independent experiments (*: P<0.05, unpaired t-test).
(F) Expression of CD25 vs. Ox40 in CD25+ Foxp3− TCRβhi thymocytes from mice of the indicated genotype. Data is representative of two mice of each genotype analyzed two independent experiments.
(G) Bar graph (left) shows relative expression (MFI) of GITR in CD25+ Foxp3− TCRβhi thymocytes from mice of the indicated genotype. Data is expressed relative to wild type cells analyzed in the same experiment, set to 100, and is from 8 to 10 mice of each genotype analyzed in six independent experiments (****: P< 10−4, unpaired t-test). A representative experiment is shown on the right.
(H) Expression of CD8 vs. CD4 by Foxp3+ (top) or Foxp3− (bottom) TCRβhi thymocytes of the indicated genotype. Data is summarized in (I).
(I) Percentage (thymus, top) and absolute number (spleen, bottom) of Foxp3+ cells in indicated populations from control (open squares), Thpokfl/fl Cd4-Cre (filled circles), and Thpokfl/fl Cd4Silfl/+Cd4-Cre (open triangles) mice. Each symbol represents an individual mouse; data is from two independent experiments. Statistical significance was determined by one-way ANOVA followed by a Tukey’s multiple comparisons test (***: P<0.001 ****: P<0.0001).
Most thymic Tregs are thought to differentiate from CD25+ Foxp3− precursors. We found that the percentage of such precursors was reduced in Thpok-deficient thymi (Fig. 1E). Expression of TNF-family receptors Ox40 and GITR is important for intrathymic Treg differentiation (36). We found that expression of both molecules was reduced in Thpok-deficient CD25+ Foxp3− precursors, and that this effect was amplified by the additional deletion of LRF, which on its own had no effect (Fig. 1FG and data not shown). The fact that such cells do not express Foxp3 indicates that the impact of Thpok on Ox40 or GITR expression is not mediated by its effect on Foxp3 expression. We conclude from these experiments that Thpok is involved in the generation of thymic Treg precursors, prior to Foxp3 expression.
In addition to thymus-generated Tregs, conventional naïve CD4+ T cells can be induced to express Foxp3+ and acquire regulatory functions when signaled by TGFβ (11, 12). Thus, we examined if post-thymic Thpok disruption would affect TGFβ-induced Foxp3+ expression and Treg differentiation. We purified CD4+ T cells from Thpokfl/fl mice carrying a previously described CD2-Cre transgene that becomes active in post-thymic CD4+ T cells (17). Thpok disruption did not impair, and in fact slightly enhanced, TGFβ-induced Foxp3 up-regulation (Fig. S1A). Thus, unlike for thymic Treg differentiation, Thpok is not needed for induced Treg differentiation.
Thpok requirement for thymic Treg differentiation is independent of its effect of CD4 expression
Thpok promotes CD4 expression in MHC II-signaled thymocytes, by antagonizing the expression or function of Runx molecules (37, 38). Because Treg differentiation requires TCR signaling and because CD4 promotes MHC II-induced TCR signaling, we considered that termination of CD4 expression in Thpok-deficient thymocytes could be responsible for their impaired Treg differentiation. Consistent with this idea, most of the few Tregs that developed in Cd4-Cre Thpokfl/fl mice expressed CD4, unlike conventional MHC II-restricted thymocytes that become CD8+ T cells (Fig. 1H). To evaluate this possibility, we generated Cd4-Cre Thpokfl/fl Cd4Silfl/+ mice, in which Cre excises floxed alleles of Thpok and of the Cd4 silencer (Cd4Sil), a cis-regulatory element mediating Cd4 silencing in differentiating CD8+-lineage thymocytes (39, 40). Even though silencer inactivation maintained CD4 expression, it failed to rescue Treg cell populations (Fig. 1HI). Thus, the impact of Thpok on Treg differentiation is not mediated solely by its effect on CD4 expression.
Thpok and LRF redundantly promote Treg homeostasis
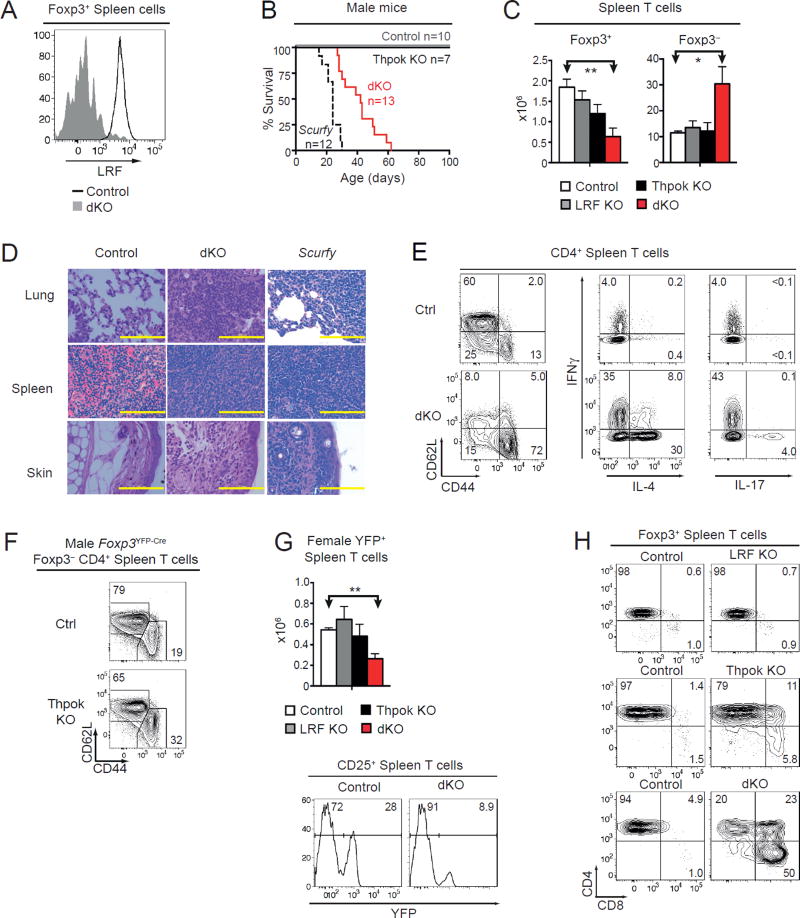
While these results demonstrated the importance of Thpok for Treg development, Cd4-Cre disruption could not address whether Thpok was needed in post-thymic Tregs, because Thpok is essential for conventional CD4+ T cell function (17, 18, 41). To overcome this obstacle, we generated Thpokfl/fl mice carrying a Foxp3YFP-Cre knockin allele (Thpok KO mice), which delete Thpok in Treg but not conventional T cells (24). Additionally, because LRF is expressed in Tregs (Fig. 2A), we generated Foxp3YFP-Cre Lrffl/fl and Thpokfl/fl Lrffl/fl mice (LRF KO and dKO mice, respectively) to evaluate the functional overlap between these two factors. Animals carrying Foxp3YFP-Cre on an otherwise wild-type background were used as controls.
Figure 2. Treg expression of Thpok or LRF is critical for immune homeostasis.
(A) LRF expression in Foxp3+ spleen cells (black line). Foxp3+ cells from dKO mice serve as a negative staining control (grey filled). Data is representative of three mice of each genotype analyzed in three independent experiments.
(B) Survival of Foxp3YFP-Cre control (grey line), Thpok KO (plain black line), dKO (red line), and Scurfy (dashed black line) male mice. Number of mice is indicated in plot. Median survival was 42 days for dKO and 24 days for Scurfy mice.
(C) Absolute number of spleen TCRβ+ Foxp3+ (left), Foxp3− (right) cells from control (open bars), LRF KO (grey bars), Thpok KO (black bars), or dKO (red bars) male Foxp3YFP-Cre mice. Data is from at least three animals per genotype, analyzed in at least three independent experiments (control vs. dKO: P<0.05 [*] and P<0.01 [**]; other differences not significant).
(D) Hematoxylin and eosin stains on paraffin-embedded organs comparing Foxp3YFP-Cre control, dKO, and Scurfy mice, all male. Yellow scale bar is 100µm. Data is representative of five separate samples from mice of each genotype stained in two experiments. At time of analysis, mice were 4–6 weeks (dKO) or 2–4 weeks (Scurfy) old. Control mice were 6–8 weeks old.
(E) (Left) CD44 vs. CD62L expression in Foxp3− CD4+ TCRβ+ spleen cells from control or dKO male mice. (Middle and right) Expression IL-4 (middle) or IL-17 (right) vs. IFNγ in CD4+ TCRβ+ spleen cells from control and dKO male mice after a 3-hour in vitro stimulation and intracellular cytokine staining. Data is representative of three mice of each genotype analyzed in three independent experiments.
(F) Expression of CD44 vs. CD62L on Foxp3− CD4+ TCRβ+ spleen cells from control (top) or Thpok KO (bottom) Foxp3YFP-Cre male mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
(G) (Top) Absolute number of spleen YFP+ TCRβ+ cells from indicated Foxp3YFP-Cre mice female mice. Data is from at least three animals per genotype, analyzed in at least three independent experiments (control vs. dKO: P<0.01 [**]; other differences not significant). (Bottom) Expression of YFP in CD25+ spleen T cells from control and dKO female mice.
(H) CD8 vs. CD4 expression in Foxp3+ TCRβ+ cells from control, LRF KO (top), Thpok KO (middle), or dKO (bottom) male mice. Data is representative of at least three mouse sets analyzed in at least three independent experiments.
Because Foxp3 is located on chromosome X, all male Tregs express Cre and delete floxed alleles, so that immune homeostasis depends on their ability to control wild-type effectors. Both Thpok and LRF KO male mice remained healthy and had normal numbers of conventional and regulatory T cells (Fig. 2BC). In contrast, Treg-specific deletion of both Thpok and Lrf in male mice resulted in an inflammatory disease characterized by growth retardation, widespread skin lesions, and inflammation of the lung, spleen and skin (Fig. 2CD). This condition was similar to that of Scurfy mice, which carry a spontaneous loss-of-function Foxp3 mutation (10); although inflammation appeared later in dKO than in Scurfy mice, it resulted in death within two months of age (Fig. 2B). Most conventional T cells in dKO mice were activated (CD44hi CD62L−) and produced cytokines, including IFNγ, IL-4 and IL-17 (Fig. 2E); deletion of either Thpok or Lrf had no such effect (Fig. 2C, F, and data not shown). Thus, Treg expression of Thpok or LRF is needed for immune tolerance.
In contrast to male mice, heterozygous (Foxp3+/YFP-Cre) Thpokfl/fl Lrffl/fl female mice (female dKO mice) generated Tregs expressing either the Foxp3YFP-Cre (deleting) or Foxp3 (non-deleting) allele, as a result of random X chromosome inactivation during development. These animals maintained immune homeostasis and did not develop disease (data not shown), indicating that Thpok-and LRF-deficient Tregs do not acquire dominant inflammatory properties.
The number of spleen Foxp3+ cells was lower in dKO than control male mice (Fig. 2C). The same was true of the numbers of YFP+ (i.e. Cre-expressing) spleen Tregs in female dKO mice, in which the ratio of YFP+ vs. YFP− cells was lower than in control animals (Fig. 2G). This indicated a cell-intrinsic effect of the double disruption on Treg homeostasis. dKO Tregs were mostly CD4+CD8+ and CD4−CD8+, with a minor CD4+CD8− component (Fig. 2H). Deletion of Thpok alone, but not of Lrf, modestly affected CD4 and CD8 expression (Fig. 2H). Thus, Thpok and LRF redundantly support the CD4+CD8− expression pattern of Tregs, as they do for conventional CD4+ T cells (16, 42).
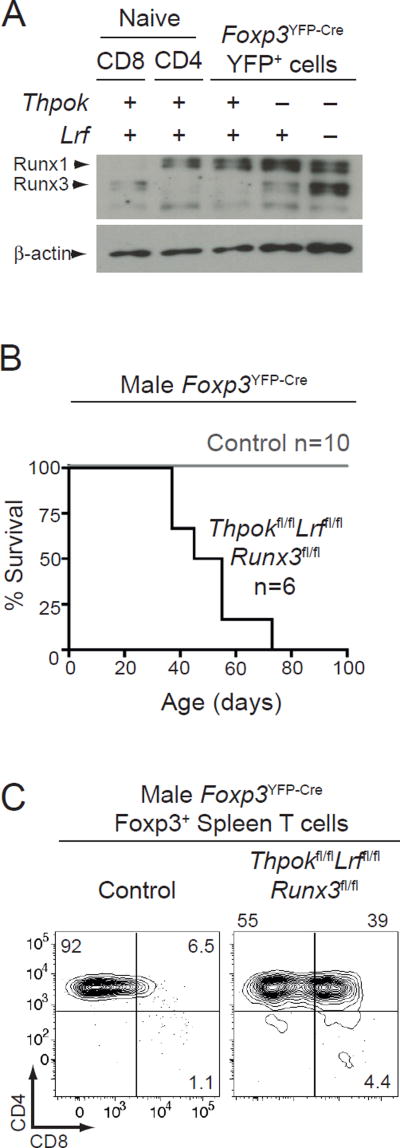
Thpok and LRF affect Treg functions independently of Runx3 repression
In thymocytes and conventional T cells, Thpok acts by inhibiting the expression and function of the transcription factor Runx3 (16, 39, 42). dKO Tregs had increased levels of Runx3, but not of its homolog Runx1 which is expressed in wild-type Tregs (Fig. 3A). To address whether Runx3 de-repression underpinned the dysfunction of dKO Tregs, we examined Foxp3YFP-Cre Runx3fl/fl Thpokfl/fl Lrffl/fl male mice, in which all Tregs lack Runx3, Thpok and LRF. The incidence and progression of inflammatory disease was similar in these and dKO mice (Figs. 3B and 2B). Of note, CD4 expression was restored by Runx3 disruption (Fig. 3C); this indicated that the impact of Thpok and LRF on Tregs is not mediated by their effect on CD4 expression, and thereby MHC II-induced TCR signaling (43, 44).
Figure 3. Runx3 independent impact of Thpok and LRF on Treg function.
(A) Immunoblot analysis of Runx protein expression in sorted CD44lo CD25− naive CD4+ and CD8+ cells from WT mice and in YFP+ Tregs from control, Thpok KO or dKO male mice. Each lane is from 105 cells, and expression of β-actin is shown as a loading control. Data is representative of two biological replicates for each sample.
(B) Survival curve of control (grey line) or Thpokfl/flLrffl/fl Runx3fl/fl (black line) Foxp3YFP-Cre male mice. Number of mice is indicated in plot. Median survival for Foxp3YFP-Cre Thpokfl/flLrffl/fl Runx3fl/fl is 50 days. Data from control mice is the same as in Fig. 2B and shown for comparison purposes.
(C) CD8 vs. CD4 expression is shown on Foxp3+ T cells from control or Thpokfl/fl Lrffl/fl Runx3fl/flFoxp3YFP-Cre male mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
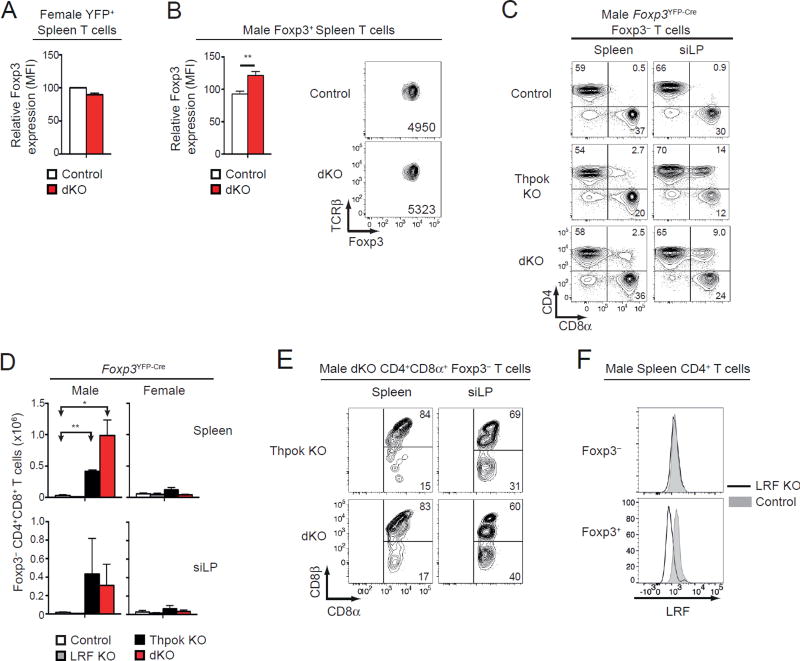
Effect of Thpok and LRF on Treg stability
It was reported that environmental cues in the small intestine, at the interface of the epithelial layer and of the lamina propria (siLP, the main site of immune activity at steady state), repress Thpok, resulting in Foxp3 silencing and conversion of Foxp3+ CD4+ Tregs into Foxp3− CD4+ CD8α+CD8β− ‘ex Treg’ intra-epithelial lymphocytes (IELs) (20); accordingly, inducible Thpok disruption resulted in the generation of Foxp3− CD4+ CD8+ ex-Tregs (20). Thus, we examined the impact of Thpok and LRF on Foxp3 expression. Foxp3+ levels were not reduced by deletion of Thpok, LRF or both, and were minimally increased in male dKO cells (Fig. 4AB). It was possible that disruption of Thpok and LRF resulted in an all-or-none Foxp3 silencing, while having no impact on Foxp3 expression levels in cells that continue to express this gene; such ‘binary’ shut-down has previously been observed in mice lacking the Foxp3 CNS2 cis-regulatory element (45, 46). Because Thpok represses CD8 expression, Foxp3 silencing caused by Thpok disruption would be accompanied by expression of CD8 in the resulting Foxp3− CD4+ cells. Consistent with this idea and in line with previous results (20), male Thpok KO mice had Foxp3− CD4+CD8+ cells, especially in the siLP (Fig. 4CD). Most of these cells expressed both CD8α and CD8β (Fig. 4E), unlike previously reported CD4+CD8α+ CD8β− ex Treg IELs (20). Foxp3− CD4+CD8+ cells were also found in dKO male mice (Fig. 4C–E).
Figure 4. Impact of Thpok and LRF on Treg stability.
(A, B) Bar graphs show relative expression (mean fluorescence intensity, MFI) of intra-cellular Foxp3 protein in YFP+ spleen cells from female mice (A) or in Foxp3+ spleen cells from male mice (B) of the indicated genotype; data is expressed relative to WT set to 100 within each experiment, and is from three (A) or five (B) mice of each genotype analyzed in three (A) or five (B) independent experiments (**: P<0.01). Right panel in (B) shows a representative experiment; actual MFI of intra-cellular Foxp3 staining is indicated in each plot.
(C) Expression of CD8α vs. CD4 on Foxp3− spleen (left) or siLP (right) TCRβ+ cells from control (top), Thpok KO (middle) or dKO (bottom) male mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
(D) Absolute numbers of TCRβ+ Foxp3− (left, male Foxp3YFP-Cre mice) or TCRβ+ YFP− (right, female Foxp3+/YFP-Cre mice) CD4+CD8α+ cells in the spleen (top) or siLP (bottom) of mice of the indicated genotype. Data is from at least three animals per genotype analyzed in at least three independent experiments (control vs. Thpok KO or dKO: P<0.05 [*] and P<0.01 [**]).
(E) Contour plots of CD8α vs. CD8β expression on CD4+CD8α+ Foxp3− T cells from the spleen (left) or siLP (right) of indicated male mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
(F) Expression of intra-cellular LRF in Foxp3− (top) and Foxp3+ (bottom) CD4+ spleen T cells from LRF KO (Lrffl/fl Foxp3YFP-Cre, plain line) and control (Lrf+/+ Foxp3YFP-Cre, grey-shaded) male mice. Data is representative of two mice.
In line with a recent report (47), we considered that, rather than being ex Tregs, Foxp3−CD4+CD8+ cells from Thpok KO or dKO male mice could be bona fide conventional CD4+ T cells that had deleted Thpok alleles because of ectopic Foxp3YFP-Cre activity. Although this possibility cannot be easily distinguished from Foxp3 silencing after Thpok inactivation in Tregs, we reasoned that ectopic Foxp3YFP-Cre expression would also inactivate Lrffl alleles in conventional CD4+ T cells from LRF KO (Lrffl/fl Foxp3YFP-Cre) male mice. Because LRF deletion per se has no detectable impact on Treg or conventional CD4+ T cell homeostasis, and can readily be detected by intra-cellular staining and flow cytometry, we used this approach to evaluate the extent of ectopic Foxp3YFP-Cre activity in conventional CD4+ T cells. Despite complete Lrf inactivation in Tregs (Fig. 4F, bottom), conventional (Foxp3−) CD4+ T cells from LRF KO male mice did not show evidence of Lrf deletion (Fig. 4F, top). This indicates that the Foxp3YFP-Cre allele had little if any ectopic activity, and supports the conclusion that Foxp3− CD4+CD8+ cells in Thpok KO male mice are indicative of Foxp3 silencing following Thpok inactivation.
In contrast to their male counterparts, we did not observe Foxp3− CD4+CD8+ cells in Thpok KO or dKO female mice (Fig. 4D). Although it is possible that impaired immune homeostasis in dKO male mice contributes to destabilize Foxp3 expression, this is unlikely to be the case in Thpok KO male mice, which show little if any evidence of inflammation. Rather, we favor the possibility that Thpok stabilizes Foxp3 expression in a cell-intrinsic manner, but that, in female Thpok KO or dKO mice, the expansion of Foxp3− ex Tregs is restrained by functionally competent Tregs (expressing the wild-type Foxp3 allele). Altogether, these observations are consistent with the idea that Thpok non-redundantly stabilizes Foxp3 expression.
The absence of detectable inflammatory disease in Thpok KO male mice suggested that the stabilization of Foxp3 expression by Thpok was not required for systemic immune homeostasis. However, it was possible that Thpok-deficient Tregs, while able to maintain immune homeostasis at steady state, would be overcome upon infectious challenge. To address this possibility, we evaluated the response to Leishmania major, an infection in which Treg cells normally restrain Th1 effectors and prevent immunopathology (48). Although Thpok KO male mice showed a trend towards increased IFNγ production by effector T cells in the draining lymph node (dLN, Fig. S1B), neither the number of Treg cells in the dLN nor disease course were reproducibly affected (Fig. S1CD). Thus, Treg disruption of Thpok, regardless of its effect on Foxp3 expression, only minimally, if at all, impairs immune homeostasis. This prompted us to consider additional functions of Thpok and LRF in Tregs.
Thpok and LRF promote Treg survival
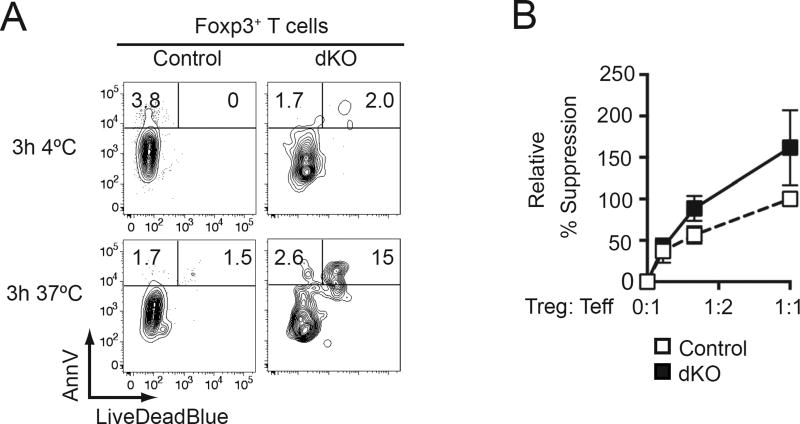
The reduced numbers of Tregs in dKO mice suggested that Thpok and LRF supported Treg survival. To assess this possibility, we stained Tregs with a dye assessing cell death and with annexin V, which marks cells undergoing apoptosis. We found a larger fraction of dying and dead cells among dKO than control Tregs (Fig. 5A), supporting the conclusion that Thpok and LRF promoted Treg survival.
Figure 5. Thpok and LRF promote Treg survival.
(A) LiveDeadBlue vs. Annexin V (AnnV) staining in male control (left) or dKO (right) spleen Foxp3+ cells cultured in medium as indicated. Data is representative of two mice of each genotype in two independent experiments.
(B) Graph indicates percent suppression of wild-type effector T cell proliferation by female dKO (filled squares) or control (open squares) Tregs at indicated Treg: Teff ratios with suppression by control Tregs at a 1:1 ratio set to 100. Data is from at least four animals per genotype analyzed in at least four independent experiments.
We next examined the impact of Thpok and LRF on Treg function. We previously showed that Thpok was dispensable for Tregs to suppress effector T cell proliferation in vitro (18), an observation consistent with the conserved immune homeostasis in Thpok KO mice. Even though the severe inflammation in dKO male mice suggested that the combined disruption of Thpok and LRF would affect Treg function, dKO Tregs efficiently restrained effector T cells in a conventional in vitro suppression assay (Fig. 5B).
Impact of Thpok and LRF on Treg gene expression
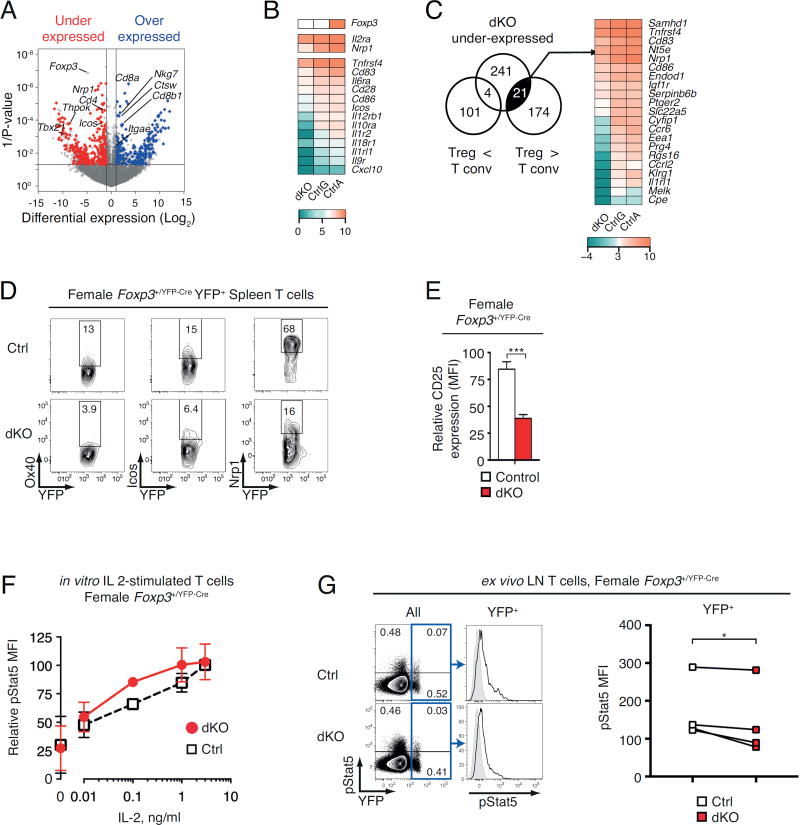
The number of Tregs in dKO mice was insufficient to conduct in vivo functional assays [e.g. prevention of inflammatory colitis (49) or rescue of the Scurfy phenotype (5)]. Thus, we sought insight into the impact of Thpok and LRF on Treg function by examining how they affect gene expression, using deep sequencing of reverse-transcribed mRNA (RNAseq). To avoid the confounding effects of inflammation in male mice, these analyses were performed on female cells. We compared YFP+ dKO Tregs with (i) CD25+ YFP− ‘animal control’ cells, obtained from the same female mice as the YFP+ Tregs but expressing the Foxp3+ allele, and (ii) YFP+ Tregs from Foxp3+/ YFP-Cre Thpok+/+ Lrf+/+ female mice (‘genetic controls’ which express the Foxp3YFP-Cre allele). Cre activity in dKO cells resulted in a complete loss of reads from floxed Thpok and Lrf exons (Fig. S2A) (19, 23). While we found lower Foxp3 read numbers in dKO than in Foxp3+ animal control cells (Fig. 6AB), this was a consequence of the Foxp3YFP-Cre knockin strategy. Indeed, there was no difference between dKO and Foxp3YFP-Cre genetic control cells, which both carry the Foxp3YFP-Cre knockin allele, consistent with the conserved Foxp3+ expression in dKO cells (Fig. 6B and 4A).
Figure 6. Impact of Thpok and LRF on Treg gene expression.
(A) Volcano plot of RNAseq data displays ratios of dKO over WT expression (x-axis, reads per million, Log2 values) vs. P-value (y-axis, -Log10 values); each symbol represents a distinct locus. Colored symbols display over-expressed (right, blue) and under-expressed (left, red) genes, defined by a 2-fold or greater change in expression with a 0.05 or lesser P-value. Relevant genes are indicated.
(B) Heat map shows expression of indicated genes in dKO, CtrlG and CtrlA cells (Log2 values, color scale at bottom).
(C) Venn diagram shows overlap between (i) genes under-expressed in dKO relative to control Tregs and (ii) genes under- or over-expressed in Treg relative to conventional T cells (50). Heat map (right) shows expression of 21 genes, defined in the left Venn diagrams, in dKO, CtrlG and CtrlA (Log2 values, color scale at bottom).
(D) Expression of YFP vs. Ox40, Icos or Nrp1 on YFP+ TCRβ+ spleen cells from control (top) and dKO (bottom) female Foxp3+/YFP-Cre mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
(E) Relative expression (MFI) of CD25 in YFP+ TCRβ+ spleen cells from Foxp3+/YFP-Cre female control (open bar) or dKO (filled red bar) mice; data is expressed relative to control set to 100 within each experiment, and is from five mice of each genotype analyzed in five independent experiments (***: P<0.001).
(F) Relative fluorescence (MFI) of pStat5 in YFP+ TCRβ+ spleen cells from Foxp3+/YFP-Cre female control (open squares) or dKO (filled red circles) mice after in vitro stimulation with IL-2 at the indicated concentrations; data is expressed relative to pStat5 MFI in control cells treated with 3 ng/ml IL-2, set to 100 within each experiment, and is from two independent experiments (except 0.1 ng/ml, one experiment). Error bars indicate SD.
(G) (Left) Expression of YFP vs. pStat5 on ex vivo gated LN T cells from control (Thpok+/+ Lrf+/+) or dKO (Thpokfl/fl Lrffl/fl) Foxp3+/YFP-Cre female mice; cells were fixed immediately after LN disruption. Histogram overlays (middle) show pStat5 fluorescence on YFP+ cells as gated in blue boxes on left plots. Grey-shaded histograms shows isotype control staining. Data is representative of four pairs of mice analyzed in four separate experiments summarized on the right plot (*: P<0.05, one-tailed paired t-test).
Among 13702 genes with detectable RNA reads, we found 497 outliers defined by a 2-fold or greater significant expression change (P-value < 0.05); of those, 233 were over-expressed and 266 under-expressed in dKO cells relative to both animal and genetic controls (Fig. 6A and Tables S1, S2). We first examined whether these outlier genes were related to previously reported Thpok- or Foxp3-dependent ‘signatures’ (17, 50). Aside from Cd4, Cd8, and Runx3, there was little overlap between outlier genes in dKO Tregs and the Thpok signature previously identified in Th1 effector cells (Fig. S2BC). Unexpectedly, the intersection with the Treg signature was similarly limited (Fig. 6C), and notably included Nrp1 and Tnfrsf4 (encoding Ox40). In addition to these intersections with Thpok and Foxp3+ signatures, the under expressed set comprised multiple genes important for immune response (Fig. 6B, D, E and S2D), including Icos and Il2ra (encoding CD25, the IL-2 receptor α chain). Accordingly, parsing the under expressed gene list with the metascape search tool (51) revealed significant enrichment with IL-2 and Stat5 targets, indicating that Thpok and LRF redundantly support IL 2-mediated gene expression in Tregs (Fig. S2EF). Of note, no consistent association emerged from the over-expressed gene list (data not shown). Thus, the impact of Thpok and LRF on Treg gene expression is distinct from that of Foxp3, or from their impact in conventional T cells.
To examine if the reduced expression of IL-2 target genes resulted from impaired transduction of IL-2 signals, we assessed phosphorylation of the IL-2 signaling relay Stat5. We found no evidence of impaired Stat5 phosphorylation in YFP+ Tregs from female dKO mice after in vitro IL-2 stimulation (Fig. 6F). In contrast, we found reduced levels of phosphorylated Stat5 (pStat5) in ex vivo Tregs fixed immediately after disruption of peripheral LN to assess in vivo IL-2 signaling (Fig. 6G) (29). Of note, the difference in in vivo Stat5 phosphorylation between wild-type and dKO Tregs varied among experiments. Together with the conserved IL-2-induced Stat5 phosphorylation in vitro, this suggests that the impact of Thpok and LRF on IL-2 target genes could be indirect, e.g. resulting from impaired access to IL-2 producing cells, rather than reflecting a cell-intrinsic impairment of IL-2 signal transduction. Consistent with this scenario, Thpok and LRF disruption affected the expression of genes involved in cell migration (Fig. S2G).
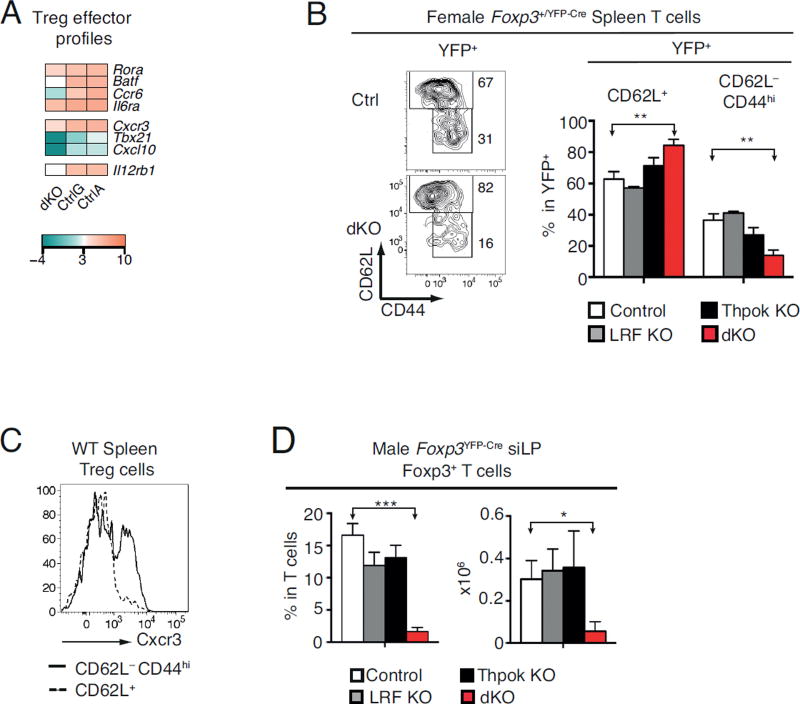
Despite the reduced survival of dKO Tregs, there was little effect of Thpok and LRF disruption on expression of cell-death related genes (Fig. S2D). In contrast, dKO cells had reduced expression of genes associated with cell migration and effector differentiation, including Tbx21 (encoding T-bet) and Cxcr3, which characterize a population of ‘Th1-like’ Tregs (52), and Batf, Rora, Il6ra, and Ccr6, characteristic of ‘Th17-like’ Tregs (Fig. 7A, Fig. S2D, G) (53, 54). Consistent with this impact, the frequency of CD44hi CD62L− effector Tregs, which normally includes all Cxcr3+ Tregs, was significantly reduced among YFP+ dKO Tregs in female mice (Fig. 7BC). Additionally, the number of siLP Tregs, most of which are effector Tregs, was strongly reduced in dKO male mice (Fig. 7D). In contrast, it was not affected by the deletion of Thpok only, unlike what was observed after inducible Thpok disruption (20). This difference may be due at least in part to the inclusion of CD8β-expressing Foxp3+ Tregs in data shown in Fig. 7D, unlike in that previous study (20). Additionally, it is possible that compensatory mechanisms maintain Treg numbers at steady-state despite Thpok disruption, but not after acute induced deletion (20). Altogether, we conclude from these findings that Thpok and LRF contribute to the differentiation or competitive fitness of effector Tregs.
Figure 7. Impact of Thpok and LRF on effector Tregs.
(A) Heat map shows expression of select Treg effector genes in dKO, CtrlG and CtrlA cells (Log2 values, color scale at bottom).
(B) Contour plots (left) show CD44 vs. CD62L expression on YFP+ (Cre-expressing) TCRβ+ spleen Tregs from control (top) or dKO (bottom) female Foxp3+/YFP-Cre mice. Bar plots (right) show percentage of CD62L+ or CD62L− CD44hi cells in YFP+ TCRβ+ spleen Tregs in control (open bars), LRF KO (grey bars), Thpok KO (black bars), or dKO (red bars) female Foxp3+/YFP-Cre mice. Data is from at least three animals per genotype and analyzed in at least three independent experiments (control vs. dKO: P<0.01 [**]; other differences not significant).
(C) Cxcr3 expression on CD62L+ (dashed line) or CD62L− CD44hi (solid line) YFP+ cells in female Foxp3+/YFP-Cre control mice. Data is representative of three mice of each genotype analyzed in three independent experiments.
(D) Percentage (left) and absolute number (right) of siLP Foxp3+ TCRβ+ cells in control (open bars), LRF KO (grey bars), Thpok KO (black bars), or dKO (red bars) male mice. Data is from at least three animals per genotype and analyzed in at least three independent experiments.
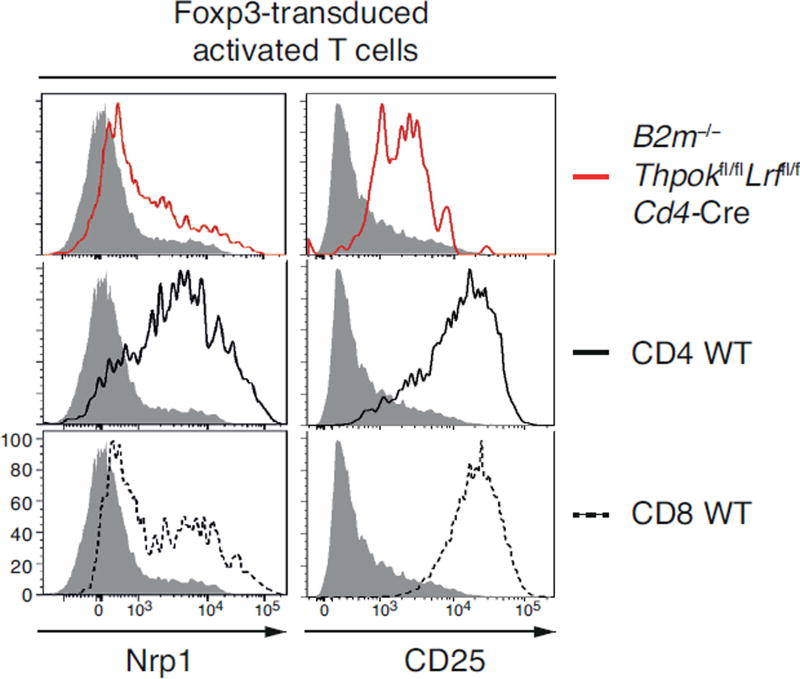
To independently document the impact of Thpok and LRF on Treg-specific gene expression, we examined whether these factors are needed to allow Foxp3-induced expression of CD25 and Nrp1, two genes characteristic of Tregs (13, 55, 56) and under-expressed in dKO cells (Fig. 6DE). We retrovirally transduced Foxp3 in conventional T cells, either Thpok- and LRF-sufficient (CD4+ and CD8+ T cells from wild-type mice) or Thpok- and LRF-deficient (CD8+ T cells from B2m−/− Thpokfl/fl Lrffl/fl Cd4-Cre mice, which all are MHC II-restricted). Whereas Foxp3 transduction up-regulated Nrp1 and CD25 in wild-type CD4+ and CD8+ T cells, this effect was blunted by disruption of Thpok and LRF (Fig. 8). Thus, Thpok and LRF are important for Foxp3-induced expression of Treg-specific genes.
Figure 8. Thpok and LRF support Foxp3-induced gene expression.
Red-line histograms (top plots) show Nrp1 (left) and CD25 (right) expression in Foxp3-transduced MHC II-restricted CD8+ T cells from B2m−/− Thpokfl/fl Lrffl/fl Cd4-Cre mice (in which MHC II-restricted thymocytes differentiate into CD8+ T cells and MHC I-restricted T cells fail to develop). Plain and dashed black-line histograms (middle and bottom plots) show Nrp1 and CD25 expression on Foxp3-transduced wild-type CD4+ and CD8+ T cells. Grey-filled histograms show Nrp1 and CD25 expression on MHC II-restricted CD8+ T cells, from B2m−/− Thpokfl/fl Lrffl/fl Cd4-Cre mice, transduced with an empty retroviral vector. Data is representative of two (Nrp1) or four (CD25) independent experiments.
In summary, we demonstrate that the transcription factor Thpok is needed for the thymic differentiation of Treg cells, and, redundantly with its homolog LRF, for Treg-mediated immune homeostasis. Both factors act in Tregs by supporting Foxp3-directed gene expression, notably by enabling expression of IL-2-dependent genes and the development of effector Treg populations.
Discussion
Here, we report that overlapping functions of the transcription factors Thpok and LRF are critical for the differentiation and homeostasis of Foxp3+ Treg cells, as shown by the severe breach of tolerance resulting from their Treg-specific disruption. Additionally, we show that these factors support intrathymic Treg development.
Although there are CD8+ Foxp3− Treg cells (57), Foxp3+ Tregs are invariably CD4+. Why this is the case has remained unclear, notably because the Foxp3 locus is epigenetically open in CD4+CD8+ (double positive, DP) thymocytes (58), which serve as precursors of both CD4+ and CD8+ cells (59). Thpok is required for the differentiation of conventional MHC II-restricted thymocytes into CD4+ T cells, but not for their positive selection. That is, Thpok-deficient MHC II-restricted thymocytes differentiate into CD8+ instead of CD4+ T cells (14, 16); importantly, despite such lineage ‘redirection’, the number and repertoire of MHC II-restricted T cells are not detectably affected by Thpok disruption (18, 19, 60). In contrast, we show here that Thpok is needed for the development of thymic Treg precursors, and not solely for their CD4+-lineage differentiation. Because the development of thymic Tregs is thought to require high-intensity TCR signaling, we considered that Thpok promoted Treg development by supporting expression of CD4, which contributes to TCR signaling. Unexpectedly, this was not the case, as the requirement for Thpok is not relieved by enforced expression of CD4.
Given the role of Thpok in stabilizing Foxp3 expression in intestinal Tregs (20), it is possible that Thpok contributes to Foxp3 expression in thymocytes. However, the fact that Thpok is needed for the proper differentiation of CD25+ Foxp3− thymic Treg precursors indicates that it serves at least in part independently of a potential direct effect on Foxp3. Chromatin immunoprecipitation analyses, which we could not perform with currently available reagents, will help address this issue. Of note, TNF-family receptors, including GITR and Ox40, promote Treg differentiation in the thymus at least in part by facilitating IL-2 signaling (36). We found expression of both receptors to be Thpok-dependent in thymocytes. Thus, it is possible that, through its effect on TNF-family receptor expression, Thpok serves in thymocytes to facilitate IL-2-mediated gene expression. Additionally, it is possible that Thpok promotes Treg thymocyte differentiation by antagonizing the activity of Runx molecules; however, the functional overlap between Runx1 and Runx3 (61), together with the dependence of Tregs on Runx activity (62, 63), create a major challenge to experimental investigations of this hypothesis.
Contrary to the importance of Thpok per se for thymic Treg development, we found that Thpok and LRF served redundantly in post-thymic Tregs to support the functions and homeostasis of these cells. Previous studies have identified components of the transcriptional network supporting Treg differentiation and functions. These include factors enabling Foxp3-mediated gene expression (64), factors enforcing Foxp3 expression and Treg fitness, including Gata3 (65, 66), Nur77-family members (67–69), the chromatin modifier Ezh2 (70), or the pathway activated by PI-3 kinase activity and targeting Foxo transcription factors and the metabolic regulator mTOR (reviewed in 71, 72). Our study supports the conclusion that the main function of Thpok and LRF in post-thymic Tregs is to support Foxp3-mediated gene expression rather than to maintain expression of Foxp3 itself.
This conclusion appears at odds with our observation, and a previous report (20), that Thpok contributes to stabilize Foxp3 expression, with little functional overlap with LRF. It was previously reported that, through this activity, Thpok prevents the conversion of siLP Foxp3+ CD4+CD8− Tregs into Foxp3− CD4+ CD8α+ CD8β− IELs (20). However, despite this impact on Foxp3 expression, Thpok is not needed for Tregs to enforce systemic immune homeostasis or for their responses to infection. Even in the gut mucosal environment, Thpok is dispensable for the prevention of intestinal inflammation in a model of antigen-induced colitis (20). Of note, disruption of the Foxp3 CNS2 cis-regulatory element, which destabilizes Foxp3 expression but does not affect the function of remaining Foxp3+ Treg cells, has a much milder impact on immune homeostasis than disruption of Thpok and LRF (45, 46).
The functions of Thpok and LRF in Tregs differ in three important respects from those in conventional CD4+ T cells. First, the functional overlap between the two factors is broader in Tregs than in conventional effectors T cells. Whereas Thpok per se is largely dispensable for Treg functions, it is needed for proper Th1 and Th2 responses with little compensation, if any, by LRF (17). Second, the functional targets of Thpok and LRF differ in Treg and conventional CD4+ effector T cells. Whereas in conventional CD4+ T cells Thpok inhibits the expression of cytotoxic genes (17, 27), many of these genes, including that for Granzyme B, are normally expressed in Tregs despite Thpok expression (73, 74). Conversely, whereas Thpok promotes expression of genes typical of helper differentiation in conventional CD4+ T cells, including Cd40lg, S1pr1 and Btla (17), these genes are not affected by inactivation of Thpok and LRF in Tregs. Genome-wide analyses of Thpok DNA binding by chromatin immunoprecipitation will assist in determining if such differences reflect distinct repertoires of Thpok binding sites among T cell subsets. Third, the key functions of Thpok and LRF in Tregs are not mediated by inhibition of Runx3 expression or activity. This conclusion is in line with the ability of Foxp3 itself to antagonize Runx activity and notably restrain Runx-mediated expression of IL-2 and IFNγ (75). Indeed, Runx activity is critical in Tregs, as demonstrated by Treg-specific disruption of Cbfβ, an obligate Runx cofactor (62, 63). Accordingly, Foxp3YFP-Cre-mediated disruption of Thpok, LRF and Cbfβ failed to generate functional Tregs (A.C. and R.B., unpublished observations).
Although additional investigations will be needed to gain a full mechanistic understanding of the functions of Thpok and LRF in Tregs, our findings indicate that these factors redundantly promote Treg survival, and suggest that they are especially important for the homeostasis of effector Tregs, a subset whose importance is increasingly appreciated (76–78). Effector Tregs express transcription factors, including T-bet, Batf and RORα, that in conventional helper T cells direct expression of inflammatory cytokines, but they do not express the corresponding cytokines (e.g. IFNγ or IL-17)(52, 76). Their chemokine receptor expression matches that of conventional effector T cells and targets them to inflammation sites, where they could serve as ‘sinks’ for inflammatory cytokines (e.g. IL-6 or IL-12) for which they carry receptors. Accordingly, there is evidence that effector Treg cells serve to control specific conventional T cell responses (79, 80). Effector Tregs are abundant at mucosal interfaces and in tissues, whereas naïve Treg cells, which do not express effector transcription factors, predominate in peripheral lymphoid organs at steady state.
Although Thpok and LRF are not strictly required for effector Treg differentiation, our findings are most consistent with a scenario whereby these factors redundantly promote the survival of Tregs when they acquire ‘effector’ functions. In this perspective, Tregs lacking Thpok and LRF are eliminated in a competitive setting (in female mice in the experimental systems used in the present study), resulting in the preponderance of wild-type Tregs which ensure immune homeostasis. In contrast, in a non-competitive setting (in male mice in the experimental systems used in the present study), Thpok and LRF deficiency results in a loss of Treg function and rupture of immune tolerance. The paucity of dKO Treg cells in siLP, where most Tregs have effector functions, is consistent with this scenario.
In summary, the severe breach of tolerance resulting from the Treg-specific disruption of Thpok and LRF shows that these transcription factors are critical components of the network controlling the differentiation and homeostasis of Foxp3+ Tregs. Together with our previous finding that both Thpok and LRF are needed for effector differentiation of conventional CD4+ T cells (17), these results demonstrate that the Thpok-LRF ‘node’ supports both the helper and regulatory arms of the MHC II-restricted response, and therefore uncovers a previously unrecognized organizational principle of the adaptive immune system.
Supplementary Material
Acknowledgments
We thank K. Beacht for technical assistance, S. Banerjee and F. Livak for flow sorting advice, T.A. Lewis, G. Jackson for expert mouse technical support, T. Ciucci, J. Grainger, O. Harrison for useful discussions, and J. Ashwell, T. Ciucci, P. Love, and A. Sher for reading the manuscript.
Funding Sources
Supported by the Intramural Research Programs of the National Cancer Institute, Center for Cancer Research, and of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. M.M.G. was supported by the German Research Foundation (Ga 1818/2-1).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annual review of immunology. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annual review of immunology. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 3.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 6.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science (New York, N.Y.) 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 7.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nature immunology. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 8.Ramsdell F, Ziegler SF. FOXP3 and scurfy: how it all began. Nature reviews. Immunology. 2014;14:343–349. doi: 10.1038/nri3650. [DOI] [PubMed] [Google Scholar]
- 9.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 10.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 11.Lee HM, Hsieh CS. Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. Journal of immunology (Baltimore, Md. : 1950) 2009;183:2261–2266. doi: 10.4049/jimmunol.0901304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malek TR. The biology of interleukin-2. Annual review of immunology. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 14.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 15.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature immunology. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. Journal of immunology (Baltimore, Md. : 1950) 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, Bosselut R. A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology. 2014;15:947–956. doi: 10.1038/ni.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpenter AC, Grainger JR, Xiong Y, Kanno Y, Chu HH, Wang L, Naik S, dos Santos L, Wei L, Jenkins MK, O'Shea JJ, Belkaid Y, Bosselut R. The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity. 2012;37:622–633. doi: 10.1016/j.immuni.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, Mucida D. Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science (New York, N.Y.) 2016 doi: 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 23.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science (New York, N.Y.) 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, Muller W, Rudensky AY. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. The Journal of experimental medicine. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung RK, Thomson K, Gallimore A, Jones E, Van den Broek M, Sierro S, Alsheikhly AR, McMichael A, Rahemtulla A. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nature immunology. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nature immunology. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol. 1991;138:1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 29.Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, Campbell DJ. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. The Journal of experimental medicine. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suffia IJ, Reckling SK, Piccirillo CA, Goldszmid RS, Belkaid Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. The Journal of experimental medicine. 2006;203:777–788. doi: 10.1084/jem.20052056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. The Journal of experimental medicine. 1998;188:1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bautista JL, Lio CW, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh CS. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nature immunology. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmud SA, Manlove LS, Schmitz HM, Xing Y, Wang Y, Owen DL, Schenkel JM, Boomer JS, Green JM, Yagita H, Chi H, Hogquist KA, Farrar MA. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nature immunology. 2014;15:473–481. doi: 10.1038/ni.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. Journal of immunology (Baltimore, Md. : 1950) 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- 38.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nature immunology. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 39.Taniuchi I, Ellmeier W. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Advances in immunology. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 40.Gialitakis M, Sellars M, Littman DR. The epigenetic landscape of lineage choice: lessons from the heritability of CD4 and CD8 expression. Current topics in microbiology and immunology. 2012;356:165–188. doi: 10.1007/82_2011_175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nature immunology. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vacchio MS, Bosselut R. What Happens in the Thymus Does Not Stay in the Thymus: How T Cells Recycle the CD4+-CD8+ Lineage Commitment Transcriptional Circuitry To Control Their Function. Journal of immunology (Baltimore, Md. : 1950) 2016;196:4848–4856. doi: 10.4049/jimmunol.1600415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nature immunology. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vahl JC, Drees C, Heger K, Heink S, Fischer JC, Nedjic J, Ohkura N, Morikawa H, Poeck H, Schallenberg S, Riess D, Hein MY, Buch T, Polic B, Schonle A, Zeiser R, Schmitt-Graff A, Kretschmer K, Klein L, Korn T, Sakaguchi S, Schmidt-Supprian M. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41:722–736. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 2014;158:734–748. doi: 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky Alexander Y. Control of the Inheritance of Regulatory T Cell Identity by a cis Element in the Foxp3 Locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franckaert D, Dooley J, Roos E, Floess S, Huehn J, Luche H, Fehling HJ, Liston A, Linterman MA, Schlenner SM. Promiscuous Foxp3-cre activity reveals a differential requirement for CD28 in Foxp3(+) and Foxp3(−) T cells. Immunology and cell biology. 2015;93:417–423. doi: 10.1038/icb.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belkaid Y, Mendez S, Lira R, Kadambi N, Milon G, Sacks D. A natural model of Leishmania major infection reveals a prolonged “silent” phase of parasite amplification in the skin before the onset of lesion formation and immunity. Journal of immunology (Baltimore, Md. : 1950) 2000;165:969–977. doi: 10.4049/jimmunol.165.2.969. [DOI] [PubMed] [Google Scholar]
- 49.Read S, Powrie F. Induction of inflammatory bowel disease in immunodeficient mice by depletion of regulatory T cells. Current protocols in immunology. 2001 doi: 10.1002/0471142735.im1513s30. Chapter 15: Unit 15.13. [DOI] [PubMed] [Google Scholar]
- 50.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 51.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yanguez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, Konig R, Stertz S, Garcia-Sastre A, Chanda SK. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell host & microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S, Fagarasan S, Mielke LA, Afshar-Sterle S, Masters SL, Nakae S, Saito H, Wentworth JM, Li P, Liao W, Leonard WJ, Smyth GK, Shi W, Nutt SL, Koyasu S, Kallies A. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nature immunology. 2015;16:276–285. doi: 10.1038/ni.3085. [DOI] [PubMed] [Google Scholar]
- 54.Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C. CCR6 regulates the migration of inflammatory and regulatory T cells. Journal of immunology (Baltimore, Md. : 1950) 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, Yang Y, Floess S, Huehn J, Oh S, Li MO, Niec RE, Rudensky AY, Dustin ML, Littman DR, Lafaille JJ. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. The Journal of experimental medicine. 2012;209:1723–1742. s1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, Ruminski P, Weiss D, Von Schack D, Bluestone JA. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. The Journal of experimental medicine. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holderried TA, Lang PA, Kim HJ, Cantor H. Genetic disruption of CD8+ Treg activity enhances the immune response to viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:21089–21094. doi: 10.1073/pnas.1320999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nature immunology. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keefe R, Dave V, Allman D, Wiest D, Kappes DJ. Regulation of lineage commitment distinct from positive selection. Science (New York, N.Y.) 1999;286:1149–1153. doi: 10.1126/science.286.5442.1149. [DOI] [PubMed] [Google Scholar]
- 61.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nature reviews. Immunology. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nature immunology. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nature immunology. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, Paul WE, Bosselut R, Wei G, Zhao K, Oukka M, Zhu J, Belkaid Y. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. The Journal of clinical investigation. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sekiya T, Kondo T, Shichita T, Morita R, Ichinose H, Yoshimura A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. The Journal of experimental medicine. 2015;212:1623–1640. doi: 10.1084/jem.20142088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H, Metzger D, Chambon P, Yoshimura A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nature immunology. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 69.Fassett MS, Jiang W, D'Alise AM, Mathis D, Benoist C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3891–3896. doi: 10.1073/pnas.1200090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DuPage M, Chopra G, Quiros J, Rosenthal WL, Morar MM, Holohan D, Zhang R, Turka L, Marson A, Bluestone JA. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity. 2015;42:227–238. doi: 10.1016/j.immuni.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nature reviews. Immunology. 2016;16:149–163. doi: 10.1038/nri.2015.18. [DOI] [PubMed] [Google Scholar]
- 72.Sawant DV, Vignali DAA. Once a Treg, always a Treg? Immunological reviews. 2014;259:173–191. doi: 10.1111/imr.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. Journal of immunology (Baltimore, Md. : 1950) 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 74.Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, Ley TJ. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 75.Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 76.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends in immunology. 2013;34:74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Gratz IK, Campbell DJ. Organ-specific and memory treg cells: specificity, development, function, and maintenance. Frontiers in immunology. 2014;5:333. doi: 10.3389/fimmu.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nature immunology. 2013;14:1007–1013. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science (New York, N.Y.) 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.