Abstract
Genome surveillance and repair, termed the DNA damage response (DDR), functions within chromatin. Chromatin-based DDR mechanisms sustain genome and epigenome integrity, defects that can disrupt cellular homeostasis and contribute to human diseases. An important chromatin DDR pathway is acetylation signalling which is controlled by histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes, which regulate acetylated lysines within proteins. Acetylated proteins, including histones, can modulate chromatin structure and provide molecular signals that are bound by acetyl-lysine binders, including bromodomain (BRD) proteins. Acetylation signalling regulates several DDR pathways, as exemplified by the preponderance of HATs, HDACs and BRD proteins that localize at DNA breaks to modify chromatin for lesion repair. Here, we explore the involvement of acetylation signalling in the DDR, focusing on the involvement of BRD proteins in promoting chromatin remodelling to repair DNA double-strand breaks. BRD proteins have widespread DDR functions including chromatin remodelling, chromatin modification and transcriptional regulation. We discuss mechanistically how BRD proteins read acetylation signals within chromatin to trigger DDR and chromatin activities to facilitate genome–epigenome maintenance. Thus, DDR pathways involving BRD proteins represent key participants in pathways that preserve genome–epigenome integrity to safeguard normal genome and cellular functions.
This article is part of the themed issue ‘Chromatin modifiers and remodellers in DNA repair and signalling’.
Keywords: chromatin, DNA damage, genome integrity, acetylation, bromodomain
1. Introduction
Genome stability and integrity are crucial for maintaining cellular and organismal homeostasis [1,2]. To ensure the stability of the genome, cells use specialized DNA damage response (DDR) pathways that manage different types of DNA lesions by promoting DNA damage signalling and repair, activities that counteract the potentially adverse effects of DNA damage [2]. DNA double-strand breaks (DSBs) represent a particularly deleterious type of DNA lesion. An inability to repair DSBs can trigger chromosome loss and cell death, while misrepaired DSBs can lead to mutations and chromosomal aberrations such as translocations. Collectively, defects in DSB repair can result in genome instability. The significance of the DDR is highlighted by the fact that dysfunctional DDR pathways can promote human diseases including cancer, ageing and neurodegenerative disorders [2]. In mammalian cells, DSBs are repaired by two dominant pathways, canonical non-homologous end joining (c-NHEJ) and homologous recombination (HR) [3,4]. C-NHEJ involves the direct re-ligation of two broken DNA ends together without a DNA template, whereas HR engages a homologous DNA sequence as a template to repair DNA (figure 1). Alternatively, other DSB repair pathways including alternative NHEJ (alt-NHEJ) or single-strand annealing (SSA) may be used to repair DSBs although these pathways are considered more mutagenic [5,6]. How the repair of a DSB is channelled into one of these specific pathways is a fundamental question. Several factors are known to influence DSB repair choice, including cell cycle phase, DNA end resection, chromatin structure and transcription [3,7–9]. Moreover, multiple DSB repair pathways may function collectively to maximize the accuracy and efficiency of DSB repair.
Figure 1.
Acetylation signalling and DNA double-strand break repair pathways by non-homologous end joining (NHEJ) or homologous recombination (HR).
Nuclear DNA in eukaryotes is organized into chromatin [10,11], which regulates access to DNA-templated processes, including transcription, replication and repair [12,13]. Chromatin exists in highly diverse states, which can be dynamically regulated via post-translational modifications (PTM) of histones by histone-modifying enzymes and through the reorganization of nucleosomes by ATP-dependent chromatin remodellers. Histone modifications regulate chromatin structure and function by altering protein–protein and DNA–protein chromatin interactions and by changing the association of proteins containing specialized PTM binding domains within modified chromatin. Dynamic alterations in histone PTMs including phosphorylation, acetylation, methylation, ubiquitylation and ribosylation have been associated with the DDR [13–18]. We refer you to additional reviews in this special issue that provide a comprehensive view of PTMs and their reader proteins in the DDR [19,20]. Upon DNA damage, the histone variant H2AX is rapidly phosphorylated on Ser-139 in its C-terminus, forming γH2AX, by the phosphatidylinositol 3-kinase-like kinases ATM, DNA-PK and ATR [21]. This modification spreads for long distances on chromatin surrounding the damage site, which provides a binding signal for the mediator protein MDC1. γH2AX orchestrates downstream damage signalling and recruitment of DNA repair proteins to DSBs [13,22–24]. In addition to histone PTMs, several complexes from distinct chromatin-remodelling families including SWI/SNF [25–27], CHD [27–31], ISWI [27,32–34] and INO80 [35–40] localize to DNA damage sites, indicating the involvement of chromatin reorganization in DNA repair. Chromatin-remodelling complexes utilize ATP hydrolysis to regulate nucleosome dynamics, including the mobilization and eviction of nucleosomes, which can modulate the local chromatin structure to allow DDR proteins to access the lesion to promote DNA repair activities within chromatin [41,42].
Acetylation signalling represents a key pathway for the detection, signalling and repair of DSBs. Acetylation is catalysed by histone acetyltransferase (HAT) enzymes through the transfer of an acetyl group (–COCH3) to lysine residues within proteins including histones (figure 1). Acetyl groups from lysines are removed by histone deacetylase (HDAC) enzymes, allowing the dynamic regulation of this histone mark (figure 1). The evidence for the importance of acetylation in DNA damage signalling and repair of DSBs is supported by the finding that half of all mammalian HAT and HDAC enzymes are recruited to DNA damage [26,43–45]. Additionally, dynamic changes in acetylation states of histones [17,43,46,47] and non-histone proteins [47,48] are observed following DNA damage. Histone acetylation can profoundly alter chromatin structure [49] but also impacts the chromatin association of proteins that bind acetylated lysines. Several domains bind this PTM, including the YEATS and bromodomain (BRD). BRDs are found in 42 proteins in mammalian cells [50] and these proteins are often components of various chromatin-associated complexes, including ATP-dependent chromatin remodellers, HAT-containing complexes and transcriptional regulators. Numerous studies have established BRD proteins as key transcriptional and DDR factors, with around one-third being reported to respond to DNA damage [27,44]. Several BRD proteins play crucial roles in cellular responses to DNA damage, including DNA damage signalling, transcriptional regulation and DSB repair [26,27,32,33,51–53] (table 1). The significance of acetylation pathways in cancer and its therapy is underscored by the fact that acetylation modifiers and readers are frequently mutated or aberrantly expressed in cancer [44,66–68]. Chromatin remodellers in general are associated with DNA repair and transcription in cancer and ageing, topics that are specifically reviewed by Yasui and co-workers in this issue [69]. Moreover, numerous studies have determined that these pathways represent attractive targets for small molecule inhibitors either as single agents or to sensitize cancer cells to chemo- or radiotherapy as a combination therapy [44,70]. For example, HDAC inhibitors (HDACi) represent one of only a few epigenetic drugs that are FDA approved as a treatment for certain cancers. Preclinical studies have demonstrated the utility of these inhibitors in combination with DNA damaging agents, owing to the finding that HDACi alter DNA repair [43,44,71]. In addition, small molecule inhibitors targeting BRD proteins, including BRD4 and CBP/p300, have been developed and have shown promise as therapeutic agents [72–74]. Understanding how acetylation signalling and BRD proteins mediate the DDR has the potential to inform the use of drugs that modulate these pathways as pharmacological agents in cancer and other diseases.
Table 1.
Mammalian BRD proteins in DSB repair pathways.
| complexes | sub-complexes | DDR-related BRD proteins | DSB repair pathways | DNA damage recruitment | references | |
|---|---|---|---|---|---|---|
| chromatin remodellers | ISWI | CHRAC, ACF | ACF1 (BAZ1A) | NHEJ, HR | Y | [27,32,54] |
| WICH | WSTF (BAZ1B) | — | Y | [27,54] | ||
| NURF | BPTF | — | Y | [27] | ||
| SWI/SNF | PBAF | BRD7 | — | — | [44] | |
| PBAF | BAF180 (PBRM1) | NHEJ | Y | [25] | ||
| BAF | BRM (SMARCA2) | NHEJ | Y | [26] | ||
| PBAF, BAF | BRG1 (SMARCA4) | NHEJ, HR | Y | [25,55–57] | ||
| INO80 | NuA4 | BRD8 | — | — | [44] | |
| NuRD | ZMYND8 | HR | Y | [27,58] | ||
| BRD-containing HATs | p300 | NHEJ, HR | Y | [26,59,60] | ||
| CBP | NHEJ, HR | Y | [26,59,60] | |||
| SAGA, ATAC | GCN5 | — | Y | [27] | ||
| ATAC | PCAF | — | Y | [27] | ||
| others | p-TEFb | BRD4 | NHEJ | — | [61] | |
| TRIM24 | — | Y | [27] | |||
| KAP1 (TRIM28) | NHEJ, HR | Y | [62–64] | |||
| TRIM33 | — | Y | [27,65] | |||
| non-DDR-related | ASH1L, ATAD2, ATAD2B, BAZ2A, BAZ2B, BRD1, BRD2, BRD3, BRDT, BRD9, BRPF1, BRPF3, BRWD1, BRWD3, CECR2, MLL, PHIP, SP100, SP110, SP140, SP140L, TAF1, TAF1L, TRIM66, ZMYND11 | |||||
In this review, we focus on our current understanding of how BRD proteins participate in DSB repair in human cells. We hope to provide a mechanistic framework for how BRD proteins promote chromatin-based DDR processes involved in DSB repair, which could point towards further studies to define how chromatin and the DDR detect, signal and repair DSBs. Obtaining a more thorough molecular understanding of acetylation signalling and the DDR within the context of chromatin will help to illuminate how DNA-templated processes occur while maintaining both genome and epigenome integrity.
2. Bromodomain proteins and non-homologous end joining
Although NHEJ is considered to be potentially error-prone compared with HR, c-NHEJ has been shown to be the major DSB repair pathway in mammalian cells, including in S and G2 phases of the cell cycle [75,76]. As mentioned earlier, DNA end resection is one of the crucial factors that determines repair pathway choice between NHEJ and HR. 53BP1 restricts end resection and HR, while BRCA1 promotes DNA end resection and HR [3,77]. For example, HR is restored in BRCA1-deficient cells upon 53BP1 loss, indicating that 53BP1 blocks end resection [77,78]. Molecularly, 53BP1 acts as a multivalent histone PTM reader, recognizing several histone modifications including H4K20me2 and damage-induced RNF168-mediated H2AK15-Ub at DSBs [79–82]. MBTD1, a subunit of the NuA4 acetyltransferase complex, can impact 53BP1 dynamics by competing for H4K20me2 near DNA breaks [38]. In addition, TIP60 (KAT5) acetylates H4K16, as well as H2AK15, in response to DNA damage. H4K16 acetylation impairs the ability of 53BP1 to bind neighbouring H4K20me2 whereas the latter further inhibits H2AK15-Ub by RNF168 [38,46]. In the c-NHEJ pathway, the first step involves the recognition of DSBs by Ku (Ku70/80 heterodimer) at broken DNA ends, which inhibits 5′–3′ end resection [4,83]. Once Ku binds DNA ends, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is recruited, resulting in an active DNA-PK complex. This complex tethers the broken DNA ends together and serves as a scaffold for the assembly of other NHEJ factors. Finally, the two DNA ends are ligated by the DNA ligase 4-XRCC4 complex to complete repair (figure 1).
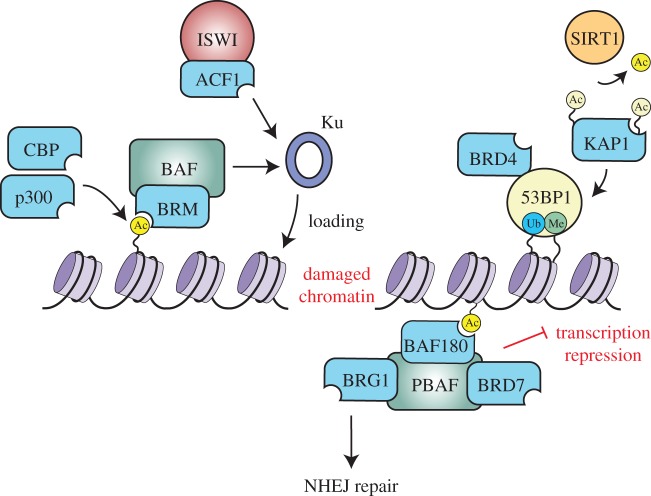
Several BRD proteins have been reported to influence c-NHEJ (table 1). The BRD protein ACF1 (BAZ1A) is an accessory subunit of the ACF and CHRAC ISWI chromatin-remodelling complexes. ACF1 accumulates at DSBs where it directly interacts with Ku and the ATPase SNF2H (SMARCA5) to facilitate their damage localization and NHEJ [32]. Depletion of ACF1 reduces NHEJ and significantly impairs Ku binding to DSBs. These observations indicate that ACF1-mediated chromatin remodelling may facilitate the recognition, and therefore repair of chromatinized DSBs by Ku [32]. Interaction between SNF2H and ACF1 increases nucleosome-remodelling efficiency [84,85], suggesting that ACF1 may interact with SNF2H at damage sites to enhance chromatin remodelling to facilitate DNA damage recognition and localization of NHEJ factors including Ku (figure 2). Although the BRD of ACF1 exhibits a binding preference towards histones H3 and H4 [84], the BRD is dispensable for damage recognition and interactions with ISWI [32]. It is conceivable that once ACF1 gains access to damaged chromatin, it then utilizes the BRD to bind acetylated substrates (i.e. histones) to facilitate repair. Alternatively, the BRD could participate in post-repair activities including the remodelling of chromatin from its damaged to repaired state or resumption of other DNA-templated processes within these chromatin regions, including transcription and replication.
Figure 2.
Acetylation signalling involving histones and BRD proteins that promote repair of DSBs by non-homologous end joining.
Human SWI/SNF chromatin-remodelling complexes include BAF and PBAF. The BAF complex contains two BRD proteins: the ATPase BRG1 (SMARCA4) or BRM (SMARCA2) and BRD9. The PBAF complex includes the BRD proteins BRG1, BAF180 (PBRM1) and BRD7. Both BRG1 and BRM are recruited to DNA damage sites [25–27,52]. BRG1 has been shown to promote the initial γH2AX formation surrounding DSBs, which requires its BRD domain to recognize GCN5-mediated H3 acetylations [52,86]. These events may influence downstream DSB repair and signalling. BRM promotes efficient c-NHEJ by facilitating the recruitment of Ku [26]. It has been speculated that BRM acts similarly to ACF1 as it can assist Ku binding by clearing chromatin obstacles at DSB ends. Multiple factors regulate the accumulation of BRM at DSB sites, which is required for the HATs p300 and CBP to acetylate H3/H4 [26]. These processes also depend on the core subunit SNF5 and the accessory subunit ARID1A or ARID1B of BAF [87], which facilitate Ku recruitment (figure 2). It is conceivable that the acetylated chromatin at DSBs modified by p300 and CBP facilitate BRM recruitment via its BRD. Furthermore, because the core subunits such as SNF5 (SMARCB1) and BAF170 stimulate the ATPase activity of BRM and BRG1 in BAF complexes [88], it is possible that other subunits of the BAF complex aid in recognizing damaged chromatin and increasing the remodelling activity for repair. The function of BRM BRD for c-NHEJ remains to be determined. The BRDs of BAF180 in the PBAF chromatin-remodelling complex are involved in transcriptional repression at DNA breaks and facilitate DNA repair by c-NHEJ [25] (figure 2). BAF180 contains six BRDs and cancer-associated BAF180 BRD mutants displayed defective DNA repair and failed to arrest transcription. BAF180 may utilize its BRDs to confine the PBAF complex to DNA breaks, thereby modulating chromatin states required for c-NHEJ. In addition to BAF180, the PBAF complex also contains two BRD proteins, BRG1 and BRD7. Identification of the targets of these BRDs could help reveal how these BRD proteins work together to respond to genotoxic stress. Whether BAF180-mediated transcription inhibition directly or indirectly affects NHEJ awaits further investigation.
Two BRD-containing HATs, p300 and CBP, are required for c-NHEJ [26] (table 1). These HATs are enriched at DSBs where they presumably participate in the DDR via acetylating histones and/or non-histone substrates. As mentioned above, these HATs can facilitate the localization of SWI/SNF complex via BRM and Ku to DSBs (figure 2). These results reveal that HATs and chromatin-remodelling complexes work in concert at DSBs to enable NHEJ factors to access damaged DNA. Chromatin-binding modules of p300 such as the BRD are important for substrate binding and target specificity. BRD binding could provide a positive feedback loop to facilitate proper HAT substrate targeting [89]. Although the function of the BRD within these HATs in c-NHEJ remains unclear, it will be interesting to determine how p300 and CBP recognize both damaged chromatin and their targets to promote c-NHEJ.
BRD4, a member of the bromodomain and extra-terminal domain (BET) family, acts as a critical regulator of transcription while also participating in DNA damage signalling and repair [51,61,90,91]. In response to DNA damage, BRD4 modulates chromatin structure by binding acetylated chromatin, thereby affecting γH2AX signalling and cell survival [51]. BRD4 also functions in repair of physiological DSBs, including class switch recombination (CSR) [61]. BRD4 is enriched in I-SceI and activation-induced cytidine deaminase (AID) induced DNA breaks. BRD4 acts as an adaptor protein to efficiently recruit key repair factors such as 53BP1 and uracil DNA glycosylase to promote CSR, which requires NHEJ (figure 2). Importantly, the BRD of BRD4 is required for recognition of the acetylated chromatin containing γH2AX, highlighting the importance of the BRD in DNA repair [51]. As BRD4 not only interacts with repair proteins but also forms the transcriptional complex with the positive transcription elongation factor (p-TEFb) [90,91], it will be interesting to understand how BRD4 orchestrates the binding of DNA repair and transcriptional regulators to promote CSR.
The transcriptional regulator KAP1 (TRIM28) contains a BRD and is an ATM kinase substrate involved in heterochromatin repair [92–94]. The BRD of KAP1 does not exhibit acetylated lysine binding [95], consistent with its functioning in heterochromatin, a chromatin state associated with low histone acetylation levels compared with euchromatin. KAP1 has also been shown to be the substrate of the deacetylase SIRT1 upon DNA damage, including an acetylation site within the BRD. It has been proposed that deacetylated KAP1 promotes NHEJ by enhancing its binding to 53BP1 and then stabilizing 53BP1 foci at DSB sites [62] (figure 2). The HAT that acetylates KAP1 and the precise function of its acetylation have yet to be elucidated. As many of these studies involving BRD proteins in NHEJ are independent from acetylation of core NHEJ factors, it is tempting to speculate that much of this regulation occurs at the level of chromatin.
3. Bromodomain proteins in homologous recombination repair
HR is a predominantly error-free DSB repair pathway that is restricted to the S/G2 phases of the cell cycle, a phase when a homologous template is readily available [3,8]. Repairing DSBs by HR requires multiple steps that have been comprehensively reviewed elsewhere [9,96] (figure 1). Briefly, HR is initiated by the MRE11-RAD50-NBS1 (MRN) complex, which detects the break and starts DNA end resection through the nuclease activity of MRE11 [97]. Following recruitment of multiple nucleases, including CtIP and EXO1, DNA resection generates 3′ single-stranded (ss) DNA overhangs [98,99]. The ensuing ssDNA overhangs are bound by the single-strand binding protein RPA, which is subsequently replaced by RAD51. RAD51-bound nucleoprotein filaments search and invade homologous sequences, leading to strand exchange and DNA synthesis to copy the sequence from an undamaged template. Finally, resolution of HR-mediated structures and ligation of the DNA strands complete the repair event [96]. Accumulating evidence suggests that mammalian chromatin-remodelling complexes as well as BRD proteins are involved in various stages of HR.
(a). Histone acetyltransferase in homologous recombination
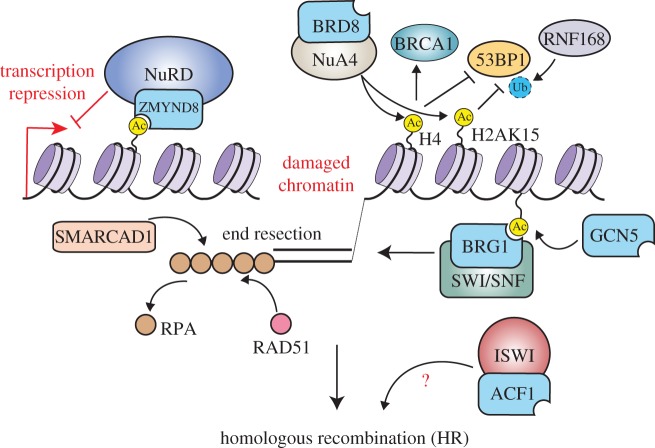
Four mammalian BRD-containing HATs (p300, CBP, GCN5 and PCAF) are recruited to DNA damage sites and regulate DSB repair [44] (table 1). Unlike BRD proteins in NHEJ [26,45], how these four HATs are involved in HR is unclear, although perturbation of acetylation signalling has been found to impact HR [45]. Currently, it is unknown if these HATs facilitate HR exclusively through their catalytic activities or also through their acetylation-binding abilities afforded by their BRDs. As HATs are well-established transcriptional activators [100], it is not surprising that defects in these enzymes can affect the transcription of DNA repair genes. For example, knockdown of p300 and CBP results in reduced mRNA and protein levels of key HR factors including BRCA1 and RAD51, with a concomitant reduction of HR repair [59]. p300 and CBP can form a complex with RBBP4, a factor known to directly bind and regulate the promoter of the RAD51 gene [60]. Mammalian GCN5 regulates the DDR and different types of repair, including nucleotide excision repair, through triggering acetylation on H3 [17,86,101]. In response to DSBs in mammalian cells, GCN5 promotes H3 acetylations for DSB repair [17,86]. Furthermore, ionizing-radiation (IR)-induced γH2AX was shown to stimulate GCN5-mediated acetylation on several H3 residues in γH2AX-containing nucleosomes. These H3 acetylation signals have been proposed to provide damage-induced docking sites for the BRD factor BRG1, the key ATPase of the SWI/SNF chromatin-remodelling complex [86] (figure 3). Consistent with these interactions being functional, siRNA-depletion of GCN5, as well as mutating the BRD of BRG1, results in radiosensitivity and defective DSB repair [86]. Although it is clear that BRD-containing HATs, including GCN5, are important for DSB repair, future investigations are necessary to delineate their DNA repair and transcriptional activities to fully understand their involvement in repairing DSBs.
Figure 3.
Roles of BRD-protein-containing chromatin-remodelling complexes in DNA damage signalling and repair by homologous recombination.
(b). Chromatin-remodelling complexes in homologous recombination
In addition to BRD-containing HATs, another group of BRD proteins involved in HR repair are ATP-dependent chromatin-remodelling complexes. All four major mammalian chromatin-remodeller families (SWI/SNF, CHD, ISWI and INO80) can promote HR repair [44]. In this review, we focus on the DDR functions of BRD proteins in these complexes. We refer readers to an additional review in this issue that comprehensively covers SMARCA- and CHD-type chromatin remodellers in the DDR [102].
(i). SWI/SNF
The core ATPase subunit of the SWI/SNF complex, BRG1, is broadly involved in DSB repair [86]. Based on the I-SceI-mediated DSB repair reporter assays DR-GFP (HR) or EJ5 (NHEJ), cells depleted of BRG1 show strong defects in HR although a reduction in NHEJ was also observed [55,56]. Qi et al. reported that loss of BRG1 caused retention of RPA-bound single-stranded DNA (ssDNA) and therefore less RAD51 binding to damaged sites. In addition, an interaction between BRG1 and the HR factor RAD52 was identified and shown to be involved in the assembly on RAD51 onto ssDNA, suggesting a role for BRG1 late in HR after DNA end resection [55]. BRG1 may function in an early step of HR too [56], because BRG1-depletion resulted in decreased chromatin-bound RPA, a phenotype consistent with impaired end resection. This study also identified the tumour suppressor retinoblastoma (RB) as a mediator for the damage recruitment of BRG1 to laser damage [56]. Taken together, these studies highlight the importance of BRG1 in HR repair. However, it is unclear whether the DDR functions of BRG1 rely on the SWI/SNF complex, as other factors within this complex are poorly characterized in the DDR. Considering the BRD domain of BRG1 is required for its damage recruitment [86], it will be important to determine whether BRD-dependent acetylation recognition promotes HR. This is especially germane given that the ability to detect visible accumulation of proteins in foci by microscopy is not always a reliable readout of function.
The SWI/SNF PBAF contains multiple BRD proteins [44], including BAF180, BRD7 and BRG1. While BAF180 promotes NHEJ together with BRG1 [25], whether any of these PBAF specific BRD proteins influence the HR function of BRG1 is unknown. SMARCAD1 is an SWI/SNF-like remodeller protein, which is classified outside this family of remodellers [103]. As shown for the yeast homologue Fun30, mammalian SMARCAD1 promotes end resection and HR repair, because SMARCAD1 loss reduces ssDNA formation and RPA binding at laser and IR-induced damage [104,105] (figure 3). In addition, mass spectrometry analysis identified a SMARCAD1 complex containing the BRD protein KAP1 that is involved in replication [106]. KAP1 is phosphorylated at DSBs by ATM at Ser824 to promote its function in DSB repair in heterochromatin [92,107]. KAP1 Ser473p regulates BRCA1 foci formation in response to the DSB-inducing agent etoposide, suggesting KAP1 may function in HR repair [63]. Conversely, KAP1 has also been proposed to repress recombination at transcriptional sites that it regulates, including ZNF genes, which may be linked to its involvement in chromosome compaction [53].
(ii). CHD
The CHD family of ATP-dependent chromatin remodellers are widely involved in the DDR [28,31]. The best-characterized complex in the mammalian CHD family is the nucleosome-remodelling and deacetylase (NuRD) complex, which regulates transcription, chromatin assembly and DNA repair [31,108]. The NuRD complex contains two histone deacetylases, HDAC1 and HDAC2, which have been linked to chromatin compaction and transcription repression [109]. NuRD also interacts with the BRD protein ZMYND8 (RACK7) to perform various cellular functions [27,58,110,111]. Numerous studies have reported the recruitment of NuRD, including its core ATPase CHD4 (Mi2β) and accessory factor ZMYND8, to DNA damage sites and involvement in HR [27,29,30,58,111–114]. Both ZMYND8 and CHD4 exhibit rapid but transient accumulations at damage sites, an involvement in very early steps of HR. ZMYND8, along with NuRD, was shown to be recruited to actively transcribed chromatin through the BRD of ZMYND8, which recognizes TIP60-mediated acetylations. ZMYND8-NuRD suppresses transcription to facilitate HR repair [27] (figure 3). These data are consistent with the reports that DSBs occurring in actively transcribing chromatin are specifically repaired by HR [7,115]. The C-terminal MYND domain of ZMYND8 interacts directly with the PPPLϕ of p66α (GATAD2A) within NuRD [58]. The PPPLϕ motif is found in p66α but not p66β (GATAD2B), which both constitute mutually exclusive NuRD sub-complexes, suggesting that ZMYND8 only regulates the damage recruitment of p66α-containing NuRD. Interestingly, a variant of ZMYND8 was shown to be recruited to damage sites independently from the BRD. This suggests that different ZMYND8 isoforms may utilize different damage recruitment mechanisms [58]. Another study provided more structural information about the chromatin recognition of ZMYND8 by its N-terminal PHD/BRD/PWWP domains [111]. Based on structural and peptide binding assays, this domain module could function as a stable unit to recognize multiple histone PTMs, including H3K14ac [111,116]. Furthermore, in addition to these chromatin-binding modules promoting damage recruitment, an additional DNA-binding region within the PHD/BRD/PWWP domain of ZMYND8 was also identified and shown to participate in damage localization [111].
ZMYND8-NuRD represents an important complex involved in HR repair as ZMYND8-NuRD deficiency results in defective RAD51 loading and low HR repair efficiency by DR-GFP report assay, although the exact step of HR that is affected by ZMYND8-NuRD loss is still unclear [27,58]. CHD4 also regulates RNF168-mediated ubiquitylation and downstream BRCA1 assembly upon damage [29]. CHD4 physically interacts with the HR factor BRIT1 and the HAT p300 to promote loading of downstream HR factors, including BRCA1, RPA and RAD51 [57,117]. The NuRD complex is also recruited to telomeres by ZNF827 to promote HR at ALT telomeres [113]. Taken together, these data suggest that the NuRD complex may regulate HR repair at several stages including by preparing the chromatin for DSB repair by repressing transcription and also by facilitating the recruitment of HR factors. Furthermore, both ZMYND8 and NuRD require the activity of the histone lysine demethylase KDM5A, as well as PARP signalling, for their damage recruitment [30,58,114,118]. How PARP promotes the recruitment of KDM5A-ZMYND8-NuRD is unclear. PARP inhibitors (PARPi) have been shown to efficiently kill HR-deficient cancer, although this appears to be due to several mechanisms, including trapping of the PARP protein at breaks sites [119–121]. Consistent with ZMYND8 and NuRD functioning in HR, cells depleted of ZMYND8 or other NuRD components are sensitive to PARPi [58,117]. It will be interesting to test whether PARPi can be used to treat cancers harbouring ZMYND8 or NuRD mutations, which are commonly found in various cancer types [108,122].
(iii). ISWI
Mammalian ISWI chromatin-remodelling complexes contain BRD proteins and participate in the DDR [42,44,123]. As the ATPase among five different mammalian ISWI complexes (WICH, NORC, RSF, ACF and CHRAC; table 1) [123], SNF2H was found to promote NHEJ and HR [32,123–125]. SNF2H functions as a downstream factor of RNF20-mediated H2B ubiquitylation and depletion of SNF2H results in defective RPA, BRCA1 and RAD51 foci formation in response to IR [125]. Supporting a role in HR, SNF2H is recruited to DNA damage together with RNF168 in a PARP-dependent manner to promote γH2AX ubiquitylation and BRCA1 accumulation [124]. As a subunit of the ISWI ACF and CHRAC complexes, the BRD protein ACF1 is also recruited to DNA damage sites and promotes both NHEJ and HR repair [32]. Although depletion of ACF1 reduces HR in the DR-GFP HR assay, how ACF1 promotes HR is still unclear [32]. It will be interesting to determine the interactors of ACF1 and its BRD domain to elucidate their HR functions, including through SNF2H and/or a particular ISWI complex. The ACF1 paralogue WSTF (BAZ1B) is another BRD protein specifically existing in the WICH complex, which is recruited to DSB sites and implicated in the DDR [33,44,54]. The N-terminus of WSTF harbours tyrosine kinase activity, which targets Tyr142 on H2AX to regulate γH2AX. Depletion of WSTF causes impaired foci formation of MDC1 and ATM Ser1981 phosphorylation [33]. The mechanistic functions of individual WTSFs or the WICH complex in the DSB signalling and repair, including the involvement of the BRD, are unclear. SNF2L (SMARCA1) is the major ATPase for the ISWI NURF and CERF complexes. The NURF complex contains the BRD protein BPTF, while CERF contains the BRD protein CECR2. Both SNF2L and BPTF are recruited to DNA damage sites [27,126]. Although there is no direct evidence for CECR2 accumulation at DNA damage, CECR2 deficiency results in radiosensitivity and DSB repair defects [127]. Numerous studies have showed that almost all ISWI complexes localize to DNA damage, although whether they act alone or in concert at all or unique DSBs is unclear [44,123]. All of the recruited ISWI complexes contain BRD proteins, suggesting that acetylation signalling is involved in the DDR functions of ISWI complexes.
(iv). INO80
INO80 chromatin-remodelling complexes are important for repairing DSBs [35–37]. In mammalian cells, INO80 contains three sub-complexes INO80, SRCAP and NuA4 (i.e. TRRAP-TIP60). NuA4 contains both chromatin remodelling and HAT subunits that localize to DNA damage and regulate HR [27,35,37–40,46]. p400, the remodelling ATPase of NuA4, is recruited to DSBs to decompact chromatin around the damage regions and to regulate RNF8-mediated chromatin ubiquitylation, which facilitates loading of BRCA1 and RAD51 for HR [40,128]. Another catalytic component of NuA4, the HAT TIP60, targets histone H4 and H2A, as well as ATM, in response to DSBs [129]. TRRAP, a key factor in this complex, also participates in DSB repair. Trrap-deficient mouse embryonic fibroblasts have impaired H4 hyperacetylation upon I-SceI break induction and reduced BRCA1 and RAD51 ionizing radiation-induced foci, resulting in defective HR [39]. In addition to promoting ZMYND8-NuRD damage recruitment as discussed above [27], acetylation on H4 by TIP60 also limits the loading of 53BP1 onto DSBs, which facilitates HR [46]. Recently, another mechanism for TIP60-mediated HR was identified [38]. In this study, TIP60 acetylates H2AK15 upon DNA damage to prevent RNF168-mediated ubiquitylation on the same residue, which antagonizes the formation of the Ub docking site for 53BP1 binding and recruitment [38,80] (figure 3). Apart from its TIP60 HAT activities, the NuA4 also contains the BRD protein BRD8 and GAS41, a YEATS protein that can bind acetylated lysines [44,130,131]. Although specific cellular functions and the acetylation-binding ability of the BRD in BRD8 is unknown, it is possible that BRD8 or GAS41 may play certain uncharacterized roles in recruiting the NuA4 complex to specific chromatin regions through acetylation recognition to promote DNA repair.
4. Other types of double-strand break repair
In addition to NHEJ and HR, several additional DSB repair pathways have been identified [5,6]. For example, DSB ends can be channelled into alternative repair pathways, including alternative end joining (alt-NHEJ) and single-strand annealing (SSA). The major form of alt-NHEJ is also known as microhomology-mediated end joining (MMEJ) [132]. In mammalian cells, MMEJ is initiated by similar end resection mechanisms to HR by MRN-CtIP to generate short 5′–3′ resected ends containing microhomology. In comparison with HR, MMEJ has a strong requirement for PARP1 signalling to help recruit MMEJ factors, which compete with HR factors to inhibit long-range resection and RPA coating on the ssDNA overhangs. After resection, the mammalian error-prone translesion synthesis polymerase Polθ (POLQ) is recruited to the break for fill-in synthesis followed by ligation by Ligase 3 [133,134]. SSA is similar to MMEJ, but usually requires a longer resected DNA end (greater than 30 nt) and is mediated by RAD52 and ERCC1, but not PARP1 and Polθ. How chromatin and chromatin regulators are involved in these pathways is still poorly understood. A recent study has reported that the NuA4 ATPase p400 promotes HR through inhibiting alt-NHEJ in normal cells, and cells with depleted p400 show increased alt-NHEJ and genome instability [135]. This study further emphasizes the importance of chromatin remodelling in regulating DSB repair pathway choice, in addition to the well-known roles of chromatin in regulating canonical NHEJ and HR repair choice [3]. The important contribution of chromatin-based mechanisms including chromatin remodelling, acetylation signalling and BRD proteins in promoting NHEJ and HR may also regulate these additional DSB repair pathways.
5. Conclusion
The network of acetylation signalling involving the ‘writers’, ‘erasers’ and ‘readers’ has emerged as a critical pathway that orchestrates DNA damage signalling and repair. The accumulating evidence reviewed here highlights the importance of this pathway, including BRD proteins, for signalling and repairing DSBs. Although many participants have been identified, additional work is necessary to further elucidate the mechanisms by which acetylation signalling link epigenetic mechanisms to the DDR and DSB repair.
Large-scale proteomic studies have identified numerous acetylated proteins including in response to DNA damage [48,136]. These include both histone and non-histone proteins, which point to potential pathways that act in addition to those described here that function within the chromatin environment. It has also been challenging to delineate clear mechanistic insights into how chromatin remodellers promote DSB repair. For example, a major question that remains is: Why are so many different chromatin-remodelling complexes involved in DSB repair? This question is simpler for transcriptional regulation given the multitude of different gene regulatory networks that must be tightly controlled by these factors. Could it also be that DSBs come in different varieties due to either the chromatin environment or type of lesion that necessitate different chromatin-remodelling complexes. It might also be that there are levels of redundancy within these pathways that are not yet fully identified for DSB repair. Given the role of chromatin remodellers in gene regulation, it cannot be ignored that perturbation of these pathways may also alter transcription and therefore impact DSB repair either directly or indirectly. Additional studies are warranted to address these important but still unanswered questions.
Obtaining a clearer molecular view of acetylation signalling in the DDR will require a better understanding of the composition of these complexes and how interactions mediate their functionality within the DDR. Many chromatin-modifying and chromatin-remodelling complexes contain multiple reader domains in addition to the BRD, including both PTM and DNA binding activities. It can be envisioned that structural studies, including cryo-electron microscopy, proteomics and biochemical analyses, will help us make new discoveries for the interplay between chromatin proteins and DNA repair [82]. As exemplified by our understanding of DNA repair reactions using purified proteins and substrates, similar advancements may require the inclusion of chromatin and synthesized ‘designer chromatin’, which contains site-specific, chemically installed PTMs within chromatin templates, into these reactions [137]. The identification of the players involved in these pathways will provide the framework on which to build these systems to interrogate chromatin-based DSB repair mechanisms, complementing other genetic and cellular studies.
Targeting DNA repair pathways and epigenetic regulators is a burgeoning field for drug development. Thus, elucidating the molecular basis of DSB repair pathways involving acetylation signalling including BRD proteins could uncover new and selective therapeutic targets. Given the numerous cancer genome studies identifying mutations within DNA repair and chromatin regulators, and the use of DNA damaging agents as frontline therapies for many cancers, understanding the interplay between these pathways is vital for informing the use of these agents within the appropriate cancer-relevant setting. Given the success of using PARPi to treat BRCA1 and BRCA2 HR-deficient cancers, there may be additional opportunities to treat cancers deficient for acetylation signalling regulators involved in DSB repair including HR.
Acknowledgements
We thank members of the Miller laboratory for useful discussions and comments for the review.
Authors' contributions
All authors contributed to the conceptual development and writing of this review.
Competing interests
We declare that no competing interests exist.
Funding
The K.M.M. laboratory is supported by the NIH National Cancer Institute (R01CA198279 and RO1CA201268) and the American Cancer Society (RSG-16–042-01-DMC).
References
- 1.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204. ( 10.1016/j.molcel.2010.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461, 1071–1078. ( 10.1038/nature08467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510. ( 10.1016/j.molcel.2012.07.029) [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211. ( 10.1146/annurev.biochem.052308.093131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceccaldi R, Rondinelli B, D'Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64. ( 10.1016/j.tcb.2015.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma P, Greenberg RA. 2016. Noncanonical views of homology-directed DNA repair. Genes Dev. 30, 1138–1154. ( 10.1101/gad.280545.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aymard F, et al. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374. ( 10.1038/nsmb.2796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hustedt N, Durocher D. 2016. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9. ( 10.1038/ncb3452) [DOI] [PubMed] [Google Scholar]
- 9.Jasin M, Rothstein R. 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 ( 10.1101/cshperspect.a012740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871. ( 10.1126/science.184.4139.868) [DOI] [PubMed] [Google Scholar]
- 11.Margueron R, Reinberg D. 2010. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296. ( 10.1038/nrg2752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128, 693–705. ( 10.1016/j.cell.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 13.Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433. ( 10.1101/gad.2021311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson SP, Durocher D. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807. ( 10.1016/j.molcel.2013.01.017) [DOI] [PubMed] [Google Scholar]
- 15.Lukas J, Lukas C, Bartek J. 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13, 1161–1169. ( 10.1038/ncb2344) [DOI] [PubMed] [Google Scholar]
- 16.Miller KM, Jackson SP. 2012. Histone marks: repairing DNA breaks within the context of chromatin. Biochem. Soc. Trans. 40, 370–376. ( 10.1042/BST20110747) [DOI] [PubMed] [Google Scholar]
- 17.Tjeertes JV, Miller KM, Jackson SP. 2009. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889. ( 10.1038/emboj.2009.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupte R, Liu Z, Kraus WL. 2017. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126. ( 10.1101/gad.291518.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD. 2017. The tale of a tail: histone H4 acetylation and the repair of DNA breaks. Phil. Trans. R. Soc. B 372, 20160284 ( 10.1098/rstb.2016.0284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson MD, Durocher D. 2017. Reading chromatin signatures after DNA double-strand breaks. Phil. Trans. R. Soc. B 372, 20160280 (doi:10.1098/rstb.2016.0280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868. ( 10.1074/jbc.273.10.5858) [DOI] [PubMed] [Google Scholar]
- 22.Rogakou EP, Boon C, Redon C, Bonner WM. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916. ( 10.1083/jcb.146.5.905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226. ( 10.1016/j.cell.2005.09.038) [DOI] [PubMed] [Google Scholar]
- 24.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457. ( 10.1038/emboj.2010.38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 55, 723–732. ( 10.1016/j.molcel.2014.06.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30, 2135–2146. ( 10.1038/onc.2010.592) [DOI] [PubMed] [Google Scholar]
- 27.Gong F, et al. 2015. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 29, 197–211. ( 10.1101/gad.252189.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall JA, Georgel PT. 2007. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 85, 463–476. ( 10.1139/O07-063) [DOI] [PubMed] [Google Scholar]
- 29.Larsen DH, et al. 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 190, 731–740. ( 10.1083/jcb.200912135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139. ( 10.1038/emboj.2010.188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley FK, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. 750, 31–44. ( 10.1016/j.mrfmmm.2013.07.008) [DOI] [PubMed] [Google Scholar]
- 32.Lan L, et al. 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 40, 976–987. ( 10.1016/j.molcel.2010.12.003) [DOI] [PubMed] [Google Scholar]
- 33.Xiao A, et al. 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62. ( 10.1038/nature07668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aydin OZ, et al. 2014. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res. 42, 8473–8485. ( 10.1093/nar/gku565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyon Y, Cote J. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin Genet. Dev. 14, 147–154. ( 10.1016/j.gde.2004.02.009) [DOI] [PubMed] [Google Scholar]
- 36.Gospodinov A, Vaissiere T, Krastev DB, Legube G, Anachkova B, Herceg Z. 2011. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol. Cell. Biol. 31, 4735–4745. ( 10.1128/MCB.06182-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gursoy-Yuzugullu O, House N, Price BD. 2016. Patching broken DNA: nucleosome dynamics and the repair of DNA breaks. J. Mol. Biol. 428, 1846–1860. ( 10.1016/j.jmb.2015.11.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacquet K, et al. 2016. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol. Cell 62, 409–421. ( 10.1016/j.molcel.2016.03.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91–99. ( 10.1038/ncb1343) [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol. 191, 31–43. ( 10.1083/jcb.201001160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. ( 10.1146/annurev.biochem.77.062706.153223) [DOI] [PubMed] [Google Scholar]
- 42.Lans H, Marteijn JA, Vermeulen W. 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5, 4 ( 10.1186/1756-8935-5-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151. ( 10.1038/nsmb.1899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong F, Chiu LY, Miller KM. 2016. Acetylation reader proteins: linking acetylation signaling to genome maintenance and cancer. PLoS Genet. 12, e1006272 ( 10.1371/journal.pgen.1006272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong F, Miller KM. 2013. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 750, 23–30. ( 10.1016/j.mrfmmm.2013.07.002) [DOI] [PubMed] [Google Scholar]
- 46.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325. ( 10.1038/nsmb.2499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennetzen MV, Larsen DH, Dinant C, Watanabe S, Bartek J, Lukas J, Andersen JS. 2013. Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response. Cell Cycle 12, 1688–1695. ( 10.4161/cc.24758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elia AE, et al. 2015. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 59, 867–881. ( 10.1016/j.molcel.2015.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. ( 10.1126/science.1124000) [DOI] [PubMed] [Google Scholar]
- 50.Filippakopoulos P, et al. 2012. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231. ( 10.1016/j.cell.2012.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Floyd SR, et al. 2013. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498, 246–250. ( 10.1038/nature12147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. 2006. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 25, 3986–3997. ( 10.1038/sj.emboj.7601291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyengar S, Farnham PJ. 2011. KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 286, 26 267–26 276. ( 10.1074/jbc.R111.252569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Molina S, Mortusewicz O, Bieber B, Auer S, Eckey M, Leonhardt H, Friedl AA, Becker PB. 2011. Role for hACF1 in the G2/M damage checkpoint. Nucleic Acids Res. 39, 8445–8456. ( 10.1093/nar/gkr435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi W, Wang R, Chen H, Wang X, Xiao T, Boldogh I, Ba X, Han L, Zeng X. 2015. BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci. 128, 317–330. ( 10.1242/jcs.159103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velez-Cruz R, Manickavinayaham S, Biswas AK, Clary RW, Premkumar T, Cole F, Johnson DG. 2016. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 30, 2500–2512. ( 10.1101/gad.288282.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi W, Chen H, Xiao T, Wang R, Li T, Han L, Zeng X. 2016. Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis 31, 193–203. ( 10.1093/mutage/gev075) [DOI] [PubMed] [Google Scholar]
- 58.Spruijt CG, et al. 2016. ZMYND8 co-localizes with NuRD on target genes and regulates poly(ADP-ribose)-dependent recruitment of GATAD2A/NuRD to sites of DNA damage. Cell Rep. 17, 783–798. ( 10.1016/j.celrep.2016.09.037) [DOI] [PubMed] [Google Scholar]
- 59.Ogiwara H, Kohno T. 2012. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS ONE 7, e52810 ( 10.1371/journal.pone.0052810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitange GJ, et al. 2016. Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 14, 2587–2598. ( 10.1016/j.celrep.2016.02.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanlie A, Yousif AS, Akiyama H, Honjo T, Begum NA. 2014. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol. Cell 55, 97–110. ( 10.1016/j.molcel.2014.05.018) [DOI] [PubMed] [Google Scholar]
- 62.Lin YH, Yuan J, Pei H, Liu T, Ann DK, Lou Z. 2015. KAP1 deacetylation by SIRT1 promotes non-homologous end-joining repair. PLoS ONE 10, e0123935 ( 10.1371/journal.pone.0123935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu C, et al. 2012. Roles of Kruppel-associated box (KRAB)-associated co-repressor KAP1 Ser-473 phosphorylation in DNA damage response. J. Biol. Chem. 287, 18 937–18 952. ( 10.1074/jbc.M111.313262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tubbs AT, et al. 2014. KAP-1 promotes resection of broken DNA ends not protected by γ-H2AX and 53BP1 in G1-phase lymphocytes. Mol. Cell. Biol. 34, 2811–2821. ( 10.1128/MCB.00441-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kulkarni A, et al. 2013. Tripartite motif-containing 33 (TRIM33) protein functions in the poly(ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with amplified in liver cancer 1 (ALC1) protein. J. Biol. Chem. 288, 32 357–32 369. ( 10.1074/jbc.M113.459164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson MA, Kouzarides T. 2012. Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27. ( 10.1016/j.cell.2012.06.013) [DOI] [PubMed] [Google Scholar]
- 67.Di Cerbo V, Schneider R. 2013. Cancers with wrong HATs: the impact of acetylation. Brief. Funct. Genomics 12, 231–243. ( 10.1093/bfgp/els065) [DOI] [PubMed] [Google Scholar]
- 68.Ropero S, Esteller M. 2007. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 1, 19–25. ( 10.1016/j.molonc.2007.01.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe R, Kanno S-i, Mohammadi Roushandeh A, Ui A, Yasui A. 2017. Nucleosome remodelling, DNA repair and transcriptional regulation build negative feedback loops in cancer and cellular ageing. Phil. Trans. R. Soc. B 372, 20160473 ( 10.1098/rstb.2016.0473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujisawa T, Filippakopoulos P. 2017. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246–262. ( 10.1038/nrm.2016.143) [DOI] [PubMed] [Google Scholar]
- 71.Groselj B, Sharma NL, Hamdy FC, Kerr M, Kiltie AE. 2013. Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. Br. J. Cancer 108, 748–754. ( 10.1038/bjc.2013.21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dawson MA, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. ( 10.1038/nature10509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Filippakopoulos P, et al. 2010. Selective inhibition of BET bromodomains. Nature 468, 1067–1073. ( 10.1038/nature09504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Picaud S, et al. 2015. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 75, 5106–5119. ( 10.1158/0008-5472.CAN-15-0236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beucher A, et al. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427. ( 10.1038/emboj.2009.276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shibata A, et al. 2011. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 30, 1079–1092. ( 10.1038/emboj.2011.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bunting SF, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. ( 10.1016/j.cell.2010.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao L, et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541. ( 10.1016/j.molcel.2009.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373. ( 10.1016/j.cell.2006.10.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fradet-Turcotte A, et al. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54. ( 10.1038/nature12318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195. ( 10.1016/j.cell.2012.08.005) [DOI] [PubMed] [Google Scholar]
- 82.Wilson MD, et al. 2016. The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103. ( 10.1038/nature18951) [DOI] [PubMed] [Google Scholar]
- 83.Fell VL, Schild-Poulter C. 2015. The Ku heterodimer: function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 763, 15–29. ( 10.1016/j.mrrev.2014.06.002) [DOI] [PubMed] [Google Scholar]
- 84.Eberharter A, Vetter I, Ferreira R, Becker PB. 2004. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 23, 4029–4039. ( 10.1038/sj.emboj.7600382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He X, Fan HY, Narlikar GJ, Kingston RE. 2006. Human ACF1 alters the remodeling strategy of SNF2h. J. Biol. Chem. 281, 28 636–28 647. ( 10.1074/jbc.M603008200) [DOI] [PubMed] [Google Scholar]
- 86.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. 2010. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445. ( 10.1038/emboj.2010.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, Yasui A. 2014. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 74, 2465–2475. ( 10.1158/0008-5472.CAN-13-3608) [DOI] [PubMed] [Google Scholar]
- 88.Phelan ML, Sif S, Narlikar GJ, Kingston RE. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253. ( 10.1016/S1097-2765(00)80315-9) [DOI] [PubMed] [Google Scholar]
- 89.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. 2013. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 20, 1040–1046. ( 10.1038/nsmb.2642) [DOI] [PubMed] [Google Scholar]
- 90.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534. ( 10.1016/j.molcel.2005.06.027) [DOI] [PubMed] [Google Scholar]
- 91.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545. ( 10.1016/j.molcel.2005.06.029) [DOI] [PubMed] [Google Scholar]
- 92.Ziv Y, et al. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876. ( 10.1038/ncb1446) [DOI] [PubMed] [Google Scholar]
- 93.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177. ( 10.1016/j.molcel.2008.05.017) [DOI] [PubMed] [Google Scholar]
- 94.Goodarzi AA, Kurka T, Jeggo PA. 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839. ( 10.1038/nsmb.2077) [DOI] [PubMed] [Google Scholar]
- 95.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ 3rd, Zhou MM. 2008. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 15, 626–633. ( 10.1038/nsmb.1416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257. ( 10.1146/annurev.biochem.77.061306.125255) [DOI] [PubMed] [Google Scholar]
- 97.Huertas P. 2010. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 17, 11–16. ( 10.1038/nsmb.1710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450, 509–514. ( 10.1038/nature06337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362. ( 10.1101/gad.2003811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee KK, Workman JL. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 8, 284–295. ( 10.1038/nrm2145) [DOI] [PubMed] [Google Scholar]
- 101.Guo R, Chen J, Mitchell DL, Johnson DG. 2011. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 39, 1390–1397. ( 10.1093/nar/gkq983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rother MB, van Attikum H. 2017. DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance. Phil. Trans. R. Soc. B 372, 20160285 ( 10.1098/rstb.2016.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okazaki N, Ikeda S, Ohara R, Shimada K, Yanagawa T, Nagase T, Ohara O, Koga H. 2008. The novel protein complex with SMARCAD1/KIAA1122 binds to the vicinity of TSS. J. Mol. Biol. 382, 257–265. ( 10.1016/j.jmb.2008.07.031) [DOI] [PubMed] [Google Scholar]
- 104.Costelloe T, et al. 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584. ( 10.1038/nature11353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580. ( 10.1038/nature11355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rowbotham SP, et al. 2011. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell 42, 285–296. ( 10.1016/j.molcel.2011.02.036) [DOI] [PubMed] [Google Scholar]
- 107.White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ 3rd. 2006. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 66, 11 594–11 599. ( 10.1158/0008-5472.CAN-06-4138) [DOI] [PubMed] [Google Scholar]
- 108.Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596. ( 10.1038/nrc3091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang XJ, Seto E. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9, 206–218. ( 10.1038/nrm2346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. 2013. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell 49, 368–378. ( 10.1016/j.molcel.2012.10.026) [DOI] [PubMed] [Google Scholar]
- 111.Savitsky P, et al. 2016. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell. Rep. 17, 2724–2737. ( 10.1016/j.celrep.2016.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190, 741–749. ( 10.1083/jcb.201001048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conomos D, Reddel RR, Pickett HA. 2014. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 21, 760–770. ( 10.1038/nsmb.2877) [DOI] [PubMed] [Google Scholar]
- 114.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. 2010. A chromatin localization screen reveals poly(ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA 107, 18 475–18 480. ( 10.1073/pnas.1012946107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pfister SX, et al. 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018. ( 10.1016/j.celrep.2014.05.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li N, et al. 2016. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell 63, 470–484. ( 10.1016/j.molcel.2016.06.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. 2012. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J. Biol. Chem. 287, 6764–6772. ( 10.1074/jbc.M111.287037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM. 2017. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 155, 94–106. ( 10.1083/jcb.201611135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. 2008. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 8, 193–204. ( 10.1038/nrc2342) [DOI] [PubMed] [Google Scholar]
- 120.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. 2012. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599. ( 10.1158/0008-5472.CAN-12-2753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O'Connor MJ. 2015. Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560. ( 10.1016/j.molcel.2015.10.040) [DOI] [PubMed] [Google Scholar]
- 122.Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, Green MR, Cantor SB. 2015. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 29, 489–494. ( 10.1101/gad.256214.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aydin OZ, Vermeulen W, Lans H. 2014. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13, 3016–3025. ( 10.4161/15384101.2014.956551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smeenk G, et al. 2013. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 126, 889–903. ( 10.1242/jcs.109413) [DOI] [PubMed] [Google Scholar]
- 125.Nakamura K, et al. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528. ( 10.1016/j.molcel.2011.02.002) [DOI] [PubMed] [Google Scholar]
- 126.Erdel F, Rippe K. 2011. Binding kinetics of human ISWI chromatin-remodelers to DNA repair sites elucidate their target location mechanism. Nucleus 2, 105–112. ( 10.4161/nucl.2.2.15209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee SK, Park EJ, Lee HS, Lee YS, Kwon J. 2012. Genome-wide screen of human bromodomain-containing proteins identifies Cecr2 as a novel DNA damage response protein. Mol. Cells 34, 85–91. ( 10.1007/s10059-012-0112-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y. 2012. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J. Cell Biol. 199, 1067–1081. ( 10.1083/jcb.201205059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun Y, Jiang X, Price BD. 2010. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9, 930–936. ( 10.4161/cc.9.5.10931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schulze JM, Wang AY, Kobor MS. 2009. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 87, 65–75. ( 10.1139/O08-111) [DOI] [PubMed] [Google Scholar]
- 131.Shanle EK, et al. 2015. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 29, 1795–1800. ( 10.1101/gad.269977.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sfeir A, Symington LS. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714. ( 10.1016/j.tibs.2015.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. 2015. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. ( 10.1038/nature14157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ceccaldi R, et al. 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262. ( 10.1038/nature14184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taty-Taty GC, Chailleux C, Quaranta M, So A, Guirouilh-Barbat J, Lopez BS, Bertrand P, Trouche D, Canitrot Y. 2016. Control of alternative end joining by the chromatin remodeler p400 ATPase. Nucleic Acids Res. 44, 1657–1668. ( 10.1093/nar/gkv1202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. ( 10.1126/science.1175371) [DOI] [PubMed] [Google Scholar]
- 137.Fierz B, Muir TW. 2012. Chromatin as an expansive canvas for chemical biology. Nat. Chem. Biol. 8, 417–427. ( 10.1038/nchembio.938) [DOI] [PMC free article] [PubMed] [Google Scholar]