Abstract
Viral hepatitis is a major cause of morbidity and mortality, affecting hundreds of millions of people worldwide. Hepatitis-causing viruses initiate disease by establishing both acute and chronic infections, and several of these viruses are specifically associated with the development of hepatocellular carcinoma. Consequently, intense research efforts have been focusing on increasing our understanding of hepatitis virus biology and on improving antiviral therapy and vaccination strategies. Although valuable information on viral hepatitis emerged from careful epidemiological studies on sporadic outbreaks in humans, experimental models using cell culture, rodent and non-human primates were essential in advancing the field. Through the use of these experimental models, improvement in both the treatment and prevention of viral hepatitis has progressed rapidly; however, agents of viral hepatitis are still among the most common pathogens infecting humans. In this Review, we describe the important part that these experimental models have played in the study of viral hepatitis and led to monumental advances in our understanding and treatment of these pathogens. Ongoing developments in experimental models are also described.
Over the past century, major advances have occurred in the study of viral hepatitis. Specifically, advances in our understanding of the epidemiology, diagnosis, pathogenesis, treatment and prevention of viral hepatitis have all been achieved. In addition, viral hepatitis A, B, C, E and Delta are well defined1. With regards to prevention, safe and effective vaccines for hepatitis A and B are available, but not yet for hepatitis C, although major efforts to this end are currently underway2. By contrast, effective hepatitis E virus (HEV) vaccines have been developed but are not yet widely available3. For chronic viral hepatitis, therapy for HBV infection is effective in achieving viral suppression, leading to undetectable HBV DNA levels whilst stopping necroinflammation and often resulting in regression of fibrosis4,5. Importantly, historical advances in therapy for HCV infection have led to interferon-free, curative, all-oral combinations with direct-acting antiviral agents (DAAs)6,7. These new treatment regimens can be used with or without ribavirin and the end goal of short-term, pangenotypic curative regimens for hepatitis C with minimal adverse effects is quickly approaching. In this Review, we highlight the expanding model systems to study viral hepatitis, focusing on available models of HCV and HBV infection as examples, which have contributed greatly to the advances in this field.
HBV and HCV biology
Overall, it has been challenging to develop models to study viral hepatitis and advances have been achieved through the generation of complex biological systems. A complete and exhaustive discussion pertaining to molecular virology and of the precise details under lying these models is beyond the scope of this Review but will be briefly discussed herein. We recommend more targeted references for precise virological and model-specific information. For hepatitis B, we recommend published papers specifically in the areas of in vitro culture systems and virology8,9, animal models10,11 and those that address these topics more broadly12,13. Similarly, for hepatitis C, we recommend published papers specifically in the areas of in vitro culture systems and virology14,15, animal models16,17 and those that address these topics more broadly18,19.
HBV is a DNA virus with a partially double-stranded genome; it predominantly infects hepatocytes and, after entry, its relaxed circular genome is released from the virion at which points its genome translocates to the nucleus (FIG. 1). There, its genome is modified to the covalently closed circular (cccDNA) form that exists stably as an extrachromosomal viral genome. Viral RNA is made from the DNA genome and is translated into the major proteins that make up the virion. HBV surface antigens are extremely potent antigens that have facilitated the development of effective vaccines20. Clearly, a major hurdle is to cure HBV with finite therapy. This step will be achieved by facilitating the loss of HBV surface antigen (HBsAg) and by eliminating cccDNA, which is thought to be a key element of chronic HBV infection13. Thus, if this feat can be achieved, hepatitis Delta virus (HDV) prevalence will also decrease because HDV requires the HBV envelope protein for the assembly of infectious virus particles. Consequently, novel models to study HBV infection are desperately needed to investigate clearance of HBV cccDNA, which exists in the nucleus as a minichromosome and persists even in the absence of active viral replication21. Unfortunately, cccDNA is not eliminated by the potent suppressive therapies that are currently available8.
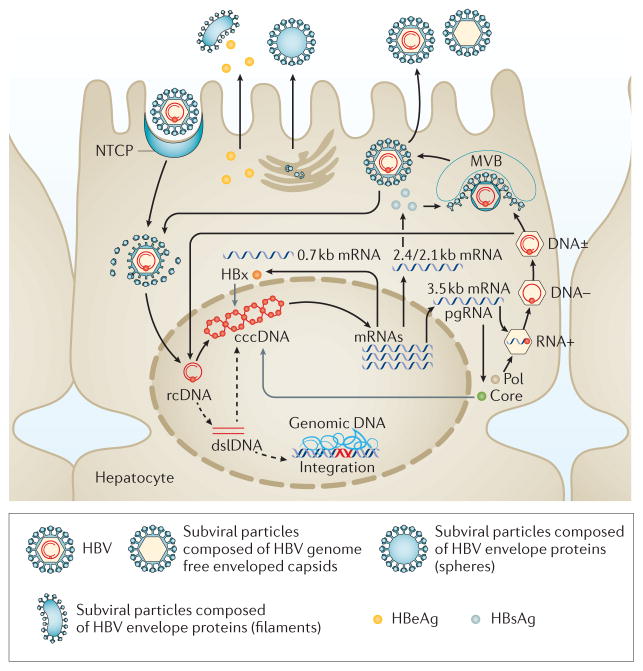
Figure 1. HBV biology.
The life cycle of HBV, including attachment, entry, uncoating, trafficking to nucleus, covalently closed circular (cccDNA) formation, integration, transcription, translation, encapsidation and secretion, is depicted. Initially, HBV particles are taken up by hepatocytes through mechanisms that involve NTCP (sodium/bile acid cotransporter, also known as SLC10A1). After internalization, viral capsids are released and subsequently directed to the nucleus where the HBV genomes are liberated. In the nucleus, relaxed circular DNA (rcDNA) genomes are converted into cccDNA that can persist in the nucleus of infected cells as a minichromosome and which serves as template for viral RNA transcription. Double-stranded linear DNA (dslDNA) is also produced that can be integrated into the cellular genome or also converted into cccDNA. Viral mRNAs are transported to the cytoplasm where they are translated into viral proteins and together with the viral polymerase, the pregenomic RNA (pgRNA) is encapsidated and reverse transcribed within the nucleocapsid into progeny rcDNA. Mature nucleocapsids are then either directed to the multivesicular body pathway for envelopment with HBV envelope proteins or re-directed to the nucleus to establish a cccDNA pool. HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBx, hepatitis B x protein; MVB, multivesicular body. Adapted with permission from Nature Publishing Group © Revill, P. et al. Nat. Rev. Gastroenterol. Hepatol. http://dx.doi.org/10.1038/nrgastro.2016.7 (2016).
HCV is a single-stranded positive-sense RNA virus; it completes its virus life cycle in the hepatocyte cytoplasm where its RNA genome is translated into the structural and nonstructural viral proteins (FIG. 2). Although antibody responses are generated that can neutralize these viral proteins, the extraordinary ability of the virus to mutate facilitates the generation of escape mutants that evade this host defence strategy. As a result, individuals who are cured of HCV with potent all-oral antiviral agents, some at great expense, can be reinfected. Thus, for HCV infection, a durable vaccine has been elusive and is desperately needed2.
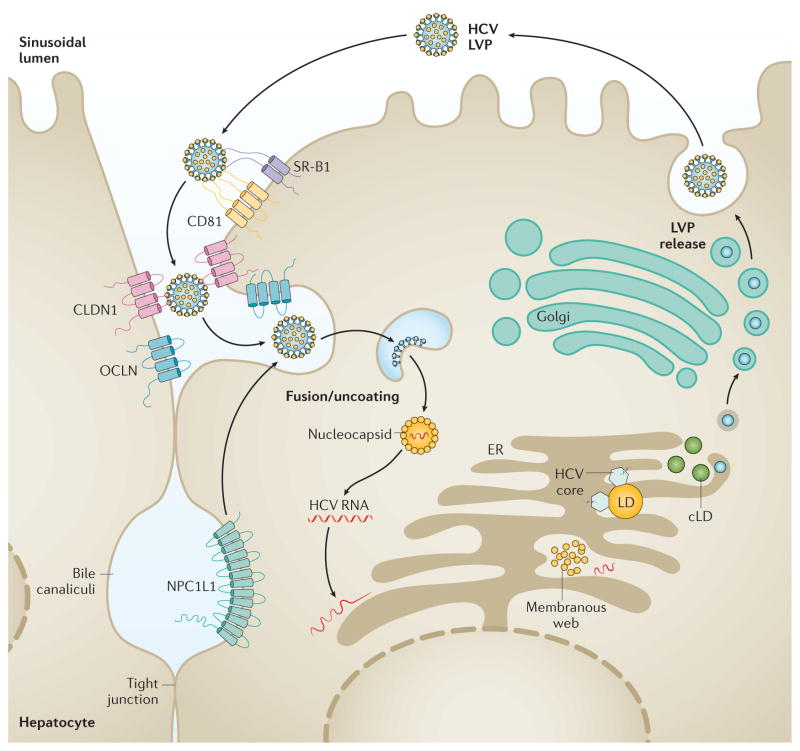
Figure 2. HCV biology.
The life cycle of HCV, including attachment, entry, uncoating, encapsidation and secretion, is depicted. Interaction of extracellular HCV with cellular surface receptors initiates the entry process, which can also occur from direct cell-to-cell transmission. After pH-dependent fusion and uncoating, the incoming HCV genome is translated and the resulting polyprotein processed. Replication takes place in ER-derived membrane spherules (membranous web). In the assembly and secretion release process, the core protein is transferred from the lipid droplets to form nucleocapsids that, assisted by NS5A, are loaded with RNA. The replicase proteins can bind to the HCV genomic RNA during transfer from replication to packaging in close proximity, but eventually are removed from the maturing nucleocapsids, the intracellular sites of which might converge. HCV virion morphogenesis is coupled to the VLDL pathway, and particles are produced as lipoviral particles (LVPs). CLDN1, claudin-1; cLD, cytoplasmic lipid droplets; ER, endoplasmic reticulum; LD, lipid droplets; NPC1L1, Niemann-Pick C1-like protein 1; OCLN, occludin; SR-B1, scavenger receptor class B member 1. Adapted with permission from Nature Publishing Group © Scheel, T. K. H. & Rice, C. M. Nat. Med, 19, 837–849 (2013).
However, even with the eradication of HBV and HCV, progressive liver disease in some infected individuals and the risk of hepatocellular carcinoma (HCC) remains a clinical challenge. Specifically, in the USA, the majority of HCV-infected individuals fall within a birth cohort born between 1945 and 1965 having acquired the virus several decades ago22; over many years, chronic HCV infection can result in progressive liver fibrosis and cirrhosis that predisposes to the development of HCC, and the ageing HCV-infected population is at substantial risk of the development of this deadly tumour. Even with cure, HCV-infected individuals with pre-existing advanced liver disease remain at increased risk of decompensation and other poor clinical outcomes including HCC23. Consequently, additional models are needed to understand progression of liver disease even after curing chronic viral hepatitis.
The early days
Modern viral hepatitis research began in 1963 with the discovery of the ‘Australian Antigen’ — now designated as HBsAg — by Baruch Blumberg and Harvey Alter24. Initially, much of the work to study viral hepatitis relied on biochemical methods, microscopy and primate models to characterize the agents of viral hepatitis. In addition, controversial experiments were carried out in populations in prisons25 and individuals with intellectual disabilities at the Willowbrook State School in Staten Island, New York, USA26. These initial studies were successful in characterizing hepatitis A virus (HAV)27, discovered by using electron microscopy on infected patient stool samples, and HBV, first identified by seroimmunoassay28. The identification and characterization of HAV and HBV subsequently led to the identification of HCV, formerly known as non-A, non-B hepatitis29. HDV was also identified by assessing liver samples from patients who were known to be positive for HBV but displayed peculiar viral protein staining patterns in the nucleus, and chimpanzees were utilized to demonstrate coexistence of HBV and HDV30–32. Similarly, HEV was characterized by using clinical samples from patients infected as a result of sporadic HEV infection epidemics33,34. Although effective in defining these aetiological agents of viral hepatitis, more sophisticated molecular studies were needed to develop targeted antivirals and vaccines for these viruses. The difficulty in generating an easily accessible animal model to study these pathogens prompted researchers to develop simpler systems that relied upon cultured cells for studies on viral hepatitis.
Hepatoma cell lines
As for most viruses, tumour-derived cells lines have been useful in increasing our understanding of viral hepatitis. General molecular biology methods, which are presented in detail in the forthcoming section, have been used to overexpress viral proteins and to generate cell lines that continuously express hepatitis viral genomes. Specifically, the HBV DNA and HCV RNA genomes and plasmids encoding viral proteins have been delivered intracellularly through various transfection techniques and viral transduction systems. In addition, with the generation of cell lines that express the viral genomes, small-molecule screens have been carried out to identify agents that could suppress viral replication35–37. Overall, the use of transformed cell lines has relied on the fact that several of them harbour defects in intrinsic innate immune antiviral pathways in cells that would normally restrict expression of viral genomes and proteins38,39. These cell lines, although abnormal, enabled these viruses to be studied without major interference from host defence signalling pathways.
The first attempts at culturing HBV and HCV in cell lines relied on using serum from infected patients to introduce the virus into an in vitro model40. Even though investigators were able to detect viral replication in these models, the resulting infection was at a very low level and was challenging to detect using standard methods including western blot, immunofluorescence and PCR41. For HCV, much more success was achieved by in vitro transcription of HCV RNA from a DNA construct containing the viral genome and then transfecting the viral RNA into a cell line14. In addition, inoculation of these in vitro transcribed viral genomes into chimpanzees subsequently led to HCV infection42. For HBV, selection of cell lines that stably incorporated into the HBV DNA genome, in multiple chromosomal sites, was found to be a successful approach43. A summary of hepatoma cell line models to study HBV and HCV is presented in TABLE 1 and shown in FIGS 3 and 4.
Table 1.
Commonly used cell culture models for the study of HBV and HCV
| Cell biology | HuH-7 cell | HepG2 cell | HepaRG cell | Stem-cell-derived hepatocytes | Primary human hepatocytes |
|---|---|---|---|---|---|
| Functional innate immunity | Minimal | Moderate | Fully functional | Fully functional | Fully functional |
| Length of propagation | Indefinite | Indefinite | Indefinite | <1 month | <1 month |
| Polarized | No | Yes | Yes | Yes | Yes |
| Immortalized | Yes | Yes | Yes | No | No |
| Transformed | Yes | Yes | No | No | No |
| Availability | High | High | Medium | Low | Low |
| Lot variability* | Minimal | Minimal | Moderate | Moderate | High |
| Supports HBV infection | No, requires NTCP | No, requires NTCP | Yes, requires differentiation | Yes | Yes |
| Supports HCV infection | Yes | No, requires CD81 and miR-122 | Yes, requires differentiation | Yes | Yes |
miR-122, microRNA 122; NTCP, sodium/bile acid cotransporter (also known as SLC10A1).
Lot variability is susceptibility to variation in experimental results obtained between different laboratories.
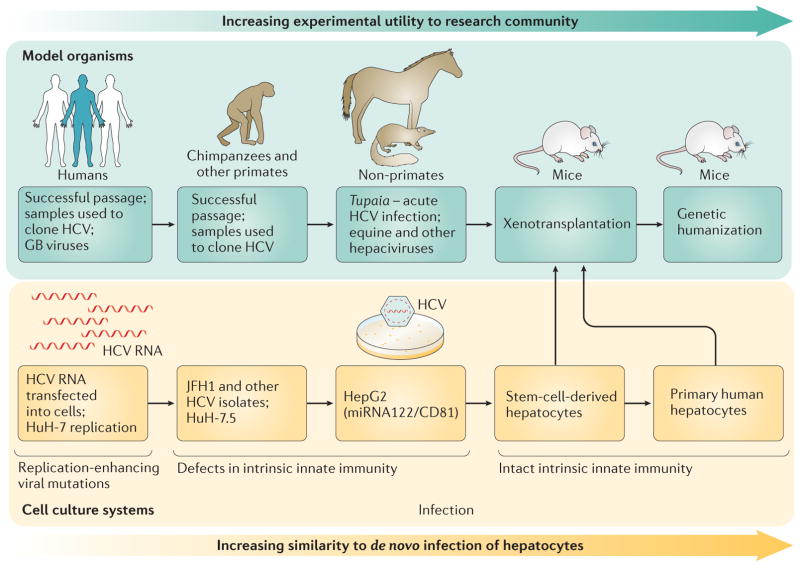
Figure 3. The evolving models to study the HCV life cycle and pathogenesis.
The use of model organisms has shifted from the use of primarily humans and primates to that of mouse models that are ‘humanized’ through xenotransplantation or genetic modification. Cell systems that initially relied upon the use of hepatoma-derived cell lines and transfected RNA genomes have transitioned to in vitro infection of primary hepatocytes and stem-cell-derived hepatocyte-like cells with functional cell intrinsic innate antiviral responses. JFH1, Japanese fulminant hepatitis 1.
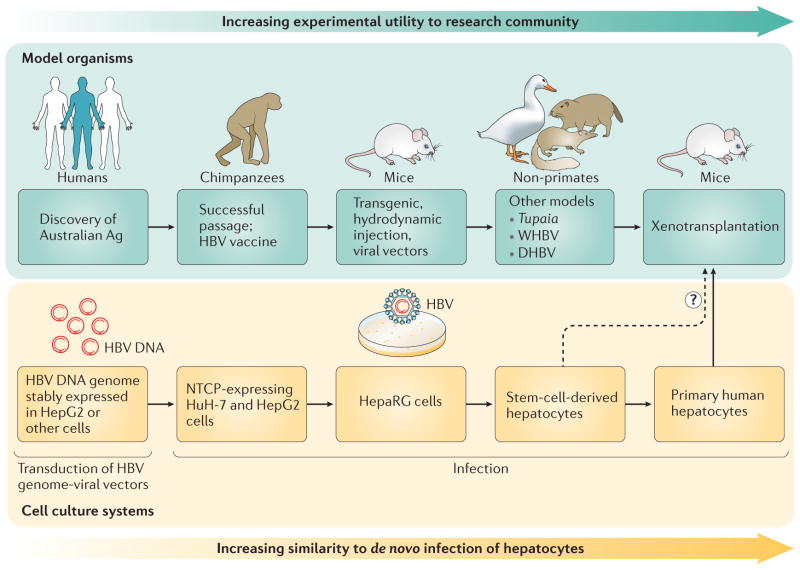
Figure 4. The evolving models to study the HBV life cycle and pathogenesis.
The use of model organisms has shifted from the use of primarily humans and primates to alternative models including Tupaia and woodchuck (WHBV) and duck (DHBV) hepatitis viruses. Xenotransplantation in mouse models has more recently been developed with animals that are susceptible to HBV infection. Cell systems that initially relied upon the use of hepatoma-derived cell lines and transfected or virally transduced DNA genomes have transitioned to infectious systems including primary hepatocytes and stem-cell-derived hepatocyte-like cells with functional cell intrinsic innate antiviral responses. Ag, antigen; DHBV, duck hepatitis B virus; NTCP, sodium/bile acid cotransporter (also known as SLC10A1); WHBV, woodchuck hepatitis B virus.
HuH-7 cells
The HuH-7 cell line, created in 1982, was derived from a well-differentiated HCC tumour that was removed from a 57-year-old Japanese man who underwent a liver resection for the tumour44. This cell line was useful in studies of HBV45 but has been more commonly used for in vitro models to study HCV. Initially, the HuH-7 cell line was successful in propagating luciferase-containing subgenomic mutants that only encoded HCV nonstructural proteins46. As a result, this cell line, described in detail in the next section, was utilized as the primary substrate for the generation of a replicon that would continuously express and replicate the HCV RNA genome, without the integration of a DNA intermediate in the cellular genome.
Replicon
The development of a cell line stably expressing the HCV RNA genome was a landmark achievement in the field46, and was rapidly duplicated by other groups47. Many of the latest DAAs that have been tested in patients with hepatitis C were initially discovered through drug screens using this model for HCV genotype 1 infection, and use of this model is one of the reasons the new DAAs can efficiently target this HCV genotype35. Interestingly, it was clearly demonstrated that the DNA from the replicon plasmid used to generate the HCV RNA did not insert into the genome as was done for HBV-overexpressing cells46. This model was also particularly useful because in addition to a selection cassette, a reporter (for example, luciferase or green fluorescent protein) could be inserted into the replicon construct in several different configurations (for example, monocistronic or bicistronic) to easily monitor genome replication by luminescence or fluorescence microscopy15. Two main reasons led to the successful establishment of this model for HCV replication: first, mutations in the viral genome occurred during the selection process that favoured replication in cell culture; and second, the presence of cells that were more permissive to hepatitis viral replication owing to defects in innate immune pathways that usually restrict virus growth14,15. Understanding the role of these two aspects of replicon creation enabled the subsequent development of cell culture models for HCV infection and propagation.
One of the advances that contributed to development of the HCV in vitro infectious model involved curing replicon cells of the HCV genome through treatment with interferon. The removal of the HCV RNA genome from these cells resulted in the selection and development of cell lines (for example, HuH-7.5 and HuH-7-lunet) that subsequently harboured more notable defects in innate immune antiviral pathways, involving genes such as RIG-I, than that observed in the parental HuH-7 cell line48,49. These cells lines were subsequently used to establish more replicon cell lines using additional HCV genotypes (such as genotypes 3 and 4)50,51 and to develop the first HCV in vitro model that could fully propagate infection. Furthermore, in a study published in 2015 (REF. 52), the HuH-7.5 cell line was utilized to support wild-type replicons for all HCV genotypes and even infection with clinical isolates through the overexpression of the SEC14L2 gene (and subsequent protein), which can promote HCV infection by enhancing vitamin-E-mediated protection against lipid peroxidation. Overexpression of this gene can also confer other hepatoma cell lines, including HuH-7 and Hep3B–miR122 albeit at lower levels, with the ability to support multiple HCV replicons52.
JFH1 strain and use of ribozymes
Although the replicon was a monumental breakthrough in the study of HCV and proved to be useful as a tool for high-throughput screening for antiviral drugs, it still failed to be quickly translated into a fully infectious in vitro system. Seminal studies demonstrated that mutations in the HCV genome, which conferred increased replication in vitro and were needed to establish the replicon cell lines, prevented the production of infectious virions in chimpanzees53. Thus, it was assumed that the best method to develop an in vitro model that fully recapitulates HCV infection would rely on wildtype HCV genomes. As replication-favouring mutations were not helpful toward this end, cell lines that were highly permissive for replication were also used. One of the first successful approaches to generating an in vitro model of HCV infection involved the JFH1 genotype 2a clone. This viral strain was generated after isolation from a Japanese patient who developed fulminant hepatitis54. Upon transfection of the HuH-7.5 cell line (which is more permissive for HCV replication48) with this cloned viral genome, it was found that infectious HCV virions were produced that could be used to infect chimpanzees and propagate the virus in this specific human hepatoma cell line55. Importantly, several other groups contributed to and confirmed this exciting achievement in HCV virology56,57. In 2006, additional infectious clones were developed for HCV genotype 1a58, and several others genotypes including 1b, 3a and 4a59,60; yet none of these replicate as robustly as the JFH1-based viruses in vitro.
A second, less frequently utilized in vitro model was also generated by an alternative approach61. In this case, the HCV cDNA, initially from genotype 1b and later extended to other genotypes, was inserted between two ribozymes that were designed to generate the naturally occurring 5. and 3. ends of the viral genome. Transfection of this construct into HuH-7 cells resulted in the production of infectious virus that can be propagated in chimpanzees62. This system has been extended to facilitate production of infectious virus from other genotypes including JFH1 (REF. 62).
With regards to HBV-related experimentation, the HuH-7 cell line has been useful but it has had less of a prominent role than that seen with HCV. Similar to HCV, this cell line was initially used to over express viral proteins from the HBV genome45. One of the important uses of HuH-7 cells to study HBV was the transfection of linear, full-length HBV cDNA into this cell line63,64. Because this model enabled the establishment of cccDNA during the HBV life cycle, this approach was used to study more complex aspects of HBV biology, including those related to epigenetic modification of the HBV episomal DNA65. In 2012, with the identification of the putative HBV entry receptor, NTCP, encoding sodium/bile acid cotransporter (also known as SLC10A1), a HuH-7 cell line stably expressing this gene was used as a model that can support HBV infection66.
HepG2 cells
Another cell line that has been used to study agents of viral hepatitis is the HepG2 cell line. This cell line was derived from the liver tissue of a 15-year-old white male patient who had a well-differentiated hepatoblastoma. In contrast to the HuH-7 cell line, the HepG2 cell line is polarized and has proved to be much more useful for the study of HBV than HCV. Initial breakthroughs in HBV research were achieved as the HBV genome could be continually expressed in HepG2 cells. This feat was achieved by selecting for stably transfected HepG2 cells carrying the HBV genome through growth in selection media43,67. Although these transfected cells were able to support HBV replication and formation of cccDNA, albeit at a low level, they were unable to be readily infected by HBV virions, now known to be due to the absence of expression of the NTCP gene. Similar approaches have been used to generate additional cell lines that were selected for carrying the HBV genome and they are designated as the HepG2 H1.3 and HepAD38 cell68–70. These cell lines continue to be used to study aspects of the HBV life cycle, including late steps of HBV replication and clearance of cccDNA by host defence pathways71. An additional model that was subsequently developed involved HepG2 cells transduced with baculovirus and adenovirus constructs, used as delivery vehicles, to efficiently express the HBV genome in these cells72,73. These transduction models were an improvement over transiently transfected or stable cell lines because HBV gene expression and replication far exceeded that found in HepG2.2.15, and these models are more reproducible than the transfected cells. With the discovery of the putative receptor for HBV (NTCP), HepG2 cells that stably express NTCP are now available to study HBV infection with infectious virions66. This model has facilitated the routine and reliable study of the entire HBV life cycle in a renewable cell culture platform.
An advance in the past decade has also enabled HepG2 cells to be more useful for studies of HCV. HepG2 cells had been observed to lack miRNA 122 (miR-122), which is necessary to support the HCV lifecycle. Specifically, miR-122 might support HCV RNA stability, replication and translation through several distinct mechanisms by directly binding homologous sequences in the untranslated region of the virus genome74,75. In addition, HepG2 cells also demonstrate very low expression of CD81 (a known entry factor for HCV)76,77. Consequently, it was demonstrated that HepG2 cells that are selected to stably express both the CD81 receptor and miR-122 can be infected by HCV and support viral replication78,79. Interestingly, owing to the differences in antiviral innate immunity within this cell line when compared with HuH-7 cells, this model has proven to be useful in the study of host defence pathways and to validate results obtained in primary human hepatocytes (PHH). Specifically, the genetically altered cell line was capable of producing large amounts of type III IFNλ in response to HCV infection, which has also been observed in primary human hepatocytes78,80.
HepaRG cells
An additional cell line has proven to be useful for studies of both HBV81 and HCV82: the HepaRG cell line. This cell line was developed from cells obtained from a resection of tumour from an HCV-infected individual with liver cancer, although the infection was not sustained in vitro after isolation of these cells. HepaRG cells are bipotent hepatic progenitor cells that can differentiate into both biliary and hepatocyte-like cells and can divide indefinitely81. To be fully capable of supporting infection with HBV and HCV, these cells must also be treated with dimethyl sulfoxide to foster additional maturation into more differentiated hepatocyte-like cells. According to head-to-head comparisons, HepaRG cells have many similarities to PHHs, which are considered to be the gold standard for hepatocyte studies in vitro, in terms of drug metabolism and ability to support infection by HBV and HCV83,84. Initially, this cell line was demonstrated to be capable of supporting HBV infection and replication81. Unfortunately, the use of polyethylene glycol is needed to facilitate viral entry by increasing interactions between the HBV virion and the cell membrane but the overall infection rate is low with minimal cell-to-cell virus spread85. Later, investigators were able to use this cell line as a substrate for HCV infection82. In this study82, the investigators were able to utilize serum-derived HCV genotype 3 to validate susceptibility to HCV infection55.
Other cell lines
Other cell lines that have been utilized for studies specifically related to viral hepatitis include PH5CH8 and Hep3B cells. Hep3B cells contain a mutation in the P53 gene that renders the protein inactive; the cell line has thus been used to study the interaction between p53 and both HCV and HBV in the context of a non-functional p53 protein86,87. PH5CH8 cell lines have been utilized for additional studies because it has a functional Toll-like receptor 3 system (necessary for recognition of double-stranded RNA associated with some viral infections) that is present in PHHs, but absent in HepG2 and HuH-7 cells88–90. Consequently, specific studies focused on RNA-sensing pathways and their role in HCV infection have been facilitated through its use.
Primary human hepatocytes
As previously mentioned, PHHs are considered the gold standard for laboratory studies of hepatocyte function. These normal cells are obtained from patients undergoing liver resection (usually for a hepatic tumour) and they are isolated from the outer edges of the tumour margin. In addition, PHHs can also be routinely obtained from the fetal livers of aborted embryos and can also serve as a substrate for infection91. Given that these fetal hepatocytes (often a combination of both hepatocytes and hepatoblasts depending on the gestational age of the donor fetus) are utilized and discarded, their use is much less controversial than that of embryonic stem cells, which offer the prospect for long-term biomedical applications such as regenerative medicine92. Unlike HepaRG cells, PHHs once plated do not divide and they have a limited lifespan in tissue culture, usually between 1 and 2 weeks, although fetal hepatocytes can be stable for several weeks91. Once in culture, these cells rapidly dedifferentiate, concomitantly downregulating biological characteristics found in mature hepatocytes93. Although these cells have a limited lifespan in culture, they readily support infection by HCV and HBV and are thus useful for the study of viral hepatitis. However, viral replication is usually limited when compared with levels seen in cell lines such as HuH-7.5; these PHHs presumably have intact host defence pathways that combat the infection80. For HCV, many studies have demonstrated productive infection of PHHs after culture, including cells that are obtained from HCV-positive patients who continue to harbour replicating virus as opposed to de novo infections80,94–96. Many studies have also demonstrated productive infection of PHHs with serum-derived and cell-culture-derived HBV97–100. In addition, primary hepatocytes isolated from other species including chimpanzees and Tupaia have proven to be capable of supporting infection by HBV101 and related hepadnaviruses that infect woodchucks and ducks102,103.
To extend the length of viability in culture and to maintain the differentiation status of PHHs, several approaches have been developed each with some success. These approaches include the growth of PHHs with additional so-called feeder cells such as mouse embryonic fibroblast cell lines (for example, STO) and non parenchymal liver cells including Kupffer and endothelial cells that support hepatocyte viability104. Hepatocytes have also been cultured in 3D organoids105. In their ‘natural’ environment, hepatocytes and other epithelial cells exist and function in a highly structured system. The organization of cells into an ordered microenvironment facilitates optimal viability by facilitating efficient nutrient uptake and waste removal. In addition, complex biological interactions with tissue matrix proteins and surrounding non parenchymal cells and vessels promotes functional polarization. Unfortunately, these critical properties of the tissue microenvironment are absent in standard in vitro cell culture systems. To provide more physiological culture conditions, micro patterning approaches facilitate the creation of a tissue-like microenvironment. This step is largely achieved by imposing a defined cell adhesion pattern that promotes the formation of islands of cells. Current micropatterning approaches also utilize feeder cells to produce growth factors and provide adhesion molecules and extracellular matrix components for cell attachment. For HCV, this approach has led to the establishment of infection of PHHs in culture for several weeks while the cells maintain functional characteristics of mature hepatocytes104. Although micropatterning of hepatocytes offer several advantages over traditional 2D cell culture, organization of hepatocytes into 3D organoids also presents additional benefits. This approach includes the ability to facilitate the organization of all three of the major nonparenchymal cells in the liver (for example, hepatic stellate cells, Kupffer cells and endothelial cells) in self-assembled networks with hepatocytes and to allow the laminar-flow of fresh media through channels, using sophisticated perfusion apparatuses, over cells that are plated in scaffolds. The design and optimization of these devices, also called bioreactors, represent an exciting area of future use for models of viral hepatitis106,107. In addition, traditional approaches to immortalize primary hepatocytes by introducing oncogenes, such as human papilloma virus encoded E6 and E7, continue to be utilized with increasing success to extend the length of utility of PHHs in culture108.
Stem cells
Progress in the propagation of stem cells in culture has led to exciting models that can be used to study viral hepatitis. Methods are now available to differentiate stem cells toward the endodermal lineage and then forward to foster development into mature hepatocytes109. Stem cells of embryonic origin are a more controversial source because of the need to derive these cells from discarded embryos and because of the possibility of long-term biomedical applications (including cloning)92. Induced pluripotent stem cells (iPSCs) are a newly developed source of hepatocytes that can be used for studies on viral hepatitis. These cells, once generated, can be a reliable source of cells that can be differentiated into mature hepatocytes. The advantages of stem-cell-derived hepatocytes over PHHs include the ability to obtain an unlimited supply of primary hepatocytes and that these cells are less variable when compared with PHHs that are obtained from different donors, who can vary by gender, age, exposure to medications and/or chemicals, genetic polymorphisms and the presence or absence of underlying liver disease110,111. These human stem-cell-derived hepatocytes have proven to be a useful substrate for infection by JFH1 and HCV obtained from infected patient serum112–114 and cells from other species such as pigtail macaques have also been utilized for HCV infection and propagation115. An interesting finding in one particular study utilizing stem-cell-derived hepatocytes was that the ability to support HCV infection was dependent upon the subsequent expression of miR-122 that was induced during the differentiation process. A similar approach has been used to successfully support infection by HBV116. Unfortunately, stem-cell-derived hepatoctyes do not reach the phenotype of fully functional mature hepatocytes; however, various approaches have been taken to overcome hurdles in function and other drawbacks of this model, including the use of small molecules that promote differentiation, and manipulation of the microenvironment used in the in vitro culture systems117,118. An exciting prospect for this line of inquiry has also been developed through the use of stem cells to develop vascularized and functional human liver in vitro119. This model might offer opportunities to study viral hepatitis in a 3D system with a functional vasculature. A summary of in vitro models to study HBV and HCV is presented in TABLE 1.
Animal models
Mouse models
Mouse models were initially developed using animals that overexpressed HBV viral proteins or the full-length replication competent viral genome so that the immune response to HBV viral antigens could be studied in an in vivo setting120. Subsequently, complete replication of HBV was accomplished using hydrodynamic injection of the HBV genome into both immunocompetent and immunodeficient mice, demonstrating an important role of the cellular immune system in controlling HBV infection in vivo121. An improvement of this method was achieved using adenoviral delivery of the HBV genome to the liver of mice, which established chronic infection and facilitated studies on adaptive immune responses and antiviral drug testing73,122.
With respect to HCV, as the virus has an RNA genome, it was much more difficult to develop stable mouse models16. Initial efforts to overexpress HCV proteins in mice were achieved much later than the corresponding HBV models123. As is the case with HBV, mouse hepatocytes are a poor substrate for natural HCV infection because of differences in multiple host factors present in human hepatocytes124. One method that has been successful in overcoming this species-specific barrier is through the generation of humanized mice through xenotransplantation of PHHs. Through this method, PHHs can be delivered into a mouse to subsequently repopulate the mouse liver. For this process to be successful, the mouse needs to be immunodeficient so as not to reject the transplanted human hepatocytes and the normal mouse hepatocytes need to be replaced. The replacement of the mouse hepatocytes can be specific ally achieved by genetically inducing liver injury in the mouse, thereby creating a growth advantage for the engrafted human hepatocytes. The first successful utilization of this approach was achieved using the SCID (severe combined immunodeficiency) mice carrying a urokinase plasminogen activator transgene that is activated by a liver-specific albumin promoter (Alb-uPA)125. Subsequent expression of the transgene results in acute hepatotoxicity that can be rescued by the engraftment of injected PHHs, which are then the substrate for HCV infection.
Unfortunately, the frailty of these mice has limited the widespread use of this model. Utilizing a promoter that is activated later in mouse development, such as the major urinary protein promoter, the development of a healthier and more robust animal model (MUP-uPA) proved to be effective to support both HCV and HBV infection126. An additional hepatotoxic model uses Fah-knockout mice for which the liver remains healthy with the administration of the small molecule 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione also called NTBC or nitisinone. Upon discontinuation of this agent, mouse hepatocytes specifically accumulate toxic metabolites and PHHs can be introduced into the mouse liver, facilitating infection by both HCV and HBV127. A more complex mouse model (AFC8) has been developed that offers both a humanized liver and rudimentary immune system. This model utilizes the caspase 8 transgene that can be activated with administration of FK506 to cause mouse hepatocyte apoptosis128. When PHHs were infected with HCV in this model, these animals developed liver fibrosis as a result of the immune response to HCV128. Similar approaches have also been used to create chimeric mice that support HBV infection and also develop liver fibrosis129. Similarly, HDV infection has also been achieved in this model130. Excitingly, humanized mice have now been developed that utilize iPSC-derived human hepatocytes (iHeps) to repopulate the livers of MUP-uPA mice to support HCV infection and this approach might be amenable to support infection with other hepatitis viruses131.
Another approach for developing humanized mice is to genetically introduce human genes to express human proteins required for HCV infection. This approach was facilitated by the discovery of specific human viral entry factors for HCV, most importantly CD81 and occludin, that are needed to enable infection of mouse hepatocytes77,132,133. By genetically engineering mice with human homologues of these two HCV entry factors, viral entry was first reported in vivo in an important proof-of-concept study134 and HCV infection was success fully demonstrated in a subsequent study135; however, viral replication was quite low and no liver disease was observed in this model135. Interestingly, a similar approach was utilized in mice with a different genetic background, and in this model sustained viraemia and infectivity was achieved for >12 months post-infection with HCV, alongside the development of fibrosis and subsequent cirrhosis136. Expression of the NTCP gene in mice has also been developed and this model has been successfully utilized to support infection with HDV but not with HBV, for which it is more difficult to achieve productive HBV infection, possibly owing to other human host factors being needed137.
Primate models
Historically, the study of viral hepatitis has greatly benefited from the use of non-human primates as a model of both acute and chronic infection beginning with landmark experiments conducted in the 1960s138. These initial experiments were performed as a result of strong evidence that hepatitis viruses could be accidentally transmitted from primates to humans139. Although there was already strong evidence supporting the existence of both HAV and HBV from epidemiological studies26, experiments performed in chimpanzees substantiated the existence of HCV in the 1970s140,141.
Until the past decade and the advent of humanized mice susceptible to infection with hepatitis viruses, the only acceptable model for vaccine development was the chimpanzee. However, the limited supply of these animals and their associated high cost has precluded their routine use. In addition, although the US Institute of Medicine report, published in 2012, on the use of chimpanzees for biomedical research has deemed these animals suitable for vaccine studies142, increased scrutiny with regards to the ethical implications of these studies has dampened their continued use and many nonhuman primate facilities have since stopped supporting chimpanzee research143. Regardless, chimpanzees, owing to their high genetic similarity with humans, have proven to be the most useful model for the study of viral hepatitis mainly because they are susceptible to infection with hepatitis viruses. However, they do not develop a similar spectrum of liver disease as humans do144,145. Another interesting caveat is that these primates are monomorphic for several of the polymorphisms in the IL28B gene that have been linked to HCV clinical outcomes146,147; thus, chimpanzees might not be suitable for this line of investigation. Specific questions that have been addressed through the use of this model have included vaccine efficacy148, antiviral responses (including both innate and adaptive responses149), and the evaluation of novel antiviral strategies150. A summary of these in vivo models to study HBV and HCV is presented in TABLE 2.
Table 2.
In vivo models for the study of HBV and HCV
| Applicability of model | Chimpanzees | Tupaia | Mouse models | |||
|---|---|---|---|---|---|---|
| Genetically humanized model |
Stem-cell- derived human hepatocytes* |
Human immune system*‡ |
Mouse immune system*‡ |
|||
| HBV infection | Yes | Yes | Not reported | Not reported | Yes | Yes |
| HCV infection | Yes | Yes | Yes | Yes | Yes | Yes |
| Liver disease | Yes, but mild | Yes, but transient | Possible | No | Possible | No |
| Development of immunotherapeutics | Yes | Yes | No | No | Possible | No |
| Antiviral testing | Yes | Yes | Yes | Yes | Yes | Yes |
| Vaccine testing | Yes | No | No | No | Possible | No |
| Availability | Very low | Low | Moderate | Low | Very low | Moderate |
Not including HBV-related or HCV-related viruses in other animals models.
Xenotransplantation.
Engrafted with primary human hepatocytes.
Other models for Hepadnavirus
Although chimpanzees have had a large contribution in terms of findings for studies of viral hepatitis, other animal models have been successfully developed. Specifically for HBV, owing to the lack of robust cell culture and mouse models historically and due to the high cost of chimpanzees, several models have been developed and have provided useful insights into the virus life cycle. Because of the known genetic similarity to primates, the tree shrew (Tupaia belangeri) has been used151. Specific beneficial aspects of this organism for studying HBV include the ability to support HBV entry and release from hepatocytes and establishment of chronic infection and cccDNA pools152,153.
Additional models of hepadnavirus infection that have been invaluable to understanding HBV biology include the duck hepatitis B virus (DHBV) and woodchuck hepatitis B virus (WHBV). Both of these animal models have proven useful because the biology of infection in each of these is similar to that seen in human HBV infection13. Subsequently, these models have been used for studies to increase our understanding of all aspects of HBV, including the virus life cycle, immune responses and for antiviral development. Specifically, WHBV has been informative towards our understanding of the liver disease and carcinogenesis as a result of hepadnavirus infection and the resulting pathology that is elicited154. DHBV has proven to be more useful in our understanding of the viral life cycle of hepadnaviruses mainly due to the fact that these animals are relatively easy to handle, can be infected easily and develop high viraemia21,155. However, there are numerous differences between these viral orthologues and HBV, such as the spectrum of comparable liver disease, transcriptional regulation of the viral genomes and substantial DNA sequence divergence156,157. The difficulty in working with these species, which are not routinely found in biomedical laboratories, limits their utility to the general viral hepatitis research community. In addition, the lack of specific research tools to study immune responses in these outbred models is also a major hurdle152.
Other models for Hepacivirus
Similar to that observed for hepadnaviruses, additional animal models of hepaciviruses have been discovered158,159. However, many of these models have only just emerged and have yet to yield a substantial amount of insight for the study of HCV as has been achieved using animal models for HBV. However, it is anticipated that these models could offer new insight into HCV virology and pathogenesis as well as insight into the evolution of this virus. The pigtail macaque might be a useful model to study HCV as cells from this primate have been shown to be capable of supporting HCV infection in vitro115. GB viruses, closely related to HCV phylogenetically, are non-human primate hepaciviruses160. The GBV-B virus has been shown to cause hepatitis in experimentally infected tamarins (genus Saguinus)161,162. Specifically, infection of New World primates with GBV-B recapitulates the phenotypes of acute viral clearance and chronic pathological disease, and this model has been used for drug testing163–165. In this regard, GBV-B seems to be a viable surrogate model for the study of HCV infection with possible utility in the testing of novel vaccine strategies. In the past few years, newly discovered hepaciviruses in bats166, dogs158 and horses (most similar to HCV)159, might also provide unique models for study. However, research on these newly discovered viruses is only just beginning and which of these animal models will prove to be most relevant to HCV remains to be seen.
Future directions
General
The mechanisms underlying how hepatitis viruses manifest as chronic infection and cause liver disease remain to be fully characterized. The means by which the genomes of these viruses persist intracellularly despite the presence of a functional innate immune response are still poorly understood. Precise insights pertaining to the strategies by which the agents of viral hepatitis are specifically able to subvert these antiviral responses and establish chronic infection are needed. Studies in PHHs and stem-cell-derived hepatocytes (iHeps) might be particularly useful in the case of HCV for which polymorphisms in the IL28B locus, an established determinant in spontaneous viral clearance146, can be specifically studied in normal cells from hepatocytes with different genotypes. In addition, improvements in these in vitro models utilizing normal hepatocytes are needed to facilitate robust infection from serum-derived HCV to facilitate studies with all genotypes; notably, major progress in this area has now been achieved52. Identifying the molecular pathways that are perturbed by chronic infection with HBV and HCV to facilitate the development of HCC is also an urgent area of unmet need. For HCV, the use of models that develop liver fibrosis and cirrhosis is important128,136 because HCC usually develops in patients with cirrhosis167. These models might be particularly useful for the identification of biomarkers to predict the development of HCC because even after achieving sustained virologic response, patients with cirrhosis because of HCV infection are still at an increased risk of developing HCC23. Although mouse models of HBV infection have been established that develop liver disease129, they need to be improved and further refined to capture the full spectrum and pathogenesis of HCC development167.
HBV
With regards to HBV, studies to increase the development of strategies to eradicate cccDNA from infected cells are currently underway71, but improved models are needed. Although HBsAg levels might be an indirect measure of cccDNA levels13, a more precise and easily accessible biomarker is needed to monitor the endogenous cccDNA pool in infected patients. Unfortunately, the lack of robust laboratory models that accurately recapitulate in vivo HBV infection has hampered the study of cccDNA persistence and removal152. With the discovery of the NTCP gene, it is anticipated that a mouse model will be developed that will support HBV infection, as has been achieved for HDV137, to aid in careful studies on the establishment and clearance of cccDNA pools. Such a model would also aid in the discovery of novel antiviral agents and new therapeutic strategies, including the use of CRISPR/Cas technology to inhibit HBV DNA replication168–170 and to purge the cccDNA reservoir in infected individuals by eliciting target specific mutations and deletions in the virus genome. As cccDNA quantification requires access to liver tissue, human studies are difficult and reliable animal models would be much more amenable to study due to the ease of access for obtaining liver tissue.
HCV
With regards to HCV, an effective vaccine is needed to prevent further spread of the virus and reinfection in those who are cured. Although chimpanzee models are useful to aid in the development of vaccines, their cost is prohibitive for routine studies and tight restrictions have been placed to limit their use. Humanized mouse models might represent a reasonable alternative in the future to evaluate vaccine efficacy, but additional effort is needed to replicate human-specific immune responses to HCV17 and facilitate their widespread use in the vaccine development community2.
Conclusions
Since the discovery of the aetiological agents of viral hepatitis B and C24,29, development of highly effective HBV vaccines and therapeutic breakthroughs that now enable successful treatment with a durable cure for HCV infection have been achieved. These efforts will promote the eradication of these viruses. Although chronic HBV infection can now be treated with potent suppressive treatment regimens, this viral illness still necessitates improvement in therapeutic intervention to facilitate a durable cure as has now been achieved for HCV. Similarly, vaccines are needed to prevent the spread of HCV infection and also to prevent reinfection in those who have achieved a sustained virologic response. In addition, even with cure, chronically infected patients are still at an increased risk of developing HCC, which remains a cancer type associated with high mortality23. Continued effort is, therefore, needed toward developing new models to study viral hepatitis to further our understanding of the mechanisms by which these viruses manifest as chronic infection and the molecular pathways that are ultimately involved in the development of liver disease and HCC.
Key points.
Early work on viral hepatitis relied on biochemical methods and microscopy, but the field greatly benefited from the use of primates and other animal models of both acute and chronic infection
Use of tumour-derived cells lines has increased our understanding of viral hepatitis; small molecule screens were performed in cell lines expressing viral genomes to identify agents that could suppress viral replication
Primary human hepatocytes are considered the most biologically relevant in vitro model for viral hepatitis infections; advances in propagation of stem cells in culture have led to exciting new stem-cell-derived hepatocyte models
Hepatocytes and other epithelial cells exist and function in a highly structured system and in vitro models are moving to those that incorporate multiple nonparenchymal cells and 3D lattices to preserve full hepatocyte function
Mouse models were initially limited by species-specific barriers, but transgenic animals can used to study immune responses and species-specific barriers can be overcome through the generation of humanized mice
Further progress in recapitulating human immune responses in mouse models might facilitate the development of an HCV vaccine, with in vitro models of HBV cccDNA persistence aiding discovery of anti-HBV small molecules
Acknowledgments
E.T. acknowledges funding support from the Miami Center for AIDS Research, Leonard M. Miller School of Medicine, USA (P30AI073961) and a the Miami CTSI KL2 program.
Footnotes
Author contributions
Both authors made equal contributions to all aspects of this manuscript.
Competing interests statement
The authors declare no competing interests.
References
- 1.Thomas E, Yoneda M, Schiff ER. Viral hepatitis: past and future of HBV and HDV. Cold Spring Harb Perspect Med. 2015;5:a021345. doi: 10.1101/cshperspect.a021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, et al. Long-term efficacy of a hepatitis E vaccine. N Engl J Med. 2015;372:914–922. doi: 10.1056/NEJMoa1406011. [DOI] [PubMed] [Google Scholar]
- 4.Wang JL, et al. Histological outcome for chronic hepatitis B patients treated with entecavir versus lamivudine-based therapy. World J Gastroenterol. 2015;21:9598–9606. doi: 10.3748/wjg.v21.i32.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai NC, et al. Viral suppression and cirrhosis regression with tenofovir disoproxil fumarate in Asians with chronic hepatitis B. Dig Dis Sci. 2015;60:260–268. doi: 10.1007/s10620-014-3336-7. [DOI] [PubMed] [Google Scholar]
- 6.Liang TJ, Ghany MG. Therapy of hepatitis C — back to the future. N Engl J Med. 2014;370:2043–2047. doi: 10.1056/NEJMe1403619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang TJ, Ghany MG. Current future therapies hepatitis C virus infection. N Engl J Med. 2013;368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;1:24–30. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan H, et al. NTCP opens the door for hepatitis B virus infection. Antiviral Res. 2015;121:24–30. doi: 10.1016/j.antiviral.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, et al. Modeling hepatitis B virus infection, immunopathology and therapy in mice. Antiviral Res. 2015;121:1–8. doi: 10.1016/j.antiviral.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason WS. Animal models and the molecular biology of hepadnavirus infection. Cold Spring Harb Perspect Med. 2015;5:a021352. doi: 10.1101/cshperspect.a021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang TJ, et al. Present and future therapies of hepatitis B: from discovery to cure. Hepatology. 2015;62:1893–1908. doi: 10.1002/hep.28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeisel MB, et al. Towards an HBV cure: state-of-the-art and unresolved questions-report of the ANRS workshop on HBV cure. Gut. 2015;64:1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 14.Murray CL, Rice CM. Turning hepatitis C into a real virus. Annu Rev Microbiol. 2011;65:307–327. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann V, Bartenschlager R. On the history of hepatitis C virus cell culture systems. J Med Chem. 2014;57:1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 16.Bukh J. Animal models for the study of hepatitis C. Virus infection and related liver disease. Gastroenterology. 2012;142:1279–1287. e3. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 17.von Schaewen M, Ploss A. Murine models of hepatitis C: what can we look forward to? Antiviral Res. 2014;104:15–22. doi: 10.1016/j.antiviral.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagaye S, et al. Major models of HCV infection. Future Virol. 2013;8:493–506. [Google Scholar]
- 19.Catanese MT, Dorner M. Advances in experimental systems to study hepatitis C virus in vitro and in vivo. Virology. 2015;479:221–233. doi: 10.1016/j.virol.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. doi: 10.1186/1743-422X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith BD, et al. Recommendations for the identification of chronic hepatitis C virus infection among persons born during 1945–1965. MMWR Recomm Rep. 2012;61:1–32. [PubMed] [Google Scholar]
- 23.Kanwal F, et al. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C Virus Infection. Gastroenterology. 2011;140:1182–1188. e1. doi: 10.1053/j.gastro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumberg BS, Alter HJ, Visnich S. A ‘new’ antigen leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 25.Melnick JL, Boggs JD. Human volunteer and tissue culture studies of viral hepatitis. Can Med Assoc J. 1972;106:461–467. [PMC free article] [PubMed] [Google Scholar]
- 26.Krugman S, Giles J, Hammond J. Evidence for two distinctive clinical, epidemiological, and immunological types of infection. JAMA. 1967;200:365–373. doi: 10.1001/jama.200.5.365. [DOI] [PubMed] [Google Scholar]
- 27.Feinstone SM, Kapikian AZ, Purceli RH. Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science. 1973;182:1026–1028. doi: 10.1126/science.182.4116.1026. [DOI] [PubMed] [Google Scholar]
- 28.Feinstone SM, et al. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med. 1975;292:767–770. doi: 10.1056/NEJM197504102921502. [DOI] [PubMed] [Google Scholar]
- 29.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 30.Rizzetto M, et al. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang KS, et al. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 32.Rizzetto M. Hepatitis D: clinical features and therapy. Dig Dis. 2010;28:139–143. doi: 10.1159/000282077. [DOI] [PubMed] [Google Scholar]
- 33.Khuroo MS. Study of an epidemic of non-A, non-B hepatitis. Possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med. 1980;68:818–824. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- 34.Reyes GR, et al. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 35.Woerz I, Lohmann V, Bartenschlager R. Hepatitis C virus replicons: dinosaurs still business? J Viral Hepatitis. 2009;16:1–9. [Google Scholar]
- 36.Bartenschlager R. Hepatitis C virus replicons: potential role for drug development. Nat Rev Drug Discov. 2002;1:911–916. doi: 10.1038/nrd942. [DOI] [PubMed] [Google Scholar]
- 37.Gao M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–U108. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sumpter R, Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naik S, Russell SJ. Engineering oncolytic viruses to exploit tumor specific defects in innate immune signaling pathways. Expert Opin Biol Ther. 2009;9:1163–1176. doi: 10.1517/14712590903170653. [DOI] [PubMed] [Google Scholar]
- 40.Brighton WD, Taylor PE, Zuckerma AJ. Changes induced by hepatitis serum in cultured liver cells. Nat New Biol. 1971;232:57–58. doi: 10.1038/newbio232057a0. [DOI] [PubMed] [Google Scholar]
- 41.Bartenschlager R, Lohmann V. Replication of the hepatitis C virus. Best Pract Res Clin Gastroenterol. 2000;14:241–254. doi: 10.1053/bega.1999.0073. [DOI] [PubMed] [Google Scholar]
- 42.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 43.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakabayashi H, et al. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 45.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 47.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 48.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friebe P, et al. Kissing-loop interaction in the 3. end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saeed M, et al. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology. 2013;144:56–U137. doi: 10.1053/j.gastro.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 51.Saeed M, et al. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother. 2012;56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saeed M, et al. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature. 2015;524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bukh J, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci USA. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato T, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 55.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 57.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi M, et al. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gottwein JM, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li YP, et al. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J Virol. 2015;89:811–823. doi: 10.1128/JVI.02877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heller T, et al. An in vitro model of hepatitis C virion production. Proc Natl Acad Sci USA. 2005;102:2579–2583. doi: 10.1073/pnas.0409666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato T, et al. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang CM, et al. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yaginuma K, Shirataka Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pollicino T, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Yan H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLIFE. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sureau C, et al. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 68.Jost S, et al. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J Virol. 2007;81:10588–10596. doi: 10.1128/JVI.02489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Protzer U, et al. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology. 2007;133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 70.Ladner SK, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lucifora J, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delaney WE, 4th, Isom H. C Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28:1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- 73.Sprinzl MF, et al. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J Virol. 2001;75:5108–5118. doi: 10.1128/JVI.75.11.5108-5118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sedano CD, Sarnow P. Interaction of host cell microRNAs with the HCV RNA genome during infection of liver cells. Semin Liver Dis. 2015;35:75–80. doi: 10.1055/s-0034-1397351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jopling CL, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 76.Rice CM, et al. Is CD81 the key to hepatitis C virus entry? Hepatology. 1999;29:990–992. doi: 10.1002/hep.510290356. [DOI] [PubMed] [Google Scholar]
- 77.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 78.Israelow B, et al. HepG2 cells mount effective antiviral interferon-lambda based innate immune response hepatitis C virus infection. Hepatology. 2014;60:1170–1179. doi: 10.1002/hep.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Narbus CM, et al. HepG2 cells expressing microRNA miR-122 support entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas E, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978–988. doi: 10.1053/j.gastro.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gripon P, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ndongo-Thiam N, et al. Long-term propagation of serum hepatitis C virus (HCV) with production of enveloped HCV particles in human HepaRG hepatocytes. Hepatology. 2011;54:406–417. doi: 10.1002/hep.24386. [DOI] [PubMed] [Google Scholar]
- 83.Guillouzo A, et al. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 84.Andersson TB, Kanebratt KP, Kenna JG. The HepaRG cell line: a unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin Drug Metab Toxicol. 2012;8:909–920. doi: 10.1517/17425255.2012.685159. [DOI] [PubMed] [Google Scholar]
- 85.Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759–1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 86.Park US, et al. Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1→S transition via a p53-independent pathway in human hepatoma cells. Oncogene. 2000;19:3384–3394. doi: 10.1038/sj.onc.1203674. [DOI] [PubMed] [Google Scholar]
- 87.Lan KH, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- 88.Naka K, et al. Hepatitis C virus NS5B delays cell cycle progression by inducing interferon-β via Toll-like receptor 3 signaling pathway without replicating viral genomes. Virology. 2006;346:348–362. doi: 10.1016/j.virol.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 89.Yu SY, et al. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKε and DDX3. J Virol. 2010;91:2080–2090. doi: 10.1099/vir.0.020552-0. [DOI] [PubMed] [Google Scholar]
- 90.Li K, et al. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-β production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 91.Andrus L, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hyun I. Policy: regulate embryos made for research. Nature. 2014;509:27–28. doi: 10.1038/509027a. [DOI] [PubMed] [Google Scholar]
- 93.Elaut G, et al. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab. 2006;7:629–660. doi: 10.2174/138920006778017759. [DOI] [PubMed] [Google Scholar]
- 94.Podevin P, et al. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139:1355–1364. doi: 10.1053/j.gastro.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 95.Fournier C, et al. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J Virol. 1998;79:2367–2374. doi: 10.1099/0022-1317-79-10-2367. [DOI] [PubMed] [Google Scholar]
- 96.O’Connell JF, et al. Primary human hepatocyte culture for the study of HCV. Methods Mol Med. 1999;19:495–500. doi: 10.1385/0-89603-521-2:495. [DOI] [PubMed] [Google Scholar]
- 97.Rijntjes PJ, Moshage HJ, Yap SH. In vitro infection of primary cultures of cryopreserved adult human hepatocytes with hepatitis-B virus. Virus Res. 1988;10:95–109. doi: 10.1016/0168-1702(88)90060-3. [DOI] [PubMed] [Google Scholar]
- 98.Gripon P, et al. Hepatitis-B virus-infection of adult human hepatocytes cultured in the presence of dimethyl-sulfoxide. J Virol. 1988;62:4136–4143. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ochiya T, et al. An in vitro system for infection with hepatitis-B virus that uses primary human-fetal hepatocytes. Proc Natl Acad Sci USA. 1989;86:1875–1879. doi: 10.1073/pnas.86.6.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Galle PR, et al. In-vitro experimental-infection of primary human hepatocytes with hepatitis-B virus. Gastroenterology. 1994;106:664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 101.Kock J, et al. Efficient infection of primary Tupaia hepatocytes with purified human and woolly monkey hepatitis B virus. J Virol. 2001;75:5084–5089. doi: 10.1128/JVI.75.11.5084-5089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nair S, et al. Differential gene expression analysis of in vitro duck hepatitis B virus infected primary duck hepatocyte cultures. Virol J. 2011;8:363. doi: 10.1186/1743-422X-8-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dandri M, et al. Woodchuck hepatocytes remain permissive for hepadnavirus infection and mouse liver repopulation after cryopreservation. Hepatology. 2001;34:824–833. doi: 10.1053/jhep.2001.28189. [DOI] [PubMed] [Google Scholar]
- 104.Ploss A, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci USA. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sivaraman A, et al. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569–591. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 106.Lu YH, et al. A novel 3D liver organoid system for elucidation of hepatic glucose metabolism. Biotechnol Bioeng. 2012;109:595–604. doi: 10.1002/bit.23349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xia L, et al. Laminar-flow immediate-overlay hepatocyte sandwich perfusion system for drug hepatotoxicity testing. Biomaterials. 2009;30:5927–5936. doi: 10.1016/j.biomaterials.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 108.Levy G, et al. Long-term culture and expansion of primary human hepatocytes. Nat Biotechnol. 2015;33:1264–1271. doi: 10.1038/nbt.3377. [DOI] [PubMed] [Google Scholar]
- 109.Cai J, et al. Stem Book. Harvard Stem Cell Institute; 2008. [Google Scholar]
- 110.Gomez-Lechon MJ, et al. Human hepatocytes in primary culture: the choice to investigate drug metabolism in man. Curr Drug Metab. 2004;5:443–462. doi: 10.2174/1389200043335414. [DOI] [PubMed] [Google Scholar]
- 111.Sheahan T, et al. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe. 2014;15:190–202. doi: 10.1016/j.chom.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu XF, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2014;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwartz RE, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roelandt P, et al. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 115.Sourisseau M, et al. Hepatic cells derived from induced pluripotent stem cells of pigtail macaques support hepatitis C virus infection. Gastroenterology. 2012;145:966–969. e7. doi: 10.1053/j.gastro.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shlomai A, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci USA. 2014;111:12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwartz RE, et al. Engineering the microenvironment of differentiating IPS derived hepatocyte-like cells enables acquisition of an adult phenotype. Gastroenterology. 2013;144:S1025–S1025. [Google Scholar]
- 118.Shan J, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–577. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 1985;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 120.Chisari FV, et al. A transgenic mouse model of the chronic hepatitis-B surface-antigen carrier state. Science. 1985;230:1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- 121.Yang PL, et al. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeissig S, et al. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med. 2013;18:1060–1068. doi: 10.1038/nm.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kawamura T, et al. Transgenic expression of hepatitis C virus structural proteins in the mouse. Hepatology. 1997;25:1014–1021. doi: 10.1002/hep.510250437. [DOI] [PubMed] [Google Scholar]
- 124.Sandmann L, Ploss A. Barriers of hepatitis C virus interspecies transmission. Virology. 2013;435:70–80. doi: 10.1016/j.virol.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mercer DF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 126.Tesfaye A, et al. Chimeric mouse model for the infection of hepatitis B and C viruses. PLoS ONE. 2013;8:e77298. doi: 10.1371/journal.pone.0077298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bissig KD, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Washburn ML, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–1344. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bility MT, et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog. 2014;11:e1004718. doi: 10.1371/journal.ppat.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Giersch K, et al. Persistent hepatitis D virus mono-infection in humanized mice is efficiently converted by hepatitis B virus to a productive co-infection. J Hepatol. 2014;60:538–544. doi: 10.1016/j.jhep.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 131.Carpentier A, et al. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J Clin Invest. 2014;124:4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cormier EG, et al. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci USA. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–246. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dorner M, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen JZ, et al. Persistent hepatitis C virus infections and hepatopathological manifestations in immune-competent humanized mice. Cell Res. 2014;24:1050–1066. doi: 10.1038/cr.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He WH, et al. Hepatitis D virus infection of mice expressing human sodium taurocholate co-transporting polypeptide. PLoS Pathog. 2015;11:e1004840. doi: 10.1371/journal.ppat.1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deinhardt F, et al. Studies of liver function tests in chimpanzees after inoculation with human infectious hepatitis virus. Am J Hyg. 1962;75:311–321. doi: 10.1093/oxfordjournals.aje.a120252. [DOI] [PubMed] [Google Scholar]
- 139.Hillis WD. An outbreak of infectious hepatitis among chimpanzee handlers at a United States air force base. Am J Hyg. 1962;73:316–328. doi: 10.1093/oxfordjournals.aje.a120191. [DOI] [PubMed] [Google Scholar]
- 140.Tabor E, et al. Transmission of non-A, non-B hepatitis from man to chimpanzee. Lancet. 1978;1:463–466. doi: 10.1016/s0140-6736(78)90132-0. [DOI] [PubMed] [Google Scholar]
- 141.Alter HJ, et al. Transmissible agent in non-A, non-B hepatitis. Lancet. 1978;1:459–463. doi: 10.1016/s0140-6736(78)90131-9. [DOI] [PubMed] [Google Scholar]
- 142.Kahn J. Raising the bar: the implications of the IOM report on the use of chimpanzees in research. Hastings Cent Rep. 2012;42:S27–S30. doi: 10.1002/hast.105. [DOI] [PubMed] [Google Scholar]
- 143.Altevogt BM, et al. Guiding limited use of chimpanzees in research. Science. 2012;335:41–42. doi: 10.1126/science.1217521. [DOI] [PubMed] [Google Scholar]
- 144.Wieland SF. The chimpanzee model for hepatitis B virus infection. Cold Spring Harb Perspect Med. 2015;5:a021469. doi: 10.1101/cshperspect.a021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 146.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Prokunina-Olsson L, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Choo QL, et al. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bigger CB, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tsukiyama-Kohara K, Kohara M. Tupaia belangeri as an experimental animal model for viral infection. Exp Anim. 2014;63:367–374. doi: 10.1538/expanim.63.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dandri M, Lutgehetmann M, Petersen J. Experimental models and therapeutic approaches for HBV. Semin Immunopathol. 2013;35:7–21. doi: 10.1007/s00281-012-0335-7. [DOI] [PubMed] [Google Scholar]
- 153.Walter E, et al. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24:1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 154.Summers J, Smolec JM, Snyder R. Virus similar to human hepatitis-B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mason WS, et al. Asymmetric replication of duck hepatitis B virus DNA in liver cells: free minus-strand DNA. Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Di Q, et al. Major differences between WHV and HBV in the regulation of transcription. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 157.Sprengel R, et al. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 158.Kapoor A, et al. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lyons S, et al. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. J Gen Virol. 2014;95:1701–1711. doi: 10.1099/vir.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 160.Stapleton JT, et al. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Deinhard F, et al. Studies on transmission of human viral hepatitis to marmoset monkeys. 1. Transmission of disease serial passages and description of liver lesions. J Exp Med. 1985;125:673–688. doi: 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simons JN, et al. Identification of 2 flavivirus-like genomes in the Gb hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Manickam C, Reeves RK. Modeling HCV disease in animals: virology, immunology and pathogenesis of HCV and GBV-B infections. Front Microbiol. 2014;56:690. doi: 10.3389/fmicb.2014.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McPhee F, et al. Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032) Antimicrob Agents Chemother. 2012;56:5387–5396. doi: 10.1128/AAC.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chavez D, Guerra B, Lanford RE. Antiviral activity and host gene induction by tamarin and marmoset interferon-α and interferon-γ in the GBV-B primary hepatocyte culture model. Virology. 2009;390:186–196. doi: 10.1016/j.virol.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Quan PL, et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci USA. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ben Ari Z, Weitzman E, Safran M. Oncogenic viruses and hepatocellular carcinoma. Clin Liver Dis. 2013;19:341–360. doi: 10.1016/j.cld.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 168.Kennedy EM, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ramanan V, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Seeger C, Sohn JA. Targeting hepatitis B virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2015;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]