Unlike canonical transport, in which cargo is attached to molecular motors, CENP-F promotes the transport of cargoes such as mitochondria by directly attaching them to the tips of growing and shrinking microtubules.
Abstract
Dynamic microtubule ends exert pulling and pushing forces on intracellular membranes and organelles. However, the mechanical linkage of microtubule tips to their cargoes is poorly understood. CENP-F is a nonmotor microtubule-binding protein that participates in microtubule binding at kinetochores and in the mitotic redistribution of the mitochondrial network. CENP-F–driven mitochondrial transport is linked to growing microtubule tips, but the underlying molecular mechanisms are unknown. Here we show that CENP-F tracks growing microtubule ends in living cells. In vitro reconstitution demonstrates that microtubule tips can transport mitochondria and CENP-F–coated artificial cargoes over micrometer-long distances during both growing and shrinking phases. Based on these and previous observations, we suggest that CENP-F might act as a transporter of mitochondria and other cellular cargoes by attaching them to dynamic microtubule ends during both polymerization and depolymerization of tubulin.
INTRODUCTION
Mitochondria form a highly dynamic network. Their intracellular transport and distribution is achieved through interaction with the cytoskeleton (Okamoto and Shaw, 2005). In animal cells, microtubules are the main tracks for mitochondrial transport. The classical model for organelle transport states that adaptor proteins on the surface of the organelle recruit molecular motors such as kinesins and dynein to transport mitochondria along the lattice of microtubules. In the case of mitochondria, the outer membrane protein Miro binds to both the kinesin KIF5b and the dynein–dynactin complex through the cytosolic adaptor Milton (Trak1 and Trak2), thus fulfilling the requirement for mitochondrial transport (Stowers et al., 2002; Guo et al., 2005; Birsa et al., 2013). Recently we discovered that at the end of mitosis, Miro also binds to centromere protein F (CENP-F), thereby influencing mitochondrial distribution in daughter cells (Kanfer et al., 2015). CENP-F is a nonmotor microtubule-binding protein that is expressed with strict cell cycle dependence (Zhu et al., 1995; Feng et al., 2006). Absent in G1, it accumulates in the nucleus and at the mitochondria during the S and G2 phases. During early mitosis (prophase), CENP-F localizes to the nuclear envelope and participates in its disassembly (Bolhy et al., 2011). In prometaphase and until anaphase, CENP-F localizes to the outer kinetochore and participates in linking kinetochores to spindle microtubules (Liao et al., 1995). During late mitosis and early G1, CENP-F is recruited by Miro to the mitochondrial network to participate in its proper distribution (Kanfer et al., 2015).
Unlike classical mitochondrial movement, which happens along the microtubule lattice, CENP-F/Miro–driven movements are linked to growing microtubule tips. Coordinated events of microtubule growth and mitochondrial movements are observed in wild-type cells but disappear in CENP-F–deficient cells. These observations together suggest that, in addition to using microtubules as passive tracks for motor-based transport, mitochondrial distribution in mitosis is linked to microtubule dynamics. Mitochondrial movements dependent on microtubule dynamics but not motor proteins have been observed in fission yeast, suggesting ancient evolutionary origins of this transport mode (Yaffe et al., 2003; Li et al., 2015).
Both growth and shrinkage of microtubules produce mechanical force. The force generated by growing microtubules can be observed in vitro when microtubules are grown toward a wall (Kerssemakers et al., 2006). These forces can also be harnessed by one factor—the tubulin polymerase XMAP215—to move a microbead in a reconstituted in vitro system (Trushko et al., 2013). The force generated by microtubule shrinking can be captured by a number of proteins in vitro (Lombillo et al., 1995; Asbury et al., 2006; McIntosh et al., 2008; Grissom et al., 2009; Welburn et al., 2009). Most of these proteins were found on the kinetochores, suggesting that microtubule depolymerization is a determinant of genome segregation. Among these, CENP-F was recently shown to follow microtubule depolymerization and transmit piconewton amounts of mechanical force to move a microbead in vitro (Volkov et al., 2015). This raises the question of whether CENP-F–dependent mitochondrial movements can be directly driven by microtubule dynamics without involving molecular motors.
RESULTS
CENP-F tracks growing microtubule tips in vivo
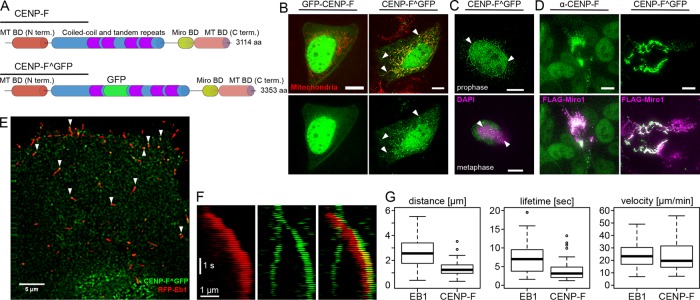
We sought to image CENP-F–based mitochondrial movements by time-lapse microscopy. However, ectopic expression of N- or C-terminal green fluorescent protein (GFP) fusions of CENP-F did not recapitulate its endogenous localization. This was possibly due to interference of GFP with CENP-F’s microtubule-binding domains, which are located on both termini of the protein. To circumvent this, we used an internal tagging strategy to insert GFP after amino acid 1529 between two coiled-coiled regions of CENP-F (Figure 1A). Ectopic expression of this internally tagged CENP-F construct (hereafter referred to as CENP-F^GFP) in live cells faithfully recapitulated the localization of endogenous CENP-F, as it localized to mitochondria, nucleus, nuclear envelope, and kinetochores, depending on the cell cycle, as expected (Figure 1, B and C). Moreover, overexpression of Miro1 led to a complete recruitment of CENP-F on mitochondria, as observed for the endogenous protein (Figure 1D; Kanfer et al. 2015). We previously reported by immunofluorescence in fixed cells that CENP-F localizes to EB1-labeled microtubule tips (Kanfer et al., 2015). This localization pattern was not readily visible in live cells, however, probably because it was masked by free CENP-F^GFP. We therefore turned to total internal reflection fluorescence (TIRF) microscopy to image only microtubules in proximity to the coverslip. In these conditions, we observed many CENP-F^GFP foci localizing to growing microtubules (Figure 1E). Moreover, these foci moved together with EB1 comets in the direction of microtubule growth (Figure 1F and Supplemental Video S1), indicating that CENP-F tracks growing microtubules. When imaged in fixed cells by superresolution microscopy, endogenous CENP-F localized ∼100 nm ahead of the EB1 comet (Kanfer et al., 2015). Although imaged at conventional resolution in live cells, CENP-F^GFP showed the same pattern as endogenous CENP-F, indicating a localization at the extreme tips of microtubules (Figure 1E). CENP-F tracked microtubules for shorter distances and shorter times on average than EB1, but the speed of CENP-F–positive comets was no different from that of CENP-F–negative, EB1-positive comets (Figure 1G).
FIGURE 1:
CENP-F tracks growing microtubules in vivo. (A) Top, domain organization of CENP-F. Bottom, position of the internal GFP tag (CENP-F^GFP). (B) Live images of U2OS cells coexpressing N-terminally GFP-tagged CENP-F (GFP-CENP-F, left) or internally GFP-tagged CENP-F (CENP-F^GFP, right) and a mitochondrial marker (mtBFP, red). Arrowheads show mitochondrial localization. Scale bars, 10 µm (C) Top, localization of CENP-F^GFP to the nuclear envelope (arrowheads) in early prophase. DNA is counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Bottom, localization of CENP-F^GFP to the kinetochores (arrowheads). Condensed chromatin is counterstained using an anti-phosphohistone H3 (H3P). Scale bars, 10 µm. (D) Cells overexpressing FLAG-Miro (magenta) and stained for endogenous CENP-F (left, green) or cotransfected with CENP-F^GFP (right, green) show recruitment of both endogenous and transfected CENP-F onto the mitochondrial network without any obvious localization elsewhere. Scale bars, 10 µm. (E) TIRF images of live U2OS cells coexpressing CENP-F^GFP (green) and RFP-EB1 (red). Arrowheads point at CENP-F foci found at extreme tips of growing microtubules. Scale bar, 5 µm. (F) Kymograph of the live images in E. Scale bars, 1 µm and 1 s. (G) Quantification of the distance traveled (top), lifetime (middle), and speed of comets labeled with EB1 (left) and CENP-F (right). Note that all comets bearing CENPF^GFP signal were also positive for EB1, whereas the reverse was not true.
Although we were able to visualize CENP-F^GFP on mitochondria and growing microtubule tips, we were not able to record CENP-F^GFP moving with growing microtubule tips attached to mitochondria. One possible explanation is that, although the use of TIRF microscopy is necessary to visualize microtubule-bound CENP-F, CENP-F–driven mitochondria–microtubule interactions might happen outside of the evanescent field of total internal reflection. This effect might be aggravated by the cell cycle regulation CENP-F/Miro interaction. Indeed, CENP-F is most present at the mitochondria during late mitosis (Kanfer et al., 2015), a phase when cells adopt a rounded, nonadherent morphology not compatible with TIRF imaging.
Mitochondria can be transported by dynamic microtubules in vitro
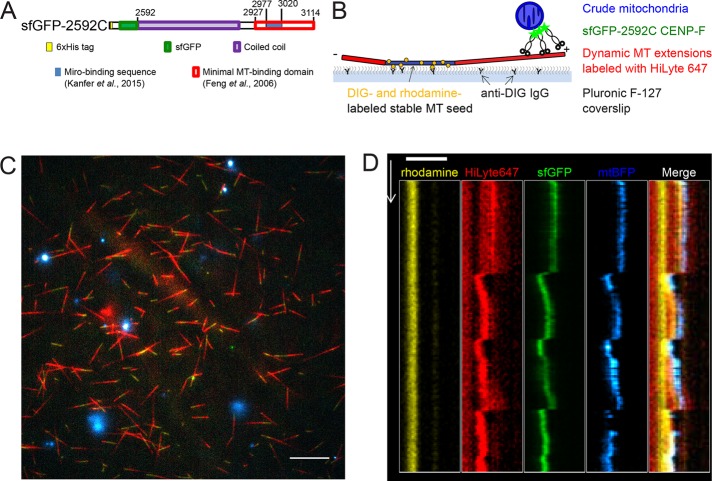
Because of these technical limitations, we asked whether mitochondria could be transported by dynamic microtubules in vitro. We prepared crude mitochondria from U2OS cells overexpressing Miro1 and a mitochondria-targeted fluorescent marker (mtBFP). These mitochondria were incubated with a previously characterized recombinant fragment of CENP-F (sfGFP-2592; Volkov et al., 2015) encompassing the Miro-binding (Kanfer et al., 2015) and C-terminal microtubule-binding domains fused to a superfolder GFP (sfGFP; Pédelacq et al., 2006) and added to dynamic microtubules. Dynamic microtubule extensions were grown on coverslips using fluorescently labeled GTP-tubulin and microtubule seeds, which were stabilized by the GTP analogue guanosine-5’-[(a,b)-methyleno]triphosphate (GMPCPP; which is essentially nonhydrolyzable within the time course of our experiments) and anchored to the coverslip via anti-digoxigenin (DIG) immunoglobulin G (IgG; Figure 2B). Using TIRF microscopy, we observed that mitochondria harboring both blue (mtBFP) and green (CENP-F-sfGFP-2592C) fluorescence bound to the microtubule extensions (Figure 2C) and moved with the tips of both growing and shortening microtubules (Figure 2D and Supplemental Video S2). Mitochondrial particles moved with shrinking microtubules over 2.4 ± 0.4 μm (n = 16, mean ± SEM) and with growing microtubules over 0.8 ± 0.1 μm (n = 11). Thus microtubule dynamics–linked mitochondrial transport can be reconstituted in an in vitro setup.
FIGURE 2:
In vitro reconstitution of mitochondria microtubule tracking using CENP-F fragment. (A) Domain organization in recombinant sfGFP-2592C fragment of human CENP-F. Numbers show amino acid position in the full-length human CENP-F. (B) Experimental setup. GMPCPP-stabilized, rhodamine- and DIG-labeled microtubule seeds are immobilized on a coverslip by anti-DIG antibodies. Dynamic microtubules are extended from these seeds by incubation with HiLyte-647–labeled tubulin and GTP. Crude mitochondria expressing a fluorescent marker (mtBFP) are incubated with recombinant sfGFP-2592C and added to the flow chamber. (C) Field of view showing dynamic microtubules (red) growing from coverslip-attached seeds (yellow) and purified BFP-mitochondria (cyan) in the presence of sfGFP-2592C (green). Scale bar, 10 µm. (D) Kymograph showing microtubule seed (yellow), dynamic microtubule extension (red), sfGFP-2592C (green), and a mitochondrial particle (blue). Scale bar, 2 µm (horizontal), 60 s (vertical).
Is CENP-F required for the phenomenon? To address this question, we used clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 engineering to generate a cell line in which a homozygous frameshift mutation interrupted CENP-F after amino acid 2991, truncating the protein within the Miro-binding domain (Supplemental Figure S1A). In these cells, the truncated CENP-F failed to localize to mitochondria (Supplemental Figure S1B). To perform the mitochondria tip-tracking assay in the absence of CENP-F, we purified crude mitochondria from these cells after Miro1 overexpression and omitted recombinant CENP-F-sfGFP-2592C. In these conditions, we could still observe mitochondria movement in concert with microtubule growth and shrinkage (Supplemental Figure S1C).
We conclude that mitochondrial movement can be coupled to microtubule dynamics in vitro but that CENP-F is not necessary for this to occur.
CENP-F can couple cargo transport to microtubule dynamics in vitro
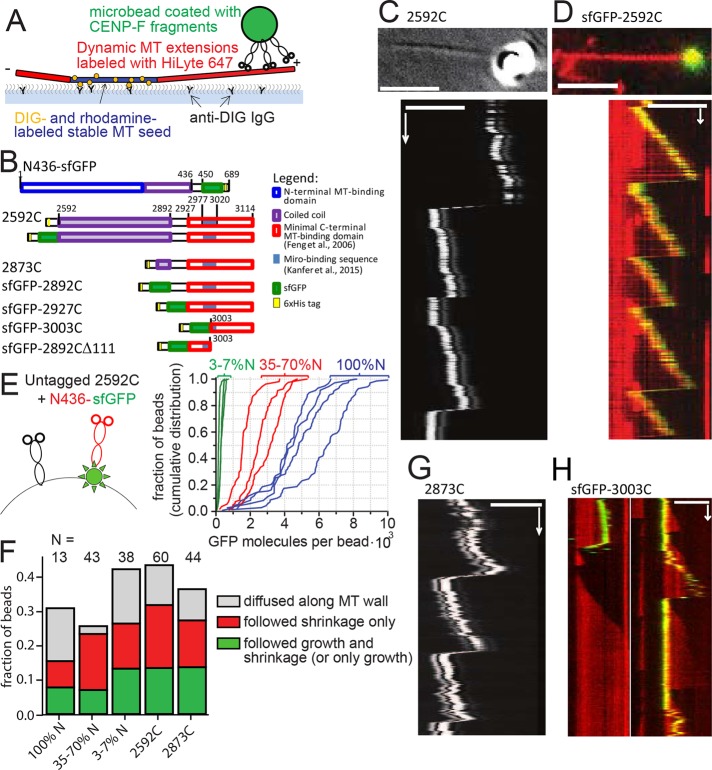
To assess whether CENP-F is sufficient to promote cargo movement on dynamic microtubules, we turned to an entirely reconstituted system in which mitochondria were mimicked by 1-μm glass microbeads coated with recombinant CENP-F fragments (Figure 3, A and B). We used a laser trap to bring beads coated with nonfluorescent 2592C fragment into the vicinity of the tips of dynamic microtubules, released the beads, and followed beads and microtubule dynamics using differential interference contrast (DIC) optics (Figure 3C). Using this assay, we could record beads following both microtubule growth and shrinkage (Supplemental Video S3). Sixty of 209 beads coated with 2592C successfully bound to microtubules (Table 1). Among the bound beads, 56% did not show any movement, 12% diffused along the microtubule lattice, and 32% moved with microtubule dynamics; 18% followed only microtubule shortening, 2% (one bead) followed only microtubule growth, and 12% stayed attached to the microtubule tip during both growth and shortening through a total of 12 catastrophe events. The ability to follow microtubule-tip dynamics over multiple cycles of growth and shortening was also observed for beads coated with GFP-tagged version of the C-terminal fragment (sfGFP-2592C), indicating that the sfGFP tag did not impede movement with microtubule growth (Figure 3D, Table 1, and Supplemental Video S4).
FIGURE 3:
Transport of CENP-F–coated beads by dynamic MT tips in vitro. (A) Experimental setup is analogous to Figure 2B, except that CENP-F–coated beads are used instead of mitochondria. (B) Domain organization of the C-terminal fragments of CENP-F used. Numbers show amino acid position in the full-length human CENP-F. (C) Top, DIC image showing a 1-µm glass bead coated with 2592C on a growing microtubule tip (averaged from 20 sequential frames). Bottom, kymograph showing movement of the same bead with the microtubule tip. (D) Top, 1-µm glass bead coated with sfGFP-2592C (green) on a dynamic microtubule labeled with HiLyte-647 (red). Bottom, kymograph showing movement of the same bead. (E) Mixtures of untagged 2592C and sfGFP-tagged N436 were added to the beads (left; not to scale). The graph shows cumulative distributions of individual bead fluorescence for three molar ratios of 2592C and N436-sfGFP: 1:10 (green) 1:1 (red), and only N436-sfGFP (blue). Intensity of a single GFP molecule was determined from the bleaching curves of individual N436-sfGFP molecules as described in Materials and Methods. (F) Fates of the beads that were successfully bound to dynamic microtubules, depending on the bead coating. (G) Kymograph showing the movement of a 1-µm glass bead coated with 2873C and attached to the tip of a dynamic microtubule by means of an optical trap; the bead and microtubule dynamics were imaged using DIC optics. (H) Beads coated with sfGFP-3003C (green) were allowed to bind spontaneously to dynamic microtubules grown using HiLyte-627–labeled tubulin (red). Images show two representative kymographs. Scale bars, 60 s (vertical); 5 µm (horizontal).
TABLE 1:
Quantification of binding and motility of beads coated with CENP-F fragments.
| Construct | 2592C (laser trap assay) | 2873C (laser trap assay) | sfGFP-2592C | sfGFP-2592C + 100 mM KCl | sfGFP-2892C | sfGFP-2927C | sfGFP-3003C | sfGFP-2892CΔ111 |
|---|---|---|---|---|---|---|---|---|
| Moved with growth | 8 | 6 | 5 | 0 | 12 | 14 | 7 | 0 |
| Moved with shortening | 18 | 12 | 21 | 1 | 34 | 14 | 9 | 2 |
| Total bound beads | 60 | 44 | 36 | 14 | 60 | 29 | 27 | 10 |
| Total beads | 209 | 164 | 97 | 233 | 358 | 300 | 567 | 424 |
| Fraction of beads binding | 0.29 | 0.27 | 0.37 | 0.06 | 0.17 | 0.1 | 0.05 | 0.02 |
Human CENP-F contains two distinct microtubule-binding sites, which bind preferentially to different tubulin oligomers (Volkov et al., 2015). To mimic a possible complementing action of N- and C-terminal microtubule-binding sites of the whole-length CENP-F, we added various mixtures of nonfluorescent 2592C (C) and N436-sfGFP (N) to the beads, resulting in the dilution of GFP signal on the beads as a function of C:N ratio (Figure 3E). No synergistic improvement of microtubule binding was observed at any concentration. Beads coated with 35–100% N436-sfGFP moved with microtubule growth in 7% of cases, and addition of more 2592C increased this fraction to 13% (Figure 3F). Thus the C-terminal 2592C CENP-F fragment alone was sufficient to provide tip-tracking ability to microbeads.
We then wondered what structural features in the C-terminal fragment of CENP-F were responsible for tip tracking. To address this point, we made a series of truncations in the construct and assessed their ability to attach beads to microtubule tips. The first feature we addressed was the long coiled-coil domain present in the 2592C fragment. Long coil-coiled domains have been shown to play important roles by positioning the cargo optimally for force transmission (Volkov et al., 2013). In the case of CENP-F, the long coiled-coil might mediate end-on attachment of the beads, allowing the growing microtubule tip to exert a pushing force on the cargo. This conformation would be incompatible with mitochondrial transport, however, because the Miro-binding domain of CENP-F is situated beyond the coiled-coil domain, proximal to the microtubule-binding domain. Beads coated with a fragment of CENP-F lacking most of the coiled-coil region (2873C, Figure 3B) could still move with microtubule growth (Figure 3G), and their behavior was indistinguishable from beads coated with 2592C (Figure 3F). Therefore coiled-coil–mediated end-on attachment of the cargo is unlikely. Consistent with this idea, the forces generated during bead movement were too small to be detected by our setup (<0.2 pN, 67 beads, 232 attempts). This low force is incompatible with an end-on attachment that allows microtubules to develop 2–4 pN of pushing force (Kerssemakers et al., 2006). We therefore conclude that CENP-F–coated microbeads are more likely to be dragged behind the growing microtubule tip than pushed in front of it.
We generated two more fragments of CENP-F, one containing the previously reported microtubule-binding domain (2927–3114), and a shorter one that did not encompass the Miro-binding domain (3003–3114). Both fragments were able to promote cargo movement with growing and shrinking microtubules, although they bound microtubules more rarely than longer fragments (Table 1). As a control, a fragment missing the terminal 111 amino acids of CENP-F (2892CΔ111) induced binding of only 2% of beads to the microtubules, and we did not observe any beads moving with microtubule growth (Table 1). Therefore microtubule-tracking properties are intrinsic to the CENP-F C-terminal microtubule-binding site; neither the coiled-coil domain nor the Miro-binding sequence is necessary for motility.
CENP-F binds tightly to the growing microtubule tip
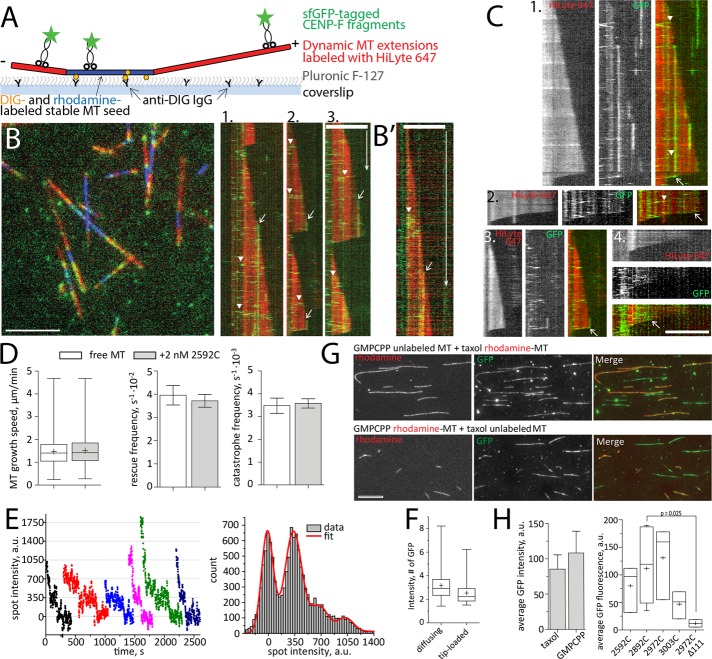
Many microtubule plus tip–tracking proteins do so by association with GTP-tubulin–binding EB proteins (reviewed in Akhmanova and Steinmetz, 2015). CENP-F, however appeared to track microtubules through a different mechanism because 1) it did not colocalize with the EB1 comet in vivo but appeared to bind microtubules closer to their tips (Kanfer et al., 2015), and 2) CENP-F–coated beads followed shrinking microtubules after the loss of the GTP-tubulin cap. This prompted us to test whether the microtubule-binding domains of CENP-F had tip-tracking capability in isolation. To assess whether single molecules or oligomers of CENP-F could follow microtubule growth and/or shrinkage, we used in vitro reconstitution and TIRF microscopy of recombinant sfGFP-2592C fragments (Figure 4A).
FIGURE 4:
Interaction of CENP-F with dynamic MTs in vitro. (A) Experimental setup. (B) Left, dynamic MT extensions (red) growing from stable MT seeds (blue) were imaged in TIRF in the presence of 2 nM sfGFP-2592C (green). Right, kymographs showing fast-diffusing molecules of sfGFP-2592C loaded along the microtubule lattice (arrowheads) and immobile spots loaded at the growing microtubule tips (arrows). (B′) A diffusing lattice-loaded complex (arrowhead) and tip-loaded immobile complex (arrow) imaged with a faster acquisition rate. (C) Kymographs showing tip-loading complexes that move with microtubule disassembly (arrows). Arrowheads indicate molecules diffusing along the microtubule lattice. (D) Microtubule growth speed, rescue frequency, and catastrophe frequency in the absence (white bars) and presence (gray bars) of 2 nM 2592C. Data are summarized from 531 growth and shrinkage events for free microtubules and 610 such events in the presence of CENP-F obtained in three independent experiments. Here and elsewhere, bar graphs show mean ± SEM, box-and-whiskers graphs show 25–75% (box), minimum and maximum (whiskers), median (horizontal line), and mean (+). (E) Bleaching curves of GFP-tagged CENP-F fragments (left) and the summary histogram of all spot intensities during photobleaching (right). The histogram is fitted with the equidistant Gaussian function (red curve). (F) Intensity of 109 immobile tip-loaded and 166 diffusing lattice-loaded molecules of sfGFP-2592C in dynamic microtubule assay. (G) Flow chambers with a mixture of unlabeled GMPCPP-stabilized and rhodamine-labeled, Taxol-stabilized microtubules (top) or vice versa (bottom) were incubated with sfGFP-2592C. The graph shows quantification of the microtubule-associated GFP fluorescence. The values are summarized from 432 Taxol-stabilized microtubules and 1018 GMPCPP-stabilized microtubules measured in four independent experiments. (H) Microtubule-associated GFP intensity in the presence of truncated fragments of CENP-F C-terminus. Data are from three independent experiments for sfGFP-2592C, sfGFP-2927C, and sfGFP-3003C; 11 experiments for sfGFP-2892C and two experiments for sfGFP-2892CΔ111. The p value is reported based on a Mann–Whitney–Wilcoxon rank sum test. Scale bars, 60 s (vertical), 5 µm (horizontal).
Using dynamic microtubules, we did not observe single CENP-F molecules moving at growing microtubule tips. Instead, we observed two distinct behaviors of CENP-F fragments on microtubules (Figure 4B): 1) some GFP-tagged molecules bound the microtubule lattice and diffused rapidly along microtubules before dissociating (Figure 4B, arrowheads), and 2) some GFP-tagged molecules loaded at the growing microtubule tip but did not follow the growth (Figure 4B, arrows). Instead, they remained stable at their loading site and stayed bound until microtubule shortening (Supplemental Video S5). As reported previously (Volkov et al., 2015), CENP-F fragments, including the tip-loading immobile spots, were occasionally able to follow microtubule shrinkage (Figure 4C, arrows). Thus individual microtubule-binding fragments of CENP-F did not appear to have intrinsic capability to follow growing microtubule tips in our in vitro system. Previous studies showed that CENP-F fragment influences microtubule dynamics (Moynihan et al., 2009; Pfaltzgraff et al., 2016). We therefore quantified the growth and shrinkage speed of microtubule in the presence of sfGFP-2592C. Consistent with our in vivo data (Figure 1F), there were no significant changes in microtubule growth rates or in the frequencies of rescues and catastrophes (Figure 4D) at nanomolar concentrations of CENP-F. Therefore CENP-F binding at the growing microtubule tip does not affect the addition of incoming tubulin dimers, suggesting that CENP-F and tubulin are not competing for the same site on microtubules.
The observed lower mobility of tip-loaded CENP-F complexes might be a result of these complexes being oligomerized and thus forming more bonds with the microtubule than the lattice-loaded ones. To test this possibility, we compared the brightness of the unbleached GFP signal of the tip-loaded complexes with the ones that loaded on the microtubule lattice. Tip-loaded complexes were not brighter than lattice-loaded complexes and were constituted of oligomers of two to eight molecules, as reported previously (Volkov et al., 2015; Figure 4, E and F), suggesting that difference in mobility was not associated with a difference in oligomer size. To test whether the nucleotide associated with tubulin played any role in the binding of CENP-F, we compared the brightness of GFP signal associated with Taxol-stabilized microtubules (mimicking GDP-tubulin) to the brightness of GFP signal on the GMPCPP-stabilized microtubules (mimicking GTP-tubulin; Figure 4, G and H). No difference was observed, suggesting that CENP-F binding to the tubulin-GTP was unlikely to result in a tighter binding of CENP-F to the GTP-tubulin cap at the growing microtubule tips. Truncated fragments (Figure 3B) bound to stabilized microtubules with similar affinities, except for the 2892CΔ111 fragment, whose binding to microtubules was barely detectable (Figure 4H).
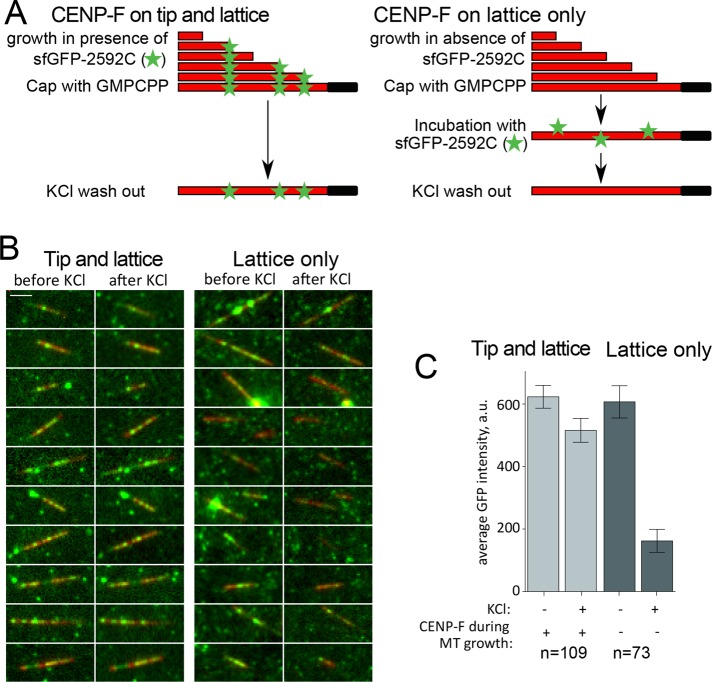
Finally, to assess further the differences in binding of lattice- versus tip-loaded complexes, we tested their stability in high-salt concentrations. To allow CENP-F loading specifically to the microtubule lattice, we grew microtubules from coverslip-attached seeds in the absence of sfGFP-2592C and stabilized them by addition of a GMPCPP-tubulin cap before washing out excess tubulin. These stable microtubules were incubated in the presence of sfGFP-2592C before being washed with 100 mM KCl (Figure 5A, lattice only). To achieve CENP-F loading on both MT tip and lattice, we used the same experimental setup but included sfGFP-2592C during the polymerization phase (Figure 5A, tip and lattice). As reported previously, 100 mM KCl removed the majority of microtubule lattice-bound complexes (Volkov et al., 2015; Figure 5, B and C, lattice only). However, if sfGFP-2592C was present during microtubule growth, KCl wash did not result in the dissociation of microtubule-bound complexes (Figure 5, B and C, tip and lattice). Thus CENP-F oligomers loaded at the growing microtubule tip formed more stable bonds with microtubules than lattice-loaded oligomers of the same size.
FIGURE 5:
Stability of lattice- and tip-loaded molecules of sfGFP-2592C in the presence of KCl. (A) Experimental setup showing the sequence of incubations to induce a mixture of lattice- and tip-binding molecules or only lattice-binding ones. Microtubules are grown in the presence (left) or absence (right) of 2 nM sfGFP-2592C and then stabilized by addition of a GMPCPP-tubulin cap. Stabilized microtubules are then incubated in the same concentration of sfGFP-2592C and washed in 100 mM KCl. The GFP signal remaining on the microtubules is then quantified. Scale bar, 5 µm. (B) Images of individual microtubules (red) grown in the presence (left) or absence of sfGFP-2592C (right) before and after KCl wash. (C) Average microtubule-associated GFP intensity for the conditions in B.
DISCUSSION
The spatial and temporal distributions of many cellular structures, including mitochondria, endoplasmic reticulum, secretory vesicles, and chromosomes, are dependent on microtubule-based transport. Although chromosome movement has long been linked to microtubule dynamics, mitochondria are believed to use microtubules as passive tracks for kinesin- and dynein-mediated motility in higher eukaryotes (Nangaku et al., 1994; Pilling et al., 2006; Birsa et al., 2013). Here we present direct evidence that dynamic microtubule tips can transport cargo by a motor-independent mechanism mediated by the C-terminal microtubule-binding domain of CENP-F; a glass bead coated with 2592C can follow both growing and shortening microtubule tips (Figure 3).
Disassembling microtubules produce a power stroke due to protofilament bending—a conformational change that allows protofilaments to pull on an attached cargo (Grishchuk et al., 2005). However, what makes a cargo move with growing microtubule tips, which are less bent and therefore structurally much less different from the wall of the microtubule? Several mechanisms have been suggested. First, the microtubule-binding molecules might have increased affinity for the GTP cap at the growing microtubule tip, as shown for the EB family (Zhang et al., 2015; Duellberg et al., 2016). Second, the cargo might be brought to the microtubule tip by the ATP-dependent motility of a kinesin domain and track the growing microtubule tip due to the activity of an additional microtubule-binding domain, as shown for the mitotic kinesin CENP-E (Gudimchuk et al., 2013).
Considering that CENP-F does not localize to EB1 comets (Kanfer et al., 2015), that the C-terminal microtubule-binding site of CENP-F does not have an increased affinity for GTP-tubulin (Figure 4H), and that molecular motors are absent in our in vitro system, the tip-tracking mechanism promoted by CENP-F appears to involve a third mechanism, more akin to the one that allows the microtubule polymerase XMAP215 to transport microbeads against an external load (Trushko et al., 2013). Indeed, the localization patterns of both CENP-F and XMAP215 at the extreme tips of microtubules are virtually indistinguishable in vivo (Nakamura et al., 2012; Kanfer et al., 2015), suggesting that both factors may bind similar determinants. Three major differences remain, however; first, XMAP215 is not known to transport any cargo; second, CENP-F fragments alone, unlike XMAP215 (Brouhard et al., 2008), cannot track growing microtubule tips, despite being preferentially loaded there; and third, unlike XMAP215, CENP-F binding does not influence microtubule dynamics.
The behavior of free CENP-F fragments in vitro is different from the behavior of full-length CENP-F in vivo. We do not know the behavior of full-length CENPF in vitro. Nonetheless, the behavior of CENP-F fragments loading onto the growing microtubule tips and staying associated there without tip tracking is unprecedented. It suggests that CENP-F microtubule-binding domains bind structurally differently to microtubule walls and tips and that this difference remains even after the tip becomes a wall. Because CENP-F fragments loaded at the growing microtubule tip were less mobile and less sensitive to high-salt wash than the lattice-loaded ones, we suggest that terminal tubulin dimers at the growing microtubule tip expose molecular interfaces that promote higher affinity of CENP-F. Diffusion of CENP-F–coated cargo along the microtubule will then be biased toward these terminal high-affinity sites, producing net movement with growing microtubule tip. The difference in behavior between free and bead-coupled CENP-F fragments suggests that CENP-F might couple organelle movement to the microtubule growth by a novel, cargo-dependent mechanism that involves collective action of multiple CENP-F molecules.
MATERIALS AND METHODS
DNA constructs
The CENP-F^GFP construct was cloned by Gap-repair cloning in yeast. First, the full-length CENP-F was amplified as two separate fragments (1–1529 and 1530–3114) from a GFP–CENP-F construct (Gurden et al., 2010) using primers 1 and 2 for 1–1529 and 3 and 4 for 1530–3114 (Table 2). These primers introduced regions homologous with yeast vector pRS426 on both termini of CENP-F. GFP was amplified from the same vector using primers 5 and 6. These primers introduced regions homologous with CENP-F fragments generated in the previous step. The yeast episomal vector pRS426 (Sikorski and Hieter, 1989) was linearized by EcoRI digestion. For homologous recombination assembly, all four DNA fragments (CENP-F 1–1529, CENP-F 1530–3114, GFP, and linearized pRS426) were cotransformed into yeast cells (BY4741) using standard procedures. Yeast cells were subsequently plated on SD–URA plates to select for successful recombination events. The resulting vector carrying full-length CENP-F^GFP was rescued from yeast and amplified in DH5α Escherichia coli strain. Finally, CENP-F^GFP was excised using BamHI and NotI digestion and cloned into pcDNA5/frt/to mammalian expression vector digested with the same enzymes. The final CENP-F^GFP construct was verified by sequencing.
TABLE 2:
Oligonucleotides used in this study.
| Number | Name | Sequence |
|---|---|---|
| 1 | fw_Cenpf1_BamHI | ccccctcgaggtcgacggtatcgataagcttgataGGATCCGCGATGAGCTGGGC |
| 2 | rev_Cenpf1 | GCTGCAAAATACTTCATCTACACTGCAC |
| 3 | fw_cenpf2 | AGTCTGCAGGAGGAGAATCTGACCAGGAAAGAAACC |
| 4 | re_cenpf2_NotI | cggccgctctagaactagtggatcccccgggctgcgcggccgcTCACTGGACCTTACAGTTCT |
| 5 | Fw_GFP | CAAACCAGTGCAGTGTAGATGAAGTATTTTGCAGCatggtgagcaagggcgagga |
| 6 | rev_GFP | GTTTCTTTCCTGGTCAGATTCTCCTCCTGCAGACTcttgtacagctcgtccatgc |
| 7 | sfGFP_fwd | GGACATATGcgtaaaggcgaagagctgt |
| 8 | sfGFP_rev | CAAGGATCCtttgtacagttcatccataccatgc |
| 9 | 2892C_fwd | GCTGGATCCcaatctaaacaagattcccgagg |
| 10 | 2892_2927_3003C_rev | TTACTCGAGTGCGtcactggaccttacagttct |
| 11 | 2927C_fwd | GCTGGATCCaataaagcttcaggcaagagg |
| 12 | 3003C_fwd | GCTGGATCcctgcgaagaacaaccatg |
| 13 | 2927CD111_fwd | GCTGGATCCcaatctaaacaagattcccgagg |
| 14 | 2927CD111_rev | TTACTCGAGTGCGtcacaggatatatgggctagtc |
| 15 | CENP-F_CRISPR1 | GAAATTAATACGACTCACTATAGGGGGATGTCAGCAAACCCTAAGTTTTAGAGCTAGAAATAGC |
| 16 | CENP-F_CRISPR1 | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
Fragments of human CENP-F were generated by PCR using 2592C fragment as the template and primers 9–14 (Table 2) inserted into pET28a vector using XhoI and BamHI sites and verified by sequencing. sfGFP was amplified by PCR using primers 7 and 8 and inserted into pET28a vector using NdeI and BamHI.
Cell culture and transfection
U2OS cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal calf serum and maintained at 80% confluency. All transfections were performed using Lipofectamine 3000 (ThermoFisher Scientific). To image EB1, cells were transfected with 0.5 µg of EB1-RFP plasmid (a gift from Tim Mitchison and Jennifer Tirnauer, Harvard Medical School; Addgene plasmid 39323). For CENP-F imaging, cells were transfected with 1.5 µg of CENP-F^GFP plasmid. Cells were analyzed 48 h posttransfection. Immunofluorescence was performed using the Ab5 antibody (Abcam) as described previously (Kanfer et al., 2015).
TIRF live imaging
CENP-F^GFP and RFP-EB1 were imaged live at 37°C using a DeltaVision OMX 3D-SIM Super-Resolution system controlled by DV-OMX software (Applied Precision) in Ring-TIRF mode. Images were captured with an ApoN 60×/1.4 numerical aperture (NA) oil TIRF, using 1.514 immersion oil. The different channels were acquired on two different cameras, which were aligned using a special camera alignment objective. Images were acquired every 50–100 ms for a maximum of 2 min. All experiments were repeated a minimum of three times, with consistent results.
Protein purification
Reagents were purchased from Sigma-Aldrich unless stated otherwise. Tubulin and CENP-F fragments were purified as described (Volkov et al., 2015). In brief, tubulin was purified from cow brains by cycles of polymerization in 0.33 M 1,4-piperazinediethanesulfonic acid (PIPES) and depolymerization in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) and 1 mM CaCl2 as described (Castoldi and Popov, 2003). Cycled tubulin was then polymerized and additionally labeled with succinimidyl esters of 5-carboxytetramethylrhodamine (ThermoFisher), HiLyte-647 (Anaspec), or DIG (ThermoFisher) as described (Hyman et al., 1991). CENP-F fragments were expressed in Rosetta cells under induction by 100 mM isopropyl-β-d-thiogalactoside for 2 h at 37°C. Cells were lysed with ultrasound or B-PER reagent (2592C and 2873C; ThermoFisher), and CENP-F fragments were purified on Ni–nitriloacetic acid (NTA)–agarose beads (Qiagen), desalted, and then applied on a prepacked HiTrap column (GE Healthcare). Protein was eluted from HiTrap columns with a linear gradient from 0.1 to 1.0 M NaCl in sodium phosphate buffer at pH 7.0 (or 7.2 for sfGFP-2892C and sfGFP-2927C). sfGFP-3003C and sfGFP-2892CΔ111 were purified using single-step Ni-NTA purification. Peak fractions were determined by SDS–PAGE, aliquoted in 30% glycerol, snap-frozen in liquid nitrogen, and stored at −80°C.
In vitro reconstitution of microtubule dynamics
Flow chambers were constructed as described previously (Volkov et al., 2014). Glass coverslips were cleaned in oxygen plasma (PlasmaCleaner PDC-32G) for 3 min at 400 mTorr and then immersed in a solution of 0.05% dichlorodimethylsilane in trichloroethylene for 1 h and washed in ethanol with sonication. Microtubule seeds were assembled by incubating 75 µM unlabeled tubulin, 17 µM DIG-labeled tubulin, and 1 mM GMPCPP (Jena) with or without addition of 5 µM rhodamine-labeled tubulin for 15 min at 35°C. Polymerized tubulin was sedimented at 25,000 × g for 15 min and resuspended in BRB-80 (80 mM PIPES, pH 6.9, 4 mM MgCl2, and 1 mM ethylene glycol tetraacetic acid [EGTA]). After assembling the chamber using double-stick tape and silanized coverslips, the coverslip surface was incubated with BRB80 supplemented with 30 µg/ml anti-DIG IgG (Roche), then 1% Pluronic F-127, and then GMPCPP-stabilized seeds diluted 1:1600. Unbound material was washed out with 10 chamber volumes of BRB80 after each step. For laser trap experiments, the motility buffer contained BRB80 supplemented with 0.4 mg/ml casein, 1 mM dithiothreitol (DTT), 12 µM unlabeled tubulin, and 1.7 mM GTP. For TIRF microscopy, this buffer additionally contained a glucose oxidase–catalase oxygen-scavenging cascade in the presence of 10 mM DTT, and 5% of tubulin was labeled with HiLyte-647. Immediately before the start of the experiment, 15–20 µl of motility buffer was warmed at 35°C for 25 s and introduced into the chamber using a syringe or peristaltic pump. All experiments were carried out at 32°C.
In vitro reconstitution of mitochondria transport by microtubules
Crude mitochondria were isolated (Wieckowski et al., 2009) from U2OS cells, which stably expressed mtBFP and were treated with CENP-F RNAi as described (Kanfer et al., 2015). The mitochondria were incubated with 50 nM sfGFP-2592C in 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, supplemented with 250 mM sorbitol and 1 mM EGTA for 40 min at 4°C, washed twice in the same buffer by centrifugation at 10,000 × g for 10 min at 4°C, and resuspended in TIRF motility buffer supplemented with 1 mM ATP. Images were acquired using a Leica DMI6000B microscope equipped with a TIRF 100×/1.47 NA oil HCX PlanApo objective and an ORCA-Flash 4.0 scientific complementary metal-oxide semiconductor (sCMOS) camera.
CRISPR targeting of CENP-F in U2OS cells
To generate CENP-F mutant cells, CRISPR/Cas9 was used to target the Miro-binding domain of CENP-F (guide RNA target sequence GGGATGTCAGCAAACCCTAAGGG). To prepare the guide RNA, a double-strand DNA template was PCR-assembled using two partially overlapping oligonucleotides (15 and 16, Table 2). The resulting DNA contains a minimal T7 promoter sequence, an invariant Cas9-scaffold sequence, and the CENP-F target sequence. DNA was transcribed in vitro using a MEGAshortscript T7 Transcription Kit and cleaned using a MEGAclear Transcription Clean-Up Kit. Cas9 RNPs were assembled by incubating 120 pmol of Cas9 protein (a kind gift from Martin Jinek, University of Zurich) with 100 pmol of guide RNA in 15 μl of Neon R buffer (Neon electroporation system, Invitrogen) at room temperature for 15 min. We harvested 200,000 cells and resuspended them in 5 μl of Neon R buffer. Cas9 ribonuclear proteins were added to the cells, and the mixture was electroporated using 10-μl Neon tips. Electroporation parameters were set to 1400 V, 15 ms and four pulses. Three days after electroporation, cells were single-cloned and further expanded. The targeted CENP-F locus was sequenced to check for mutation.
Optical trap
The optical trap was custom built using an AxioImager Z1 microscope (Carl Zeiss), a Spectra Physics 5W Nd:YaG 1064-nm trapping laser, a custom-built quadrant photodetector (QPD), a 100-mW, 830-nm Qioptiq iFlexx 2000 tracking laser, and a piezoelectric stage (Physik Instrumente P-561.3DD). Calibration, bead positioning, and force measurement were performed using software written in-house using LabVIEW 9 as described previously (Volkov et al., 2013). DIC imaging was performed using an EC Plan-Neofluar NA 1.3 objective (Zeiss) and a Cascade II electron-multiplying charge-coupled device camera (Roper Scientific).
Experiments with CENP-F–coated beads
Glass 1-µm beads coated with carboxyl groups (Bangs Laboratories) were suspended as 1% (wt/vol) by sonication and then activated with 50 mg/ml Sulfo-NHS and 50 mg/ml EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; ThermoFisher) by incubation for 30 min at 25°C in 25 mM MES, pH 5.0, supplemented with 0.05% Tween-20 (MES buffer). Unbound reagents were washed three times by sedimenting the beads for 1 min at 13,000 × g. Beads were then resuspended in 3 mg/ml streptavidin (Pierce) and incubated for 2 h at 4°C with rocking. Reaction was stopped by addition of 20 mM glycine, and the beads were washed three times in MES buffer and stored for several months with rocking. To attach 2592C, N436, and 2873C fragments, the streptavidin-coated beads (0.1% wt/vol) were functionalized with biotinylated anti-Penta His IgG (Qiagen) in phosphate-buffered saline supplemented with 2 mM DTT, 0.4 mg/ml casein, and 0.5 mg/ml bovine serum albumin, washed three times, and then incubated with the CENP-F fragment, followed by three washes in the same buffer and resuspended in motility buffer. Fragments tagged with sfGFP were attached similarly using biotinylated anti-GFP IgG (Rockland).
For experiments with nonfluorescent 2592C and 2873C fragments, individual beads were captured in a laser trap with a spring constant of (5–10) × 10-3 pN/nm and brought into the vicinity of a growing microtubule tip. The bead was then released by shutting the trapping beam. Because our laser trap is built on an upright microscope, the beads that failed to attach sank fast out of objective focus due to the bead weight. Attached beads remained close to the coverslip and displayed arc-like motion (Supplemental Video S2). Motions of the successfully attached beads were recorded every 1–2 s. In an attempt to measure microtubule pushing force acting on the beads, the successfully attached beads were then trapped once again, and bead displacement from the trap center was recorded using a QPD until microtubule disassembly as judged by DIC.
For TIRF microscopy of beads coated with sfGFP-tagged fragments, the flow chambers with coverslip-attached seeds were first incubated with tubulin in motility buffer to allow microtubule extensions, and then beads resuspended in the same buffer were introduced. The fact that our TIRF microscopes were inverted allowed the beads to sink to the chamber floor with anchored microtubules and bind to microtubules spontaneously. Images were acquired using a Nikon Eclipse Ti microscope equipped with a Plan Apochromat 100×/1.49 NA objective, a TIRF laser LU-N4 module, and an Andor iXon+ camera.
Single-molecule experiments
To test whether CENP-F was tracking microtubule growth, the TIRF motility buffer was supplemented with 0.5–5 nM sfGFP-2592C. Microtubule growth and CENP-F motility were monitored using DeltaVision OMX microscope in TIRF mode equipped with an ApoN 60×/1.49 NA oil TIRF objective and four sCMOS OMX V4 cameras. Additional experiments to quantify intensity of tip-loaded and lattice-loaded complexes were performed using a Nikon Eclipse Ti TIRF microscope (as described earlier).
CENP-F decoration of stable microtubules
To test for nucleotide preference of CENP-F binding to microtubules, DIG-labeled, rhodamine-labeled, and unlabeled tubulins were polymerized in the presence of 1 mM of GMPCPP or 10 µM Taxol for 15 min at 35°C. Microtubule polymer was separated by centrifugation at 25,000 × g and resuspended in BRB80. Taxol-stabilized, rhodamine-labeled microtubules were mixed with GMPCPP-stabilized unlabeled microtubules, or Taxol-stabilized unlabeled microtubules were mixed with GMPCPP-stabilized, rhodamine-labeled microtubules. These mixtures of microtubules were introduced into the flow chamber with silanized coverslips coated with anti-tubulin IgG (Serotec) and Pluronic F-127. After unbound microtubules were washed away, 2 nM sfGFP-2592C in BRB80 supplemented with 0.4 mg/ml casein and a glucose oxidase–catalase oxygen-scavenging cascade in the presence of 10 mM DTT was perfused through the chamber.
KCl-washout experiments
DIG-labeled, GMPCPP-stabilized microtubule seeds were attached to the silanized coverslips in the flow chamber as described. The seeds were then extended by incubation with 12 µM tubulin labeled with 5% HiLyte-647 with 1.7 mM GTP and with or without 2 nM sfGFP-2592C. After 10 min of microtubule growth, the dynamic tips were stabilized by rapidly exchanging the solution to the one with the 2.2 μM tubulin concentration but in the presence of 0.5 mM GMPCPP. All tubulins were then washed out using BRB80 with 0.4 mg/ml casein and 1 mM DTT. In the control experiment, which lacked sfGFP-2592C during microtubule growth, 2 nM sfGFP-2592C was added after removing tubulin. The chamber was then washed to remove unbound sfGFP-2592C, several microtubule-containing regions on the coverslip were imaged in GFP and HiLyte-647 channels, and positions of these regions were recorded. The chamber was then washed with the same buffer containing 100 mM KCl, and the same regions on the coverslip were imaged again.
Image and data analysis
All microscopy images were processed using ImageJ. To correct for unevenness of the background in DIC images, at the beginning of an experiment, 50–100 images in an empty chamber were collected and averaged, and a constant value of 3000 a.u. was subtracted from each pixel. The resulting average background image was subtracted from each individual DIC image acquired during this experiment. Kymographs were built using a custom ImageJ script that projects a maximum intensity across a 9-pixel-wide line. Microtubule and spot intensities were processed as described earlier (Volkov et al., 2014, 2015). Intensity of a single GFP molecule was obtained from the bleaching curves (Figure 4E) measured using picomolar concentrations of N436-sfGFP adsorbed to plasma-cleaned coverslips and imaged under conditions identical to the conditions in the experimental chambers. Frequencies of microtubule rescue and catastrophes were calculated as described in Walker et al. (1988). Frequency of catastrophes was calculated by dividing the amount of events over total time of microtubule growth. Frequency of rescue was calculated by dividing the amount of events over total time of microtubule disassembling. Data are reported as mean ± SEM unless stated otherwise. p values are reported based on the two-sample t test unless stated otherwise.
Supplementary Material
Acknowledgments
We are grateful to the members of Kornmann group for discussions, Richard McIntosh (University of Colorado) and Louis Reese (Delft University of Technology) for critical reading of the manuscript, Arkady Fradkov (ZAO Evrogen, Moscow, Russia) for excellent technical assistance with the cloning of CENP-F fragments, and Martin Jinek (University of Zurich) and Michel O. Steinmetz (Paul Scherrer Institut, Villigen) for kindly sharing reagents. Fluorescence microscopy was performed in part at the Scientific Center for Optical and Electron Microscopy of the ETH Zurich. This study was supported by grants of the Swiss National Science Foundation (PP00P3_13365 to B.K.), the European Research Council (Grant 337906-OrgaNet to B.K.), the Russian Science Foundation (Grant 16-14-00-224) to F.I.A., the Russian Fund for Basic Research (15-04-04467 to F.I.A. and 14-04-00057 to V.A.V.), and a fellowship from the Dmitry Zimin Dynasty Foundation to V.A.V.
Abbreviations used:
- CENP-F
centromere protein F
- CRISPR
clustered regularly interspaced short palindromic repeats
- DIG
digoxigenin
- EB
microtubule end-binding
- GFP
green fluorescent protein
- GMPCPP
guanosine-5’-[(a,b)-methyleno]triphosphate
- IgG
immunoglobulin G
- MT
microtubule
- mtBFP
mitochondrial blue fluorescent protein
- sfGFP
superfolder GFP
- TIRF
total internal fluorescence microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-11-0756) on July 12, 2017.
REFERENCES
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsa N, Norkett R, Higgs N, Lopez-Domenech G, Kittler JT. Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Biochem Soc Trans. 2013;41:1525–1531. doi: 10.1042/BST20130234. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- Duellberg C, Cade NI, Holmes D, Surrey T. The size of the EB cap determines instantaneous microtubule stability. Elife. 2016;5:e13470. doi: 10.7554/eLife.13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudimchuk N, Vitre B, Kim Y, Kiyatkin A, Cleveland DW, Ataullakhanov FI, Grishchuk EL. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat Cell Biol. 2013;15:1079–1088. doi: 10.1038/ncb2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Gurden MDJ, Holland AJ, van Zon W, Tighe A, Vergnolle MA, Andres DA, Spielmann HP, Malumbres M, Wolthuis RM, Cleveland DW, Taylor SS. Cdc20 is required for the post-anaphase, KEN-dependent degradation of centromere protein F. J Cell Sci. 2010;123:321–330. doi: 10.1242/jcs.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison TJ. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Kanfer G, Courthéoux T, Peterka M, Meier S, Soste M, Melnik A, Reis K, Aspenström P, Peter M, Picotti P, Kornmann B. Mitotic redistribution of the mitochondrial network by Miro and Cenp-F. Nat Commun. 2015;6:8015. doi: 10.1038/ncomms9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Li T, Zheng F, Cheung M, Wang F, Fu C. Fission yeast mitochondria are distributed by dynamic microtubules in a motor-independent manner. Sci Rep. 2015;5:11023. doi: 10.1038/srep11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, Richard McIntosh J. Minus-end-directed motion of kinesin–coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KL, Pooley R, Miller PM, Kaverina I, Bader DM. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. 2009;20:4790–4803. doi: 10.1091/mbc.E09-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Grigoriev I, Nogi T, Hamaji T, Cassimeris L, Mimori-Kiyosue Y. Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS One. 2012;7:e51442. doi: 10.1371/journal.pone.0051442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Pfaltzgraff ER, Roth GM, Miller PM, Gintzig AG, Ohi R, Bader DM. Loss of CENP-F results in distinct microtubule related defects without chromosomal abnormalities. Mol Biol Cell. 2016;27:1990–1999. doi: 10.1091/mbc.E15-12-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Trushko A, Schäffer E, Howard J. The growth speed of microtubules with XMAP215-coated beads coupled to their ends is increased by tensile force. Proc Natl Acad Sci USA. 2013;110:14670–14675. doi: 10.1073/pnas.1218053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VA, Grissom PM, Arzhanik VK, Zaytsev AV, Renganathan K, McClure-Begley T, Old WM, Ahn N, Richard McIntosh J, McIntosh JR. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J Cell Biol. 2015;209:813–828. doi: 10.1083/jcb.201408083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VA, Zaytsev AV, Grishchuk EL. Preparation of segmented microtubules to study motions driven by the disassembling microtubule ends. J Vis Exp. 2014:85, e51150. doi: 10.3791/51150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VA, Zaytsev AV, Gudimchuk N, Grissom PM, Gintsburg AL, Ataullakhanov FI, McIntosh JR, Grishchuk EL. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc Natl Acad Sci USA. 2013;110:7708–7713. doi: 10.1073/pnas.1305821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Stuurman N, Vale RD. Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc Natl Acad Sci USA. 2003;100:11424–11428. doi: 10.1073/pnas.1534703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Mancini MA, Chang KH, Liu CY, Chen CF, Shan B, Jones D, Yang-Feng TL, Lee WH. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.