Abstract
Chromatin on the inactive X chromosome (Xi) of female mammals is enriched for the histone variant macroH2A that can be detected at interphase as a distinct nuclear structure referred to as a macro chromatin body (MCB). Green fluorescent protein-tagged and Myc epitope-tagged macroH2A readily form an MCB in the nuclei of transfected female, but not male, cells. Using targeted disruptions, we have identified two macrochromatin domains within macroH2A that are independently capable of MCB formation and association with the Xi. Complete removal of the non-histone C-terminal tail does not reduce the efficiency of association of the variant histone domain of macroH2A with the Xi, indicating that the histone portion alone can target the Xi. The non-histone domain by itself is incapable of MCB formation. However, when directed to the nucleosome by fusion to core histone H2A or H2B, the non-histone tail forms an MCB that appears identical to that of the endogenous protein. Mutagenesis of the non-histone portion of macroH2A localized the region required for MCB formation and targeting to the Xi to an ∼190 amino acid region.
INTRODUCTION
The levels of X-linked gene expression between male and female mammals are normalized by rendering all but one X chromosome largely transcriptionally silent in female cells (1). X chromosome inactivation occurs early in development and the choice of which chromosome to inactivate is random (2). The inactive X chromosome (Xi) is heterochromatic and can be observed as a staining mass (Barr body) at the periphery of interphase nuclei (3). Several general features of gene silencing characterize the Xi, including DNA hypermethylation (4,5), histone H3 and H4 hypoacetylation (6–8) and late replication in S phase (9,10).
In addition, chromatin of the Xi demonstrates a number of unique features that distinguish it from other regions of heterochromatin. One such feature is the colocalization with the Xi of a large untranslated RNA, the X inactive specific transcript (XIST) (11–13). XIST is expressed exclusively from the Xi and associates in cis along the length of the chromosome. Another characteristic of the Xi is its distinct nucleosome composition. Four variants of the core histone H2A have a differential distribution in chromatin of the Xi. Two variants, macroH2A1 and macroH2A2, are enriched on the Xi (14–16), while the variants H2A-Bbd, and to a lesser extent H2A.Z, are deficient on the Xi (15,17).
MacroH2A1 and macroH2A2 are unusual histone H2A variants that have an extensive C-terminal tail that comprises nearly two-thirds of the protein. Originally identified through its association with the nucleosome (18), macroH2A is highly localized in female cells as a distinct nuclear body, referred to as a macro chromatin body (MCB), which is coincident with the Xi and the Barr body (14). Two distinct macroH2A genes encode the macroH2A1 and macroH2A2 proteins which are 80% identical and are both enriched on the Xi chromatin (15,16). In undifferentiated embryonic stem cells, macroH2A is sequestered at the centrosome (19) and is relocated to the Xi after the counting and choice of which X chromosome to inactivate has been made (19,20). This implies that macroH2A is not directly involved in the early stages of X inactivation. Despite a physical association with XIST RNA (21), a role for macroH2A in the maintenance of X inactivation appears redundant, as deletion of XIST after inactivation results in loss of macroH2A association with the Xi, yet gene silencing is apparently not affected (22). This may reflect generally the highly redundant nature of X inactivation, as removal of a single silencing feature does not lead to reactivation of gene expression from the Xi (4,22–27). Therefore, while the precise role of macroH2A in the X inactivation process is unclear, macroH2A is a key component of Xi chromatin and may be a critical player in imprinted X inactivation of pre-implantation embryos (28).
Expression of a green fluorescent protein (GFP)-tagged macroH2A1 or macroH2A2 in cells in culture demonstrates an MCB that is indistinguishable from the endogenous proteins (15,17,29). In this manuscript we describe an assay for MCB formation and the disruption of macroH2A to identify functional domains of macroH2A that are critical for the formation of an MCB and its targeting to the Xi.
MATERIALS AND METHODS
Mammalian expression constructs
GFP-tagged and Myc epitope-tagged versions of human macroH2A1, macroH2A2, H2B and H2A were generated as described previously (15). Full-length human cDNA clones of HP1-α (IMAGE 627533) and HMG14 (IMAGE 645015) were obtained from Research Genetics. The complete coding sequence from each clone was PCR-amplified with primers incorporating restriction enzyme recognition sites and subcloned into pcDNA3.1-CT-GFP (Invitrogen) using standard techniques (30). To ensure sequence integrity, subclones were sequenced with a fluorescence labeled dye-terminator cycle sequencing kit according to the manufacturer’s instructions (PRISM Ready DyeDeoxy Terminator Premix from Applied Biosystems Inc.) and electrophoresed on an ABI 373 (Perkin-Elmer). Chimeric constructs of H2A-macroH2A1, H2B-macroH2A1, H2B-macroH2A2, HP1-α-macroH2A1 and HMG14-macroH2A1 were generated by PCR amplification of the regions described (Fig. 1) with primers incorporating restriction enzyme recognition sites, and subcloned into pcDNA3.1-CT-GFP. Myc-tagged cDNAs were generated by transfer of inserts from GFP-tagged constructs into pcDNA-3.1-CT-Myc/His (Invitrogen). An SV40 nuclear localization sequence (NLS)-tagged macroH2A1 tail was generated using a PKKKRKV AgeI–KpnI adapter (top strand 5′-CCG GGG CCC AAG AAG AAG CGG AAG GTA C-3′, bottom strand 5′-CTT CCG CTT CTT CTT GGG CC-3′) and subcloned into the AgeI–KpnI sites between the GFP and macroH2A1 tail.
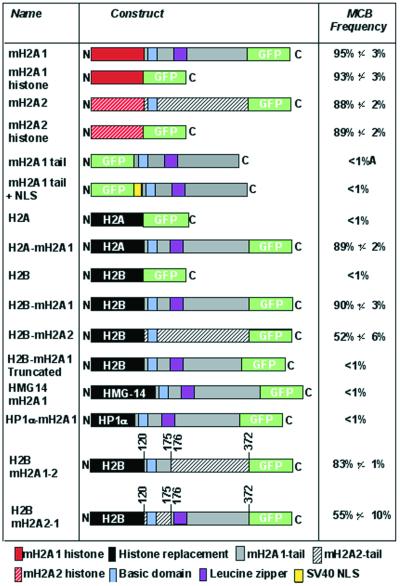
Figure 1.
Schematic representation of macroH2A constructs and their ability to form an MCB in female cells. The N-terminus is represented by an N and the C-terminus is represented by a C. Histone substitutions are in black. The frequency of MCB formation is given to the right for each construct 96 h post-transfection into 46,XX cells. Data represent mean ± SD for two independent experiments. A, macroH2A1 non-histone tail domain was distributed throughout the cytoplasm and nucleus.
Mutagenesis
Fusion proteins of macroH2A1 and macroH2A2 were generated by PCR amplification with primers incorporating restriction enzyme recognition sites and subcloned into pcDNA3.1-CT-GFP. Constructs were sequenced to ensure sequence integrity as described above.
Transposon mutagenesis was achieved in vitro using the EZ::TNTM<NotI/KAN-3> transposon (Epicentre Technologies). The non-histone region of macroH2A1 (amino acids 119–371) was subcloned into pGEM-Zf(+) (Promega) and used as target DNA for mutagenesis. Transposon insertion was carried out according to the manufacturers’ instructions using 80 ng of transposon and 200 ng of macroH2A1-pGEM. The site of insertion and maintenance of the open reading frame was confirmed by DNA sequencing as described above. The macroH2A1 tails of 35 clones containing independent 57 bp insertion sites were subcloned with histone H2B into pcDNA3.1-CT-GFP (Invitrogen).
Site-directed deletions of the macroH2A1 non-histone domain were generated using the ExSiteTM PCR-based site directed mutagenesis kit (Stratagene). Primers were designed in accordance with the manufacturers’ recommendations, and mutagenesis was performed using 1.5 µg of H2B-mH2A1.2-CT-GFP as above. The site of deletion and maintenance of the open reading frame was confirmed by DNA sequencing as described above.
Site-directed mutants of the histone domain of macroH2A1 were generated using the QuickChangeTM site-directed mutagenesis kit (Stratagene). Primers were designed in accordance with the manufacturers’ recommendations. Target DNA for mutagenesis was the macroH2A1 histone domain (amino acids 1–123) subcloned into pBluescript-sk(–) (Stratagene). Mutagenesis was performed using 25 ng of target DNA and site changes were confirmed by DNA sequencing. Mutated clones were then subcloned into pcDNA3.1-CT-GFP for expression analysis.
Cell culture and transfection
Cell lines used included GM04626 and GM00254, both 47,XXX primary fibroblast strains (National Institute of General Medical Sciences Cell Repository, Camden, NJ); hTERT-RPE1, a 46,XX telomerase-immortalized cell line derived from a retinal pigment epithelial cell line RPE-340 (31) (Clontech Laboratories, Inc. No. C4000-1); and hTERT-BJ1, a 46,XY telomerase-immortalized cell line derived from a primary foreskin fibroblast cell line (31) (Clontech Laboratories, Inc. No. C4001-1).
GM04626 and GM00254 were maintained as described previously (15,16). hTERT-RPE1 and hTERT-BJ1 were maintained according to manufacturer’s recommendations (Clontech).
Transfections were performed as described (15,17). Cells were processed 96 h post-transfection.
Immunofluorescence and fluorescence in situ hybridization (FISH)
Immunolocalization, detection of GFP-tagged proteins and FISH using X-specific probes were carried out essentially as described previously (15,17).
RESULTS
To identify regions of macroH2A1 and macroH2A2 required for MCB formation, both proteins were separated into the variant histone and non-histone tail domains and fused to GFP (Fig. 1). Female 46,XX cells were transfected with macroH2A1-GFP histone and macroH2A2-GFP histone and assessed for the ability to form MCBs. By themselves, the histone domains of macroH2A1 and macroH2A2 were each capable of forming a single MCB in the nucleus at a frequency comparable to the full-length GFP-tagged proteins (Fig. 1). To confirm that the MCB was associated with an X chromosome, Myc epitope-tagged constructs of both histones were generated, and their distribution in relation to the X chromosomes was determined by FISH. Figure 2b’ clearly shows association of the macroH2A1-Myc histone with an X chromosome. The macroH2A1-Myc histone MCB is indistinguishable from an MCB generated using a full-length macroH2A1-Myc construct (Fig. 2a’ and a”). As predicted by the ‘n – 1’ rule of X inactivation (1,2), the number of inactive X chromosomes, and consequently the number of MCBs in a nucleus, is one less than the total number of X chromosomes. The full-length macroH2A1-GFP transfected into 47,XXX cells forms two MCBs in each nucleus (Fig. 2a”), as demonstrated previously (15). Both macroH2A-Myc histone constructs also formed two MCBs in 47,XXX cells (Fig. 2b” and c”), confirming association with the Xi. Conversely, no MCBs were observed when the same constructs were transfected into 46,XY cells (Fig. 2b and c).
Figure 2.
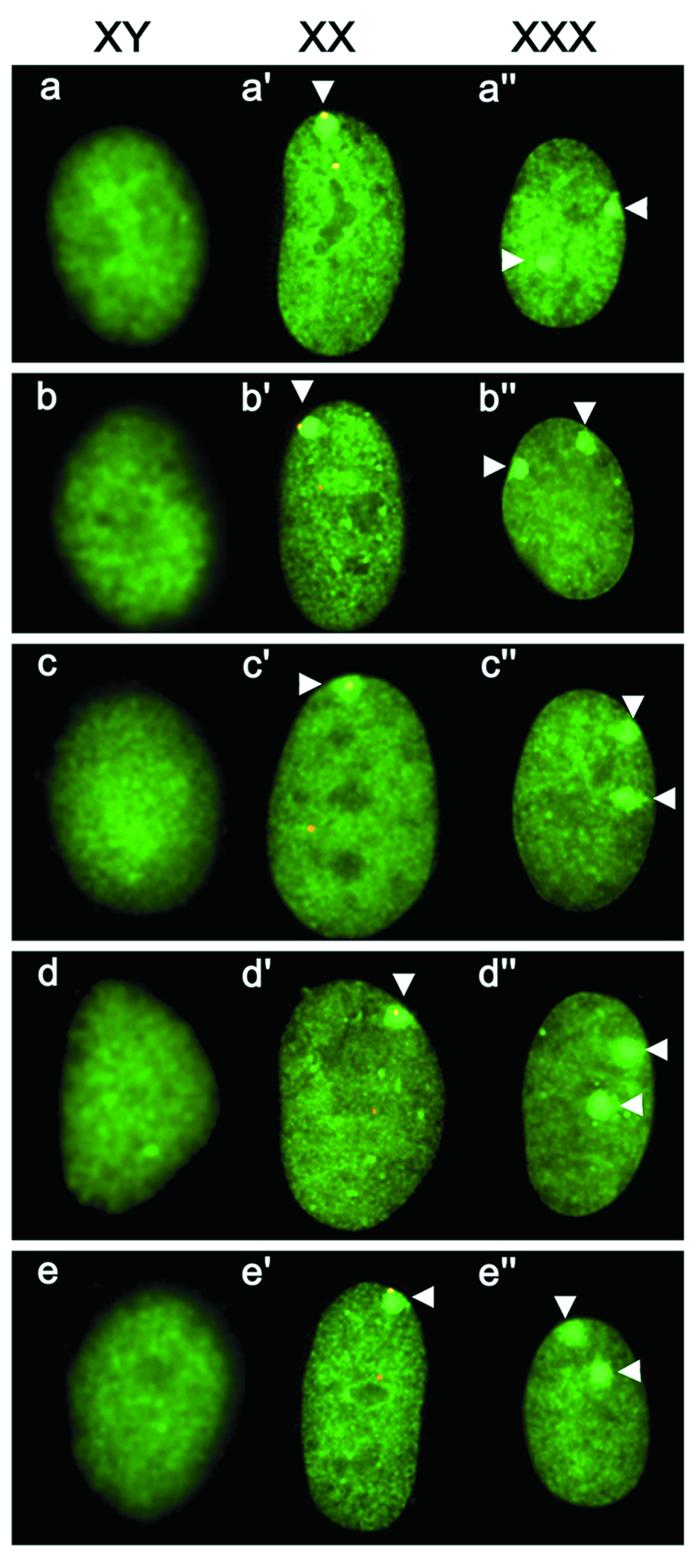
Nuclear distribution of macroH2A constructs in a variety of cell types. Karyotype of cell line is indicated at the top as 46,XY, 46,XX and 47,XXX. White arrowheads indicate the position of MCBs. For images a’–e’, the macroH2A image (FITC, green) is merged with the FISH signals for a human X alpha satellite probe (orange, rhodamine). Nuclear distribution of macroH2A1-Myc (a–a”), macroH2A1 histone domain-Myc (b–b”), macroH2A2 histone domain-Myc (c–c”), core histone H2A substituted macroH2A1-Myc (d–d”) and core histone H2B substituted macroH2A1-Myc (e–e”) 96 h post-transfection in the different cell types.
In an attempt to identify the region of the histone domain of macroH2A that targets the histone to the Xi, we generated a number of mutants and assessed MCB formation. Figure 3 summarizes analysis of the histone domain of macroH2A. The N-terminal tail of core H2A contains two lysine residues that are the site of acetylation (32). Only one of these sites is conserved in the macroH2A histones. Additionally, the N-terminal histone portion of macroH2A1 and macroH2A2 contains three conserved lysine residues that may present macroH2A-specific target sites of covalent modification. Truncated histones have been shown previously to be transferred efficiently to daughter DNA strands during DNA replication (33), indicating that the removal of the N-terminal tail of macroH2A histone is unlikely to prevent deposition into nucleosomes. Therefore, to assess the role of the tail in MCB formation, a deletion of the tail was generated, extending to the first globular domain of core histone H2A (34) that removed all possible sites of covalent modification, and fused to GFP at the N-terminus. The construct was capable of forming MCBs in 46,XX cells at a frequency indistinguishable from that observed for the complete variant histone alone. A similar deletion was made at the C-terminus, removing several unique macroH2A residues and removing the conserved site of ubiquitination (32). As with the N-terminal deletion, removal of a portion of the C-terminal tail did not lower the frequency of MCB formation in 46,XX cells.
Figure 3.
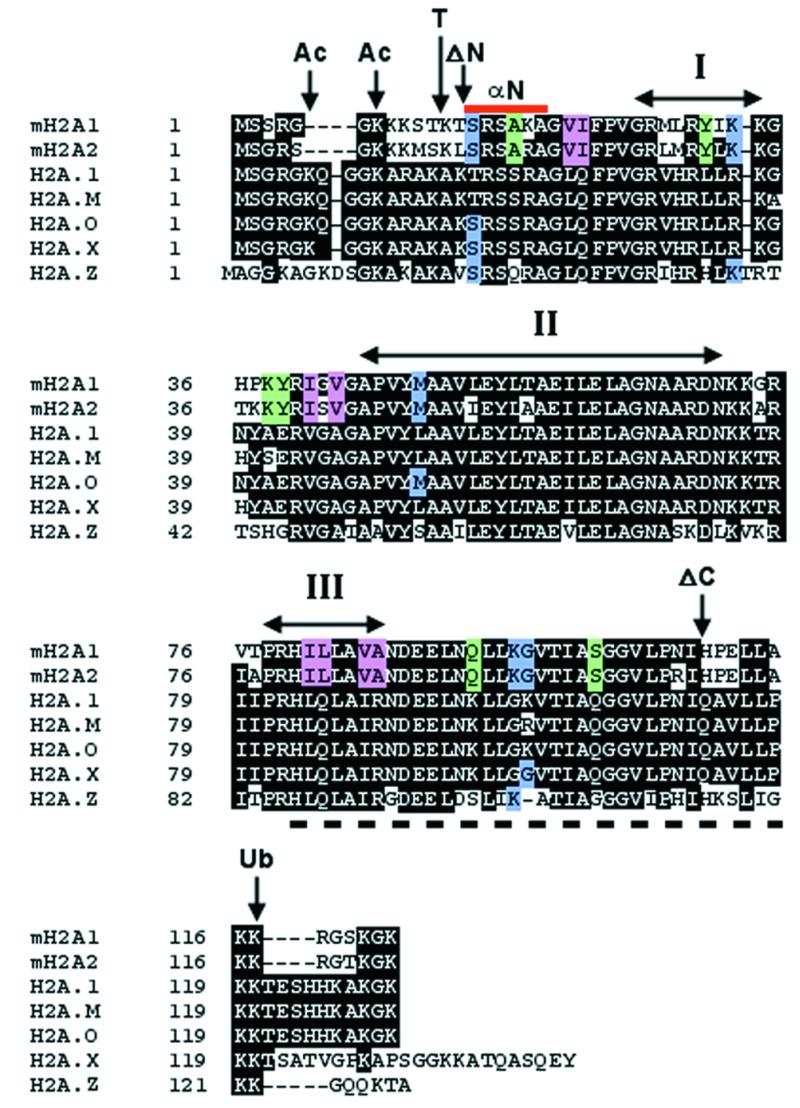
Summary of mutations made to the histone domain of macroH2A. Amino acid alignment of the histone domain of macroH2A1 and macroH2A2 with representative members of the human H2A family. Identical residues are highlighted in black. Gaps in the sequence required for alignment are represented by dashes. Sites of modification for acetylation (Ac) and ubiquitination (Ub) are indicated, as is the position of N-terminal tail removal by treatment with trypsin (T) (42). The α-helices of the histone fold domain (I–III) are indicated by the double-headed arrows (43). The positions of the N-terminal histone deletion (ΔN) and the C-terminal histone deletion (ΔC) are indicated. The region underlined by the dashed line at the C-terminus (amino acids 80–115 of macroH2A1 and macroH2A2) defines the docking domain of histone H2A-H2A interaction in the nucleosome, while the red bar at the N-terminus represents the histone fold extension (αN) (34). Amino acid residues of macroH2A1 replaced by the corresponding core H2A.1 residues are highlighted in color. Blue indicates residues shared between macroH2A1 and macroH2A2 that differ from core H2A.1 but are present in other H2A variants. Green indicates residues shared between macroH2A1 and macroH2A2 that differ from core H2A.1 but are also different in some H2A variants. Red indicates residues specific to macroH2A1 and macroH2A2.
Together, these data indicate that the information required for directing macroH2A histones to the Xi is contained within the central globular region of the histone domain. The globular region of the macroH2A histone has 19 amino acid residues that are conserved between macroH2A1 and macroH2A2, but that differ from core histone H2A. Each such residue highlighted in Figure 3 was converted to the corresponding H2A residue. No single amino acid substitution was capable of preventing MCB formation, and the MCB frequency was unaffected (88–99% for site-directed mutants compared to 88–95% for the parental construct). Although this was only tested in the context of macroH2A1, we anticipate that the histone domain of macroH2A2 would not behave differently. To reflect the ability of the globular region of the variant histone to form an MCB, we have termed this a macrochromatin domain (MCD).
To test if the non-histone tail of macroH2A1 also contained an MCD, an N-terminal GFP-tagged tail construct was generated (Fig. 1) and transfected into 46,XX cells. The GFP-tagged tail was distributed throughout the nucleus and cytoplasm (data not shown). This indicates that the non-histone tail depends upon the histone domain for complete nuclear localization. As the formation of an MCB presumably requires macroH2A to be nuclear, the SV40 T-antigen NLS (35) was inserted at the N-terminus of the tail (Fig. 1). While the NLS-tagged tail localized to the nucleus in 46,XX cells, no obvious MCBs were observed (data not shown), suggesting that nuclear localization by itself is not sufficient to target the non-histone tail to the Xi.
To test if the tail could form an MCB when directed to chromatin generally, the tail was fused to the C-terminus of two chromatin-associated proteins: human heterochromatin protein HP1-α (36) and the high mobility group protein HMG-14 (37) (Fig. 1). Both HP1-α-macroH2A1-GFP and HMG-14-macroH2A1-GFP were nuclear and had a distribution indistinguishable from wild-type HP1-α-GFP or HMG-14-GFP (data not shown). Lastly, to test if the non-histone tail could form an MCB when redirected to the nucleosome, the macroH2A1 variant histone domain was substituted with core histone H2A (Fig. 1). Core histone H2A by itself does not form an MCB and has a uniform distribution throughout the nucleus (15). However, the H2A-macroH2A1 construct did form an MCB in 46,XX cells (Fig. 2d’). This indicates that, in addition to the MCD of the macroH2A variant histone, the non-histone tail of macroH2A1 contains an independent MCD that requires nucleosomal association for formation. The MCB formed by the core H2A-macroH2A1 fusion associated with an X chromosome (Fig. 2d’) and conformed to the n – 1 rule in 47,XXX cells (Fig. 2d and d”).
To further investigate the nucleosome-dependent MCB forming ability of the macroH2A1 tail, the histone domain of macroH2A1 was replaced with core histone H2B (Fig. 1). Core H2B, like core H2A, does not form an MCB and is distributed throughout the nucleus (17). Surprisingly, H2B-macroH2A1 formed an MCB in 46,XX cells that was associated with an X chromosome (Fig. 2e’) and conformed to the n – 1 rule (Fig. 2e and e”). This suggests that MCB formation is not dependent upon the position of the non-histone tail in relation to the nucleosome core, as core H2A and core H2B are located in different positions in the nucleosome (34).
The non-histone domains of macroH2A1 and macroH2A2 are not as well conserved (60% identity) as the variant histone domains (84%) (15). To investigate whether the tail of macroH2A2 contains a MCD, a core histone H2B-replacement of macroH2A2 was generated and transfected into 46,XX cells. As with macroH2A1, H2B-macroH2A2 formed a single MCB in female cells, but the frequency of MCB formation was considerably lower than for H2B-macroH2A1 (Fig. 1). This indicates that differences in amino acid composition of the tail may influence its ability to target to the Xi and/or to form an MCB. The most diverged region between the non-histone domain of macroH2A1 and macroH2A2 is between amino acids 120 and 175 at 35% identity, while the remainder of the non-histone domains, amino acid 176–372, share 68% amino acid identity (15). To determine if this region influences MCB formation, the corresponding regions of both proteins were substituted for each other (Fig. 1). The non-histone domain of macroH2A2 containing amino acids 120–175 of macroH2A1 formed MCBs in 46,XX cells at a frequency comparable to the full macroH2A1 non-histone domain (Fig. 1). Conversely, the non-histone domain of macroH2A1 containing amino acids 120–175 of macroH2A2 formed MCBs in 46,XX cells at a frequency comparable to the full macroH2A2 non-histone domain (Fig. 1). This indicates that the amino acid differences between residues 120–175 of the two proteins significantly affect the efficiency of the non-histone MCD.
The high frequency of MCB formation of H2B-macroH2A1-GFP provides a system to identify the tail MCD by targeted mutagenesis, in an assay that is independent of any influence of the macroH2A-histone MCD. A number of C-terminal truncations of the tail were made and transfected into 46,XX cells. No MCBs were observed for any of the constructs, the smallest of which removed 30 amino acids from the C-terminus, indicating that at least the very C-terminal portion of the tail is necessary for MCB formation.
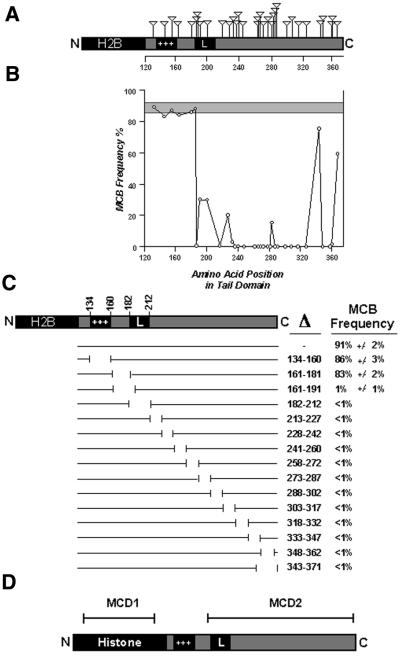
Two complementary approaches were adopted to generate internal disruptions of the tail. The first involved the insertion of 19 amino acids into the tail by in vitro transposon mutagenesis. A total of 35 different in-frame insertions were made and tested for the ability to form an MCB. Figure 4A shows the position of each insertion and the efficiency of MCB formation when each construct was transfected into 46,XX cells. Insertions into positions 130–184, which includes the basic domain, did not affect the ability to form an MCB (Fig. 4B). However, with the exception of an insertion at amino acid 344 and one at amino acid 366, all insertions between amino acid 185 and the C-terminus seriously impaired or removed MCB forming ability.
Figure 4.
Schematic representation of the transposon and deletion mutations of H2B-macroH2A1, results of the assay for MCB formation and summary of MCD positions. (A) Diagram of the H2B-macroH2A1 construct indicating the position of each of 35 insertions represented by the inverted triangle above the construct. Core histone H2B is highlighted in black, as is the basic domain (+++) and predicted leucine zipper (L). Scale beneath the tail indicates amino acid position. (B) Graphical representation of MCB-forming capability of the mH2A1 tail containing different insertions. The frequency of MCB formation is given on the y axis, and relative amino acid position in the tail is given along the x axis. The shaded region represents the range of MCB frequency observed for the non-mutated H2B-macroH2A1 construct from four independent transfections. Circles represent the means of two independent transfections. No result varied >3% either side of the mean value. MCB frequency was determined by scoring 50 random nuclei 96 h post-transfection. (C) A schematic representation of the macroH2A1 non-histone domain deletions. Gaps in lines directly under the macroH2A1 tail represent relative positions of each targeted deletion. The amino acids deleted in each construct are indicated to the right under the Δ symbol. The frequency of MCB formation for each construct is given on the far right. Data represent means ± SD for two independent experiments. (D) Schematic representation of macroH2A1 indicating the position of two independent MCDs, MCD1 and MCD2.
The second approach involved systematic removal of small adjacent sections along the length of the non-histone tail, fused to core histone H2B. Figure 4C shows the position of 14 internal deletions covering amino acids 134–363, and the percentage of 46,XX cells showing an MCB 96 h post-transfection. In agreement with the insertion data above, deletion of amino acids 134–181, including the basic domain, did not disrupt the MCD, while any deletion of amino acids 182–371 completely prevented MCB formation.
To distinguish the two separate regions of macroH2A that are essential for targeting the protein to the Xi, we have called the globular variant histone Xi-targeting region MCD1 and the non-histone Xi-targeting region MCD2, as indicated in Figure 4D. The failure of some constructs to form an MCB might not be due to the disruption of an Xi targeting signal, but could simply reflect the inability of those constructs to substitute core H2A and integrate into nucleosomes.
DISCUSSION
The enrichment of macroH2A in chromatin of the Xi results in the formation of an MCB in the nucleus. Despite evidence of a physical association of macroH2A with XIST RNA (21), colocalization with XIST (19,20) and macroH2A dependence upon XIST for localization with the Xi (22), nothing is known about the regions of macroH2A that are important for targeting to the Xi.
The variant histone domains of macroH2A1 and macroH2A2 alone are capable of forming an MCB in 46,XX cells at a frequency comparable to the full-length protein (Fig. 1), but core histones H2A or H2B do not form MCBs in 46,XX cell lines (15,17). This contrasts with the observations by Perche et al. (29) which suggest that the nucleosome density of the inactive X chromosome is higher, as core H2A, H2B and H3 were also found to be enriched at the Barr body at a level comparable to macroH2A. While the inactive X chromosome may well have a higher nucleosome density at interphase, macroH2A is clearly enriched on the inactive X chromosome at metaphase (14,20), suggesting that nucleosome density alone cannot account for the enrichment of macroH2A on chromatin of the inactive X chromosome. We do not find core H2A or H2B to be enriched on the inactive X chromosome under the conditions described in the cell lines we have tested, while macroH2A1 and macroH2A2 consistently demonstrate enrichment on the inactive X chromosome at interphase (15,17).
The macroH2A histone domain MCB looks identical to one formed by the full-length protein (Fig. 2a’), is associated with an X chromosome (Fig. 2b’ and c’) and conforms to the n – 1 rule (Fig. 2b, b”, c and c”). Therefore, the variant histone portion of macroH2A alone contains information sufficient for direction of the Xi to chromatin. MCD1 is limited to the core globular domain of the histone, as indicated in Figure 4D, and as a consequence is independent of any covalent modification of the histone tails. No single amino acid substitution at any of the 19 amino acids that differ between the globular domain of macroH2A and core H2A was sufficient to remove the capability to form an MCB. Therefore, it is likely that a combination of substitutions are required to change the structural conformation sufficiently so that the variant histone domain of macroH2A is no longer recognized during chromatin assembly as macroH2A, and consequently not targeted to the Xi.
By itself, the non-histone domain of macroH2A1 was incapable of MCB formation and distributed throughout the cell (Fig. 1). Therefore, nuclear localization of endogenous macroH2A is a function of the histone domain, probably mediated by importin β1 as demonstrated for core H2A and the variant H2A.Z (38). Indeed, MCB formation only occurred when the non-histone tail was targeted directly to the nucleosome by replacing the macroH2A histone domain with a core histone (Fig. 1). Therefore, like the macroH2A histone portion, the non-histone tail also contains an MCD (MCD2). Surprisingly, replacement of the histone domain of macroH2A with core histone H2B was as effective at H2A in directing MCB formation (Fig. 1). This indicates that, although MCB formation is dependent upon nucleosome association, the position of the tail in relation to the nucleosome is not critical, as histone H2B is located at a different position from H2A in the nucleosome (34). Replacement of the macroH2A2 histone domain with core histone H2B also formed an MCB, but the frequency was significantly lower than that observed for macroH2A1-H2B (Fig. 1). The tail domains of macroH2A1 and macroH2A2 are the least conserved portions of the proteins, sharing only 60% amino acid identity (15). Within the non-histone domains of macroH2A1 and macroH2A2, amino acids 120–175 are only 35% identical. When this region was substituted from one protein with the same interval of the other protein, the efficiency of MCB formation followed that of the protein from where amino acids 120–175 originated. Therefore, the amino acid differences between the two proteins within this interval are responsible for the differences in MCB efficiency. Although amino acids 120–175 of macroH2A2 significantly reduce MCB formation when the variant histone domain is substituted with histone H2B (Fig. 1), this interval does not influence MCD1 in the wild-type protein and hence the full-length macroH2A2 forms MCBs at a level comparable to macroH2A1 (15).
To delimit the MCD within the tail, we made targeted disruptions to the macroH2A1-H2B chimera as this construct has a higher ability to form an MCB, and changes in the frequency of MCB formation would be more obvious. The results of insertional mutagenesis (Fig. 4B) and deletion mutagenesis (Fig. 4C) are almost completely complementary. Both indicate that the basic domain of macroH2A1 is not required for MCB formation, while the remaining portion of the tail is critical (Fig. 4D), but as outlined above, this region of the non-histone domain does influence the efficiency of MCD2. The location of macroH2A in the nucleus is coincident with XIST RNA (19,20) and is physically associated, either directly or in combination with additional factors, to XIST (21). Deletion of or insertion into the basic domain shifts the relative position of the remainder of the tail in relation to the nucleosome, yet MCB formation is not affected. This suggests that if MCD2 is forming an MCB through its ability to interact with the XIST complex, then the tail must be fairly flexible. Not only can it interact when the basic domain is removed or disrupted, but the interaction can also occur when fused to histone H2B and consequently presented to the complex at a different angle from the nucleosome. Two insertions made towards the C-terminal end of the tail domain did not prevent MCB formation, but only served to reduce the frequency of MCBs (Fig. 4A and B). These same intervals, when deleted, removed the ability to form an MCB (Fig. 4C). This suggests that the amino acids in this interval are essential for the MCD, and that alteration of the spatial relationship of the residues reduces the frequency of MCB formation, probably through reduced efficiency of interaction with components of the XIST complex.
We have found that MCB frequency in 46,XX transfected cells steadily increases until ∼96 h post-transfection, whereupon the number of MCBs reaches a maximum and remains constant (data not shown). This time dependence probably reflects that macroH2A-GFP constructs are competing with endogenous macroH2A for integration into the nucleosome and that at least one round of DNA replication is required for deposition of macroH2A-GFP nucleosomes into Xi chromatin. During DNA replication, newly synthesized chromatin of the daughter DNA is formed through the random segregation of parental nucleosomes onto one daughter strand, and the assembly of a new nucleosome on the other daughter strand (39). Histone H2A is deposited onto newly synthesized (H3-H4)2 cores along with H2B through association with a number of chaperone proteins (40). The recent solving of the structure of a nucleosome core particle containing the histone H2A variant H2A.Z suggests that the H2A content of a nucleosome is homogenous for one type of H2A. Differences in the docking domains would prevent two different H2A molecules stably interacting within the nucleosome (41). The docking domains of macroH2A1 and macroH2A2 are almost identical (Fig. 3). Therefore, the two proteins are unlikely to prevent each other occupying the same nucleosome, and as a consequence nucleosomes containing combinations of the two proteins are likely (i.e. macroH2A1 with macroH2A1, or 1 with 2 or 2 with 2).
How subtle differences in the histone domain of macroH2A are recognized by chaperone proteins, such that macroH2A is directed to the Xi, is intriguing and is a critical question to address within the context of X inactivation. It is conceivable that a chaperone protein specific for macroH2A-H2B heterodimers exists that is responsible for depositing macroH2A onto newly synthesized nucleosomes on daughter strands of the Xi. Alternatively, the deposition of macroH2A may be restricted to late-replicating chromatin, and therefore targeting is a consequence of the late replication of the Xi (9,10). Understanding the inheritance of the Xi chromatin state by the daughter strands remains a key question that can then be directed to how this state is established early in development.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Lisa Chadwick for critical examination of the manuscript. This work was supported by research grant GM45441 to H.F.W. from the National Institutes of Health. B.P.C. is supported by a postdoctoral fellowship from the Rett Syndrome Research Foundation.
References
- 1.Lyon M.F. (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature, 190, 372–373. [DOI] [PubMed] [Google Scholar]
- 2.Avner P. and Heard,E. (2001) X-chromosome inactivation: counting, choice and initiation. Nat. Rev., 2, 59–67. [DOI] [PubMed] [Google Scholar]
- 3.Barr M.L. and Bertram,E.G. (1949) A morphological distinction between neurones of the male and female and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature, 163, 676–677. [DOI] [PubMed] [Google Scholar]
- 4.Mohandas T., Sparkes,R.S. and Shapiro,L.J. (1981) Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science, 211, 393–396. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer G., Tanguay,R., Steigerwald,S. and Riggs,A. (1990) In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev., 4, 1277–1287. [DOI] [PubMed] [Google Scholar]
- 6.Jeppesen P. and Turner,B.M. (1993) The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell, 74, 281–289. [DOI] [PubMed] [Google Scholar]
- 7.Boggs B.A., Connors,B., Sobel,R.E., Chinault,A.C. and Allis,C.D. (1996) Reduced levels of histone H3 acetylation on the inactive X chromosome in human females. Chromosoma, 105, 303–309. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert S.L. and Sharp,P.A. (1999) Promoter-specific hypoacetylation of X-inactivated genes. Proc. Natl Acad. Sci. USA, 96, 13825–13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert C.W., Muldal,S., Lajthal,L.G. and Rowley,J. (1962) Time-sequence of human chromosome duplication. Nature, 195, 869–873. [DOI] [PubMed] [Google Scholar]
- 10.Morishma A., Grumbach,M.M. and Taylor,J.H. (1962) Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc. Natl Acad. Sci. USA, 48, 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown C.J., Ballabio,A., Rupert,J.L., Lafreniere,R.G., Grompe,M., Tonlorenzi,R. and Willard,H.F. (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature, 349, 38–44. [DOI] [PubMed] [Google Scholar]
- 12.Brockdorff N., Ashworth,A., Kay,G.F., McCabe,V.M., Norris,D.P., Cooper,P.J., Swift,S. and Rastan,S. (1992) The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell, 71, 515–526. [DOI] [PubMed] [Google Scholar]
- 13.Brown C.J., Hendrich,B.D., Rupert,J.L., Lafreniere,R.G., Xing,Y., Lawrence,J. and Willard,H.F. (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell, 71, 527–542. [DOI] [PubMed] [Google Scholar]
- 14.Costanzi C. and Pehrson,J.R. (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature, 393, 599–601. [DOI] [PubMed] [Google Scholar]
- 15.Chadwick B.P. and Willard,H.F. (2001) Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet., 10, 1101–1113. [DOI] [PubMed] [Google Scholar]
- 16.Costanzi C. and Pehrson,J.R. (2001) MacroH2A2, a new member of the macroH2A core histone family. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 17.Chadwick B.P. and Willard,H.F. (2001) A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol., 152, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pehrson J.R. and Fried,V.A. (1992) MacroH2A, a core histone containing a large nonhistone region. Science, 257, 1398–1400. [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen T.P., Mastrangelo,M.A., Eden,A., Pehrson,J.R. and Jaenisch,R. (2000) Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol., 150, 1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mermoud J.E., Costanzi,C., Pehrson,J.R. and Brockdorff,N. (1999) Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol., 147, 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert S.L., Pehrson,J.R. and Sharp,P.A. (2000) XIST RNA associates with specific regions of the inactive X chromatin. J. Biol. Chem., 275, 36491–36494. [DOI] [PubMed] [Google Scholar]
- 22.Csankovszki G., Panning,B., Bates,B., Pehrson,J.R. and Jaenisch,R. (1999) Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet., 22, 323–324. [DOI] [PubMed] [Google Scholar]
- 23.Brown C.J. and Willard,H.F. (1994) The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature, 368, 154–156. [DOI] [PubMed] [Google Scholar]
- 24.Driscoll D. and Migeon,B. (1990) Sex differences in methylation of single-copy genes in human meiotc germ cells: implications for X chromosome inactivation, parental imprinting and origin of CpG mutations. Somat. Cell Mol. Genet., 16, 267–282. [DOI] [PubMed] [Google Scholar]
- 25.Gartler S.M. and Goldman,M.A. (1994) Reactivation of inactive X-linked genes. Dev. Genet., 15, 504–514. [DOI] [PubMed] [Google Scholar]
- 26.Rack K.A., Chelly,J., Gibbons,R.J., Rider,S., Benjamin,D., Lafreniere,R.G., Oscier,D., Hendriks,R.W., Craig,I.W., Willard,H.F., Monaco,A.P. and Buckle,V.J. (1994) Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet., 3, 1053–1059. [DOI] [PubMed] [Google Scholar]
- 27.Singer-Sam J., Goldstein,L., Dai,A., Gartler,S.M. and Riggs,A.D. (1992) A potentially critical Hpa II site of the X chromosome-linked PGK1 gene is unmethylated prior to the onset of meiosis of human oogenic cells. Proc. Natl Acad. Sci. USA, 89, 1413–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzi C., Stein,P., Worrad,D.M., Schultz,R.M. and Pehrson,J.R. (2000) Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development, 127, 2283–2289. [DOI] [PubMed] [Google Scholar]
- 29.Perche P., Vourc’h,C., Konecny,L., Souchier,C., Robert-Nicoud,M., Dimitrov,S. and Khochbin,S. (2000) Higher concentrations of histone macroH2A in the barr body are correlated with higher nucleosome density. Curr. Biol., 10, 1531–1534. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 32.Wolffe A.P. and Hayes,J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintini G., Treuner,K., Gruss,C. and Knippers,R. (1996) Role of amino-terminal histone domains in chromatin replication. Mol. Cell. Biol., 16, 2888–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 35.Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- 36.Saunders W.S., Chue,C., Goebl,M., Craig,C., Clark,R.F., Powers,J.A., Eissenberg,J.C., Elgin,S.C., Rothfield,N.F. and Earnshaw,W.C. (1993) Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J. Cell Sci., 104, 573–582. [DOI] [PubMed] [Google Scholar]
- 37.Postnikov Y.V., Trieschmann,L., Rickers,A. and Bustin,M. (1995) Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J. Mol. Biol., 252, 423–432. [DOI] [PubMed] [Google Scholar]
- 38.Johnson-Saliba M., Siddon,N.A., Clarkson,M.J., Tremethick,D.J. and Jans,D.A. (2000) Distinct importin recognition properties of histones and chromatin assembly factors. FEBS Lett., 467, 169–174. [DOI] [PubMed] [Google Scholar]
- 39.Krude T. (1999) Chromatin assembly during DNA replication in somatic cells. Eur. J. Biochem., 263, 1–5. [DOI] [PubMed] [Google Scholar]
- 40.Ridgway P. and Almouzni,G. (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci., 113, 2647–2658. [DOI] [PubMed] [Google Scholar]
- 41.Suto R.K., Clarkson,M.J., Tremethick,D.J. and Luger,K. (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol., 7, 1121–1124. [DOI] [PubMed] [Google Scholar]
- 42.Bohm L. and Crane-Robinson,C. (1984) Proteases as structural probes for chromatin: the domain structure of histones. Biosci. Rep., 4, 365–386. [DOI] [PubMed] [Google Scholar]
- 43.Argents G. and Moudrianakis,N.N. (1995) The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl Acad. Sci. USA, 92, 11170–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]