Abstract
T5 5′–3′ exonuclease is a member of a homologous group of 5′ nucleases which require divalent metal co-factors. Structural and biochemical studies suggest that single-stranded DNA substrates thread through a helical arch or hole in the protein, thus bringing the phosphodiester backbone into close proximity with the active site metal co-factors. In addition to the expected use of Mg2+, Mn2+ and Co2+ as co-factors, we found that divalent zinc, iron, nickel and copper ions also supported catalysis. Such a range of co-factor utilisation is unusual in a single enzyme. Some co-factors such as Mn2+ stimulated the cleavage of double-stranded closed-circular plasmid DNA. Such endonucleolytic cleavage of circular double-stranded DNA cannot be readily explained by the threading model proposed for the cleavage of substrates with free 5′-ends as the hole observed in the crystal structure of T5 exonuclease is too small to permit the passage of double-stranded DNA. We suggest that such a substrate may gain access to the active site of the enzyme by a process which does not involve threading.
INTRODUCTION
Bacteriophage T5 exonuclease belongs to a family of enzymes previously shown to possess both 5′–3′ exo- and structure-specific endo-nucleolytic activities (1–4). Both of these activities require a substrate with a free 5′-end. This requirement prompted Dahlberg and co-workers (3) to suggest the term 5′ nuclease to better describe their actions. These enzymes play important roles in DNA replication (5) and act on DNA flap strands that possess a 5′-single-stranded arm (Fig. 1A). Dahlberg and co-workers (3) found that 5′ nucleases are able to cut the single-stranded arm close to the site of bifurcation (the junction of single-stranded and double-stranded DNA). They postulated that the single-stranded 5′-end of the bifurcated substrate becomes threaded through the nuclease. The nuclease then slides to the branch point where endonucleolytic cleavage occurs (3) releasing the 5′-single-stranded arm. Bambara and co-workers (6) provided evidence that the homologous mammalian 5′ nuclease flap endonuclease 1 (FEN-1) may operate in a similar manner. The enzymes are thought to thread or slide along the single-stranded DNA; this sliding is not inhibited by adducts on the bases or sugar–phosphate backbone up to a certain size (streptavidin–biotin being too large), nor is it inhibited by an abasic site (7–10). Double-stranded endonuclease activity independent of a free 5′-end (i.e. cleavage of closed-circular double-stranded DNA) has not previously been demonstrated for these 5′ nucleases.
Figure 1.
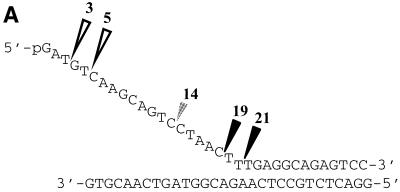
T5 exonuclease can utilise divalent metal ions other than Mg2+ and Mn2+ as co-factors. (A) The DNA pseudo-Y structure used in characterisation of the cleavage specificity of T5 5′–3′ exonuclease. The pseudo-Y structure is composed of a flap strand and a template strand. The flap strand is 5′-32P-labelled in these studies. Triangles indicate the sites of hydrolysis. The large open triangles show exonucleolytic (3 and 5mer products) cleavage. Black triangles show sites of structure-specific endonucleolytic (19 and 21mer products) cleavage. The smaller shaded triangle indicates endonucleolytic hydrolysis (14mer product) corresponding to cleavage of a transient duplex formed by this single-stranded sequence alone (42). (B) Structure-specific digest of pseudo-Y substrate by wild-type T5 exonuclease; enzyme was incubated with the substrate in the presence of either 2 mM zinc acetate (Zn2+), 5 mM nickel chloride (Ni2+), 2 mM cobalt chloride (Co2+), 1 mM ferrous ascorbate (Fe2+) or 10 mM magnesium chloride (Mg2+) for 5 and 30 min at 37°C, as described previously (2). Lane C1: control lane, pseudo-Y substrate incubated with 10 mM magnesium chloride (Mg2+) for 30 min in the absence of enzyme; identical results were obtained in similar controls using the other listed metal co-factors (data not shown). Lane C2: control lane, pseudo-Y substrate incubated with 600 nM enzyme for 30 min in the absence of metal co-factor. Lanes ‘M’: marker lanes, single nucleotide ladder produced by digestion of substrate oligonucleotide with snake venom phosphodiesterase.
Further evidence supporting a threading/sliding model exists. When Taq polymerase was tested on a substrate with a very long single-stranded 5′-arm (268 nt in the 5′ overhang) it was able to cut at the single-strand/double-stranded junction at 72°C in the absence of KCl (3) but not under conditions which promoted transient folding of the long 5′ overhang. The endonuclease activity of FEN-1 is inhibited by annealing an oligonucleotide to the 5′-end of a single-stranded arm thus blocking the threading action (6). This strongly suggests that the free 5′-end of the single-stranded arm can become threaded through the enzyme. It may either cut immediately acting as an exonuclease, or slide to the point of bifurcation to act as a structure-specific endonuclease.
Recent structural studies have provided physical evidence supporting the threading hypothesis. T5 5′–3′ exonuclease (11) contains a hole large enough for the passage of single- but not double-stranded DNA. Thus, the enzyme would become arrested at the junction of double- and single-stranded DNA. The hole is formed by an inverted V-shaped helical archway set astride an active site rich in conserved acidic residues. These residues form the binding site for essential divalent metal co-factors. The structures of two archaeal 5′ nucleases also show a hole or channel and are similar to the T5 structure (11–14). The reported crystal structures show significant variation in the position and spacing of the two divalent metal co-factors present in the active sites. The distances range from ∼8 Å in T5 enzyme to 5 Å in the Methanococcus enzyme. Indeed, the 5′–3′ exonuclease domain of Taq polymerase appears to have a third metal binding site showing a preference for Zn2+ ions. This suggests some flexibility of the ligand geometry within the nuclease active sites.
During initial attempts to carry out hydroxy-radical footprinting on T5 exonuclease/DNA complexes we made the chance discovery that ferrous ion (Fe2+) was able to act as co-factor in the cleavage of oligonucleotide flap structures (Fig. 1A). This prompted us to investigate the ability of various divalent cations to act as co-factors with this 5′ nuclease. The experiments described here show that T5 exonuclease is able to make use of an unusually wide range of divalent metal co-factors. This implies some degree of flexibility in the active site. Furthermore, some metal ions (such as Mn2+ ions) can promote double-stranded endonucleolytic activity not previously reported for this class of enzymes. The proposed DNA threading mechanism for 5′ nuclease action cannot explain such an activity. The mechanistic implications of these observations are discussed.
MATERIALS AND METHODS
Production of enzymes
Escherichia coli M72(λ) were transformed with the tightly regulated expression vectors pJONEX4 (15) carrying the wild-type (15) or 2CS-exo [C115S, C265S (2)] T5 exonuclease coding sequences. Cells were grown to mid-log phase in 1–2 l of rich media (containing 40 g tryptone, 25 g yeast extract, 5 g NaCl, 100 mg ampicillin/l) at 28°C. Expression from the λPL promoter was induced by shifting the culture to 42°C for 2 h. Cells were recovered by centrifugation at 13 000 g for 20 min at 4°C. Cell pellets were washed with 1% (w/v) NaCl, centrifuged as above, then frozen in liquid nitrogen for storage at –80°C. After lysing the cells, wild-type protein was purified to homogeneity from the soluble fraction by a combination of ion exchange and affinity chromatography as described (16). The 2CS-exo mutant protein was purified from the cell pellet after lysis and was purified as described previously (2,15). Protein concentration was determined by the method of Bradford (17).
Protein purification by preparative SDS–PAGE
Approximately 200 µg wild-type exonuclease was loaded onto an oversized lane of an SDS–PAGE gel and electrophoresis carried out as normal. After electrophoresis, the marker lane included for subsequent alignment was cut away from the gel and stained with Coomassie blue stain. The remainder of the gel was placed into an elution chamber assembly of a Mini-Whole Gel Eluter (Bio-Rad, USA). The protein band corresponding to the 33 kDa T5 exonuclease was electro-eluted from the gel by electrophoresis at 75 mA for 30 min in 50 mM Tris-Bicine pH 8.3 containing 2 mM DTT, 0.2% Triton X-100 and 3 M urea. The eluted protein was diluted and concentrated (with the aid of Amicon 10 K Microsep Microconcentrators) in 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 2 mM DTT, 200 mM NaCl, 5% glycerol, 25 µg/ml PMSF and 25 µg/ml acetylated BSA. The purified protein was made 50% in glycerol and stored at –20°C (16).
Double-stranded endonucleolytic activity assay
Endonucleolytic activity was examined using a pUC19 plasmid as a covalently-closed circular (double-stranded) substrate. Reactions contained 16 nM substrate DNA, in 25 mM potassium glycinate (pH 9.3), 0.1 mg/ml acetylated BSA, 1 mM DTT (unless indicated otherwise in the figure legends). Divalent cations, monovalent salts (all of the highest purity from BDH Merck) and enzyme were added to each reaction as described in the relevant Results section. After addition of the metal ion the reactions were mixed carefully but not centrifuged, in order to avoid precipitation of the metal ion or metal-induced precipitation of the DNA or of the enzyme. The reactions were incubated at 37°C. Aliquots of the reactions were stopped by addition of 10 µl of the reaction mix to 10 µl of agarose gel loading buffer (50% glycerol, 50 mM EDTA, 140 µg/ml bromophenol blue). The products of the reaction were analysed on a 1% agarose gel buffered with 1× TAE. The gel and buffer contained ethidium bromide at 0.2 µg/ml. The gels were run for 1.5 h at 5 V cm–1. The DNA was visualised with a UV transilluminator. Quantitative densitometry of the images obtained were processed using NIH Image v1.61 software (National Institute of Health, USA).
Binding (EMSA) and structure-specific cleavage of oligonucleotide substrates
Electrophoretic mobility shift assays (EMSA) were carried out in the absence of co-factor on 5′-32P-end-labelled pseudo-Y and blunt-ended duplex substrates as described (2). The pseudo-Y structure was made by annealing the two oligonucleotides 5′-GATGTCAAGCAGTCCTAACTTTGAGGCAGAGTCC and 5′-GGACTCTGCCTCAAGACGGTAGTCAACGTG. This produces a structure which is fully base paired at one end only (Fig. 1A). The double-stranded blunt-ended substrate was composed of 5′-CTTGAGGCAGAGTCC and 5′-GGACTCTGCCTCAAG. Reactions were performed in 25 mM potassium glycinate pH 9.3, 1 mM EDTA, 5% glycerol, 1 mM DTT and 0.1 mg/ml acetylated BSA. The concentration of KCl added to the reaction mix was varied to assess the influence of increasing concentrations of KCl on protein–DNA interaction. The reactions were incubated on ice for 10 min and analysed on a 17% native acrylamide gel, in 50 mM Tris-Bicine pH 8.3, 1 mM EDTA and 1 mM DTT at 4°C for 2 h at 15 V cm–1. The gel was analysed using a Molecular Imager and Molecular Analyst v2.0.1 software (Bio-Rad, USA).
Structure-specific DNA cleavage was examined using 5′-32P-end-labelled pseudo-Y substrate as described (2) with the specified co-factor in 25 mM potassium glycinate pH 9.3, 100 mM KCl, 0.5 mg/ml acetylated BSA, 2% glycerol and 0.2 mM EDTA. None of the reactions contained more than 0.2 mM DTT. Pseudo-Y substrate was prepared as described above. The reactions were incubated at 37°C and 10 µl aliquots taken at intervals during the reaction. All reactions were stopped by addition to 10 µl stop mix (95% formamide, 15 mM EDTA, 200 µg/ml bromophenol blue). The products of the reactions were analysed on a 7 M urea–15% polyacrylamide gel, buffered in 1× TBE at 50 W for 2 h. The gel was visualised with a Molecular Imager and Molecular Analyst v2.0.1 software (Bio-Rad, USA). The length of the digestion products was determined by comparison to an SVPD (snake venom phosphodiesterase) digestion of the labelled substrate.
Exonuclease activity assay
This UV spectroscopy-based assay demonstrates the presence of (predominantly) exonucleolytically derived, acid soluble degradation products. The release of acid-soluble nucleotides from high molecular weight DNA (herring sperm Type XIV, Sigma) was determined with a standard UV spectrophotometric assay essentially as described (16). The 600 µl assay mixture contained 2 mM Type XIV DNA (in nucleotides), 25 mM potassium glycinate, pH 9.3, 1 µg enzyme and divalent cation (as specified in the Results section). Curves were plotted from the data obtained (from at least triplicate experiments) and estimates of the initial velocity were calculated.
RESULTS
Exonucleolytic activity and co-factor promiscuity
The optimal co-factors for (exonucleolytic) activity are Mg2+ or Mn2+ (18). However, we found that other divalent cations could also be used as co-factors in an assay of 5′ nuclease activity based on structure-specific cleavage of oligonucleotides annealed to form a pseudo-Y structure (Fig. 1). These included Co2+, Fe2+, Zn2+ and Ni2+. Cleavage was also supported by Cu2+ but at a vastly reduced rate in comparison to that seen with the other co-factors (data not shown). The substrate used for this assay had a defined structure, a pseudo-Y, and the products of degradation were similar to those obtained from structure-specific cleavage using Mg2+ as co-factor; a summary showing the percentage of endo- and exo-nucleolytic cleavage products is given in Table 1. In separate experiments, no cleavage of substrate was observed in reactions containing enzyme but no divalent metal ion. Similarly, no cleavage of substrate was observed in reactions containing any divalent metal ion but no enzyme (Fig. 1B and data not shown). Besides Mn2+ (data not shown) and Mg2+, the co-factors Co2+ and Ni2+ were the most efficient.
Table 1. Structure-specific digest of pseudo-Y substrate by wild-type T5 exonuclease.
| Co-factor |
Zn2+ |
|
|
|
Ni2+ |
|
|
|
Co2+ |
|
|
|
Fe2+ |
|
|
|
Mg2+ |
|
|
|
| [Enzyme]/nM | 600 | 60 | 600 | 60 | 60 | 6 | 600 | 60 | 6 | 0.6 | ||||||||||
| Time/min | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 | 5 | 30 |
| Substrate remaining (34mer) | 1 | 1 | 31 | 29 | 2 | 0 | 3 | 0 | 6 | 2 | 34 | 11 | 4 | 3 | 67 | 66 | 5 | 1 | 57 | 12 |
| Endo products (14, 19 and 21mer) | 23 | 21 | 8 | 9 | 48 | 40 | 60 | 59 | 30 | 29 | 18 | 16 | 17 | 18 | 9 | 10 | 12 | 11 | 13 | 20 |
| Exo products (3 and 5mer) | 50 | 54 | 51 | 53 | 23 | 30 | 15 | 17 | 43 | 52 | 34 | 56 | 59 | 54 | 14 | 14 | 70 | 80 | 21 | 51 |
A summary of percentage endo- and exo-nucleolytic products obtained after incubation of substrate with enzyme in the presence of different co-factors as illustrated in Figure 1, as determined by quantitative phosphoimager analysis. The percentages have been expressed to the nearest whole percentage.
Given the low level of hydrolysis seen using the unusual co-factors, we examined the possibility that the observed activity could be due to contamination of the metal ions with e.g. Mg2+ or Mn2+. We determined that the minimum concentration of Mg2+ required to yield detectable hydrolysis (of the pseudo-Y substrate) was 100 µM (data not shown). Thus, the metal salts used as alternative co-factors would have to have been contaminated with at least 2% Mg2+ to produce the activity seen; the stated maximum level of Mg2+ contamination of the zinc acetate used was 0.005% (BDH, AnalaR).
In a separate assay of exonuclease activity based on quantification of acid soluble nucleotide released from a high molecular weight DNA substrate, the rate of the Co2+ assisted reaction was ∼35 and 65% that of the rate obtained using Mg2+ and Mn2+, respectively (summarised in Table 2). Exonucleolytic activity, though very low, was detectable with Zn2+ as co-factor, while no detectable activity was apparent with Cu2+ or Ni2+ (<1%). The use of this particular assay with Fe2+ was not possible because the acid precipitation step produced an opaque solution in this case. Assays containing either Mg2+ or Mn2+ as principle co-factor could be inhibited by competition with high concentrations (≥5 mM) of Zn2+ and Cu2+, possibly due to precipitation of protein or substrate (19). Conversely, when Zn2+ or Cu2+ (5 and 10 mM, respectively) were used as principle co-factor, the addition of increasing Mn2+ did not result in recovery of activity (data not shown).
Table 2. Exonucleolytic activity of wild-type T5 5′–3′ nuclease.
| Divalent cation (mM) | Specific activitya | ||||
| |
Mg2+ |
Mn2+ |
Co2+ |
Zn2+ |
Cu2+ |
| 0.5 | nd | 11.8 | nd | nd | nd |
| 1 | 18.0 | 13.0 | 3.1 | 0.5 | 0.1 |
| 2 | 20.4 | 10.8 | 8.4 | 1.6 | 0.2 |
| 5 | 23.8 | 6.5 | 6.8 | 0.1 | 0.2 |
| 10 | 23.9 | 4.5 | 2.5 | 0.0 | 0.2 |
Exonucleolytic digestion of high molecular weight DNA was assayed spectrophotometrically (16). Specific activity for each reaction was determined from at least three separate assays.
aSpecific activity: 1 U is defined as 1 nmol of released nucleotides per min per µg of protein in the standard spectrophotometric assay at 37°C.
nd, not determined.
Degradation of covalently-closed circular DNA
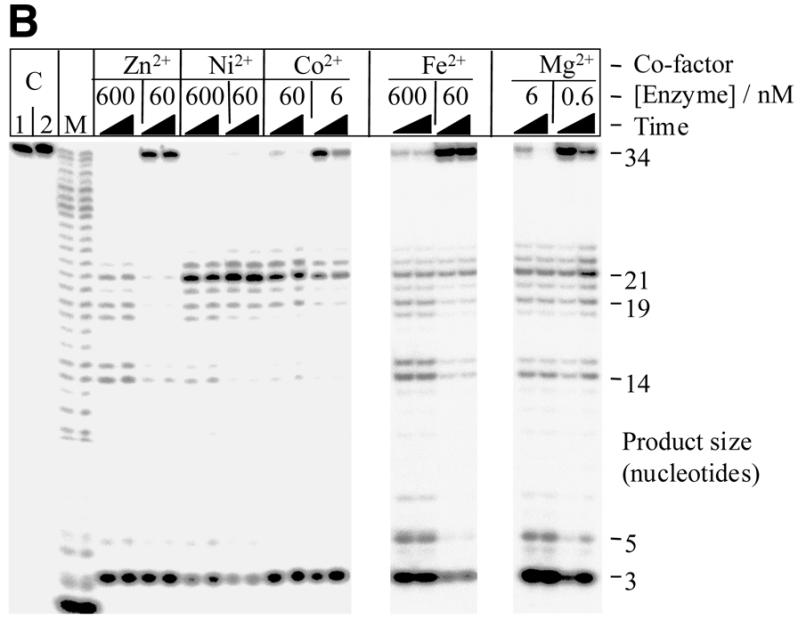
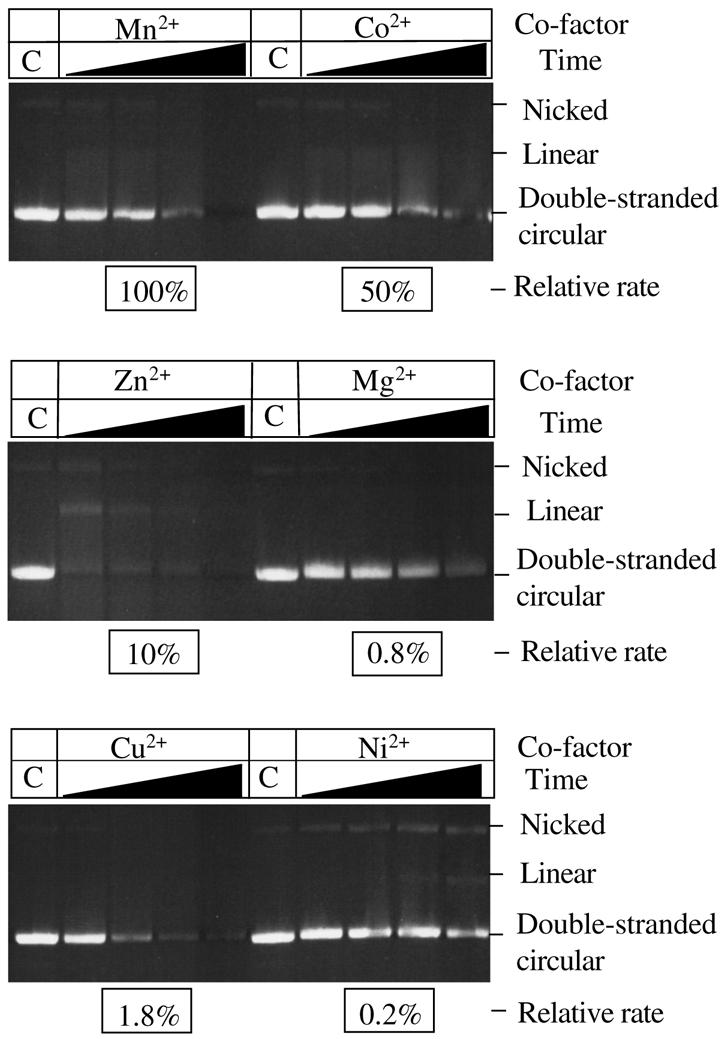
The degradation of covalently-closed circular double-stranded DNA by T5 nuclease was supported by several co-factors tested (Fig. 2). Degradation was most efficient in the presence of Mn2+, followed by Co2+ and Zn2+ as shown by the loss of the double-stranded circular DNA substrate. Degradation of the covalently-closed circular DNA occurred through both nicked and linear intermediates. This purely endonucleolytic activity was also supported, albeit to a lesser extent, by Mg2+ (at low monovalent salt concentrations) and most surprisingly by Cu2+. Some linearisation of plasmid DNA was observed with Ni2+ as a co-factor. Quantitative densitometry of the gels further demonstrated the relative influence of co-factor on degradation of double-stranded circular DNA. Using reactions with co-factor Mn2+ as the standard, relative rates of percentage loss of double-stranded circular DNA were determined for reactions containing other divalent metal ions as co-factor; the enzyme concentration used per reaction was taken into consideration. As previously indicated, degradation was most efficient in the presence of Mn2+, followed by Co2+ and Zn2+ (Fig. 2). The effectiveness of Fe2+ as a co-factor in this system could not be tested, as the substrate precipitated. In the absence of co-factor there was no apparent single-stranded or double-stranded endonuclease activity (15). Several lines of evidence suggest that the ability of T5 5′–3′ exonuclease to degrade covalently-closed double-stranded circular (plasmid) DNA under certain conditions was not due to a contaminating nuclease. The double-stranded endonuclease activity was present in enzyme preparations purified by different methods (from soluble cell-free extracts or by denaturation/renaturation protocols from inclusion body material, data not shown) and in enzyme separated and renatured from an SDS–PAGE gel (Fig. 3); albeit a higher concentration of enzyme was used to account for loss of activity during purification and renaturation.
Figure 2.
Degradation of covalently-closed double-stranded circular DNA by wild-type T5 exonuclease in the presence of different divalent cations. The reactions contained 16 nM substrate DNA, in 25 mM potassium glycinate (pH 9.3), 0.1 mg/ml acetylated BSA and metal ion as indicated. In reactions containing either Mn2+ or Co2+, 40 nM wild-type T5 enzyme was used, in all other reactions a higher concentration (3 µM) of enzyme was necessary to demonstrate cleavage of the substrate on the same time-scale. Reactions were incubated at 37°C and aliquots taken at 5, 15, 30 and 60 min time points and quenched with excess EDTA. Lanes marked ‘C’ represent 60 min time point for control reactions containing the appropriate divalent metal ion co-factor but no enzyme. Relative rate: relative percentage loss of double-stranded circular DNA in comparison to degradation in the presence of Mn2+; rates have been determined using quantitative densitometry of gels and have also been corrected for the enzyme concentration used per reaction. Data from at least two experiments.
Figure 3.
T5 exonuclease purified by preparative PAGE retains nuclease activity on double-stranded covalently-closed circular DNA. The reactions were performed in 25 mM potassium glycinate pH 9.3, 50 mM NaCl, 0.1 mg/ml acetylated BSA and 1 mM DTT at 37°C. Reaction A contained 2 mM manganese chloride; reaction B contained no divalent metal co-factor. Both reactions contained 0.15 µM renatured enzyme. Aliquots were taken at various time points as indicated. Three replicate experiments were performed and the rate of percentage loss of double-stranded circular DNA was 1.4% min–1 as determined using quantitative densitometry.
Influence of potassium chloride on double-stranded endonuclease activity
Potassium chloride was shown to inhibit degradation of covalently-closed double-stranded circular DNA. An enzyme concentration of 40 nM, with 1 mM Mn2+, was used in these reactions. In the absence of KCl, substrate degradation was obvious but not complete until prolonged incubation, thus allowing evaluation of the effect of increasing KCl concentration on the reaction. Some inhibition was evident with 50 mM KCl and was almost complete at 100 mM KCl (Fig. 4A). Quantitative densitometry was used to determine the percentage rate of loss of double-stranded circular DNA and compared to that observed in the absence of KCl (i.e. 0 mM KCl); these results have been expressed as relative rates (Fig. 4A). It is interesting to note that the initial relative rate for degradation in the presence of 25 mM KCl was increased over that seen in the absence of KCl; concentrations of KCl of 25–150 mM are known to increase the exonuclease activity of T5 exonuclease as previously demonstrated (16). However higher concentrations of KCl (≥50 mM) inhibited the rate of degradation.
Figure 4.

(A) Influence of increasing KCl concentration on the cleavage of double-stranded covalently-closed circular DNA by T5 exonuclease. The reactions contained 16 nM substrate DNA, in 25 mM potassium-glycinate pH 9.3, 0.1 mg/ml acetylated BSA, 1 mM DTT, 1 mM MnCl2. KCl and 40 nM enzyme were added as indicated. Aliquots were taken at 5, 15, 30 and 60 min time points. Lanes marked ‘C’ represent 60 min time points for control reactions without enzyme. Relative rate: relative percentage loss of double-stranded circular DNA in comparison to degradation in the absence of KCl (i.e. 0 mM KCl). Three replicate experiments were performed and rates were determined using quantitative densitometry. (B) Increased KCl concentration stimulated binding to both pseudo-Y and blunt ended duplex substrates. Lanes marked ‘+’ contained substrate incubated with T5 5′–3′ exonuclease; enzyme concentration was 3 µM in the reactions containing blunt duplex and 12 nM with the pseudo-Y substrate. Lanes marked ‘ss’ contained the labelled oligonucleotide alone i.e. not annealed to the complementary oligonucleotide.
Increasing the KCl concentration was shown to increase binding of the exonuclease to oligonucleotide substrates, particularly blunt double-stranded substrates (Fig. 4B) as shown in EMSAs At low concentrations of KCl, dissociation of the protein–DNA substrate complex during electrophoresis was observed as indicated by the diffuse bands and altered mobility; high concentrations of KCl appear to stabilise the interaction between the protein and substrate as indicated by the tighter, slowly migrating band observed in the presence of 100 mM KCl. Thus, the inhibition of plasmid degradation observed is unlikely to be due to weakening of protein–substrate interactions.
A mercury compound stimulates endonucleolytic activity
The covalently-closed double-stranded plasmid substrate was incubated with p-hydroxymercuribenzoate (PHMB), Mg2+ as co-factor and mutant T5 exonuclease lacking cysteine residues (2CS-exo). 2CS-exo is a mercury tolerant mutant of the T5 exonuclease, in which both cysteine residues are replaced by serine residues (2). No cleavage of the substrate was apparent when PHMB alone was added; a small amount of cleavage was observed in the presence of Mg2+ alone. In the presence of both metal salts the substrate was completely degraded by 2CS-exo (Fig. 5). The rate of percentage loss of substrate DNA for reactions containing Mg2+ with either wild-type or 2CS-exo T5 were similar (Fig. 5), however the rate almost doubled when reactions contained 2CS-exo T5 with both Mg2+ and PHMB. A lower concentration of enzyme was used in these reactions (refer to Fig. 2) to facilitate demonstration of the influence of PHMB on cleavage; this allowed us to observe the early course of the reaction. In separate experiments no loss of closed-circular plasmid DNA was observed in reactions containing enzyme but no metal ions. Similarly, no loss of closed-circular plasmid DNA was observed in reactions containing metal ions but no enzyme. This type of qualitative analysis can detect both endonuclease activity (as formation of nicked and linear molecules) as well as exonuclease activity (loss of ethidium fluorescence). No degradation of the substrate was apparent when wild-type exonuclease was used in the presence of both metal salts i.e. PHMB inhibits both exo- and endonuclease activity of the wild-type nuclease but neither activity is inhibited in the cysteine-free mutant. This result strongly suggests that both activities reside on the T5 5′ nuclease.
Figure 5.
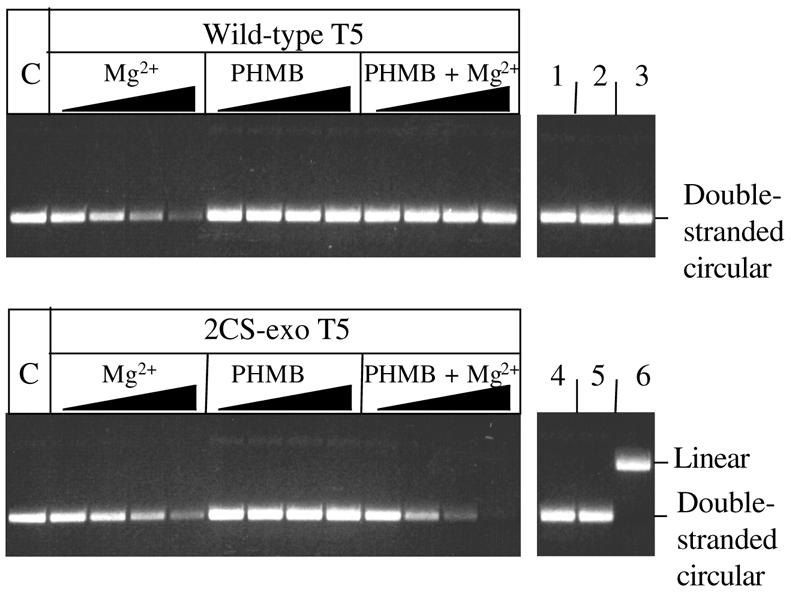
Stimulation of endonuclease activity by PHMB. The reactions contained 16 nM substrate (double-stranded covalently closed-circular) DNA, in 25 mM potassium-glycinate pH 9.3, 0.1 mg/ml acetylated BSA, 25 mM KCl. Metal ion [10 mM magnesium chloride (Mg2+) or 2 mM PHMB] and 1.5 µM enzyme (wild-type or 2CS-exo T5) were added as indicated. The DNA was incubated with metal ions for 5 min before addition of the enzyme. Aliquots were taken at 5, 15, 30 and 60 min time points. Lanes marked ‘C’ represent untreated substrate DNA. Lanes numbered 1–5 represent 60 min time points for control reactions: lanes 1, 2 and 3 contain Mg2+, PHMB, both Mg2+ and PHMB, respectively, but no enzyme; lanes 4 and 5 contain wild-type and 2CS-exo, respectively, but no Mg2+ nor PHMB. Lane 6 shows linearised plasmid. The rate of percentage loss of double-stranded circular DNA was 1.3, 1.1 and 2.5% min–1 for reactions containing wild-type/Mg2+, 2CS-exo/Mg2+ and 2CS-exo/Mg2+/PHMB, respectively. Rates were determined using quantitative densitometry from at least triplicate experiments.
DISCUSSION
Co-factor tolerance
The 5′ nucleases require divalent cations for activity and T5 5′–3′ exonuclease was reported to be able to use Mg2+, Mn2+ or Co2+ as co-factors (18). The work presented here shows that T5 exonuclease is able to utilise an unusually wide range of divalent cations as co-factors. In an assay using a pseudo-Y substrate several divalent cations were able to effect structure-specific endonucleolytic cleavage as well as exonucleolytic attack. In decreasing order of effectiveness (as judged by the amount of remaining labelled 34mer substrate), the co-factors tested were Mg2+ ≈ Mn2+ > Co2+ ≈ Ni2+ > Zn2+ ≥ Fe2+ >> Cu2+. Similar patterns of structure-specific endonucleolytic cleavage were observed with the active co-factors i.e. cleavage of the 5′-single-stranded arm within the hinge region of the substrate (producing products of 19 and 21 nt) and exonucleolytic cleavage producing a trimer. Crystallographic studies on the 5′ nucleases show that they share very similar structures (14,20). All appear to possess a cluster of conserved acidic residues able to bind two divalent metal ions and sequence alignments identify these conserved regions (4,21,22). The structures show significant variation with respect to the distances between the bound manganese ions ranging from 5 to 10 Å (13,23). Extrapolation from crystallographic data involving manganese ions alone to describe other co-factors has been shown to require caution (24). Some, or all, of the co-ordination of the divalent cations could be indirect, for instance binding through an intermediate water molecule has been directly observed in two 5′ nuclease structures (13,25). Both Mg2+ and Mn2+ occupy sites with octahedrally co-ordinated ligands. Cobalt(II) can adopt octahedral or tetrahedral co-ordination geometries, but is generally a better replacement for Zn2+ at tetrahedrally co-ordinated sites than it is for Mg2+ (26). Given the co-ordination geometry requirements of all metals which supported activity, we suggest that substantial conformational variation/flexibility within the active site is possible, as suggested for exonuclease III from E.coli (27). Direct interaction between the carboxylate groups and the cations would involve minimal metal–ligand distances ranging from 1.82 Å (Cu2+) to 2.08 Å (Mn2+) (28).
Double-stranded endonuclease activity
The T5 exonuclease (15) and the Taq polymerase 5′ nuclease (29) domain have previously been shown to cleave single-stranded circular DNA endonucleolytically. We found that double-stranded plasmid DNA could also be cleaved by T5 exonuclease in the presence of certain co-factors. This truly endonucleolytic reaction was most effective in the presence of Mn2+, Co2+ or Zn2+ although some activity was detectable with Mg2+, Cu2+ and Ni2+. The presence of KCl inhibited double-stranded endonucleolytic (ds-endo) activity on covalently-closed circular DNA despite the observation that KCl increases the exonuclease activity of T5 enzyme (16) and affinity for oligonucleotide substrates (Fig. 4B).
The double-stranded covalently-closed circular substrate was degraded through both nicked and linearised intermediates by most of the co-factors tested. The endonucleolytic activity could involve nicking of only one strand or cleavage of both strands simultaneously. The sole mechanism of endonucleolytic cleavage cannot be through simultaneous cleavage of both strands, otherwise a nicked intermediate could not be formed. We were unable to discover reaction conditions under which the substrate is cleaved exclusively through a linear intermediate, suggesting that concerted cleavage of both strands does not occur.
It appears that the T5 nuclease cleaves some form of localised disruption of the double-helix in covalently-closed circular double-stranded DNA caused by interaction with divalent metal cations. This disruption can be reversed by KCl. Low monovalent salt concentrations tend to cause the supercoiled DNA to melt locally as repulsion between the duplex DNA strands is increased (30). If endonucleolytic cleavage depends on disruption of the DNA structure, this would explain why Mn2+ causes much more efficient endonucleolytic cleavage of the plasmid DNA than does Mg2+. Both metal ions may be similarly effective as co-factors, but Mn2+ tends to destabilise the double helix more than Mg2+. This would similarly explain why Zn2+ is a more effective co-factor for ds-endo cleavage than Mg2+, when on all other substrates the opposite is true, as Zn2+ is also capable of altering DNA structure (31). The relative affinity of divalent cations for the backbone phosphate decreases, compared to affinity for the nucleotide base, in the order Mg2+ > Co2+ > Ni2+ > Mn2+ > Zn2+ > Cu2+/Hg2+ (31,32). Alternative explanations are also possible, e.g. Halford and co-workers found that Mn2+ stimulates the formation of tighter enzyme–DNA complexes than does Mg2+ (33), allowing normally very slow reactions to be observed. This could explain why Mn2+ often alters the activity of Mg2+-requiring enzymes (34).
Plasmid DNA was incubated with both magnesium chloride and a mercury compound (PHMB) in order to test the hypothesis that T5 exonuclease-mediated cleavage of covalently-closed circular DNA requires disruption of the double helix. Mercury compounds disrupt the DNA double helix (35). The double-cysteine mutant 2CS-exo was used to demonstrate cleavage of plasmid DNA in the presence of PHMB, as this mutant is resistant to inactivation by PHMB (2). The double-stranded circular DNA was much more efficiently cleaved by T5 5′–3′ exonuclease in the presence of both PHMB and Mg2+ than in the presence of magnesium ions alone.
Stimulation of ds-endo activity by PHMB and Zn2+ may be through different mechanisms; mercury compounds can cause large changes in DNA conformation, to the extent of inducing a transition between right-handed and left-handed helical screwness (35). Divalent zinc ions tend to interact preferentially with N7 of guanine in double-helical DNA and the context of the guanine base has been shown to be important in the interaction with Zn2+. This interaction of Zn2+ with guanine did not cause a major perturbation of the DNA structure (36). If T5 5′–3′ exonuclease is endonucleolytically cleaving DNA in response to a particular DNA conformation then it could be used as a probe for structural change; in a similar manner, the reduction in activity of staphylococcal nuclease has been used as a probe for Z-DNA formation (35).
There is no possibility that this double-stranded endonuclease activity was due to the co-purification of an endonuclease with a requirement for Mn2+ as a co-factor; such an endonuclease would nick the plasmid, creating a suitable substrate for subsequent exonuclease activity. The endonuclease activity of the wild-type protein was shown to be sensitive to inhibition by a mercury derivative, as was the exonuclease activity. However, the 2CS-exo mutant retained exonuclease and endonuclease activities in the presence of PHMB (Fig. 5) implying that activity is not due to a contaminating endonuclease. Furthermore, T5 5′–3′ exonuclease retained the same activities after purification and renaturation from an SDS–PAGE gel (Fig. 3).
The ability to utilise such a wide range of co-factors is unusual but not without precedent; TaqI endonuclease can utilise Mg2+, Mn2+, Fe2+, Co2+, Ca2+, Ni2+ and Zn2+ as co-factors (37), BamHI can use Mg2+, Mn2+, Co2+, Zn2+ and Cd2+, but is inhibited by Ca2+ (38). DNase I has been shown to use Mg2+, Mn2+, Co2+, Ca2+ and Zn2+ (39), and E9 DNase is reported to be able to utilise Mg2+, Mn2+, Co2+ and Ni2+ (40). Results presented here show that T5 5′–3′ exonuclease can utilise an equally wide, if not wider, range of co-factors: Mg2+, Mn2+, Co2+, Ni2+, Fe2+, Zn2+ and Cu2+.
Implications for threading model
The T5 helical arch is flexible (20) but forms a hole only large enough for the passage of single-stranded DNA. Biophysical studies support the threading mechanism in T5 exonuclease. Substrates with free 5′-ends bound the enzyme with much higher affinity than blunt-ended or single-stranded substrates (2). This, and the flap endonuclease activity of the enzyme, suggests that the protein loads on to the 5′-end and tracks along to the duplex region which prevents further threading. Published crystal structures of 5′–3′ exonucleases show the region corresponding to the helical arch is disordered or present in varying conformations (13,23,25,41,42). How can the cleavage of covalently-closed double-stranded DNA by T5 5′–3′ exonuclease be reconciled with the published structure, showing the active site beneath a helical arch, through which single-stranded DNA is thought to be threaded (11)? We suggest that the helical arch could fold (or collapse), away from the active site (Fig. 6) in order to accommodate double-stranded DNA lacking a free 5′-end. There is little steric hindrance to such a process. This would fully expose a region of positive potential containing the active site (11) to which the circular DNA would then have ready access. A similar mechanism has been proposed for endonucleolytic cleavage by related 5′ nucleases (10,29). Elegant work by Bornath et al. (10) showed that flap substrates with bulky 5′-ends (which would preclude threading) were nonetheless cleaved endonucleolytically. Whether the purely endonucleolytic cleavage of circular plasmid DNA occurs at melted bubbles, bifurcations, cruciforms or by direct contact of duplex DNA with the active site remains unclear. In any case, it seems unlikely that cleavage of closed-circular DNA involves threading of a duplex through the helical arch.
Figure 6.
Proposed DNA threading model. (A) The crystal structure of T5 exonuclease (helical arch region in cyan, remainder in grey) was docked with a DNA flap structure. The 5′-end of the displaced strand (blue) is threaded through the helical arch. The flap structure is composed of DNA template strand (yellow), invading strand (green) and the displaced strand. (B) Backbone structure of T5 exonuclease showing position of the two divalent metal ions (red spheres) docked with a ladder representation of a DNA flap structure. (C) The model rotated in the absence of threaded DNA. The helical arch (cyan) could potentially fold away from the metal ions, allowing greater access to the active site. This region has been observed to be mobile in one T5 exonuclease structure [(42), PDB ID code 1xo1].
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Jane Grasby, Martin Nicklin and Thomas Ceska for helpful discussions and advice. This work was supported by the Wellcome Trust (grant number 052123), a Biotechnology and Biological Research Council studentship (to S.J.G) and a White Roses Consortium studentship (to M.F).
References
- 1.Harrington J.J. and Lieber,M.R. (1994) The characterisation of a mammalian DNA structure-specific endonuclease. EMBO J., 13, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garforth S.J. and Sayers,J.R. (1997) Structure-specific DNA binding by bacteriophage T5 5′–3′ exonuclease. Nucleic Acids Res., 25, 3801–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyamichev V., Brow,M.A.D. and Dahlberg,J.E. (1993) Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science, 260, 778–783. [DOI] [PubMed] [Google Scholar]
- 4.Ceska T. and Sayers,J.R. (1998) Structure-specific cleavage by 5′ nucleases. Trends Biochem. Sci., 23, 319–367. [DOI] [PubMed] [Google Scholar]
- 5.Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- 6.Murante R.S., Rust,L. and Bambara,R.A. (1995) Calf 5′ to 3′-exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem., 270, 30377–30383. [DOI] [PubMed] [Google Scholar]
- 7.Barnes C.J., Wahl,A.F., Shen,B.H., Park,M.S. and Bambara,R.A. (1996) Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′–3′-exonuclease/endonuclease RAD2 homologue 1 or flap endonuclease 1. J. Biol. Chem., 271, 29624–29631. [DOI] [PubMed] [Google Scholar]
- 8.Price A. (1992) Action of Escherichia coli and human 5′–3′ exonuclease functions at incised apurinic apyrimidinic sites in DNA. FEBS Lett., 300, 101–104. [DOI] [PubMed] [Google Scholar]
- 9.Demott M.S., Shen,B.H., Park,M.S., Bambara,R.A. and Zigman,S. (1996) Human RAD2 homolog 1 5′- to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J. Biol. Chem., 271, 30068–30076. [DOI] [PubMed] [Google Scholar]
- 10.Bornath C.J., Ranalli,T.A., Henricksen,L.A., Wahl,A.F. and Bambara,R.A. (1999) Effect of flap modifications on human FEN1 cleavage. Biochemistry, 38, 13347–13354. [DOI] [PubMed] [Google Scholar]
- 11.Ceska T.A., Sayers,J.R., Stier,G. and Suck,D. (1996) A helical arch allowing single-stranded DNA to thread through T5 5′- exonuclease. Nature, 382, 90–93. [DOI] [PubMed] [Google Scholar]
- 12.Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: Coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- 13.Hwang K.Y., Baek,K., Kim,H.Y. and Cho,Y. (1998) The crystal structure of Flap Endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol., 5, 707–713. [DOI] [PubMed] [Google Scholar]
- 14.Sayers J.R. and Artymiuk,P.J. (1998) Flexible loops and helical arches. Nat. Struct. Biol., 5, 668–670. [DOI] [PubMed] [Google Scholar]
- 15.Sayers J.R. and Eckstein,F. (1991) A single-strand specific endonuclease activity copurifies with overexpressed T5 D15 exonuclease. Nucleic Acids Res., 19, 4127–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayers J.R. and Eckstein,F. (1990) Properties of overexpressed phage T5 D15 exonuclease. J. Biol. Chem., 265, 18311–18317. [PubMed] [Google Scholar]
- 17.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 18.Paul A.V. and Lehman,I.R. (1966) The Deoxyribonucleases of Escherichia coli. J. Biol. Chem., 241, 3441–3451. [PubMed] [Google Scholar]
- 19.Kejnovsky E. and Kypr,J. (1997) DNA extraction by zinc. Nucleic Acids Res., 25, 1870–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artymiuk P.J., Ceska,T.A., Suck,D. and Sayers,J.R. (1997) Prokaryotic 5′–3′ exonucleases share a common core structure with gamma-delta resolvase. Nucleic Acids Res., 25, 4224–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen B.H., Qiu,J.Z., Hosfield,D. and Tainer,J.A. (1998) Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem. Sci., 23, 171–173. [DOI] [PubMed] [Google Scholar]
- 22.Gutman P.D. and Minton,K.W. (1993) Conserved sites in the 5′–3′ exonuclease domain of Escherichia coli DNA polymerase. Nucleic Acids Res., 21, 4406–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y., Eom,S.H., Wang,J.M., Lee,D.S., Suh,S.W. and Steitz,T.A. (1995) Crystal structure of Thermus aquaticus DNA polymerase. Nature, 376, 612–616. [DOI] [PubMed] [Google Scholar]
- 24.Casareno R.L.B. and Cowan,J.A. (1996) Magnesium vs. manganese cofactors for metallonuclease enzymes. A critical evaluation of thermodynamic binding parameters and stoichiometry. Chem. Commun., 1813–1814. [Google Scholar]
- 25.Mueser T.C., Nossal,N.G. and Hyde,C.C. (1996) Structure of bacteriophage-T4 RNase-H, a 5′ to 3′ RNA–DNA and DNA–DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell, 85, 1101–1112. [DOI] [PubMed] [Google Scholar]
- 26.Maret W. and Vallee,B.L. (1993) Cobalt as probe and label of proteins. Methods Enzymol., 226, 52–71. [DOI] [PubMed] [Google Scholar]
- 27.Cowan J. (1998) Magnesium activation of nuclease enzymes – the importance of water. Inorg. Chim. Acta, 275–276, 24–27. [Google Scholar]
- 28.Carrell C.J., Carrell,H.L., Erlebacher,J. and Glusker,J.P. (1988) Structural aspects of metal ion-carboxylate interactions. J. Am. Chem. Soc., 110, 8651–8656. [Google Scholar]
- 29.Lyamichev V., Brow,M.A.D., Varvel,V.E. and Dahlberg,J.E. (1999) Comparison of the 5′ nuclease activities of Taq DNA polymerase and its isolated nuclease domain. Proc. Natl Acad. Sci. USA, 96, 6143–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp K.A. and Honig,B. (1995) Salt effects on nucleic acids. Curr. Opin. Struct. Biol., 5, 323–328. [DOI] [PubMed] [Google Scholar]
- 31.Saenger W. (1988) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY.
- 32.Nakano S., Fujimoto,M., Hara,H. and Sugimoto,N. (1999) Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res., 27, 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanford N.P., Halford,S.E. and Baldwin,G.S. (1999) DNA cleavage by the EcoRV restriction endonuclease: pH dependence and proton transfers in catalysis. J. Mol. Biol., 288, 105–116. [DOI] [PubMed] [Google Scholar]
- 34.Eldeiry W.S., So,A.G. and Downey,K.M. (1988) Mechanisms of error discrimination by Escherichia coli DNA- Polymerase-I. Biochemistry, 27, 546–553. [DOI] [PubMed] [Google Scholar]
- 35.Gruenwedel D.W. and Cruikshank,M.K. (1990) Mercury-induced DNA polymorphism: Probing the conformation of Hg(II)-DNA via staphylococcal nuclease digestion and circular dichroism measurements. Biochemistry, 29, 2110–2116. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Balbas M.A., Jimenez-Garcia,E. and Azorin,F. (1995) Zinc(II) ions selectively interact with DNA sequences present at the TFIIIA binding site of the Xenopus 5S-RNA gene. Nucleic Acids Res., 23, 2464–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao W. and Barany,F. (1998) Identification of TaqI endonuclease active site residues by Fe2+-mediated oxidative cleavage. J. Biol. Chem., 273, 33002–33010. [DOI] [PubMed] [Google Scholar]
- 38.Viadiu H. and Aggarwal,A.K. (1998) The role of metals in catalysis by the restriction endonuclease BamHI. Nat. Struct. Biol., 5, 910–916. [DOI] [PubMed] [Google Scholar]
- 39.Price P.A. (1975) The essential role of Ca2+ in the activity of bovine pancreatic deoxyribonuclease. J. Biol. Chem., 250, 1981–1986. [PubMed] [Google Scholar]
- 40.Pommer A.J., Wallis,R., Moore,G.R., James,R. and Kleanthous,C. (1998) Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem. J., 334, 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rumbaugh J.A., Murante,R.S., Shi,S. and Bambara,R.A. (1997) Creation and removal of embedded ribonucleotides in chromosomal DNA during mammalian Okazaki fragment processing. J. Biol. Chem., 272, 22591–22599. [DOI] [PubMed] [Google Scholar]
- 42.Garforth S.J., Ceska,T.A., Suck,D. and Sayers,J.R. (1999) Mutagenesis of conserved lysine residues in bacteriophage T5 5′–3′ exonuclease suggests separate mechanisms of endo- and exonucleolytic cleavage. Proc. Natl Acad. Sci. USA, 96, 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]