Abstract
The genus Camelus is an interesting model to study adaptive evolution in the mitochondrial genome, as the three extant Old World camel species inhabit hot and low-altitude as well as cold and high-altitude deserts. We sequenced 24 camel mitogenomes and combined them with three previously published sequences to study the role of natural selection under different environmental pressure, and to advance our understanding of the evolutionary history of the genus Camelus. We confirmed the heterogeneity of divergence across different components of the electron transport system. Lineage-specific analysis of mitochondrial protein evolution revealed a significant effect of purifying selection in the concatenated protein-coding genes in domestic Bactrian camels. The estimated dN/dS < 1 in the concatenated protein-coding genes suggested purifying selection as driving force for shaping mitogenome diversity in camels. Additional analyses of the functional divergence in amino acid changes between species-specific lineages indicated fixed substitutions in various genes, with radical effects on the physicochemical properties of the protein products. The evolutionary time estimates revealed a divergence between domestic and wild Bactrian camels around 1.1 [0.58–1.8] million years ago (mya). This has major implications for the conservation and management of the critically endangered wild species, Camelus ferus.
Introduction
The genus Camelus is an interesting model to study the role of natural selection in the mitogenome as a driving force for adaptive divergence. The three extant Camelus species exist in very different habitats, each encompassing environmental pressures potentially resulting in the evolution of adaptations specific to each species. Dromedaries (Camelus dromedarius) inhabit semi-arid and hot desert regions of North and East Africa, Arabian Peninsula and southwest Asia1, 2, whereas domestic Bactrian camels (Camelus bactrianus) are distributed over central (Kazakhstan, Iran) and eastern (Russia, Mongolia, China) Asia, ranging from rocky mountains to flat and cold (semi-) deserts3–5. The wild two-humped camels (Camelus ferus) once were distributed over eastern and central Asia, adapted to arid and cold plains and hills4, 6. Today, this critically endangered wild species (IUCN 2014) can only be found in the Chinese Taklimakan and Lop Noor deserts and in the Mongolian Great Gobi Strictly Protected Area “A”, with a population census indicating as few as 2,000 individuals remain7, 8.
The ancestors of Camelus emerged on the North American continent 35–40 mya. After splitting into New World (Lamini) and Old World (Camelini) camels around 16.3 mya9, 10, the latter migrated via the Bering land bridge to the eastern hemisphere (the Old World), while the ancestors of llamas and alpacas spread to South America. Although the differentiation of the two domestic Old World camel species, dromedary and Bactrian camel, has long been established and estimated at around 4.4–8 mya9, 10, the wild two-humped camel (C. ferus) has been recognized only lately as a separate species11–14. However, a reliable estimate for the divergence time between domestic and wild Bactrian camels has been missing. Contrary to the last remaining wild relatives of the domestic Bactrian camel, the wild ancestors of the dromedary became extinct less than one millennium after the domestic form appeared (3,000–4,000 years ago), contributing to its domestication with multiple introgressions15.
Inferring the evolutionary and demographic history of wild and domestic species using mitochondrial DNA (mtDNA) has been established for a long time16. The relatively small mitogenome (16–17 kb) encodes a suite of 13 proteins, which interact with the nuclear DNA to build the oxidative phosphorylation (OXPHOS) pathway. Although these proteins are involved in key processes in the cells, the evolution of mtDNA has long been considered nearly neutral, and many studies have neglected the direct impact of positive and/or negative selection in shaping mitogenome diversity. A growing number of studies, however, favor the view that variation in the mitogenome is not selectively neutral (e.g., refs 17–21). Purifying selection has been implicated as a predominant force in shaping mitogenome evolution by limiting the accumulation of deleterious mutations at evolutionary constrained sites (e.g., cytochrome oxidase subunits (COX), NADH dehydrogenase subunits (ND), and cytochrome b (CYTB))17, 22–25.
Positive selection has been associated with shaping the mitogenome diversity in various species, such as snakes26, salmonids27, killer whales28, and across the mammalian phylogeny29, 30. Positive selection in the mitochondrial genes (e.g., COX, ATPase complexes, CYTB) has been shown to be involved in adaptation to different environmental pressures such as high altitude31–33 or temperature18, 34–36.
The most common approach in detecting selection in protein-coding genes involves estimating the rates of nonsynonymous (dN) and synonymous (dS) nucleotide substitutions and calculating the rates ratio as ω = dN/dS. In the McDonald-Kreitman (MK) test, the neutrality index (NI), or the excess of polymorphisms within species compared with the fixed substitutions between species, is used as an indicator to investigate the role of selection in shaping mitochondrial divergence between species37.
In the case of mtDNA, its high mutation rate can create intensive background noise hiding potential positively selected sites of adaptive value. Moreover, the commonly used methods such as dN/dS ratio and McDonald-Kreitman test are biased towards moderately conserved protein-coding genes38. The accurate diagnosis of individual amino acid changes with adaptive or non-adaptive effects is mathematically difficult because multiple nonsynonymous changes are required to satisfy the condition of dN > dS in a statistically significant manner39. One way to circumvent this problem is to analyze specific amino acid changes with adaptive values. Insights about selection acting on specific mitochondrial genes can be gained from branch-site tests on single codons40 and also by estimating lineage-specific deviations of dN/dS ratios in single genes41. Additional methods target the detection of positive selection at the mitochondrial level by comparing patterns of physicochemical changes of amino acids in the context of their effects on protein structure42–44. The technique of targeting changes in amino acid properties should be more sensitive than the dN/dS approach45, 46, and results in a high degree of resolution for the diagnosis of individual amino acid changes with adaptive values.
In this study, we screened for signatures of natural selection and divergence in 27 mitogenomes of the three different Old World camelid species. Specifically, we used a hierarchical approach and investigated (i) mitogenome evolution in the genus Camelus under a neutral model versus a model of natural selection; (ii) the evolution of mitochondrial protein-coding genes focusing on lineage-specific evolutionary changes; (iii) selection pressure on single sites in protein-coding genes within the Camelus lineage using a codon-based analysis; and (iv) changes in the physicochemical properties of functional proteins caused by amino acid replacements.
Finally, to gain knowledge about the evolutionary history of Old World camelids we estimated divergence times between domestic Bactrian camels and their wild, critically endangered relatives as well as between one- and two-humped camels using a Bayesian approach. Our study contributes to the understanding of evolutionary processes in Old World camels, and we provide support for the conservation and management of the critically endangered wild species Camelus ferus.
Results
Mitochondrial gene diversity and differentiation among Camelus
We analyzed 27 complete mitogenomes of Old World camels (Table S1), including three sequences from GenBank (C. dromedarius: NC_009849.1, C. bactrianus: NC_009628.2, C. ferus: NC_009629.2) to investigate signals of selection in Camelus possibly due to adaptation to different environmental regimes. With a mean read coverage of 20X [10–26X] after correcting for a maximal read coverage of 30, the alignment of all mitogenomes displayed a total length of 16,390 bp excluding repetitive regions and contained 1,347 variable sites (1,262 parsimony-informative, 85 singletons). We calculated the mitogenome diversity within each of the three Old World camel species and found higher haplotype (H d) and nucleotide diversity (π) in dromedaries (H d = 1.0, π = 0.00169) compared to domestic (H d = 0.952, π = 0.00132) and wild (H d = 0.822, π = 0.00131) Bactrian camels, respectively (Table 1). Tajima’s D47 (−2.003, P < 0.01) as well as Fu and Li’s F48 (−2.580, P < 0.02) were significantly negative in dromedaries, whereas in wild camels they were positive (2.331, P < 0.05 and 1.772; P < 0.02, respectively). For the domestic Bactrian camels we could not reject the hypothesis of selective neutrality for Tajima’s D (−0.004, P > 0.10) and Fu and Li’s F (0.298, P > 0.10) (Table 1). The tests for past population growth based on the number of pairwise nucleotide differences considering a small sample size, i.e., Ramos-Onsins and Rozas R2 statistic49, as well as Fu’s Fs (−1.206, P < 0.002) resulted in a significant expansion signal only for dromedaries (Table 1).
Table 1.
Mitogenome diversity in the three Old World camel species.
| Species | n | Lengtha (bp) | S | h | H d (SD) | k | θπ (SD) | θS (SD) | Tajima’s D | Fu and Li’s F test | Ramos-Onsins and Rozas (R2) statistic | Fu’s Fs statistic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. dromedarius | 10 | 16375 | 131 | 10 | 1.0 (0.045) | 27.644 | 0.00169 (0.00097) | 0.00283 (0.00116) | −2.00336** | −2,57981** | 0,2330 ns | −1,206** |
| C. bactrianus | 7 | 16385 | 53 | 6 | 0.952 (0.096) | 21.619 | 0.00132 (0.00028) | 0.00132 (0.00061) | −0.00364 ns | 0.29791 ns | 0,1555 ns | 1,466 ns |
| C. ferus | 10 | 16383 | 41 | 5 | 0.822 (0.097) | 21.444 | 0.00131 (0.00017) | 0.00088 (0.00038) | 2.33046* | 1.77232** | 0,2547 ns | 6,667 ns |
aAlignment excluding repetitive parts of the CR, gaps and missing data; n = number of samples; S = segregating sites; h = number of haplotypes; H d = haplotype diversity; k = average number of nucleotide differences; θπ = theta estimator based on the mean number of nucleotide differences; θS = Waterson’s theta estimator based on the number of segregating sites; SD = standard deviation values. Statistical significance: *P < 0.05; **P < 0.02, ns = not significant.
The average number of nucleotide differences between species (n) and the net nucleotide substitutions per site (Da) between the pairwise species comparisons were calculated as n = 520.9/588.9 and Da = 0.069/0.070 (dromedary vs. domestic/wild Bactrian camel) and n = 160.7 and Da = 0.018 (domestic vs. wild Bactrian camel) (Tables S2–4). We observed substantial differentiation in protein-coding genes between the pairwise comparisons of Old World camels (dromedary vs. Bactrian camel, dromedary vs. wild camel and Bactrian vs. wild camel). This resulted in 862, 874 and 223 fixed synonymous and nonsynonymous substitutions (Ds + Dn), leading to a permanent replacement (Dn) of 84, 90 and 25 amino acids, respectively (Tables S2–4). In all pairwise comparisons, net divergence (Da) varied substantially between genes; however, it was lower in the two rRNA subunits (12S, 16S) and in the concatenated tRNAs in comparison with protein-coding genes (Tables S2–4).
To investigate the heterogeneity of divergence across gene classes with different functions we examined the divergence level in protein-coding genes classified into different mitochondrial components of the electron transport system (ETS) complexes. In each pairwise comparison among the three species, we observed the highest differentiation in ETS III (Da (db) = 0.092, Da (df) = 0.099, Da (bf) = 0.023) and the lowest in ETS IV (Da (db) = 0.068, Da (df) = 0.069, Da (bf) = 0.017). The Da (bf) between domestic and wild Bactrian camels showed low differentiation among all ETS complexes (Da ETS I = 0.022, Da ETS III = 0.023, Da ETS IV = 0.017, Da ETS V = 0.018) (Table S5).
Signals of purifying selection in the genus Camelus
Mitogenome protein evolution under a neutral model versus a model of natural selection
The global MK test37 performed pairwise on the concatenated protein-coding genes (11,379 bp) in the three Old World camel species showed an excess of amino acid polymorphism in dromedary and domestic Bactrian camels (Pn/Ps = 0.21) as well as a deficiency of interspecific fixed amino acid replacements (Dn/Ds = 0.10). This resulted in a NI of 2.0 (P = 0.006) for dromedary and domestic Bactrian camels, indicative of purifying selection. A similar NI of 2.17 (P = 0.035) was detected in the pairwise comparison between domestic and wild Bactrian camels. In contrast, between dromedary and wild Bactrian camels we could not identify any signal of positive or negative selection (NI = 1.41, P = 0.244). Moreover, we compared the one- and two-humped camels, resulting in NI = 1.46 (P = 0.041) (Table S6).
Next, we investigated lineage-specific mtDNA protein evolution by performing the “extended MK test”50 for both concatenated protein-coding genes as well as each gene separately. In dromedaries, the NI indicated an excess of nucleotide polymorphism in ND6 in respect to its divergence from the domestic (NI = 13.33, P = 0.009) and wild Bactrian camels (NI = 14.66, P = 0.007), suggesting purifying selection against mildly deleterious mutations in this gene (Tables S7 and 8). In domestic Bactrian camels, we detected significant effects of purifying selection in concatenated protein-coding genes with respect to dromedaries (NI = 3.47, P = 0.001) and wild Bactrian camels (NI = 2.97, P = 0.0083) (Tables S9 and 10). The excess of replacement polymorphism was not significant in the wild Bactrian camel lineage (Tables S11 and 12). The significant results of the MK test (global and extended) indicating signals of purifying selection in the mitogenomes of the genus Camelus are summarized in Table 2.
Table 2.
Summary of the significant results of the global and extended MK tests indicating purifying selection in Camelus mitogenomes.
| Selection test | mtDNA gene | NI (P) | |
|---|---|---|---|
| dromedary vs. domestic Bactrian camel | g-MKT | PC genes | 2 (0.006**) |
| e-MKT | ND6 | 13.33 (0.009**) | |
| dromedary vs. wild Bactrian camel | e-MKT | ND6 | 14.66 (0.007**) |
| domestic Bactrian camel vs. dromedary | e-MKT | PC genes | 3.47 (0.001**) |
| domestic vs. wild Bactrian camel | e-MKT | PC genes | 2.17 (0.035*) |
| domestic vs. wild Bactrian camel | e-MKT | PC genes | 2.97 (0.008**) |
| one- vs. two-humped camels | g-MKT | PC genes | 1.48 (0.041*) |
The two-tailed P-values of Fisher’s exact tests are shown. P-values with asterisk are significant with the following significant levels *P < 0.05; **P < 0.01. PC = concatenated protein coding genes, g-MKT = global MKT, e-MKT = extended MKT, NI = neutrality index.
Evolution of mitochondrial protein-coding genes focusing on the lineage-specific evolutionary changes
To test for selective pressure acting on a particular Camelus lineage, we used a maximum likelihood (ML) approach and examined variation in the ratio of dN/dS (ω) on the codon-based alignments of ten different mammalian species including Old World camels (C. dromedarius, C. bactrianus, C. ferus), New World camels (Lama glama, L. guanicoe, Vicugna pacos, V. vicugna), Bos taurus, Pantholops hodgsonii and Homo sapiens (see Table S1 for accession numbers). In the background model, the uniform dN/dS (ω) ratio across the reconstructed phylogeny (HKY + G + I) of concatenated protein-coding genes was estimated at ω = 0.035. In the species-specific model, which was a significant improvement over the background model (likelihood ratio test [LRT], χ2 = 179.7, df = 64, P < 0.001), the highest dN/dS ratio (ω = 0.063) was estimated in wild camels followed by dromedaries (ω = 0.049) and domestic Bactrian camels (ω = 0.041) (Fig. 2). The two-humped camel lineage displayed an ω of 0.025 preceding its separation into the domestic and wild Bactrian camels.
Figure 2.
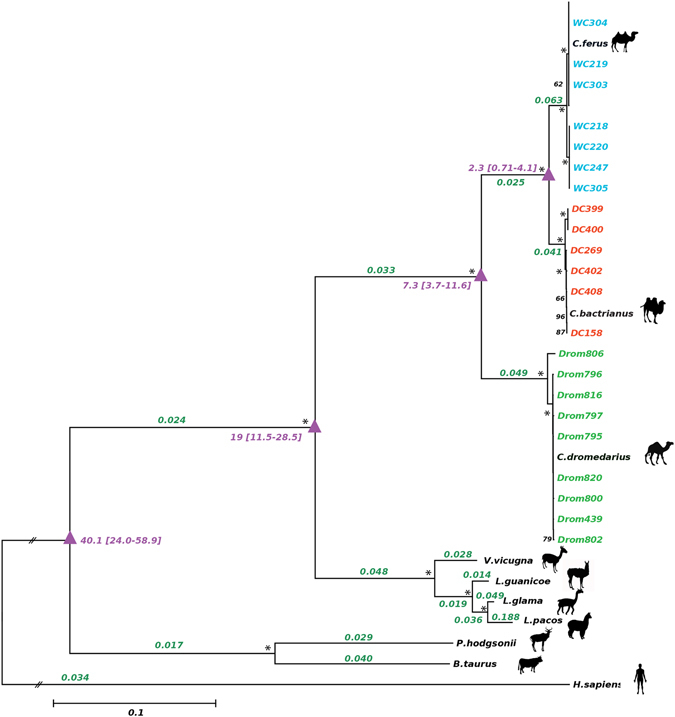
The Maximum likelihood phylogenetic tree inferred from 11,379 bp concatenated protein-coding genes from 10 different species. The HKY substitution model with a gamma distribution and proportion of invariant sites (+GI) was used to model evolutionary rate differences. Lineage-specific dN/dS values for concatenated protein-coding genes are indicated in green over each branch. The estimated divergence time (mya; using first and second protein-coding positions) of each evolutionary lineage is shown in purple color, with confidence intervals in brackets. The statistical support for each node is based on 100 bootstrap replicates, with values (in black) on nodes indicating bootstrap supports above 50%; an asterisk indicates a value of 100%. Branch lengths are scaled as the number of base pair changes per site, with the exception of the human outgroup sequence which is rescaled for aesthetic purposes. The figures depicted on the phylogenetic tree are adopted from https://openclipart.org, and are licensed under Creative Commons Zero 1.0 License.
Selection pressure on single sites in protein-coding genes within the Camelus lineage using a codon-based analysis
Using the branch-site model A (foreground lineage vs. background lineages) implemented in CODEML (PAML)51, the Bayes empirical Bayes (BEB) analysis indicated a total of 18 sites under positive selection in the entire Camelus lineage (Table S13). We detected 4 sites (in ND2, ATP6), 10 sites (in ND1, ND2, COX3, ND3, ND4, ND5) and 12 sites (in ND2, ATP8, ND4, ND5, CYTB) under positive selection in dromedaries, Bactrian camels and wild camels, respectively (Table S13). However, the significance of the detected sites could not be confirmed in the subsequent LRTs (Table S13).
Changes in the physicochemical properties of functional proteins by amino acid replacements
The analysis of functional divergence in amino acid changes was performed using TreeSAAP software52, which takes into account the magnitude of the impact of the amino acid replacement on local physicochemical properties of the protein. Changes with a magnitude ≥6 and P < 0.001 were considered as indicating positive directional selection for a given physiochemical property52. From a total of 84 (dromedary vs. Bactrian camel), 90 (dromedary vs. wild camel) and 25 (domestic vs. wild Bactrian camel) fixed nonsynonymous substitutions between Camelus species, we found evidence of positively selected sites in ATP8 (1 site), ATP6 (2 sites), ND3 (2 sites), ND5 (15 sites) and ND6 genes (4 sites) (Table S14). In these protein-coding genes, we observed 12, 14 and 12 ancestral amino acids in dromedary, domestic and wild Bactrian camels, respectively (Table S14). These amino acid changes affect physicochemical properties such as alpha-helical tendencies (P α), equilibrium constant (ionization of COOH) (pK′), polar requirement (P r), surrounding hydrophobicity (H p), power to be at the middle of alpha-helix (αm), isoelectric point (pHi), and solvent accessible reduction ratio (Ra).
Divergence time estimations among Camelidae
We used a Bayesian approach to estimate the 95% credibility intervals (CIs) of divergence times between the camelid species (Fig. 2 and Table 3). We performed five independent runs of MULTIDIVTIME53, all of which indicated good convergence (all corrected scale reduction factors = 1.0). Furthermore, the multivariate potential scale reduction factor (MPSRF)54, 55 was 1.001 and 1.01 for analyses including all sites and only first and second positions, respectively, supporting that the results across all runs had reached a stationary distribution. The mean and 95% CI for the divergence between Camelidae and Bovidae was estimated at 41.4 [24.7–60.6] mya. Within the Camelidae, the mean divergence time between the Camelini and Lamini tribes was 15.8 [CI 9.2–23.2] mya. The mean divergence between one-humped and two-humped camels occurred around 5.3 [CI 2.9–7.7] mya, and domestic and wild Bactrian camels split at about 1.1 [CI 0.58–1.8] mya. The estimates for divergence times when restricted to only the first and second codon positions were similar, albeit slightly higher than estimates from all sites (Table 3). For example, this analysis suggested the divergence between domestic and wild Bactrian camels had occurred 2.3 [CI 0.71–4.1] mya.
Table 3.
Divergence time estimates among Camelidae.
| Phylogenetic node | All sites | First and second protein-coding positions only |
|---|---|---|
| Camelidae & Bovidae | 41.4 [24.7–60.6] | 40.1 [24.0–58.9] |
| Camelini & Lamini | 15.8 [9.2–23.2] | 19 [11.5–28.5] |
| C. dromedarius & [C. bactrianus + C. ferus] | 5.3 [2.9–7.7] | 7.3 [3.7–11.6] |
| C. bactrianus & C. ferus | 1.1 [0.58–1.8] | 2.3 [0.71–4.1] |
Mean and 95% posterior credibility intervals (within brackets) of divergence time estimates for the four nodes of interest produced using MULTIDIVTIME. All values are given in millions of years ago (mya).
The mtDNA saturation analysis showed that the %Ti did not approach 50% until a sequence divergence of around 0.23 and 0.12 for all sites and first and second positions, respectively (Figure S1). The mean sequence divergence across all genes for Bovidae vs. Camelidae pairwise comparisons was 0.23 and 0.10 for all sites and first and second positions, respectively. This suggests that the Bovidae-Camelidae divergence time may be underestimated due to saturation, whereas shallower nodes are less likely to be affected.
Discussion
We investigated the evolutionary history of Old World camelids and studied the role of natural selection in shaping mitochondrial diversity. The three species are well-adapted to different environmental regimes as their habitats are spread over different altitudes and temperatures. Particularly, we were interested in estimating the divergence time between the domestic and wild Bactrian camel because a reliable estimate had not been obtained previously.
Diversity and differentiation in the mitochondrial genes among Old World camels
We observed higher haplotype and nucleotide diversity in the ten investigated dromedary mitogenomes compared to the seven domestic and ten wild Bactrian camels (Table 1), respectively. The geographically larger distribution of the dromedary samples over three continents and seven countries versus the domestic Bactrian camel specimen collected from Mongolia, Kazakhstan and Austria (with potential Mongolian origin) in principle could account for these differences. Similarly, the wild two-humped camel samples originated from one national park, the Strictly Protected Area “A” in the Mongolian Gobi desert (Table S1), a consequence of their small census size and limited geographic habitat in Mongolia and China.
During the process of domestication, population growth and dispersion of animals across a wider geographic range can be captured from molecular signals of sudden expansion56. In this study we obtained significant negative values of Tajima’s D, and Fu and Li’s F test in dromedaries (Table 1), which can indicate demographic expansion and/or positive selection. Evidence for recent population expansion in the context of dromedary domestication was confirmed by partial mitochondrial ND5, CYTB and CR analysis from over 600 individuals throughout their range15. On the contrary, the positive values of Tajima’s D and Fu and Li’s F test in the wild two-humped camels are indicative of a decrease in population size and/or balancing selection. These results are in agreement with a reduced genetic variation and small effective population size observed in wild Bactrian camels7, 57.
Overall, we calculated a mitogenome differentiation of 6.9% and 7% between dromedaries and domestic and wild Bactrian camels (Tables S2 and 3), respectively; whereas the sequence differences among two-humped camels showed 1.8% (Table S4). This corresponds to previous estimates (1.9%–2%) on partial mitochondrial genome sequences including ND5, CYTB and CR13, 14.
Investigating the heterogeneity of divergence across the different gene classes the rRNAs (12 S and 16S) and tRNAs, which are required for mitochondrial protein synthesis, were highly conserved. This is in agreement with general slow divergence rates of mitochondrial RNA genes observed in other mammals58–62. The observed rates of molecular evolution in different protein-coding genes of the ETS complexes reflected the differences in their functional constraints. In the ETS complexes the rates of molecular evolution followed the order CYTB > NDs > ATPs > COXs, with the majority of fixed differences observed in CYTB and the fewest in the COX genes. Functional modifications in the CYTB protein may be involved in physiological adaptation to different thermal environments25, 30, 36. The high variation observed in CYTB in the Camelus lineage could thus be explained with more specialized metabolic requirements in camels, like the adaptation to low-energy diet or living at higher altitudes in the case of Bactrian camels. A pattern of high variation in CYTB was also observed in other species with specialized metabolic requirements, such as the elephant, dugong, sloth and pangolin29. Higher rates of evolution in ND genes compared to the ATP and COX were reported previously in different species of insects, fish and mammals63, 64. As reported in other studies, nonsynonymous mutations in the ND genes are generally less deleterious than mutations in the COX genes, which oftentimes greatly alter functional properties of the protein and fitness24, 31, 65, 66. When we took into account the different sequence lengths of the ETS complexes, we observed the highest number of fixed nonsynonymous substitutions in the ATP genes in pairwise comparison between dromedary versus domestic and wild Bactrian. In all comparisons, the COX genes contained the lowest number of nonsynonymous substitutions, indicating their high evolutionary constraint in the Camelus lineage.
Signatures of purifying selection in the mitogenomes of Camelus
Mitogenome protein evolution under a model of natural selection
The pattern of intraspecific polymorphism in dromedary and interspecific divergence to domestic/wild Bactrian camel indicated the presence of purifying selection in the ND6 gene. This result showed that selection prevents the accumulation of slightly deleterious mutations in ND6, which is suggested to be one of the active subunits of the enzyme involved in OXPHOS pathways67. No evidence of selection was found in other protein-coding genes in dromedaries, probably a result of insensitivity of the extended MK test for the restricted number of polymorphisms and divergence in species with recent population expansion34. In domestic Bactrian camels, we did not detect signals of purifying selection in any particular gene; however, we observed a significant level of purifying selection over the concatenated protein-coding regions.
Evolution of mitochondrial protein-coding genes focusing on the lineage-specific evolutionary changes
The low dN/dS ratio estimated for the Camelus lineage (dN/dS = 0.033) indicated the effect of purifying selection on the mitochondrial protein-coding genes in this lineage. Purifying selection has been shown to be the main driver in shaping mitogenome diversity17, 22–25. However, it has been observed that domesticated species have a higher dN/dS ratio than their wild counterparts; a result possibly attributed to relaxed purifying selection during domestication (e.g., yak68, goat69). We observed this pattern in South American camelids, but contrary to this hypothesis, in wild two-humped camels we estimated slightly higher dN/dS ratio (dN/dS = 0.063) in comparison to the two domestic species (dromedaries dN/dS = 0.049; Bactrian camels dN/dS = 0.041). Wild camels are a sister species to the domestic Bactrian camels and have never been domesticated. Their original population size has been reduced over 80% during the last century and probably numbers less than 2,000 individuals7, 8. The current extremely small population size might be a reason for the relaxed pressure of purifying selection acting on the mitochondrial protein-coding genes, which could be explained by the stronger effect of genetic drift than natural selection in populations with smaller size70–73. Furthermore, compared with other domesticated ungulates, domesticated camels (dromedaries and Bactrian camels) have maintained unusually high levels of genetic variation and lack the extensive secondary bottlenecks experienced during breed formation15. The combination of the small population size in wild camels and the chance of more effective purifying selection in the domesticated species could be responsible for this result. Nonetheless, we conclude that purifying selection acts to maintain the stability and efficiency of the OXPHOS system, which are the requirements for aerobic cell respiration and energy production.
Selection pressure on single sites in protein-coding genes within Camelus
Despite the low dN/dS ratio values across the examined Camelus phylogeny indicating purifying selection, the BEB analysis of the branch-site model A recovered 18 codons potentially under positive selection (BEB > 50%, including one codon BEB > 95%) in seven genes (Table S13). However, their significance could not be further confirmed with the LRT. The BEB approach is not a direct estimator of positive selection and is used to allow sampling error in parameter estimates. Considering the small sample size and possibly large parameter estimate errors, there is very little power to detect individual sites under selection. Similarly, Matosiuk et al.58 identified 12 sites in roe deer lineage that may have been positively selected, which their significance could not be confirmed with subsequent LRT.
Changes in the physicochemical amino acid properties of functional proteins
We detected candidate positions for positive selection at fixed nonsynonymous substitutions in ATP8 (1 site), ATP6 (2 sites), ND3 (2 sites), ND5 (15 sites) and ND6 genes (4 sites) when comparing species-specific lineages (Table S14). The observed amino acid changes can affect physicochemical properties such as alpha-helical tendencies, equilibrium constant (ionization of COOH), polar requirement, surrounding hydrophobicity, power to be at the middle of alpha-helix, isoelectric point, and solvent accessible reduction ratio. The ancestral amino acid states are equally represented in dromedary, domestic and wild Bactrian camels (Table S14).
The detection of positive selection by comparing physicochemical changes of amino acids is very sensitive, as in highly conserved genes even single amino acid changes can be adaptive if they cause biochemically advantageous characterization. However, a single base change seems too weak to be identified as a positively selected site with the dN/dS ratio approach45, 46, 74, 75. As a reaction to this potential weakness in dN/dS, we applied a physicochemical property model in TreeSAAP and characterized positive destabilizing selection that correlated with seven biochemical functional shifts in local regions of the proteins. McClellan et al. (2005) argued that when this radical amino acid changes are favored by selection, they potentially result in local directional shifts in biochemical function, structure, or both, representing the unambiguous signature of molecular adaptation46. The observed changes in the amino acid properties can affect protein functions in different ways. First, we identified an increase or decrease in the alpha-helical tendency (P α) in the ND5 gene caused by five or three amino acid changes, respectively. Increasing the alpha-helical tendencies results in a longer, more rigid alpha helix, which makes interactions with other amino acid motifs more difficult. On the contrary, decreasing this property allows for a more flexible and open alpha helix, which increases the accessibility at protein interfaces76. Second, we detected an increase (eight amino acid changes) or decrease (three changes) in the equilibrium constant for the ionization of COOH (pK′) in ND6 and ND5, respectively, which make the region more or less water-soluble and hydrophilic. An increase in hydrophilicity is very important regarding a reduced reactive oxygen species (ROS) production and increased longevity during calorie restriction77. An increase in surrounding hydrophobicity (H p) property in ND3 gene, indicated that the regions surrounding these amino acid sites become less hydrophilic and more hydrophobic by introducing an isoleucine instead of a threonine.
Long-term divergence between domestic and wild Bactrian camels
Until recently, the critically endangered wild two-humped camels have been either considered to be feral (domestic runaways; reviewed in Peters & van den Driesch78), or classified as a subspecies of the domestic Bactrian camel, Camelus bactrianus ferus 12. However, earlier studies on partial mitochondrial genes indicated a high amount of differentiation (1.8–2%; refs 13, 14 and 79) between domestic and wild two-humped camels, and the International Commission of Nomenclature80 designated the species status “Camelus ferus” to the wild camel81. Genome-wide comparison between the two species confirmed their differentiation12, however, no divergence time was investigated. Here, we estimated the split between domestic Bactrian camels and their wild relatives to 1.1 [0.58–1.8] mya using all sites, and to 2.3 [0.71–4.1] mya using first and second protein-coding positions only (Table 3); long before domestication started around 4000–6000 ya82. With no mitochondrial haplotype sharing and a high number of fixed nucleotide polymorphisms (n = 233) between them, we conclude that wild camels are not only a separate species but also not the direct ancestors of contemporary domestic Bactrian camels. Our modern livestock consequently was domesticated from different ancestral populations, which had diverged from their wild sister species around 1.1–2.3 mya, and became extinct after founding the modern domestic Bactrian camel gene pool. A similar evolutionary history can be observed in the Przewalski and the domestic horse83, 84. Finally, our divergence time estimates between Camelidae and Bovidae 41.4 [24.7–60.6] mya as well as between Camelini and Lamini 15.8 [9.2–23.2] mya were in line with the latest genome-wide comparisons between the different species (42.7 [33.2–51.0] and 16.3 [9.4–25.3], respectively)9. Considering that most wild relatives of our domesticates (including llama and alpaca) are still alive, our results emphasize the evolutionary importance of Camelus ferus as the last wild representatives of the Old World camelids.
Methods
Sampling and sequencing of complete mtDNA
Genomic DNA was extract ed from whole blood of 24 specimens of Old World camelids (Table S1 and Fig. 1), using Master PureTM DNA purification kit for blood (Epicentre version III). This sampling was designed to span the known phylogenetic tree of Old World camelids, and to include mtDNA of species that are well adapted to hot, arid (nine dromedaries) and cold, dry (nine wild and six domestic Bactrian camels) environments. Blood samples were retrieved commensally either during routine veterinary treatment or during a radio-collaring project for Mongolian wild camels. All data sets were collected within the frames of the legal requirements of Austria and Mongolia. Capture and collaring of wild camels was conducted within a cooperation agreement between the International Takhi Group and the Mongolian Ministry of Nature, Environment and Tourism (signed on 15.02.2001 and renewed on 27.01.2011). The 500 bp paired-end Illumina library preparation and sequencing of each sample on a single lane of Illumina HiSeq 2000 platform was performed within the framework of a whole-genome re-sequencing study of Old World camelids (Fitak et al. in preparation). Three reference mitogenomes were retrieved from GenBank (Accession numbers NC_009849.1, NC_009628.2, NC_009629.2).
Figure 1.
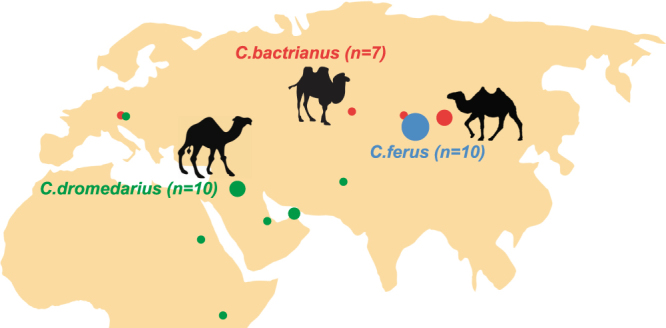
Geographical locations of the samples used in this study, including three published mitogenomes obtained from GenBank. The size of each circle is proportional to the sample size for that location. Colors correspond to the three Old World camel species, and the number of individuals per species are shown within parentheses. Detailed sample information is given in Table S1. The map and figures are adopted from https://openclipart.org and are licensed under Creative Commons Zero 1.0 License.
Read processing and alignments
The raw sequence reads for each individual were trimmed at the 3′ end to a minimum phred-scaled base quality of 20 and minimum length of 50 bp (base pairs) using POPOOLATION v1.2.285. For each species, the trimmed reads were mapped against the corresponding mitochondrial reference genome (C. dromedarius Genbank Accession no: NC_009849.1, C. bactrianus: NC_009628.2, and C. ferus: NC_009629.2), using BWA v0.786 with the following parameters (-l 200 -n 0.01 -o 1 -e 12 -d 12 -t 6). Duplicate reads were removed using Picard-tools ‘MarkDuplicates’ v.1.89 (http://www.picard.sourceforge.net) and only the filtered reads, which were properly paired and unambiguously mapped with mapping quality score >20 were used for further analyses. The consensus sequence for each sample was generated using CLC Genomics Workbench 6.5.1 (http://www.clcbio.com). The consensus mitogenomes were aligned with CodonCode Aligner v.3.7.1 (Codon Code Corporation, USA). The mitochondrial sequences of all 13 protein-coding genes, two rRNA subunits (12S, 16S) and control region of the three camel species were identified using the corresponding reference sequences of dromedary, domestic and wild Bactrian camel. The overlapping fragments of all sequences were examined to ensure complete sequence coverage. To avoid nuclear insertions of mitochondrial sequences (Numts), all amino acid sequences of protein-coding genes were screened for frameshift mutations and/or early stop codons. As additional quality control we visually screened the sequence reads for misalignments and/or large gaps using Integrative Genomics Viewer v.2.387.
Mitochondrial gene diversity and differentiation among Camelus
Summary statistics of the 27 camel mitogenomes, within and between species were calculated with DnaSP v5.10.188. Within species diversity parameters included the number of polymorphic (segregating) sites (S), number of haplotypes (h), haplotype (H d) and nucleotide (π) diversity, average number of nucleotide differences (k) and Watterson’s theta (θS) based on the number of segregating sites (Table 1). The software was also used to carry out neutrality tests including Tajima’s D47, Fu and Li’s F48, Fu’s Fs89, and Ramos-Onsins and Rozas R2 statistic49. Both D and F can detect historical population expansions and contractions, whereas Fs and R2 explicitly test past population expansion and are more powerful49. When testing for past expansions, R2 is preferred for small sample sizes (n ≤ 20) and Fs for larger sample sizes49. Significance and 95% confidence intervals of the Fs and R2 statistics were calculated with 1000 coalescent simulations. Pairwise differentiation comparisons between species comprised the average number of nucleotide differences between species (n), number of fixed synonymous (Ds) and nonsynonymous substitutions (Dn) between species, and a sliding window analysis (window size 100 bp, step size 25 bp) of the number of net nucleotide substitutions per site (Da) between species-specific lineages. The summary statistics of differentiation in pairwise comparisons between species were calculated for the protein-coding genes, rRNAs, tRNAs and the control region separately, with the overall differentiation as a sum of individual gene divergence.
Mitochondrial genome evolution and screening for natural selection
To address our main question, whether mitogenomes in the genus Camelus evolve under a model of neutrality or natural selection, we employed four different approaches (i–iv): first, we performed a global MK test37 on the concatenated protein-coding genes (11,379 bp), using DnaSP v5.10.188. We calculated the neutrality index (NI = (Pn/Ps)/(Dn/Ds); polymorphic sites/ fixed sites among species) at non-synonymous (dN) and synonymous (dS) sites in pairwise comparison between the three species. The significance of the NI values was assessed with the two-tailed Fisher’s exact test. Next, to explicitly examine which Camelus lineage had either a deficiency or excess of amino acid polymorphisms and as such might be affected by natural selection, we performed an extended MK test50 on the concatenated protein-coding genes, and each of 13 protein-coding genes separately. In the extended MK test we calculated polymorphisms within one species and the divergence (fixed SNPs) at nonsynonymous and synonymous sites in pairwise comparison.
In our second approach, we examined whether mitochondrial protein-coding genes in the Camelus lineage show any specific patterns of evolutionary rates. We assessed the variation in the ratio of dN to dS among the branches of a phylogenetic maximum likelihood (ML) tree using MEGA690, with the best-fitting model HKY + G + I according to the Bayesian Information Criterion (BIC). Using the ML method implemented in the CODEML program of PAML v.4.851, we applied the background and the branch-specific model on the 11,379 bp concatenated alignment (including gaps) of the mitochondrial protein-coding genes from ten different species (34 individuals). We included Old World (C. dromedarius, C. bactrianus, C. ferus) and New World camels (Lama glama, L. guanicoe, Vicugna pacos, V. vicugna), Bos taurus, Pantholops hodgsonii and Homo sapiens; all accession numbers are given in Table S1. While in the background model a single value of the dN/dS ratio (ω) is considered among all sites and branches, the branch-specific model allows a varying ω among the branches in the phylogeny91. We used a LRT to determine whether or not the branch-specific model was a significant improvement over the background model.
Third, we tested for selection in single sites of the protein-coding genes within the genus Camelus using the branch-site model A40 in CODEML. In this model, the ω value is allowed to vary among different branches as well as in the nucleotide sites of a particular branch (foreground lineage). We tested a null model (H0), in which the foreground branch (Camelus sp.) may have different proportions of sites under neutral selection than the background (i.e., relaxed purifying selection), and an alternative model (H1), in which the foreground branch may have a proportion of sites under positive selection. If in the alternative model a site was identified as potentially fixed due to positive selection (dN/dS > 1), its posterior probability was estimated with the Bayes empirical Bayes (BEB) approach51. For each model we retrieved the log likelihood values from which we computed the likelihood ratio tests (LRT) for the significance (P-value) of the sites detected under positive selection.
Finally, we investigated the effect of amino acid changes on the protein function, using TreeSAAP52. This program uses statistical models of molecular evolution and phylogenetic trees to predict positive selection based on amino acid physiochemical properties. TreeSAAP takes into account the magnitude of the impact of the amino acid replacements on local physiochemical properties and determines whether the observed degree of amino acid changes deviates from the neutral expectation. Radical magnitudes of changes ≥6, with P ≤ 0.001, are considered as an evidence for positive directional selection for a given physiochemical property92. To strengthen the accuracy and avoid false positive results we only used amino acid properties with the accuracy of detecting selection higher than 85% (i.e., 11 out of 31 sites were excluded) as recommended in92. In case of detecting fixed amino acid changes with effects on the physiochemical properties of a protein, we determined the ancestral state of the amino acid preceding the split of one- and two-humped camels, and domestic and wild Bactrian camels with the ML analysis in MEGA690. We constructed the phylogenetic tree for each protein coding gene in eight different species (Camelini, Lamini, and B.taurus) in MEGA as described above and used these trees as input into TreeSAAP. We restricted the sites analyzed to only the fixed nonsynonymous substitutions in pairwise comparisons in the three camel species. This analysis allowed us to investigate whether the domestic species have more amino acid changes with a predicted negative or positive impact on the respective protein due to artificial selection during domestication.
Divergence time estimation
We estimated the divergence times among Camelidae with MULTIDIVTIME53, which calculates posterior credibility intervals (CIs) of divergence times using a Bayesian approach and assuming a relaxed molecular clock with autocorrelation among rates. This method has been shown to produce posterior CIs that contain the true divergence time of simulated datasets ≥95% of the time as long as the assumption of autocorrelation among rates was not violated93. We followed the protocol outlined by Rutschmann94 for the 13 protein-coding DNA sequences. The analysis can be summarized as follows: for each gene’s alignment we determined the best-fit nucleotide substitution model and generated a ML tree topology in MEGA6 according to the procedure above. We calculated model parameters, including nucleotide frequencies, the transition/transversion rate ratio, and rate heterogeneity among sites for each gene using ‘baseml’ in PAML v4.851. Next, branch lengths and a variance-covariance matrix were estimated using the ‘estbranches’ function in MULTIDIVTIME against the ML tree topology for the 10-species dataset described above. We then ran five independent chains of MULTIDIVTIME to estimate divergence times, with each chain containing 105 burn-in steps followed by 106 steps sampled every 100th step (104 samples in total). We parameterized the MULTIDIVTIME runs using rttm = 6.5, rtrate = 0.07, and brownmean = 0.3 and with equivalent standard deviations, respectively. The time between the ingroup root and median of the tips (rttm) corresponded to 1 unit ≈ 10 million years, and was retrieved from the TimeTree database95 as the mean time of the most recent common ancestor between the families Bovidae and Camelidae (65 mya). The rate (rtrate) was calculated as rttm divided by the median branch length between the ingroup root and tips. We set brownmean to approximately 2/rttm. We included a prior to constrain the node connecting the Camelini and Lamini tribes to 8–30 mya based upon both fossil96, 97, and molecular98, 99 evidence.
Prior to estimating posterior probabilities, we assessed convergence of the independent MULTIDIVTIME chains using the R package ‘boa’ v1.1.8100 to ensure the runs reached a stationary distribution. We examined each chain and its parameters (divergence time and rate for each node) independently for convergence using the corrected scale reduction factors (CSRF) from54, 55. A chain is suggested to have converged when the CSRF is 1, or more specifically, when the 97.5% upper confidence limit of the estimate is ≤1.254. Furthermore, we also examined for convergence across all parameters and chains simultaneously using the multivariate potential scale reduction factor (MPSRF)54, 55. The MPSRF converges to 1 upon reaching stationarity when the number of steps is reasonably large. After convergence was verified, all chains were combined and the posterior density mean and 95% CIs were estimated for the parameters. We repeated the above analysis in MULTDIVTIME using only the first and second codon positions with a modified rtrate = 0.012.
Because mtDNA evolves rapidly, it is often subject to the effects of saturation (multiple mutations at a single site) – especially at deep divergence times101, 102. As a result, divergence estimates need to take into account the possible effects of saturation. To determine the potential for saturation effects in our dataset, we used the DNA saturation analysis suggested in ref. 101. Briefly, at low levels of divergence the ratio of transitions to transversions (Ti/Tv) in mtDNA is relatively high. However, as divergence increases, the chance for multiple transversions at a site increases, thus the Ti/Tv ratio approaches 1:2 when base frequencies are equal and phylogenetic information is eventually lost. Therefore, the simultaneous comparison of divergence and Ti/Tv ratio (as a proxy for saturation) serve as an indicator for phylogenetic information101. We calculated the proportion of transitions (%Ti)101 to avoid instances where no transversions occurred and compared it with pairwise sequence divergence to assess the potential for saturation. Regions of low %Ti (≤50%)101 were defined as likely affected by saturation. We performed the DNA saturation analysis separately for all sites in the protein-coding sequences and also for the first and second codon positions.
Availability of data
The complete camel mitochondrial genomes obtained in this study are deposited in Genbank with accession numbers listed below: Drom439 (Qatar): KU605072, Drom795 (Saudi Arabia): KU605073, Drom796 (Saudi Arabia): KU605074, Drom797 (Saudi Arabia): KU605075, Drom801A (Austria): KU605076, Drom802 (UAE, Dubai): KU605077, Drom806 (Kenya): KU605078, Drom816 (Sudan): KU605079, Drom820 (Pakistan): KU605080, DC158 (Austria): KU666460, DC269 (Kazakhstan): KU666461, DC399 (Mongolia): KU666462, DC400 (Mongolia):KU666463, DC402 (Mongolia): KU666464, DC408 (Mongolia): KU666465, WC214 (Mongolia): KU666451, WC216 (Mongolia): KU666452, WC218 (Mongolia): KU666453, WC219 (Mongolia): KU666454, WC220 (Mongolia): KU666455, WC247 (Mongolia): KU666456, WC303 (Mongolia): KU666457, WC304 (Mongolia): KU666458, WC305 (Mongolia): KU666459.
Electronic supplementary material
Acknowledgements
We are very grateful to the Wild Camel Protection Foundation for the support in sample collection, to W. Asaad and R. Saleh for facilitating sample collection in Syria and Jordan and to G. Gassner, First Austrian Camel Riding School, and H. Burgstaller, Graefliche Kamelheimat, for reference samples from Austria. We acknowledge general support from C. Schlötterer, Institute of Population Genetics, Vetmeduni Vienna, Austria. EM and RRF were supported by the Austrian Science Foundation (FWF) grant P24706-B25 to PAB, recipient of an APART fellowship (11506) of the Austrian Academy of Sciences. JC was partially funded by the COIN Centre of Excellence, Academy of Finland. The authors wish to acknowledge CSC – IT Center for Science, Finland, for computational resources.
Author Contributions
EM wrote the paper and performed laboratory work and bioinformatic analyses. RRF and PAB wrote the paper and performed data analysis. JC provided computational resources and revised the manuscript. AD, BC, OA, AR, PN, GS, CW, BF provided the samples and revised the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08995-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elmira Mohandesan, Email: elmira.mohandesan@univie.ac.at.
Pamela A. Burger, Email: pamela.burger@vetmeduni.ac.at
References
- 1.Bourzat D, Wilson RT. Research on the dromedary in Africa. Scientific and Technical Review. 1987;6:383–389. doi: 10.20506/rst.6.2.304. [DOI] [PubMed] [Google Scholar]
- 2.Wardeh MF. Classification of the Dromedary Camels. Journal Camel Science. 2004;1:1–7. [Google Scholar]
- 3.Faye, B. & Konuspayeva, G. The Encounter between Bactrian and Dromedary Camels in Central Asia. In: Knoll, E.-M. and Burger, P. editors. Camels in Asia and North Africa. Vienna: Interdisciplinary perspectives on their significance in past and present. Austrian Academy of Sciences Press, Wien (Austria). p. 27–33 (photos p. 248–250) (2012).
- 4.Grubb, P. Order Artiodactyla. In: Wilson, D. E., Reeder, D. M. editors. Mammal species of the world: a taxonomic and geographic reference. Baltimore: Johns Hopkins University Press. p. 637–722 (2005).
- 5.Potts D. Camel hybridization and the role of Camelus bactrianus in the Ancient Near East. Journal of the Economic and Social History of the Orient. 2004;47:143–165. doi: 10.1163/1568520041262314. [DOI] [Google Scholar]
- 6.Reading RP, Mix H, Lhagvasuren B, Blumer ES. Status of wild Bactrian camels and other large ungulates in south-western Mongolia. Oryx. 1999;33:247–255. doi: 10.1017/S0030605300030593. [DOI] [Google Scholar]
- 7.Yadamsuren, A., Dulamtseren, E. & Reading, R. P. The Conservation Status and Management of Wild Camels in Mongolia. In: Knoll, E. M., Burger, P. A. editors. Camels in Asia and North Africa. Vienna: Interdisciplinary perspectives on their significance in past and present. Austrian Academy of Sciences Press, Wien (Austria). p. 45–54 (2012).
- 8.Lei, Y., Hare, J., Guoying, Y. & Yun, C. The Status of the Wild Camel in China. In: E. M., Knoll and P. A, Burger (Eds) Camels in Asia and North Africa. Interdisciplinary perspectives on their significance in past and present. Austrian Academy of Sciences, Vienna, p.55–60 (2012).
- 9.Wu H, et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun. 2014;5 doi: 10.1038/ncomms6188. [DOI] [PubMed] [Google Scholar]
- 10.Kadwell M, et al. Genetic analysis reveals the wild ancestors of the llama and the alpaca. Proc Biol Sci. 2001;268:2575–84. doi: 10.1098/rspb.2001.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger PA. The history of Old World camelids in the light of molecular genetics. Trop Anim Health Prod. 2016;48:905–13. doi: 10.1007/s11250-016-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jirimutu., et al. Genome sequences of wild and domestic bactrian camels. Nat. Commun. 2012;3 doi: 10.1038/ncomms2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silbermayr K, et al. Isolation and characterization of nine new microsatellite loci in the domestic Bactrian camel (Camelus bactrianus) and amplification in the wild Bactrian camel (C. ferus) Mol Ecol Res. 2010;10:1106–1108. doi: 10.1111/j.1755-0998.2010.02916.x. [DOI] [Google Scholar]
- 14.Ji R, et al. Monophyletic origin of domestic bactrian camel (Camelus bactrianus) and its evolutionary relationship with the extant wild camel (Camelus bactrianus ferus) Anim. Gen. 2009;40:377–82. doi: 10.1111/j.1365-2052.2008.01848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almathen F, et al. Ancient and modern DNA reveal dynamics of domestication and cross-continental dispersal of the dromedary. Proc Natl Acad Sci USA. 2016;113:6707–6712. doi: 10.1073/pnas.1519508113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avise, J. C. Molecular Markers, Natural History, and Evolution, 2nd edn. Sinauer Associates, Sunderland, MA (2004).
- 17.Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007;23:259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst. 2005;36:621–642. doi: 10.1146/annurev.ecolsys.36.091704.175513. [DOI] [Google Scholar]
- 19.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294X.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 20.Blier PU, Dufresne F, Burton RS. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 2001;17:400–406. doi: 10.1016/S0168-9525(01)02338-1. [DOI] [PubMed] [Google Scholar]
- 21.Ballard JWO, Kreitman M. Is mitochondrial-DNA a strictly neutral marker? Trends Ecol Evol. 1995;10:485–488. doi: 10.1016/S0169-5347(00)89195-8. [DOI] [PubMed] [Google Scholar]
- 22.Kerr KC. Searching for evidence of selection in avian DNA barcodes. Mol Ecol Resour. 2001;11:1045–1055. doi: 10.1111/j.1755-0998.2011.03049.x. [DOI] [PubMed] [Google Scholar]
- 23.Rutledge LY, Patterson BR, White BN. Analysis of Canis mitochondrial DNA demonstrates high concordance between the control region and ATPase genes. BMC Evol Biol. 2010;10 doi: 10.1186/1471-2148-10-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 26.Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garvin MR, Bielawski JP, Gharrett AJ. Positive Darwinian selection in the piston that powers proton pumps in complex I of the mitochondria of Pacific salmon. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foote AD, et al. Positive selection on the killer whale mitogenome. Biol Lett. 2011;7:116–118. doi: 10.1098/rsbl.2010.0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Fonseca RR, Johnson WE, O’Brien SJ, Ramos MJ, Antunes A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishmar D, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott GR, et al. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 2011;28:351–63. doi: 10.1093/molbev/msq205. [DOI] [PubMed] [Google Scholar]
- 32.Hassanin A, Ropiquet A, Couloux A, Cruaud C. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae Antilopinae) J Mol Evol. 2009;68:293–310. doi: 10.1007/s00239-009-9208-7. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, et al. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion. 2008;8:352–357. doi: 10.1016/j.mito.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Melo-Ferreira J, et al. The elusive nature of adaptive mitochondrial DNA evolution of an arctic lineage prone to frequent introgression. Genome Biol Evol. 2014;6:886–96. doi: 10.1093/gbe/evu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva G, Lima FP, Martel P, Castilho R. Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. Proc Biol Sci. 2014;281 doi: 10.1098/rspb.2014.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontanillas P, Depraz A, Giorgi MS, Perrin N. Nonshivering thermogenesis capacity associated to mitochondrial DNA haplotypes and gender in the greater white-toothed shrew, Crocidura russula. Mol Ecol. 2005;14:661–670. doi: 10.1111/j.1365-294X.2004.02414.x. [DOI] [PubMed] [Google Scholar]
- 37.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 38.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Crandall KA, Kelsey CR, Imamichi H, Lane HC, Salzman NP. Parallel evolution of drug resistance in HIV: failure of nonsynonymous/synonymous substitution rate ratio to detect selection. Mol Biol Evol. 1999;16:372–382. doi: 10.1093/oxfordjournals.molbev.a026118. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 41.Toll-Riera M, Laurie S, Alba MM. Lineage-specific variation in intensity of natural selection in mammals. Mol Biol Evol. 2011;28:383–398. doi: 10.1093/molbev/msq206. [DOI] [PubMed] [Google Scholar]
- 42.McClellan DA, McCracken KG. Estimating the influence of selection on the variable amino acid sites of the cytochrome b protein functional domains. Mol Biol Evol. 2001;18:917–25. doi: 10.1093/oxfordjournals.molbev.a003892. [DOI] [PubMed] [Google Scholar]
- 43.Xia X, Li WH. What amino acid properties affect protein evolution? J Mol Evol. 1998;47:557–564. doi: 10.1007/PL00006412. [DOI] [PubMed] [Google Scholar]
- 44.Hughes AL, Ota T, Nei M. Positive darwinian selection promotes charge profile diversity in the antigen–binding cleft of class i major– histocompatibility–complex molecules. Mol Biol Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 45.Porter ML, Cronin TW, McClellan DA, Crandall KA. Molecular characterization of crustacean visual pigments and the evolution of pancrastacean opsins. Mol Biol Evol. 2007;24:253–268. doi: 10.1093/molbev/msl152. [DOI] [PubMed] [Google Scholar]
- 46.McClellan DA, et al. Physicochemical evolution and molecular adaptation of the cetacean and artiodactyl cytochrome b proteins. Mol Biol Evol. 2005;22:437–455. doi: 10.1093/molbev/msi028. [DOI] [PubMed] [Google Scholar]
- 47.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- 50.Messer PW, Petrov DA. Frequent adaptation and the McDonald-Kreitman test. Proc Natl Acad Sci USA. 2013;110:8615–8620. doi: 10.1073/pnas.1220835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 52.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19:671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- 53.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 54.Brooks S, Gelman A. General methods for monitoring convergence of iterative simulations. Journal of Computational and Graphical Statistics. 1998;7:434–455. [Google Scholar]
- 55.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science. 1992;7:457–511. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- 56.Bruford MW, Bradley DG, Luikart G. DNA markers reveal the complexity of livestock domestication. Nat Rev Genet. 2003;4:900–910. doi: 10.1038/nrg1203. [DOI] [PubMed] [Google Scholar]
- 57.Charruau, P. Insights from evolutionary history and population genetics for domestic and wildlife conservation – cases of the Old World camelids and cheetahs. Thesis, University of Veterinary Medicine, Vienna (Austria) (2012).
- 58.Matosiuk M, Sheremetyeva IN, Sheremetyev IS, Saveljev AP, Borkowska A. Evolutionary neutrality of mtDNA introgression: evidence from complete mitogenome analysis in roe deer. J Evol Biol. 2014;27:2483–94. doi: 10.1111/jeb.12491. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, et al. Species identification through mitochondrial rRNA genetic analysis. Sci Rep. 2014;4 doi: 10.1038/srep04089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abhyankar A, Park HB, Tonolo G, Luthman H. Comparative sequence analysis of the non-protein-coding mitochondrial DNA of inbred rat strains. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hellberg ME. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol Biol. 2006;6 doi: 10.1186/1471-2148-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilmi T, Moilanen JS, Finnilä S, Majamaa K. Sequence variation in the tRNA genes of human mitochondrial DNA. J Mol Evol. 2005;60:587–597. doi: 10.1007/s00239-003-0202-1. [DOI] [PubMed] [Google Scholar]
- 63.Ballard JW. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J Mol Evol. 2000;51:48–63. doi: 10.1007/s002390010066. [DOI] [PubMed] [Google Scholar]
- 64.Zhang F, Broughton RE. Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol. 2013;5:1781–91. doi: 10.1093/gbe/evt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blier PU, Breton S, Desrosiers V, Lemieux H. Functional conservatism in mitochondrial evolution: insight from hybridization of arctic and brook charrs. J Exp Zool B Mol Dev Evol. 2006;306:425–432. doi: 10.1002/jez.b.21089. [DOI] [PubMed] [Google Scholar]
- 66.Montooth KL, Abt DN, Hofmann JW, Rand DM. Comparative genomics of Drosophila mtDNA: Novel features of conservation and change across functional domains and lineages. J Mol Evol. 2009;69:94–114. doi: 10.1007/s00239-009-9255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brandt U. Energy converting NADH: quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 68.Colli, et al. Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability. BMC Genomics. 2015;16 doi: 10.1186/s12864-015-2342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, et al. Domestication relaxed selective constraints on the yak mitochondrial genome. Mol Biol Evol. 2010;28:15553–15556. doi: 10.1093/molbev/msq336. [DOI] [PubMed] [Google Scholar]
- 70.Furlan E, et al. Small population size and extremely low levels of genetic diversity in island populations of the platypus, Ornithorhynchus anatinus. Ecol Evol. 2012;2:844–57. doi: 10.1002/ece3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frankham, R., Ballou, J. D. & Briscoe, D. A. Introduction to conservation genetics. 2nd ed. Cambridge, U. K: Press Syndicate for the Univ. of Cambridge (2010).
- 72.Willi Y, Van, Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Ann Rev Ecol Evol Syst. 2006;37:433–458. doi: 10.1146/annurev.ecolsys.37.091305.110145. [DOI] [Google Scholar]
- 73.Wright, S. Classification of the factors of evolution. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 20, 16–24 (1955). [DOI] [PubMed]
- 74.McClellan DA. Directional Darwinian Selection in proteins. BMC Bioinformatics. 2013;14:1–8. doi: 10.1186/1471-2105-14-S13-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–73. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 76.Burkin DJ, Kim JE, Gu M, Kaufman SJ. Laminin and alpha7beta1 integrin regulate agrininduced clustering of acetylcholine receptors. J. Cell Sci. 2000;113:2877–86. doi: 10.1242/jcs.113.16.2877. [DOI] [PubMed] [Google Scholar]
- 77.Beckstead WA, Ebbert MT, Rowe MJ, McClellan DA. Evolutionary pressure on mitochondrial cytochrome b is consistent with a role of CytbI7T affecting longevity during caloric restriction. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peters J. von den Driesch. The two-humped camel (Camelus bactrianus): New light on its distribution, management and medical treatment in the past. J Zool. 1997;242:651–679. doi: 10.1111/j.1469-7998.1997.tb05819.x. [DOI] [Google Scholar]
- 79.Jianlin H, Jiexia Q, Zhenming M, Yaping Z, Wen W. Rapid communication: three unique restriction fragment length polymorphisms of EcoRI, PvuII, and ScaI digested mitochondrial DNA of Bactrian camels (Camelus bactrianus ferus) in China. J. Anim. Sci. 1999;77:2315–6. doi: 10.2527/1999.7782315x. [DOI] [PubMed] [Google Scholar]
- 80.International Commission on Zoological Nomenclature, 2003. Opinion 2027 (Case 3010): Usage of 17 specific names based on wild species which are pre-dated by or contemporary with those based on domestic animals (Lepidoptera, Osteichthyes, Mammalia): conserved. http://iczn.org/content/biodiversity-studies. Accessed on 8 Feb 2016.
- 81.Gentry A, Clutton-Brock J, Groves CP. The naming of wild animal species and their domestic derivates. J Archeol Sci. 2004;31:645–651. doi: 10.1016/j.jas.2003.10.006. [DOI] [Google Scholar]
- 82.Han, J., Quan, J. X., Han, J. L. & Men, Z. M. Genetic differentiation between Camelus bactrianus ferus and C. bactrianus inferred from mitochondrial DNA RFLPS. In: Reading RP, Enkhbileg, D., Galbaatar, T. editors. Ecology and Conservation of Wild Bactrian Camels (Camelus bactrianus ferus) Ulaanbaatar, Mongolia: Mongolian Conservation Coalition & Admon Printing. p. 65–70 (2002).
- 83.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–48. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 84.Goto H, Ryder OA, Fisher AR. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski’s horses. Genome Biol Evol. 2011;3:1096–106. doi: 10.1093/gbe/evr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kofler R, et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson JT, et al. “Integrative Genomics Viewer.”. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 89.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–25. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/PL00006320. [DOI] [PubMed] [Google Scholar]
- 92.McClellan DA, Ellison DD. Assessing and improving the accuracy of detecting protein adaptation with the TreeSAAP analytical software. Int J Bioinform Res Appl. 2010;6:120–133. doi: 10.1504/IJBRA.2010.032116. [DOI] [PubMed] [Google Scholar]
- 93.Battistuzzi FU, Filipski A, Hedges BA, Kumar S. Performance of relaxed-clock methods in estimating evolutionary divergence times and their credibility intervals. Mol Biol Evol. 2010;27:1289–1300. doi: 10.1093/molbev/msq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rutschmann, F. Bayesian molecular dating using PAML/multidivtime. A step-by-step manual. University of Zurich, Switzerland. Available at www.plant.ch/software.html (2004).
- 95.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 96.Harrison JA. Revision of the Camelinae (Arteriodactyla, Tylopoda) and description of the new genus Alforjas. Paleont. Cont. 1979;95:1–20. [Google Scholar]
- 97.Webb, S. D. Pleistocene llamas of Florida, with a brief review of the Lamini. In Pleistocene mammals of Florida (ed. Webb, S. D.). University Press of Florida, Gainesville. p. 170–214 (1974).
- 98.Cui P, et al. A complete mitochondrial genome sequence of the wild two-humped camel (Camelus bactrianus ferus): an evolutionary history of camelidae. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stanley HF, Kadwell M, Wheeler, Jane C. Molecular evolution of the family Camelidae: a mitochondrial DNA study. Proc R Soc Lond B. 1994;256:1–6. doi: 10.1098/rspb.1994.0041. [DOI] [PubMed] [Google Scholar]
- 100.Smith BJ. boa: an R package for MCMC output convergence assessment and posterior inference. Journal of Statistical Software. 2007;21:1–37. doi: 10.18637/jss.v021.i11. [DOI] [Google Scholar]
- 101.Roe AD, Sperling FAH. Patterns of evolution of mitochondrial cytochrome c oxidase I and II DNA and implications for DNA barcoding. Molecular Phylogenetics and Evolution. 2007;44:325–345. doi: 10.1016/j.ympev.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 102.Zheng U, Peng R, Kuro-o M, Zeng X. Exploring Patterns and Extent of Bias in Estimating Divergence Time from Mitochondrial DNA Sequence Data in a Particular Lineage: A Case Study of Salamanders (Order Caudata) Molecular Biology and Evolution. 2003;28:2521–2535. doi: 10.1093/molbev/msr072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


