Abstract
In our previous work we demonstrated that a small protein called affibody can be used for a cytotoxic conjugate development. The anti-HER2 affibody was armed with one moiety of a highly potent auristatin E and specifically killed HER2-positive cancer cells with a nanomolar IC50. The aim of this study was to improve the anti-HER2 affibody conjugate by increasing its size and the number of conjugated auristatin molecules. The affibody was fused to the Fc fragment of IgG1 resulting in a dimeric construct with the molecular weight of 68 kDa, referred to as ZHER2:2891-Fc, ensuring its prolonged half-life in the blood. Due to the presence of four interchain cysteines, the fusion protein could carry four drug molecules. Notably, the in vitro tests of the improved anti-HER2 conjugate revealed that it exhibits the IC50 of 130 pM for the HER2-positive SK-BR-3 cells and 98 nM for the HER2-negative MDA-MB-231 cells. High efficacy and specificity of the auristatin conjugate based on ZHER2:2891-Fc indicate that this construct is suitable for further in vivo evaluation.
Keywords: affibody, HER2, Fc fragment of IgG1, monomethyl auristatin E (MMAE)
1. Introduction
Safe and efficient delivery of cytotoxic payloads to solid tumors remains one of the main challenges for advanced cancer therapies. An antibody-drug conjugate (ADC) consists of a monoclonal antibody that recognizes tumor-associated antigens covalently linked to a highly cytotoxic agent. This composition confers a limited systematic exposure to the drug minimizing adverse effects of the therapy [1,2]. Since the 1980s, many ADCs have undergone clinical trials and currently, more than 60 are being tested [3,4,5,6]. Nevertheless, only two ADCs, brentuximab vedotin (Adcetris, Seattle Genetics) against Hodgin’s lymphoma and ado-trastuzumab emtansine (Kadcyla, Genentech) against metastatic breast cancer, were approved for marketing by the Food and Drug Administration (FDA) and are being used to treat patients.
Conventional ADCs employ monoclonal antibodies as targeting molecules. Recently, the feasibility of other molecules to deliver cytotoxic payloads or radionuclides to cancer cells has been addressed in an increasing number of studies [7,8]. Among the antibody alternatives are antibody fragments such as scFvs (28 kDa) and Fabs (55 kDa) [9,10,11,12], nanobodies (12–15 kDa) [13,14,15,16], affimers (12–14 kDa) [17], signaling molecules [18] and affibodies (6–7 kDa) [19,20,21,22,23,24]. Affibodies are engineered proteins based on a three-helix bundle Z domain that was derived from the IgG-binding B domain of protein A from Staphylococcus aureus [25]. Randomization of 13 surface-exposed residues present in the first two alpha-helices followed by phage display allowed for the generation of affibodies that recognize and bind various molecular targets with high affinity [26,27,28]. One of the most studied affibodies is a variant selected against HER2 receptor (Human Epidermal Growth Factor Receptor 2) [29,30,31]. This receptor is overexpressed in 20–25% of metastatic breast cancer cases [32]. The main advantages of affibodies over antibodies in terms of targeted therapy and molecular imaging include their small size that ensures better penetration of solid tumors, high stability, lack of disulphide bridges and low-cost manufacturing in a bacterial expression system.
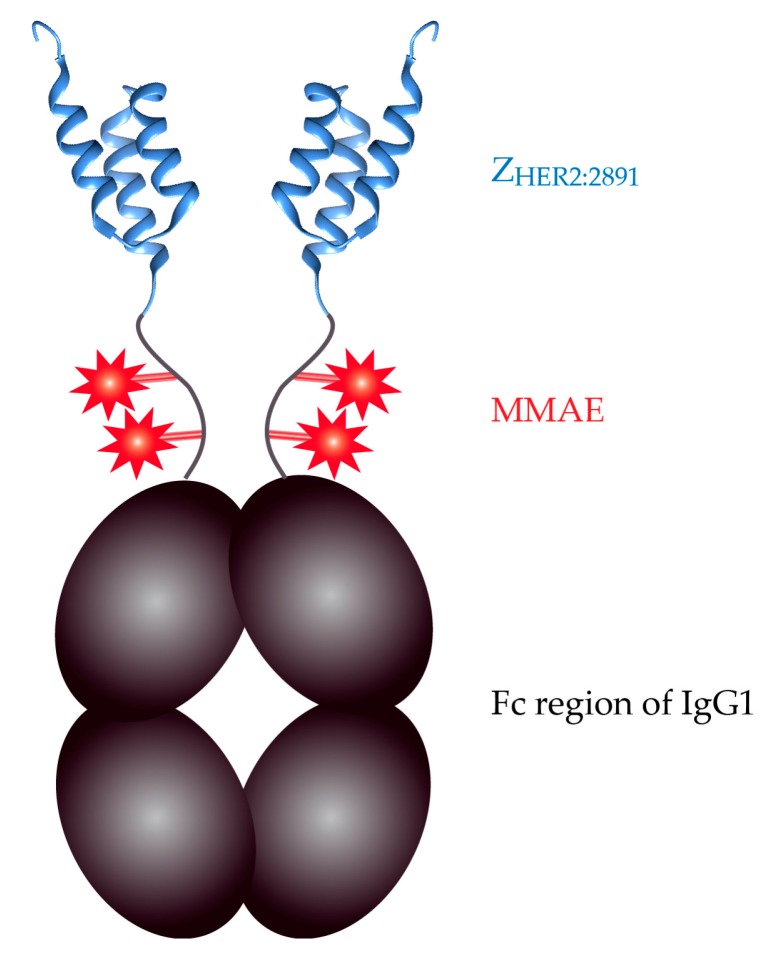
In our previous study, we demonstrated that the anti-HER2 affibody ZHER2:2891, originally designed by Feldwisch et al. (2010) efficiently delivers a highly cytotoxic antimitotic agent, auristatin E, to HER2-positive cancer cells [19]. However, the small size of this conjugate (8647 Da) would result in its fast renal clearance as the molecules smaller than 25–40 kDa are rapidly removed from circulation via kidney filtration [22,33]. In order to expand the size of our conjugate and increase its half-life in blood, we decided to fuse the ZHER2891 affibody to the Fc fragment of IgG1 [34,35,36]. The resulting ZHER2:2891-Fc fusion homodimerizes via Fc has the molecular mass of 68 kDa, which should contribute to its retention in the circulation. Additionally, the Fc fragment ensures that the conjugate will be recycled from epithelial cells back to blood vessels via interaction with FcRn [37,38]. Moreover, the presence of two interchain disulphide bonds in the Fc part allowed us to conjugate four auristatin molecules to our dimeric construct (Figure 1) whereas the previous ZHER2:2891-DCS-MMAE conjugate was loaded only with a single auristatin molecule. The cytotoxicity of our improved construct referred to as ZHER2:2891-Fc-MMAE, was evaluated on breast cancer cell lines.
Figure 1.
Scheme of the ZHER2:2891-Fc-MMAE homodimer.
2. Results
2.1. Expression and Purification of the ZHER2:2891-Fc from Mammalian Cells
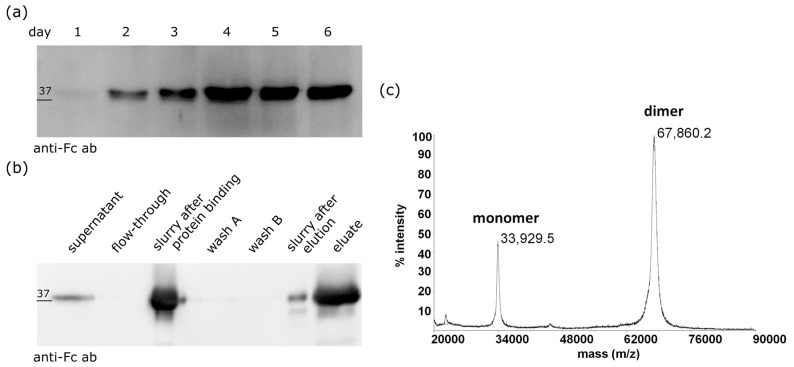
The conjugate developed here is based on the molecule named ZHER2:2891, which exhibits increased thermal stability and hydrophilicity along with lower liver uptake in animals when compared to the parental ZHER2:342 affibody [30,39]. To expand the size of our construct, we fused the ZHER2:2891 molecule to the Fc fragment of IgG1 (Figure 1). A similar fusion was previously proposed by Rönnmark et al. (2002) who fused the Tag DNA polymerase specific affibody to the Fc fragment and purified this construct from E. coli cells [40]. However, the ZHER2:2891-Fc fusion protein was expressed in Chinese Hamster Ovary CHO-S cells to ensure its proper folding and glycosylation [41,42]. The highest levels of ZHER2:2891-Fc in the culture medium were observed since day 4 following the transfection with pLEV113-ZHER2:2891-Fc (Figure 2a). Therefore, ZHER2:2891-Fc was purified on day 5 or day 6 using a single-step affinity chromatography on protein A-Sepharose. The purification process was analyzed by Western blotting with the anti-Fc antibody conjugated with HRP (Figure 2b). The ZHER2:2891-Fc fusion migrated as a 36-kDa band in SDS-PAGE in the presence of the reducing agent, β-mercaptoethanol (5%), in Laemmli sample buffer. Additionally, the identity and purity of the product were confirmed by mass spectrometry (MS) (Figure 2c). The detected m/z peaks corresponded to a covalent dimer (67,860.2 Da) and monomer (33,929.5 Da) of the ZHER2:2891-Fc after cleavage of a 19 amino acid-long secretion signal peptide (2273.7 Da). We obtained 1.5 mg of the ZHER2:2891-Fc protein from 1 liter of CHO-S culture.
Figure 2.
Expression and purification of ZHER2:2891-Fc. (a) The levels of ZHER2:2891-Fc in the CHO-S cells culture medium; (b) Western blot analysis of the purification of ZHER2:2891-Fc; (c) The mass spectrum of the purified ZHER2:2891-Fc.
2.2. Specificity of the ZHER2:2891-Fc Fusion Protein
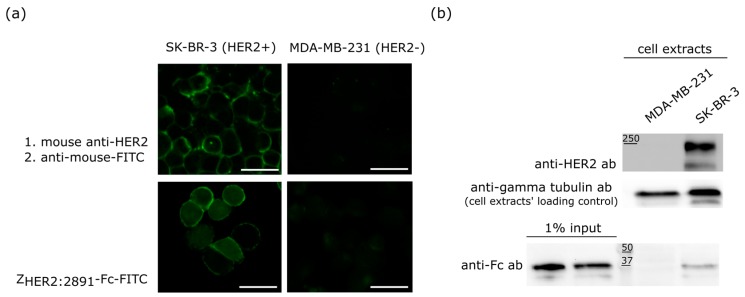
Our previous study clearly demonstrated that ZHER2:2891 selectively recognized HER2 receptor present on the surface of cancer cell lines that overexpress this receptor [19]. Since the proposed fusion contains the same targeting molecule, we verified the specificity of ZHER2:2891-Fc towards two out of four cell lines that were further used to assess cytotoxicity of monomethylauristatin E (MMAE) coupled to ZHER2:2891-Fc. We chose the SK-BR-3 and MDA-MD-231 breast cancer cells as they express very high and low, physiological levels of HER2 receptor, respectively [19,43,44]. The ZHER2:2891-Fc was labeled with FITC and used for the immunostaining experiment. The fluorescent signal was detected on the surface of SK-BR-3 cells when ZHER2:2891-Fc–FITC or anti-HER2 antibody were used to recognize HER2 receptor (Figure 3a). In the case of the HER2-negative MDA-MB-231 cells, only a weak background signal was observed. We also analyzed ZHER2:2891-Fc binding to SK-BR-3 and MDA-MB-231 cells by Western blotting with anti-Fc antibody-HRP. The band corresponding to ZHER2:2891-Fc was present only in the case of the HER2-positive SK-BR-3 cells (Figure 3b). Taken together, these biochemical and microscopic results are consistent with the analysis of ZHER2:2891 binding to different cell lines carried out in our previous study [19] and confirm that the fusion protein specifically recognizes HER2 receptor.
Figure 3.
Specificity of the ZHER2:2891-Fc binding. (a) Fluorescent microscopy images of the HER2-positive SK-BR-3 and MDA-MB-231 HER2-negative cells. The anti-HER2 antibody was detected with a FITC-labeled secondary antibody whereas the affibody was FITC labelled, scale bar = 100 µm; (b) ZHER2:2891-Fc binding to the SK-BR-3 and MDA-MB-231 cells analyzed by Western blotting. The upper panel shows HER2 levels in SK-BR-3 and MDA-MB-231cells, the middle panel severs as a loading control for the experiment, the bottom panel demonstrates that ZHER2:2891-Fc is specifically enriched at the surface of the SK-BR-3 cells.
2.3. The ZHER2:2891-Fc-MMAE Conjugate
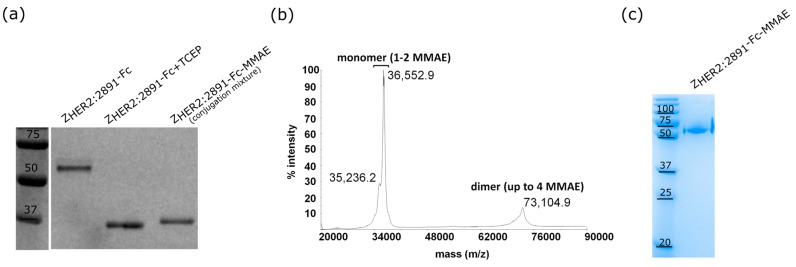
Reduction of the two interchain disulphide bonds between the Fc fragments of the fusion protein resulted in four –SH groups which reacted with the maleimide group present in the drug molecule with a high efficiency (~90%) (Figure 4a). In the MS spectrum recorded for the conjugate, two main m/z peaks were present. The major m/z peak (36,552.9 Da) corresponds to a monomeric ZHER2:2891-Fc loaded with two MMAE molecules whereas the second m/z peak (73,104.9 Da) represents a dimeric ZHER2:2891-Fc carrying up to four auristatin E (MW 1316.7 Da). Our analysis also revealed a trace of the ZHER2:2891-Fc monomer conjugated with one MMAE molecule (m/z = 35,236.2) (Figure 4b). The strong signal detected for the monomeric conjugate loaded with one or two MMAE can be attributed to the breakdown of the non-covalent conjugate dimer since the disulphide bridges were replaced with MMAE molecules. Such noncovalent dimer is prone to decomposition during sample preparation and exposure to a laser beam in the MS experiment. To confirm that the conjugate dimerizes, we subjected ZHER2:2891-Fc-MMAE to a polyacrylamide gel electrophoresis with a low concentration of SDS (0.0375%) in the running buffer under non-reducing conditions [45]. The conjugate migrated as a band of a molecular mass between 50 and 75 kDa, which demonstrates that the dimeric format of ZHER2:2891-Fc is preserved in its auristatin conjugate (Figure 4c).
Figure 4.
Conjugation of MMAE to ZHER2:2891-Fc. (a) The ZHER2:2891-Fc species before and after conjugation separated on 12% SDS-PAGE gel and subjected to Coomassie staining. The samples were heated (96 °C, 10 min) in non-reducing Laemmli buffer prior the electrophoretic separation; (b) The mass spectrum of the conjugation product; (c) The ZHER2:2891-Fc-MMAE conjugate migration on a native 12% polyacrylamide gel under non-reducing conditions stained with Coomassie [45].
2.4. In Vitro Efficacy of the ZHER2:2891-Fc-MMAE
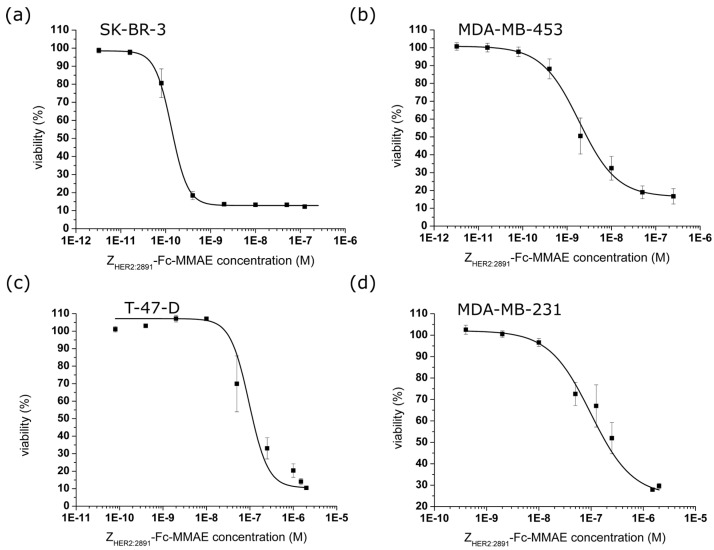
Our previous conjugate based on a single ZHER2:2891 with the C-terminal extension, referred to as drug conjugation sequence (DCS), could carry only a single MMAE moiety. The IC50 of this construct determined for SK-BR-3 cells, which overexpress high levels of HER2 receptor was ~5 nM [19]. Since the ZHER2:2891-Fc-MMAE contained two affibody molecules and was armed with four drug molecules, we had expected to observe a significant decrease in the IC50 value. The cytotoxicity assays were carried out exactly the same way as for ZHER2:2891-MMAE and free MMAE in our previous study [19]. In order to more precisely compare HER2 expression levels between used breast cancer cell lines: MDA-MB-231, T-47-D, MDA-MB-453 and SK-BR-3, we carried out a densitometric quantification on the anti-HER2 Western blots published previously [19]. According to our analysis, HER2 levels in MDA-MB-453 were three to five times higher than in T-47-D cells and about six to eight times lower than in SK-BR-3 cells. We did not detect the band corresponding to HER2 in MDA-MD-231, which confirms that this is a strong HER2-negative line. For clarity, we decided to divide the used cell lines for HER2-positive (SK-BR-3 and MDA-MB-453) and for HER2-negative cell lines (MDA-MB-231 and T-47-D) which is in accordance with literature [43,44]. These four cell lines were subjected to different concentrations of ZHER2:2891-Fc-MMAE and the cell viability was determined with the Alamar Blue assay after 96 h of the continuous exposure to the conjugate. The IC50 values corresponded to the HER2 level of tested cells. Consequently, SK-BR-3 cells exhibited the lowest IC50 value of 134 pM, the IC50 obtained for MDA-MB-453 cells was 1.9 nM whereas IC50 values for T-47-D and MDA-MB-231 were 45.7 nM and 98.2 nM, respectively (Figure 5 and Table 1). Notably, these results show that we significantly improved in vitro efficacy of our new anti-HER2 affibody-based conjugate in comparison to the previous ZHER2:2891-DCS-MMAE construct (Table 1).
Figure 5.
Cytotoxicity of the ZHER2:2891-Fc-MMAE conjugate. The conjugate was tested on four breast cancer cell lines: (a) SK-BR-3 (HER2+); (b) MDA-MB-453 (HER2+); (c) T-47-D (HER2-) and (d) MDA-MB-231 (HER2-). Error bars represent standard error of the mean (SEM).
Table 1.
The cytotoxicity of ZHER2:2891-Fc-MMAE in comparison to the previously published IC50 values obtained for the ZHER2:2891-MMAE conjugate. IC50 results are expressed as mean ± SEM.
| Cytotoxic Agent | SK-BR-3 (10) * | MDA-MB-453 (8) * | T-47-D (5) * | MDA-MB-231 (2) * | |
|---|---|---|---|---|---|
| IC50 (nM) | free MMAE [19] | 1.76 ± 0.26 (n = 3) | 1.27 ± 0.46 (n = 3) | 3.25 ± 1.06 (n =3) | 5.21 ± 1.46 (n = 3) |
| ZHER2:2891-MMAE [19] | 5.16 ± 1.09 (n = 4) | 24.83 ± 5.62 (n =3) | 135.55 ± 22.7 (n = 4) | 161.53 ± 49.9 (n = 3) | |
| ZHER2:2891-Fc-MMAE | 0.13 ± 0.01 (n = 7) | 1.87 ± 0.27 (n = 4) | 45.67 ± 7.43 (n = 3) | 98.22 ± 30.9 (n = 7) |
* The erbB2 gene expression values according to HerceptinR, a database for Herceptin resistance [46].
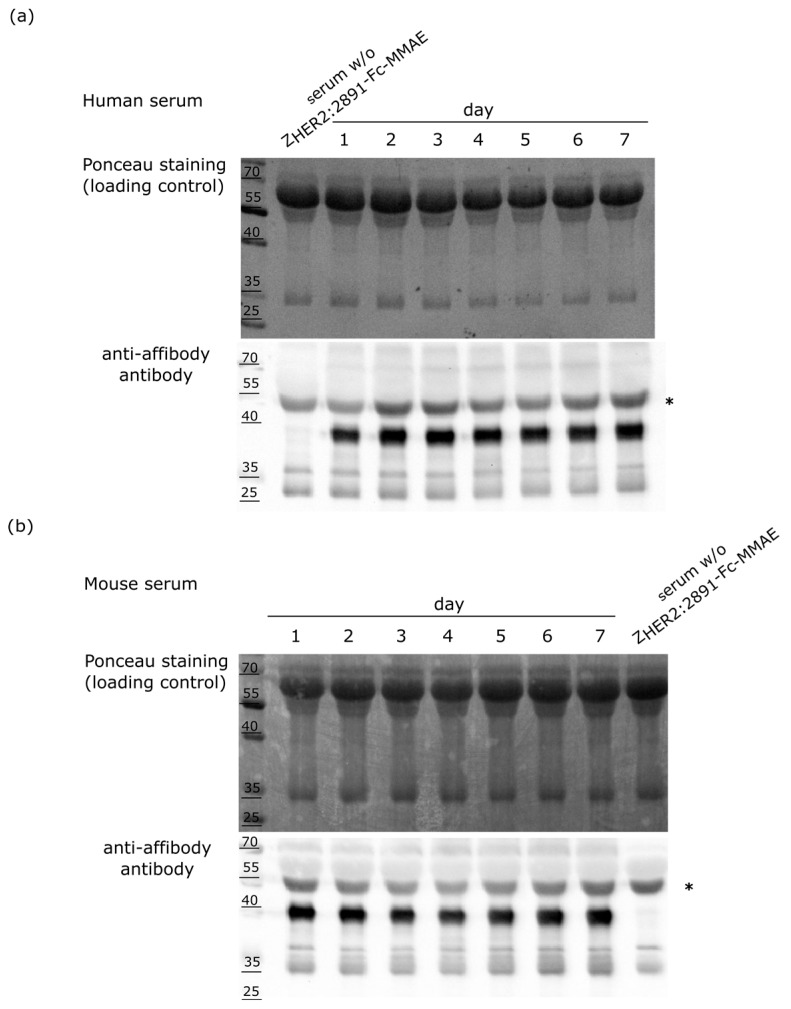
2.5. Stability of the ZHER2:2891-Fc-MMAE Conjugate in Serum
The stability of ZHER2:2891-Fc-MMAE in human and mouse sera was monitored by Western blotting with an anti-affibody antibody. The strong signal of the ZHER2:2891-Fc-MMAE conjugate was detected for each day of the 7-day incubation both in human and mouse plasma (Figure 6a,b). This indicates that in the above-mentioned conditions our construct remains relatively stable since the levels of the conjugate detected by the anti-affibody antibody were comparable between different days of the incubation.
Figure 6.
The stability of ZHER2:2891-Fc-MMAE in (a) human and (b) mouse plasma analyzed by Western blotting. The asterisk indicates a nonspecific band.
3. Discussion
Antibodies are commonly used to generate new cytotoxic conjugates. These proteins are highly specific which ensures that the drug is delivered to target cells overexpressing tumor-associated antigens. However, due to the large size of these molecules (~150 kDa), their ability to penetrate solid tumors is limited [47]. Therefore, other macromolecules are also tested as drug carriers. The anti-HER2 affibodies labeled with radioisotopes have been widely investigated for molecular imaging of HER2-positive tumors [31,48,49,50]. These efforts have recently led to successful clinical trials of the second-generation anti-HER2 affibody loaded with 111In and 68Ga [51,52,53].
The anti-HER2 affibody armed with one MMAE molecule specifically targets HER2-positive cells and destroys them with a low nanomolar IC50 [19]. Here, we improved this conjugate by fusing the Fc fragment of IgG1 to ZHER2:2891 to expand its size and increase the number of the coupled drug molecules (Figure 1). The obtained ZHER2:2891-Fc construct run on SDS-PAGE gel under reducing conditions as a ~36 kD-protein (Figure 2b and Figure 3b) while the mass of the monomeric ZHER2:2891-Fc determined by MS was 33929.5 Da (Figure 2c). The band shift towards higher molecular weight observed on the SDS-PAGE gel and the ~1100 Da discrepancy between the apparent molecular mass of the ZHER2:2891-Fc monomer (33,929.5 Da) and its calculated molecular mass (32,833 Da) suggest that similar to other Fc fusions [41,54], the Fc fragment was glycosylated in CHO-S cells. Combining the anti-HER2 affibody with Fc allowed us to expand the molecular mass of the affibody from ~7 kDa to ~70 kDa, which is still about half of the antibody size. Such size should contribute to a slower kidney removal of our construct since the kidney filtration cutoff is around 40–60 kDa [33]. Additionally, the Fc domain prolongs the serum half-life of antibodies and Fc-fusion proteins due to pH-dependent binding to the neonatal Fc receptor (FcRn) which protects the proteins from endosomal degradation in the epithelial cells. The Fc-fusion bound to FcRn is then recycled back into circulation [34,36,38]. The ability of the Fc region to improve pharmacokinetics has been verified for numerous proteins including eleven Fc fusions approved by FDA till 2017 [55].
The ZHER2:2891-Fc construct specifically recognized HER2-positive cells, which was confirmed by microscopic images and a biochemical binding assay (Figure 3). The presence of Fc allowed us to conjugate MMAE molecules to four cysteines obtained by treating ZHER2:2891-Fc with a reducing agent to break interchain disulphide bonds prior to the conjugation process, as it has been described for many ADCs [56,57,58]. According to the analysis of mass spectra, one or two MMAE molecules were coupled to a single ZHER2:2891-Fc which resulted in up to four MMAE molecules per dimeric ZHER2:2891-Fc (Figure 4b). The ZHER2:2891-Fc-MMAE dimer underwent decomposition to monomer during the MS sample preparation and measurement (Figure 4b). However, the dimeric format of the conjugate was preserved during electrophoretic separation with a low amount of SDS in the running buffer (Figure 4c). This indicates that in physiological conditions ZHER2:2891-Fc-MMAE should remain dimeric.
We obtained the following IC50 values for cells treated with the ZHER2:2891-Fc-MMAE conjugate: 130 pM for SK-BR-3 (HER2+) cells, 1.87 nM for MDA-MB-453 (HER2+) cells, 45.67 nM for T-47-D (HER2-) cells and 98.22 nM for MDA-MB-231 (HER2-) cells (Figure 5 and Table 1). This translates into a significant increase in the sensitivity of HER2-positive cells towards ZHER2:2891-Fc-MMAE in comparison with their sensitivity to ZHER2:2891-DCS-MMAE loaded with a single auristatin E molecule (the 38- and 12-fold increase in the case of SK-BR-3 and MDA-MB-453 cells, respectively). Importantly, the sensitivity of the HER2 negative cells to ZHER2:2891-Fc-MMAE was elevated to a lesser extent (Figure 5 and Table 1). The higher efficacy of the ZHER2:2891-Fc-MMAE conjugate may also be caused by an enhanced internalization of this bivalent construct, which is in accordance with the results demonstrating that cross-linking or clustering of transmembrane receptors accelerates endocytosis [59].
Recently, we have generated an auristatin conjugate based on the bivalent ZHER2:4 affibody referred to as (ZHER2:4)2DCS-MMAE [21]. ZHER2:4 is an earlier version of the anti-HER2 affibody and binds HER2 receptor with the affinity three orders of magnitude lower than ZHER2:2891 [29,30]. The (ZHER2:4)2DCS-MMAE conjugate killed SK-BR-3 cells with a similar efficacy (IC50 = ~500 pM) as ZHER2:2891-Fc-MMAE. However, this bivalent construct is highly heterogeneous since it carries from zero to three MMAE molecules. Moreover, the size of (ZHER2:4)2DCS-MMAE is below 20 kDa which makes it prone to removal through kidney clearance. In this context, the ZHER2:2891-Fc fusion provides a better platform for a cytotoxic conjugate development than single and bivalent affibodies. Interestingly, the dimeric (ZHER2:4)2DCS affibody slightly affected the SK-BR-3 cell viability during a 4-day incubation [21]. This finding is in contrast to our preliminary observation that the ZHER2:2891 affibody fused to the Fc fragment enhances viability of the HER2 positive cells (data not shown). This effect of ZHER2:2891-Fc was not observed for MDA-MB-231 cells that do not overexpress HER2 receptor. We hypothesize that the ZHER2:2891-Fc dimer induces cell proliferation by activating HER2 signaling pathways. However, this possibility needs to be further investigated.
Ado-trastuzumab emtansine (Kadcyla, Genentech), which is one of the two ADCs currently approved for clinical use by the FDA, similarly to the ZHER2:2891-Fc-MMAE, targets HER2 receptor. In this ADC cytotoxic payload, DM1 (emtansine), is attached to lysine amines via the SMCC linker [60,61,62]. This conjugate carries 3.2–3.8 DM1 molecules per antibody on average [61,63] which is close to four that is the optimal drug to antibody ratio (DAR) proposed by Hamblett et al. (2004) [57]. However, the distribution of DM1 is not uniform since there are about 40 lysines that can be modified in an antibody affecting pharmacokinetics of the conjugate [64]. Our conjugate relied on MMAE coupling to the interchain cysteines, which results in defined conjugation sites. This strategy ensures batch-to-batch consistency in terms of MMAE location. Importantly, the efficacy of trastuzumab-DM1 tested on the SK-BR-3 cell lines is comparable to the cytotoxicity of (ZHER2:4)2DCS-MMAE [61].
The Fc part of IgG1 contributes to a cell killing-activity by three mechanisms: antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent phagocytosis (ADPh) and complement-dependent cytotoxicity (CDC) [65,66,67]. Notably, a recent study by Mobergslien et al. demonstrated that a fusion of a targeting peptide, referred as WN, with the Fc fragment activated innate immune cells (NK cells, monocytes, dendritic cells). Injection of this construct to 4T1 tumor-bearing BALB/c mice inhibited tumor growth and the tumor tissue was infiltrated by NK cells and T cells [54]. These findings suggest that the cytotoxicity of the proposed ZHER2:2891-Fc-MMAE conjugate can be further enhanced by its potential to trigger the immunological response directed against HER2-positive cancer cells. This possibility will be addressed in further studies.
4. Materials and Methods
4.1. Construction of the ZHER2:2891-Fc Expression Plasmid
The nucleotide sequence of ZHER2:2891 was amplified by PCR (Polimerase Chain Reaction) from the bacterial expression plasmid pDEST-ZHER2:2891-DCS [19] using the primers A: 5′-CTCTTCTTCCTGTCAGTAACGACTGGTGTCCACTCCGCCGAAGCCAAATATGCAAAA-3′ and B: 5′-TTGCGTCCGGATTTCGGTGCCTGGCTATCAT-3′. This PCR resulted in the DNA fragment encoding the last 12 amino acids of the signaling peptide, followed by the sequence encoding of the affibody and Kpn2I (BspMII) restriction site at the 3′ of the PCR product. The second round of PCR was carried out with primer C: 5′-CTCCAAGCTTTGAACCACCATGGAATGGAGCTGGGTCTT TCTCTTCTTCCTGTCAGTAACG-3′ and B and resulted in the DNA fragment comprising HindIII restriction site at the 5′, the sequence encoding all amino acids of the signal peptide, the affibody and Kpn2I (BspMII) restriction site at the 3’ of the PCR product. The PCR product and the pLEV113-ECD_FGFR2-Fc construct [41] were digested with HindIII and Kpn2I (BspMII) and subjected to ligation using T4 ligase. The resulting construct pLEV113-ZHER2:2891-Fc was verified by sequencing, propagated in DH10α E. coli cells and purified using Plasmid Maxi Kit (Qiagen, Hilden, Germany).
4.2. Protein Expression
The ZHER2:2891-Fc expression was carried out according to Sokolowska-Wedzina et al. (2014) with minor modifications [41]. Briefly, CHO-S cells (Invitrogen, Carlsbad, CA, USA) were cultured in PowerCHO-2CD medium (Lonza, Basel, Switzerland) supplemented with 8 mM L-glutamine and 1× penicillin/streptomycin solution (Biowest, Nuaillé, France). Cells were grown at 37 °C in a shaking incubator (140 rpm) with 8% CO2. The cells were diluted with fresh medium to density of 0.3 × 106 cells/mL when the culture density reached 1–1.2 × 106 cells/mL. One day before transfection the CHO-S cells were diluted to 0.6 × 106 cells/mL. On the day of transfection culture was centrifuged and cell pellet were resuspended in ProCHO4 medium (Lonza, Basel, Switzerland) at 2 × 106 cells/mL. Appropriate amount of plasmid (1.25 µg of DNA per 1 × 106 cells/mL) was resuspended in 150 mM NaCl. The final volume of the DNA solution was 5% of the cell suspension volume. Appropriate amount of 1 mg/mL linear PEI (Polyscience, Warrington, PA, USA) was added to 150 mM NaCl aiming for 5 µg PEI per 1 × 106 cells/mL. The final volume of the PEI solution was 5% of the cell suspension. The DNA and PEI solutions were mixed together, incubated at room temperature for 10 min and then added dropwise to the cell solution. The cells were incubated for 4 h in standard conditions (37 °C, 140 rpm, 8% CO2). After this time, the culture was diluted with an equal volume of PowerCHO-2CD supplemented with 8 mM l-glutamine and 2× penicillin-streptomycin solution (Biowest, Nuaillé, France) and incubated at 31 °C. On the second day following the transfection the culture was supplemented with an additional 4 mM l-glutamine and harvested on sixth day.
4.3. Protein Purification
The purification of ZHER2:2891-Fc was carried out according to Sokolowska-Wedzina et al. (2014) with minor modifications [41]. EDTA was added to the CHO-S cell culture to a final concentration of 2 mM and the culture was centrifuged (25 min at 15,000× g, 4 °C). Supernatant was collected, centrifuged (25 min at 15,000× g, 4 °C), filtered through 0.22-micron filter units and loaded onto a Protein A Sepharose column (GE Healthcare, Chicago, IL, USA) pre-equilibrated with PBS. The unbound fraction was removed and the resin was washed with buffer A: PBS, 200 mM NaCl, 0.1% Tween 20, 2 mM EDTA, pH 7.4 and then with buffer B: PBS, 500 mM NaCl, 2 mM EDTA, pH 7.4. ZHER2:2891-Fc was eluted with 100 mM triethylamine, and neutralized with 1 M Tris–HCl, pH 7.2. Fractions with the highest protein concentrations were collected, dialyzed against PBS, and concentrated using Centriprep 10 K centrifugal filter units (Mreck Millipore, Billerica, MA, USA). Protein purity was checked by SDS–PAGE and their identity was confirmed by mass spectrometry.
4.4. Mass Spectrometry
ZHER2:2891-Fc and its conjugate were detected by MS 4800 Plus MALDI TOF/TOF Analyzer (AB Sciex, Framingham, MA, USA) with sinapic acid (Sigma-Aldrich, Saint Louis, MO, USA) 10 mg/mL 50% acetonitrile 0.1% TFA as a matrix.
4.5. Western Blotting
Western blotting was performed as described in Sochaj-Gregorczyk et al. (2016) [19]. Primary antibodies used in this study include: the goat anti-affibody antibody (Affibody AB, Solna Sweden) (conjugate stability assay), the anti-Fc antibody-HRP (KPL, Milford, MA, USA) (expression and purification analysis, ZHER2:2891-Fc binding to SK-BR-3 and MDA-MB-231 cell surface), the mouse anti-gamma-tubulin monoclonal antibody (Sigma-Aldrich, Saint Louis, MO, USA) (ZHER2:2891-Fc binding to SK-BR-3 and MDA-MB-231 cell surface) and the mouse anti-HER2 (e2-4001) monoclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA) (ZHER2:2891-Fc binding to SK-BR-3 and MDA-MB-231 cell surface). For detection of the goat and mouse primary antibodies, the secondary anti-goat antibody-HRP (Jackson ImmuoResearch, West Grove, PA, USA) and anti-mouse antibody-HRP (Jackson ImmuoResearch, West Grove, PA, USA) were used, respectively.
4.6. The ZHER2:2891-Fc-MMAE Conjugate Preparation
ZHER2:2891-Fc at the concentration of 0.5 mg/mL in PBS pH 7.2 was subjected to reduction with a 10 molar excess of tris-2-karboksyetylofosfine (TCEP) for 2 h at 37 °C. The reduced ZHER2:2891-Fc was incubated with a 16 molar excess of MC-Val-Cit-PABC-MMAE (maleimidocaprylvaline-citruline-p-amino-benzyloxycarbonyl-mono-methylauristatin E) (ChiroBlock, Wolfen, Germany) for 3 h with gentle mixing at 4 °C. The conjugation mixture was centrifuged at 20,000× g for 10 min. Unconjugated MMEA was removed using Zeba spin column (Thermo Fisher Scientific, Waltham, MA, USA). The efficiency of conjugation was monitored on 12.5% SDS-PAGE gel and by mass spectrometry. The conjugate was stored at 4 °C.
4.7. ZHER2:2891-Fc Labeling with FITC
50 µg fluorescein 5(6)-isothiocyanate (FITC) (Sigma-Aldrich, Saint Louis, MO, USA) was added to 500 µg of ZHER2:2891-Fc dissolved in PBS, pH 7.2. The sample was incubated in the dark for 3 h. In order to remove unconjugated FITC, the PD10 column (GE Healthcare, Chicago, IL, USA) was used.
4.8. Immunostaining
SK-BR-3 and MDA-MB-231 cells were allowed to grow in appropriate media on cover slips that were placed in a 6-well plates for two days. Cells were washed twice with PBS and then fixed with 4% paraformaldehyde for 10 minutes. After paraformaldehyde removal cells were washed 3 times with PBS and then incubated with PBS, 2% BSA, 0.01% Triton X-100 for 15 min at RT. Cells were washed twice with PBS, 0.01% Triton X-100 and incubated for 1 h with a mouse anti-HER2 (e2-4001) monoclonal antibody (Thermo Fisher Scientific, Waltham, MA, USA) or ZHER2:2891-Fc-FITC diluted in PBS, 0.01% Triton X-100. Following the incubation, cells were washed three times with PBS, 0.01% Triton X-100. Then a 1 h incubation with a secondary fluorescein (FITC) AffiniPure F (ab’) fragment donkey anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, USA) was carried out for the cells stained with the mouse anti-HER2 antibody. Following three washes with PBS, 0.01% Triton X-100 cover slips were mounted onto slides.
4.9. Microscopy
Microscopical analysis was performed as described in Sochaj-Gregorczyk et al. (2016) [19].
4.10. Cell Lines
The following breast cancer cell lines were used in this study: SK-BR-3, MDA-MB-453, MDA-MB-231 and T-47-D (American Type Culture Collection, Manassas, VA, USA). The MDA-MB-453, MDA-MB-231 and T-47-D cells were cultured in Dulbecco's Modified Eagle Medium, DMEM (Sigma-Aldrich, Saint Louis, MO, USA), and the SK-BR-3 in the McCoy’s 5A medium (Lonza, Basel, Switzerland). All media contained 2 mM l-glutamine and were supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) and 1 x penicillin-streptomycin solution (Biowest, Nuaillé, France). The cells were cultured at 37 °C in 5% CO2 and humidified atmosphere. Upon reaching 90% confluence, the cells were passaged using Trypsin-EDTA solution (Thermo Fisher Scientific, Waltham, MA, USA)
4.11. Cytotoxicity Assays
The early-passage cells were seeded at the density of 5000 cells per well in a 96-well plate. The cells were allowed to attach and after 24 h different concentrations of the conjugate were added to the wells. After 96-incubation of continuous exposure to the ZHER2:2891-Fc-MMAE conjugate, the medium was replaced with a fresh medium containing 10% of AlamarBlue reagent (Thermo Fisher Scientific, Waltham, MA, USA). After 4 h, formation of a fluorescent reduced form of the dye was determined using EnVision Multimode Plate Reader (PerkinElmer, Waltham, MA, USA). The IC50 values were determined using the OriginPro8 software v8.0724 (OriginLab, Northampton, MA, USA).
4.12. Monitoring Stability of the ZHER2:2891-Fc-MMAE Conjugate in Mouse and Human Sera
For the stability assay, 10 µL of conjugate (0.5–1 mg/mL solution) was added to 90 µL of mouse (Invitrogen, Carlsbad, CA, USA) or human serum (Sigma-Aldrich, Saint Louis, MO, USA) and incubated for 6 days at 37 °C. The eppendorf tubes were sealed with parafilm to minimize evaporation. The 10 µL samples were taken every 24 h, mixed with reducing Laemmli sample buffer, boiled (96 °C, 10 min) and separated on 12% SDS-PAGE gel. Upon electrotransfer the membrane was stained with Ponceus S solution and subjected to Western blotting with the goat anti-affibody antibody (Affibody AB, Solna, Sweden) and secondary anti-goat antibody-HRP (Jackson ImmuoResearch, West Grove, PA, USA).
Acknowledgments
This work was supported by the National Science Centre, Poland (grants number 2013/08/S/NZ1/00845 and 2011/02/A/NZ1/00066). The cost of publication was covered by the Polish Ministry of Science and Higher Education under the Leading National Research Centre (KNOW) Programme (2014–2018).
Author Contributions
Alicja M. Sochaj-Gregorczyk—experiments design, execution and analysis, manuscript preparation and editing; Patryk Ludzia and Emilia Kozdrowska—experiments execution and analysis; Piotr Jakimowicz—mass spectrometry analysis; Aleksandra Sokolowska-Wedzina—eksperiments design, manuscript edition; Jacek Otlewski—manuscript edition, revision and proof-reading.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zolot R.S., Basu S., Million R.P. Antibody-drug conjugates. Nat. Rev. Drug Discov. 2013;12:259–260. doi: 10.1038/nrd3980. [DOI] [PubMed] [Google Scholar]
- 2.Diamantis N., Banerji U. Antibody-drug conjugates—an emerging class of cancer treatment. Br. J. Cancer. 2016;114:362–367. doi: 10.1038/bjc.2015.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters C., Brown S. Antibody-drug conjugates as novel anti-cancer chemotherapeutics. Biosci. Rep. 2015;35:e00225. doi: 10.1042/BSR20150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senter P.D., Sievers E.L. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 5.Perez H.L., Cardarelli P.M., Deshpande S., Gangwar S., Schroeder G.M., Vite G.D., Borzilleri R.M. Antibody-drug conjugates: Current status and future directions. Drug Discov. Today. 2014;19:869–881. doi: 10.1016/j.drudis.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Beck A., Goetsch L., Dumontet C., Corvaia N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 7.Casi G., Neri D. Antibody–drug conjugates and small molecule–drug conjugates: Opportunities and challenges for the development of selective anticancer cytotoxic agents. J. Med. Chem. 2015;25:8751–8761. doi: 10.1021/acs.jmedchem.5b00457. [DOI] [PubMed] [Google Scholar]
- 8.Spiess C., Zhai Q., Carter P.J. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol. Immunol. 2015;67:95–106. doi: 10.1016/j.molimm.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Nelson A.L. Antibody fragments: Hope and hype. MAbs. 2010;2:77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junutula J.R., Bhakta S., Raab H., Ervin K.E., Eigenbrot C., Vandlen R., Scheller R.H., Lowman H.B. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. J. Immunol. Methods. 2008;332:41–52. doi: 10.1016/j.jim.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Petters E., Sokolowska-Wedzina A., Otlewski J. Selection and characterization of single chain antibody fragments specific for Hsp90 as a potential cancer targeting molecule. Int. J. Mol. Sci. 2015;16:19920–19935. doi: 10.3390/ijms160819920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandström K., Haylock A.K., Spiegelberg D., Qvarnström F., Wester K., Nestor M. A novel CD44v6 targeting antibody fragment with improved tumor-to-blood ratio. Int. J. Oncol. 2012;40:1525–1532. doi: 10.3892/ijo.2012.1352. [DOI] [PubMed] [Google Scholar]
- 13.Teillaud J.L. From whole monoclonal antibodies to single domain antibodies: Think small. Methods Mol. Biol. 2012;911:3–13. doi: 10.1007/978-1-61779-968-6_1. [DOI] [PubMed] [Google Scholar]
- 14.Cortez-Retamozo V., Backmann N., Senter P.D., Wernery U., De Baetselier P., Muyldermans S., Revets H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004;15:2853–2857. doi: 10.1158/0008-5472.CAN-03-3935. [DOI] [PubMed] [Google Scholar]
- 15.De Meyer T., Muyldermans S., Depicker A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014;32:263–270. doi: 10.1016/j.tibtech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira S., Heukers R., Sornkom J., Kok R.J., van Bergen en Henegouwen P.M. Targeting tumors with nanobodies for cancer imaging and therapy. J. Control. Release. 2013;28:607–617. doi: 10.1016/j.jconrel.2013.08.298. [DOI] [PubMed] [Google Scholar]
- 17.Tiede C., Tang A.A., Deacon S.E., Mandal U., Nettleship J.E., Owen R.L., George S.E., Harrison D.J., Owens R.J., Tomlinson D.C., et al. Adhiron: A stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng. Des. Sel. 2014;27:145–155. doi: 10.1093/protein/gzu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szlachcic A., Zakrzewska M., Lobocki M., Jakimowicz P., Otlewski J. Design and characteristics of cytotoxic fibroblast growth factor 1 conjugate for fibroblast growth factor receptor-targeted cancer therapy. Drug Des. Devel. Ther. 2016;10:2547–2560. doi: 10.2147/DDDT.S105896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sochaj-Gregorczyk A.M., Serwotka-Suszczak A.M., Otlewski J. A novel affibody-auristatin E conjugate with a potent and selective activity against HER2+ cell lines. J. Immunother. 2016;39:223–232. doi: 10.1097/CJI.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 20.Tolmachev V., Tran T.A., Rosik D., Sjöberg A., Abrahmsén L., Orlova A. Tumor targeting using affibody molecules: Interplay of affinity, target expression level, and binding site composition. J. Nucl. Med. 2012;53:953–960. doi: 10.2967/jnumed.111.101527. [DOI] [PubMed] [Google Scholar]
- 21.Serwotka-Suszczak A.M., Sochaj-Gregorczyk A.M., Pieczykolan J., Krowarsch D., Jelen F., Otlewski J. A conjugate based on anti-HER2 diaffibody and auristatin E targets HER2-positive cancer cells. Int. J. Mol. Sci. 2017;18:E401. doi: 10.3390/ijms18020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zielinski R., Lyakhov I., Hassan M., Kuban M., Shafer-Weaver K., Gandjbakhche A., Capala J. HER2-affitoxin: A potent therapeutic agent for the treatment of HER2-overexpressing tumors. Clin. Cancer Res. 2011;1:5071–5081. doi: 10.1158/1078-0432.CCR-10-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., Seijsing J., Frejd F.Y., Tolmachev V., Gräslund T. Target-specific cytotoxic effects on HER2-expressing cells by the tripartite fusion toxin ZHER2:2891-ABD-PE38X8, including a targeting affibody molecule and a half-life extension domain. Int. J. Oncol. 2015;47:601–609. doi: 10.3892/ijo.2015.3027. [DOI] [PubMed] [Google Scholar]
- 24.Zielinski R., Lyakhov I., Jacobs A., Chertov O., Kramer-Marek G., Francella N., Stephen A., Fisher R., Blumenthal R., Capala J. Affitoxin—A Novel Recombinant, HER2-Specific, Anti-Cancer Agent for Targeted Therapy of HER2-Positive Tumors. J. Immunother. 2009;32:817–825. doi: 10.1097/CJI.0b013e3181ad4d5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson B., Moks T., Jansson B., Abrahmsén L., Elmblad A., Holmgren E., Henrichson C., Jones T.A., Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 26.Nygren P.A. Alternative binding proteins: Affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008;275:2668–2676. doi: 10.1111/j.1742-4658.2008.06438.x. [DOI] [PubMed] [Google Scholar]
- 27.Löfblom J., Feldwisch J., Tolmachev V., Carlsson J., Ståhl S., Frejd F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010;584:2670–2680. doi: 10.1016/j.febslet.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Nord K., Nilsson J., Nilsson B., Uhlén M., Nygren P.A. A combinatorial library of an α-helical bacterial receptor domain. Protein Eng. 1995;8:601–608. doi: 10.1093/protein/8.6.601. [DOI] [PubMed] [Google Scholar]
- 29.Wikman M., Steffen A.-C., Gunneriusson E., Gunneriusson E., Tolmachev V., Adams G.P., Carlsson J., Ståhl S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004;17:455–462. doi: 10.1093/protein/gzh053. [DOI] [PubMed] [Google Scholar]
- 30.Feldwisch J., Tolmachev V., Lendel C., Herne N., Sjöberg A., Larsson B., Rosik D., Lindqvist E., Fant G., Höidén-Guthenberg I., Galli J., Jonasson P., Abrahmsén L. Design of an optimized scaffold for affibody molecules. J. Mol. Biol. 2010;398:232–247. doi: 10.1016/j.jmb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Orlova A., Magnusson M., Eriksson T.L., Nilsson M., Larsson B., Höidén-Guthenberg I., Widström C., Carlsson J., Tolmachev V., Ståhl S., et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66:4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 32.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasarao M., Galliford C.V., Low P.S. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015;14:203–219. doi: 10.1038/nrd4519. [DOI] [PubMed] [Google Scholar]
- 34.Beck A., Reichert J.M. Therapeutic Fc-fusion proteins and peptides as successful alternatives to antibodies. MAbs. 2011;3:415–416. doi: 10.4161/mabs.3.5.17334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kontermann R.E. Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 2011;22:868–876. doi: 10.1016/j.copbio.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Strohl W.R. Fusion proteins for half-life extension of biologics as a strategy to make biobetters. BioDrugs. 2015;29:215–239. doi: 10.1007/s40259-015-0133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitragotri S., Burke P.A., Langer R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roopenian D.C., Akilesh S. FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 39.Ahlgren S., Orlova A., Wållberg H., Hansson M., Sandström M., Lewsley R., Wennborg A., Abrahmsén L., Tolmachev V., Feldwisch J. Targeting of HER2-expressing tumors using 111In-ABY-025, a second-generation affibody molecule with a fundamentally reengineered scaffold. J. Nucl. Med. 2010;51:1131–1138. doi: 10.2967/jnumed.109.073346. [DOI] [PubMed] [Google Scholar]
- 40.Rönnmark J., Hansson M., Nguyen T., Uhlén M., Robert A., Ståhl S., Nygren P.A. Construction and characterization of affibody-Fc chimeras produced in Escherichia coli. J. Immunol. Methods. 2002;261:199–211. doi: 10.1016/S0022-1759(01)00563-4. [DOI] [PubMed] [Google Scholar]
- 41.Sokolowska-Wedzina A., Borek A., Chudzian J., Jakimowicz P., Zakrzewska M., Otlewski J. Efficient production and purification of extracellular domain of human FGFR-Fc fusion proteins from Chinese hamster ovary cells. Protein Expr. Purif. 2014;99:50–57. doi: 10.1016/j.pep.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Hossler P., Khattak S.F., Li Z.J. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–949. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- 43.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally. Cancer Cell. 2009;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holliday D.L., Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowakowski A.B., Wobig W.J., Petering D.H. Native SDS-PAGE: High resolution electrophoretic separation of proteins with retention of native properties including bound metal ions. Metallomics. 2014;6:1068–1078. doi: 10.1039/C4MT00033A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad S., Gupta S., Kumar R., Varshney G.C., Raghava G.P. Herceptin resistance database for understanding mechanism of resistance in breast cancer patients. Sci. Rep. 2014;4:4483. doi: 10.1038/srep04483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee C.M., Tannock I.F. The distribution of the therapeutic monoclonal antibodies cetuximab and trastuzumab within solid tumors. BMC Cancer. 2010;10:255. doi: 10.1186/1471-2407-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trousil S., Hoppmann S., Nguyen Q.D., Kaliszczak M., Tomasi G., Iveson P., Hiscock D., Aboagye E.O. Positron emission tomography imaging with 18F-labeled ZHER2:2891 affibody for detection of HER2 expression and pharmacodynamic response to HER2-modulating therapies. Clin. Cancer Res. 2014;20:1632–1643. doi: 10.1158/1078-0432.CCR-13-2421. [DOI] [PubMed] [Google Scholar]
- 49.Kramer-Marek G., Bernardo M., Kiesewetter D.O., Bagci U., Kuban M., Aras O., Zielinski R., Seidel J., Choyke P., Capala J. PET of HER2-Positive Pulmonary Metastases with 18F-ZHER2:342 affibody in a murine model of breast cancer: Comparison with 18F-FDG. J. Nucl. Med. 2012;53:939–946. doi: 10.2967/jnumed.111.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Zhao X., Wang S., Wang N., Han J., Jia L., Ren X. Monitoring therapeutic response of human ovarian cancer to trastuzumab by SPECT imaging with (99m)Tc-peptide-ZHER2:342. Nucl. Med. Biol. 2015;42:541–546. doi: 10.1016/j.nucmedbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 51.Sörensen J., Sandberg D., Sandström M., Wennborg A., Feldwisch J., Tolmachev V., Åström G., Lubberink M., Garske-Román U., Carlsson J., et al. First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule. J. Nucl. Med. 2014;55:730–735. doi: 10.2967/jnumed.113.131243. [DOI] [PubMed] [Google Scholar]
- 52.Sandberg D., Tolmachev V., Velikyan I., Olofsson H., Wennborg A., Feldwisch J., Carlsson J., Lindman H., Sörensen J. Intra-image referencing for simplified assessment of HER2-expression in breast cancer metastases using the affibody molecule ABY-025 with PET and SPECT. Eur. J. Nucl. Med. Mol. Imaging. 2017;44:1337–1346. doi: 10.1007/s00259-017-3650-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sörensen J., Velikyan I., Sandberg D., Wennborg A., Feldwisch J., Tolmachev V., Orlova A., Sandström M., Lubberink M., Olofsson H., et al. Measuring HER2-receptor expression in metastatic breast cancer using [68Ga]ABY-025 Affibody PET/CT. Theranostics. 2016;6:262–271. doi: 10.7150/thno.13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mobergslien A., Peng Q., Vasovic V., Sioud M. Cancer cell-binding peptide fused Fc domain activates immune effector cells and blocks tumor growth. Oncotarget. 2016;7:75940–75953. doi: 10.18632/oncotarget.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jafari R., Zolbanin N.M., Rafatpanah H., Majidi J., Kazemi T. Fc-fusion proteins in therapy: An updated view. Curr. Med. Chem. 2017;4:1–10. doi: 10.2174/0929867324666170113112759. [DOI] [PubMed] [Google Scholar]
- 56.Doronina S.O., Toki B.E., Torgov M.Y., Mendelsohn B.A., Cerveny CG., Chace D.F., DeBlanc R.L., Gearing R.P., Bovee T.D., Siegall C.B., et al. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat. Biotechnol. 2003;21:778–784. doi: 10.1038/nbt832. [DOI] [PubMed] [Google Scholar]
- 57.Hamblett K.J., Senter P.D., Chace D.F., Sun M.M., Lenox J., Cerveny C.G., Kissler K.M., Bernhardt S.X., Kopcha A.K., Zabinski R.F., et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin. Cancer Res. 2004;10:7063–7070. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 58.Sun M.M., Beam K.S., Cerveny C.G., Hamblett K.J., Blackmore R.S., Torgov M.Y., Handley F.G., Ihle N.C., Senter P.D., Alley S.C., et al. Reduction−alkylation strategies for the modification of specific monoclonal antibody disulfides. Bioconjug. Chem. 2005;16:1282–1290. doi: 10.1021/bc050201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moody P.R., Sayers E.J., Magnusson J.P., Alexander C., Borri P., Watson P., Jones A.T. Receptor crosslinking: A general method to trigger internalization and lysosomal targeting of therapeutic receptor:ligand complexes. Mol. Ther. 2015;23:1888–1898. doi: 10.1038/mt.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barginear M.F., John V., Budman D.R. Trastuzumab-DM1: A clinical update of the antibody–drug conjugate for HER2-overexpressing breast cancer. Mol. Med. 2013;18:1473–1479. doi: 10.2119/molmed.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis Phillips G.D., Li G., Dugger D.L., Crocker L.M., Parsons K.L., Mai E., Blättler W.A., Lambert J.M., Chari R.V.J., Lutz R.J., et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 62.LoRusso P.M., Weiss D., Guardino E., Girish S., Sliwkowski M.X. Trastuzumab emtansine: A unique antibody–drug conjugate in development for human epidermal growth factor receptor 2-positive cancer. Clin. Cancer Res. 2011;17:6437–6447. doi: 10.1158/1078-0432.CCR-11-0762. [DOI] [PubMed] [Google Scholar]
- 63.Kim M.T., Chen Y., Marhoul J., Jacobson F. Statistical modeling of the drug load distribution on trastuzumab emtansine (kadcyla), a lysine-linked antibody drug conjugate. Bioconjug. Chem. 2014;25:1223–1232. doi: 10.1021/bc5000109. [DOI] [PubMed] [Google Scholar]
- 64.Wang L., Amphlett G., Blättler W.A., Lambert J.M., Zhang W. Structural characterization of the maytansinoid–monoclonal antibody immunoconjugate, huN901-DM1, by mass spectrometry. Protein Sci. 2005;14:2436–2446. doi: 10.1110/ps.051478705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki M., Kato C., Kato A. Therapeutic antibodies: Their mechanisms of action and the pathological findings they induce in toxicity studies. J. Toxicol. Pathol. 2015;28:133–139. doi: 10.1293/tox.2015-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Redman J.M., Hill E.M., AlDeghaither D., Weiner L.M. Mechanisms of action of therapeutic antibodies for cancer. Mol. Immunol. 2015;67:28–45. doi: 10.1016/j.molimm.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zipfel P.F., Skerka C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]