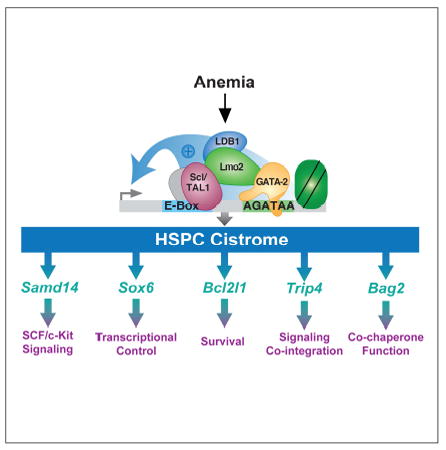
SUMMARY
An enhancer with amalgamated E-box and GATA motifs (+9.5) controls expression of the regulator of hematopoiesis GATA-2. While similar GATA-2-occupied elements are common in the genome, occupancy does not predict function, and GATA-2-dependent genetic networks are incompletely defined. A “+9.5-like” element resides in an intron of Samd14 (Samd14-Enh) encoding a sterile alpha motif (SAM) domain protein. Deletion of Samd14-Enh in mice strongly decreased Samd14 expression in bone marrow and spleen. Although steady-state hematopoiesis was normal, Samd14-Enh-/- mice died in response to severe anemia. Samd14-Enh stimulated Stem Cell Factor/c-Kit signaling, which promotes erythrocyte regeneration. Anemia activated Samd14-Enh by inducing enhancer components and enhancer chromatin accessibility. Thus, a GATA-2/anemia-regulated enhancer controls expression of a SAM domain protein that confers survival in anemia. We propose that Samd14-Enh and an ensemble of anemia-responsive enhancers are essential for erythrocyte regeneration in stress erythropoiesis, a vital process in pathologies including β-thalassemia, myelodysplastic syndrome and viral infection.
Keywords: GATA-2, erythroid, enhancer, anemia, hematopoiesis, regeneration
Graphical abstract
INTRODUCTION
Transcription factor complexes occupy vast ensembles of cis-regulatory elements to establish/maintain genetic networks controlling physiological processes such as hematopoiesis (Fujiwara et al., 2009; Li et al., 2011; Shen et al., 2012b; Wilson et al., 2010; Yu et al., 2009). In hematopoiesis, two Gata2 cis-elements (Grass et al., 2003; Grass et al., 2006; Johnson et al., 2012; Johnson et al., 2015) induce expression of the master regulatory transcription factor GATA-2 (Tsai et al., 1994). A GATA-2-occupied Gata2 intronic enhancer (+9.5) containing an E-box-8 bp spacer-AGATAA composite element (E-box-GATA element) is required for hematopoietic stem cell (HSC) emergence from hemogenic endothelium in the mouse embryo (Gao et al., 2013). A GATA-2-occupied enhancer 77 kb upstream of Gata2 (-77), which contains several GATA motifs (Grass et al., 2006), confers myelo-erythroid progenitor differentiation potential, without impacting HSC emergence (Johnson et al., 2015). Insufficient GATA-2 levels/activity resulting from GATA2 coding or +9.5 enhancer mutations underlie hematologic diseases including primary immunodeficiencies that frequently progress to myelodysplastic syndrome (MDS), acute myeloid leukemia (AML) (Dickinson et al., 2011; Hahn et al., 2011; Hsu et al., 2011; Ostergaard et al., 2011) and pediatric MDS/AML independent of immunodeficiency (Wlodarski et al., 2016). Since GATA factor occupancy of a GATA motif in chromatin does not predict GATA factor-dependent regulation of the linked gene (DeVilbiss et al., 2014; Hewitt et al., 2015; Sanalkumar et al., 2014), many questions remain unanswered regarding mechanisms conferring GATA-2 activity.
Given the essential +9.5 enhancer activity, we reasoned that a cohort of cis-elements with sequence and molecular attributes resembling the +9.5 (+9.5-like) are also critical. Prioritization of GATA-2-occupied E-box-GATA elements with attributes resembling the +9.5 enhancer, and gene editing in G1E mouse erythroid precursor cells revealed sites that regulate their endogenous loci and seemingly similar sites that do not (Hewitt et al., 2015; Tanimura et al., 2015). The functional sites included a conserved intronic enhancer (Samd14-Enh) at the largely unstudied, GATA-2-occupied Samd14 gene (Hewitt et al., 2015). SAMD14 is expressed in HSC, megakaryocyte erythrocyte progenitor (MEP) and committed erythroid progenitors (Chen and Lodish, 2014; Hewitt et al., 2015), downregulated in colorectal cancer and adenocarcinoma (Shen et al., 2012a; Sun et al., 2008), and polymorphisms associated with blood platelet volume are linked to altered SAMD14 expression (Fehrmann et al., 2011). A genome-wide screen for human genetic variants linked to hematologic phenotypes correlated a polymorphism within the SAMD14 5’ UTR with platelet distribution width (Astle et al., 2016). Samd14 is downregulated ~20 fold in myeloid progenitors lacking the Gata2 -77 enhancer (Johnson et al., 2015). shRNA-mediated downregulation of Samd14 in mouse fetal liver hematopoietic stem and progenitor cells (HSPCs) reduced stem cell factor (SCF)-induced c-Kit signaling and myelo-erythroid progenitor levels (Hewitt et al., 2015).
Conforming to a Type I feed-forward loop (Shoval and Alon, 2010), GATA-2 directly activates Kit and Samd14 transcription, and Samd14 promotes SCF-induced c-Kit receptor tyrosine kinase signaling (Hewitt et al., 2015). Given the vital GATA-2 and c-Kit functions in diverse hematopoietic cells, it is instructive to consider the biological contexts of Samd14 function. c-Kit signaling is an important determinant of erythropoiesis (Munugalavadla and Kapur, 2005; Paulson et al., 2011). Kit or SCF mutations cause macrocytic anemia (Nocka et al., 1989) and impair recovery from acute anemia (Broudy et al., 1996; Harrison and Russell, 1972). By lysing red blood cells, phenylhydrazine (PHZ) induces acute hemolytic anemia, triggering c-Kit+ erythroid stress progenitor expansion at extramedullary sites, including spleen (Lenox et al., 2005; Paulson et al., 2011). These stress progenitors express GATA-2 and the E-box-binding basic helix-loop-helix protein Scl/TAL1. In response to hypoxia, SCF and BMP-4, the stress progenitors generate erythrocytes (Harandi et al., 2010; Perry et al., 2007). Signaling-defective Kit mutant mice exhibit macrocytic anemia to varying degrees, and their response to anemic stress is impaired (Agosti et al., 2009; Perry et al., 2007). Endothelial cell-derived SCF supports stress erythropoiesis, as conditional SCF deletion in splenic endothelial cells suppresses recovery from anemia (Inra et al., 2015). Since stress erythropoiesis regenerates red blood cells in anemia (Bozzini et al., 1970), during recovery from surgery (Schlitt et al., 1995), chemotherapy (Chang et al., 2013), bone marrow transplantation (Harandi et al., 2010) and viral infection (Subramanian et al., 2008), ensuring the integrity of stress erythropoiesis mechanisms is critical.
Herein, we demonstrate that targeted deletion of Samd14-Enh in mice strongly reduced Samd14 expression in bone marrow and spleen and establish it as a GATA-2- and anemia-activated (G2A) enhancer conferring survival in severe anemia. Mechanistic analyses indicated that Samd14-Enh is the founding member of an ensemble of anemia-responsive enhancers essential for red blood cell regeneration during severe anemia, but dispensable for steady-state hematopoiesis.
RESULTS
GATA Factor-Activated Enhancer Regulates Expression of a Sterile Alpha Motif Domain Protein In Vivo
GATA-2 and Scl/TAL1 occupy Samd14-Enh 2.5 kb downstream of the Samd14 transcription start site in G1E mouse erythroid precursor cells (Figure 1A). The intronic Samd14-Enh site harbors a composite E-box-GATA element (Hewitt et al., 2016; Hoang et al., 2016; Wadman et al., 1997) resembling the +9.5 site that increases Gata2 expression in hemogenic endothelium and triggers HSPC genesis in the mouse embryo (Gao et al., 2013). Genome editing revealed that Samd14-Enh increases Samd14 transcription in G1E cells (Hewitt et al., 2015). To determine if Samd14-Enh regulates Samd14 transcription in vivo and if Samd14 functions in a Scl/TAL1-GATA-2-dependent mechanism, we used TALENs to generate a C57BL/6N mouse strain with a 33 bp deletion lacking the E-box and GATA motifs (Samd14-Enh-/-) (Figure 1B, left). Although Samd14-Enh-/- embryos were born at Mendelian ratios (Figure 1B, right), Samd14 mRNA was 23 fold lower in Samd14-Enh-/- vs. WT bone marrow (Figure 1C); Samd14 expression in brain was unaffected (Figure 1C). Samd14 primary transcripts (intron 9) were 11 fold lower (p = 0.0019) in Samd14-Enh-/- vs. WT. Thus, Samd14-Enh activates Samd14 transcription in vivo (Figure 1C). While Samd14 mRNA levels were indistinguishable in Lin-Sca1+Kit+ (LSK) cells sorted from WT and Samd14-Enh-/- bone marrow, it was reduced significantly in Samd14-Enh-/- Common Myeloid Progenitor (CMP), Granulocyte Macrophage Progenitor (GMP) and Megakaryocyte Erythrocyte Progenitor (MEP) populations vs. WT (Figure 1D).
Figure 1. Samd14-Enh Regulates Samd14 Transcription in Hematopoietic Cells and Tissues In Vivo.
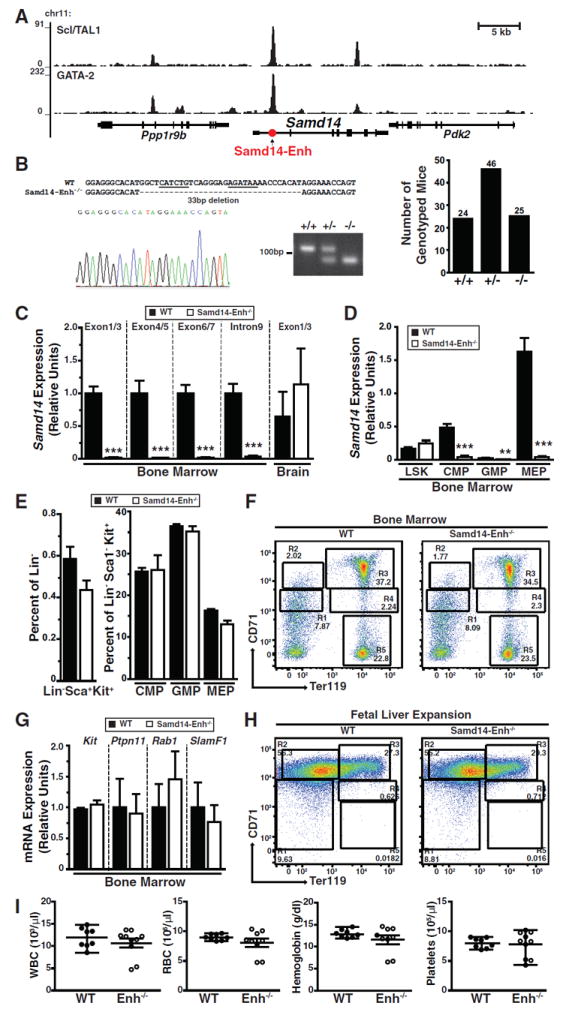
(A) ChIP-seq of GATA-2 (GSE29193) and Scl/TAL-1 (GSE36029) occupancy at Samd14 in mouse G1E proerythroblast cells (Trompouki et al., 2011). (B) Sequence validation and genotyping of a 33 base pair deletion in intron 1 of Samd14 (left). Genotypes of offspring from heterozygous matings (right). (C) Relative Samd14 mRNA and primary transcript in bone marrow and brain cerebellum from WT and Samd14-Enh-/- mice (n = 3). (D) Relative Samd14 mRNA expression in FACS-purified L-S+K+, CMP, GMP and MEP populations from bone marrow (n = 3). (E) Flow cytometric analysis of the percentage of CMP, GMP and MEP populations in WT and Samd14-Enh-/- total bone marrow, using Lin- Sca-Kit+ cells (n = 3). (F) Representative flow cytometric plot of CD71 and Ter119 staining from bone marrow of WT and Samd14-Enh-/- mice. (G) Relative expression of Kit, Ptpn11, Rab1 and SlamF1 in WT and Samd14-Enh-/- cells from bone marrow (n = 3). (H) Representative flow cytometric plot of CD71 and Ter119 staining from WT and Samd14-Enh-/- cells isolated from E14.5 fetal liver and expanded in vitro for 3 days. (I) Hematologic parameters (white blood cells (WBC), red blood cells (RBC), hemoglobin, platelets) from WT (n = 8) and Samd14-Enh-/- (n = 10) mice. Statistical significance represented by mean +/- SEM.; **p<0.01, ***p<0.001.
As Samd14 downregulation reduces mouse fetal liver myelo-erythroid progenitor CFUs (Hewitt et al., 2015), we assessed bone marrow cellularity of Samd14-Enh-/- mice. No differences were detected in the percentages of bone marrow LSK, CMP, GMP, or MEP populations (Figure 1E, Figure S1A). Using the erythroid lineage markers CD71 and Ter119, the percentages of cells at distinct stages of erythroid maturation (R1-R5 populations) (Zhang et al., 2003) were unaltered in Samd14-Enh-/- bone marrow (Figure 1F, Figure S1B). The expression of genes deregulated upon shRNA-mediated Samd14 downregulation in cultured fetal liver erythroid precursors (Kit, Ptpn11, Rab1, SlamF1) (Hewitt et al., 2015) was unaffected in Samd14-Enh-/- bone marrow (Figure 1G). In addition, cellularity and expansion potential were unaffected upon ex vivo culture of fetal liver erythroid precursors (Figure 1H, Figure S1C). In eight week old WT and Samd14-Enh-/- mice, complete blood count (CBC) hematological parameters were indistinguishable (Figure 1I, Table S1). Although Samd14-Enh strongly increased Samd14 transcription in vivo (Figure 1C), its deficiency did not impact steady-state hematopoiesis.
GATA-2- and Anemia-Activated Enhancer Confers Survival in Severe Anemia
Since Samd14-Enh was not required for steady-state hematopoiesis, we tested whether Samd14-Enh and Samd14 are important in PHZ-induced hemolytic anemia. After two daily administrations of 60 mg/kg PHZ to induce severe anemia, 15 of 16 mutant animals died before day 6; all WT mice survived (median survival = 4 days, log-rank test p = 0.0001) (Figure 2A). A single PHZ injection (100 mg/kg) in Samd14-Enh-/- mice produced anemia but was not lethal, and this dose was used for subsequent mechanistic analyses. Three days post-PHZ injection, Samd14-Enh-/- spleen weight was 15% less than WT (p = 0.044) (Figure 2B). Whereas PHZ treated Samd14-Enh-/- mice had a reduced hematocrit (Figure 2C, left) and circulating RBCs (Figure 2C, right) on days 1-4 relative to WT littermates, both cohorts of mice recovered by day 7. Prior to PHZ treatment, WT and Samd14-Enh-/- control spleens generated similar numbers of burst forming unit-erythroid (BFU-E) colonies in methylcellulose containing Epo, SCF, IL-3 and IL-6 (Figure 2D). However, Samd14-Enh-/- splenocytes generated 2.9 fold fewer (p = 0.0015) BFU-E vs. WT 48 hours post-PHZ injection and 2.9 fold fewer (p = 0.009) BFU-E vs. WT 72 hours post-PHZ injection (Figure 2D). In Epo-containing methylcellulose, spleens from vehicle-treated mice generated similar numbers of BFU-E in WT and Samd14-Enh-/- (Figure 2E). Three days post-PHZ treatment, Samd14-Enh-/- splenocytes generated 15% fewer (p = 0.048) BFU-E vs. WT; Colony Forming Unit-Erythroid (CFU-E) were unaffected (Figure 2E).
Figure 2. Samd14-Enh Confers Survival in Severe Anemia.
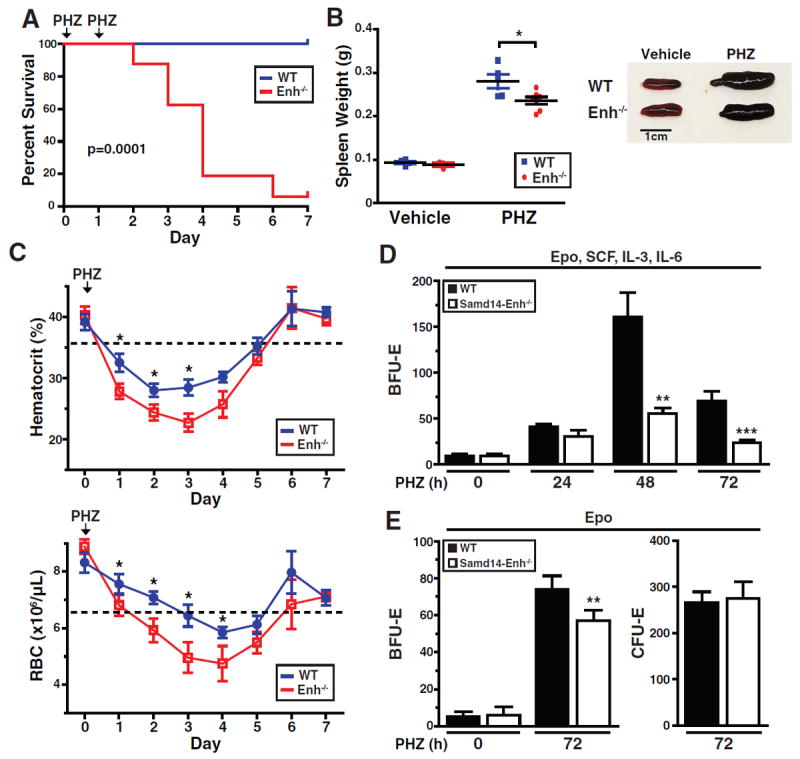
(A) Kaplan-Meier survival curve of WT and Samd14-Enh-/- mice following two doses (one per day) of 60 mg/kg PHZ (n = 16 in each group). (B) Spleen weight of WT and Samd14-Enh-/- mice 3 days after treatment with vehicle (PBS) or PHZ (100 mg/kg). (C) Hematologic parameters of WT (n = 6) and Samd14-Enh-/- (n = 6) mice following PHZ treatment (100 mg/kg). (D) Quantification of Burst Forming Unit-Erythroid (BFU-E) colonies 5 days after plating splenic cells (1 × 105) from mice treated with PHZ for 0, 24, 48, or 72 hours (h) in methylcellulose containing Epo, SCF, IL-3, and IL-6. (E) Quantification of BFU-E colonies 5 days after plating, and Colony Forming Unit-Erythroid (CFU-E) colonies 2 days after plating, 1 × 105 cells from mice treated with PHZ for 0 or 72 h in Epo-containing methylcellulose. Statistical significance represented by mean +/- SEM.; *p<0.05. **p<0.01, ***p<0.001.
Samd14-Enh increased Samd14 expression in bone marrow, and we tested whether it also regulates Samd14 in spleen in normal and stress erythropoiesis contexts. Samd14 mRNA levels were 20 fold lower in Samd14-Enh-/- vs. WT spleen (Figure 3A). PHZ increased Samd14 mRNA 36 fold (p = 0.002) in WT spleen, but not brain, and Samd14-Enh deletion abrogated this response (Figure 3A). Expression of genes flanking Samd14 (Ppp1r9b and Pdk2) was similar in WT and Samd14-Enh-/- spleen (Figure S2). PHZ increased Ppp1r9b expression similarly in WT and Samd14-Enh-/-, indicating that Samd14-Enh selectively activates Samd14 transcription. As Samd14 protein had not been described, targeted mass spectrometry was used to test whether the changes in Samd14 mRNA impact Samd14 protein levels. Analysis of three Samd14 peptides revealed that PHZ increased Samd14 levels 3.6 fold (p = 0.0001) (Figure 3B). Samd14 was 16 fold lower (p < 0.0001) in Samd14-Enh-/- vs. WT spleen. PHZ did not affect Samd14 levels in the mutant cells (Figure 3B). Since no Samd14 antibody had been reported, we generated a rabbit anti-murine Samd14 polyclonal antibody. The antibody detected a major 55 kDa band in spleen from PHZ-treated mice and weak 55 and 69 kDa bands in vehicle-treated spleen (Figure 3C). The 55 kDa protein was not detected in spleen from vehicle or PHZ-treated Samd14-Enh-/- mice (Figure 3C). An anti-Samd14 shRNA expressed in G1E cells downregulated the 55 kDa protein (Figure 3D). PHZ increased GATA-2 in WT and Samd14-Enh-/- spleen (Figure 3E).
Figure 3. Samd14-Enh Mediates Anemia-Dependent Samd14 Expression.
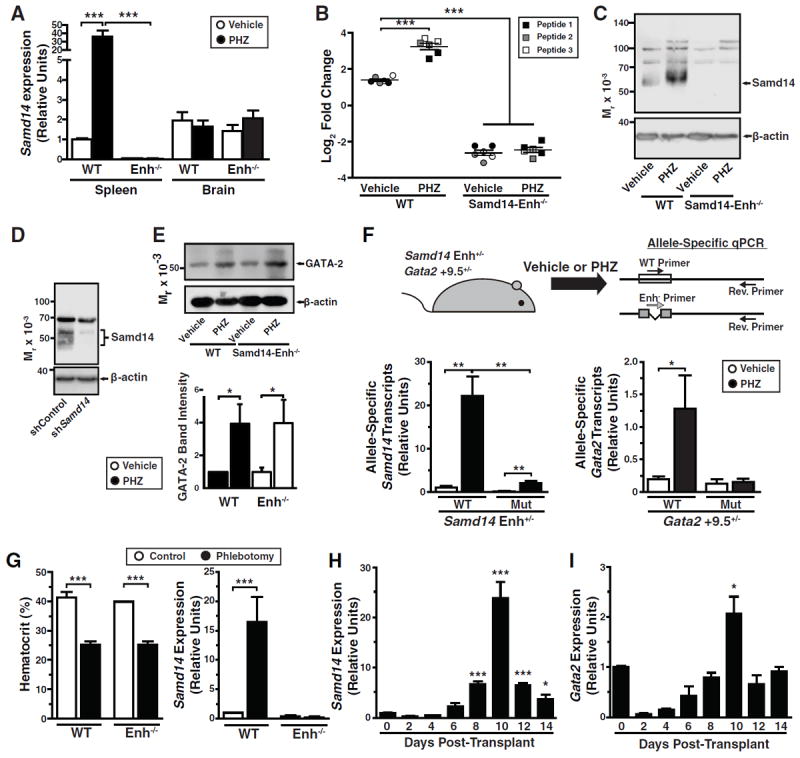
(A) Samd14 mRNA levels in WT and Samd14-Enh-/- spleen and brain from vehicle-treated and PHZ-treated animals (n = 3 in each group). (B) Targeted mass spectrometry using three distinct Samd14 peptides in WT and Samd14-Enh-/- spleen from vehicle-treated and PHZ-treated animals. Peptide 1 - SASQESTLSDDSTPPSSSPK, Peptide 2 - SLDEDEPPPSPLAR, Peptide 3 - QVDGPQLLQLDGSK. (C) Western blotting of Samd14 in WT and Samd14-Enh-/- spleen from vehicle- and PHZ-treated animals. (D) Western blot analysis of Samd14 in G1E cells retrovirally infected with an shRNA targeting Samd14 mRNA (shSamd14) relative to control shRNA (shControl). (E) Top, Western blotting of GATA-2 in WT and Samd14-Enh-/- spleen from vehicle- and PHZ-treated animals. Bottom, densitometric quantitation (n = 3). (F) (top) Experimental strategy to test allele-specific transcription changes in Samd14-Enh and Gata2 +9.5 heterozygous spleen following a 3 d PHZ treatment. Forward primers exclusively anneal to WT or mutant Enh- alleles of primary transcripts, and reverse primers anneal to both alleles. (bottom) Allele-specific qRT-PCR analysis of Samd14 and Gata2 primary transcripts from Samd14-Enh+/- and Gata2 +9.5+/- mice, respectively, in control (n = 3) and PHZ-treated (n = 4) spleen. (G) Hematocrit (left) and Samd14 mRNA (right) in WT and Samd14-Enh-/- spleen from control and phlebotomized animals (n = 4 in each group). (H) Samd14 mRNA levels in total spleen at 2, 4, 6, 8, 10, 12, and 14 days post-transplantation (n =2). (I) Gata2 mRNA levels in total spleen at 2, 4, 6, 8, 10, 12, and 14 days post-transplantation (n = 2). Statistical significance represented by mean +/- SEM.; *p<0.05 ***p<0.001. (Related to Figure 3)
Samd14 and Gata2 share nearly identical intronic E-box-GATA elements that mediate enhancer activity (Hewitt et al., 2015). As PHZ increased Samd14 and GATA-2 mRNA and protein levels, we devised an allele-specific transcription assay to test whether Samd14-Enh and Gata2 +9.5 are anemia-responsive enhancers (Figure 3F). We quantitated allele-specific primary transcripts from WT and mutant alleles in spleen from PHZ-treated (100 mg/kg) Samd14-Enh+/- mice. PHZ increased Samd14 primary transcripts from the WT allele 22 fold (p = 0.003) (Figure 3F). While mutant allele expression was higher in PHZ- vs. vehicle-treated spleen, Samd14 primary transcripts were 10 fold lower in PHZ-treated mutant vs. wild type spleen (p = 0.004) (Figure 3F). To test whether anemia-induced Gata2 expression required the +9.5 enhancer, we quantitated allele-specific primary transcripts from control and PHZ-treated spleens of Gata2 +9.5+/- mice. PHZ increased Gata2 primary transcripts from the WT allele 6.5 fold (p = 0.013); expression from the mutant allele was unaffected (Figure 3F).
We tested whether anemia-dependent increases in Samd14 expression can be generalized to distinct anemia models. Phlebotomy-induced anemia (41% decrease in hematocrit) increased Samd14 mRNA (16.5 fold, p = 0.023) in WT, but not Samd14-Enh-/-, spleen (Figure 3I). Bone marrow transplantation results in a similarly reduced hematocrit at early times post-transplant, requiring short-term radioprotective cells to generate new erythrocytes (Harandi et al., 2010). At 10 days post-transplant, Samd14 mRNA increased 24 fold (p = 0.0004) (Figure 3H), and Gata2 mRNA increased 2.1 fold (p = 0.02) in the spleen (Figure 3I). Thus, three distinct modes of inducing acute anemia elevate Samd14 expression.
To identify splenic cells in which anemia increases Samd14 expression, we used fluorescence-activated cell sorting (FACS) with anti-CD71 and -Ter119 antibodies to isolate erythroid cell populations (Figure 4A). The relative frequencies and number of CD71-Ter119-, CD71+Ter119-, and Ter119+ populations were similar in WT and Samd14-Enh-/- spleen in control and PHZ-treated mice (Figure 4B). In WT spleen, Samd14 mRNA was highest in Ter119+ and lowest in CD71-Ter119- cells (Figure 4C). Whereas PHZ did not affect Samd14 mRNA in CD71-Ter119- cells, Samd14 mRNA increased 33 fold in CD71+Ter119- cells. In Ter119+ cells, PHZ increased Samd14 mRNA 3 fold (Figure 4C). By contrast, PHZ did not affect Samd14 expression in any population in the Samd14-Enh-/- spleen. PHZ increased Gata2 expression 15-20 fold in WT and Samd14-Enh-/- CD71+Ter119- cells (Figure 4D). Gata2 expression was very low in other populations (Figure 4D). In WT and Sam14-Enh-/- mice, PHZ increased Gata1 expression in CD71+Ter119- and Ter119+ cells (Figure 4E). PHZ increased Kit expression in WT, but not Samd14-Enh-/-, CD71+Ter119- cells (Figure 4F). PHZ strongly induced Scl/TAL1 expression in CD71+Ter119- and Ter119+ cells, which was unaffected by Samd14-Enh deletion (Figure 4G). In aggregate, the mRNA, mass spectrometric and Western blotting data indicate that Samd14-Enh and anemia increase Samd14 mRNA and Samd14 protein levels in splenic erythroid cells.
Figure 4. Samd14-Enh opposes anemia-dependent apoptosis of CD71+Ter119-Kit+splenic erythroid progenitor cells.
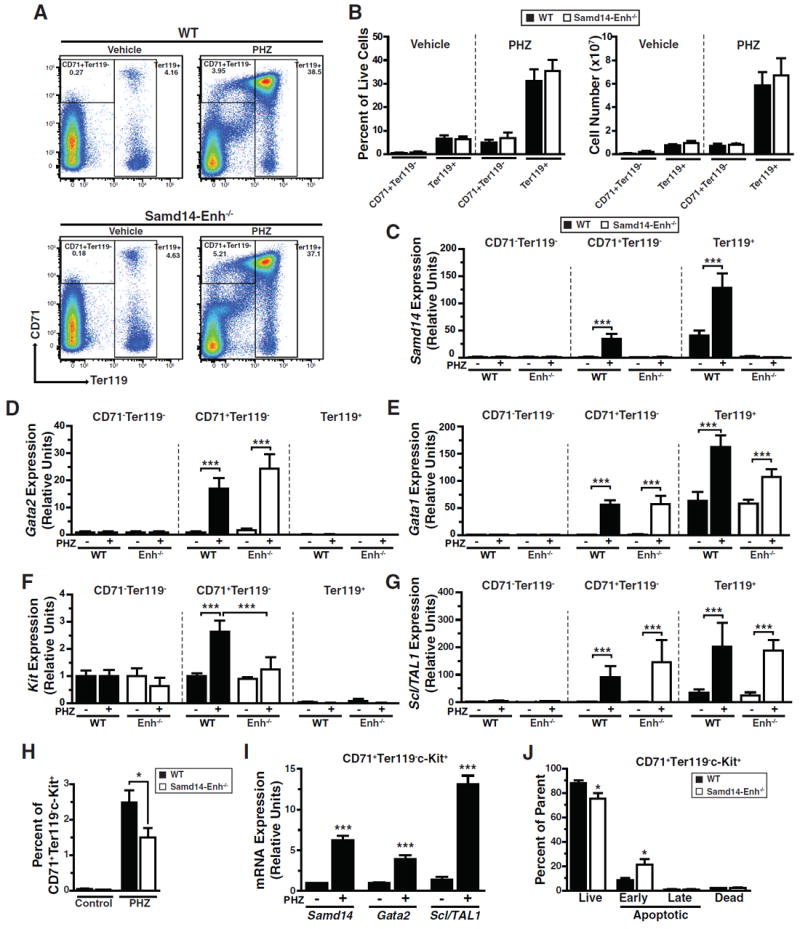
(A) Representative flow cytometric analysis of CD71 and Ter119 staining in WT and Samd14-Enh-/- spleen following PHZ treatment. (B) Quantitation of the percentage (top) and cell number (bottom) per spleen of CD71+Ter119- and Ter119+ cells in WT and Samd14-Enh-/- mice. (C) Relative Samd14 mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen (n = 3). (D) Relative Gata2 mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen (n = 3). (E) Relative Gata1 mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen (n = 3). (F) Relative Kit mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen (n = 3). (G) Relative Scl/TAL1 mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen. (H) Flow cytometric analysis of percentages of c-Kit+CD71+ cells within the Ter119- population of PHZ-treated spleen (n = 4). (I) Relative Samd14, Gata2, and Scl/TAL1 mRNA levels in FACS-purified cells from WT spleen and Samd14-Enh-/- spleen. (J) Quantitation of percentages of live (DRAQ7-AnnexinV-), early apoptotic (DRAQ7-AnnexinV+), late apoptotic (DRAQ7+AnnexinV+), and dead (DRAQ7+AnnexinV-) cells in CD71+c-Kit+Ter119- cells in PHZ-treated WT and Samd14-Enh-/- spleen (n = 4). Statistical significance represented by mean +/- SEM.; *p<0.05, ***p<0.001.
Anemia-induced Kit expression is attenuated in Samd14-Enh-/- mice (Figure 3F), and the percentage of c-Kit-expressing cells is lower in CD71+Ter119- PHZ-treated Samd14-Enh-/- spleen in comparison with WT spleen (Figure 4H, Figure S3). Within CD71+Ter119-Kit+ splenic cells Samd14, Gata2, and Scl1/TAL1 expression were upregulated following PHZ treatment (Figure 4I). To establish whether the reduced Kit+ cell numbers in Samd14-Enh-/- spleen resulted from decreased proliferation and/or increased apoptosis, we quantified the percentage of Ki67+ and AnnexinV+ cells in control and PHZ-treated spleen. In populations defined by CD71 and Ter119 markers following PHZ treatment, proliferation (Figure S4A) and apoptosis (Figure S4B) were similar between WT and Samd14-Enh-/- spleen. To evaluate a more enriched population of splenic erythroid stress progenitors, we incorporated c-Kit into the flow cytometric strategy. This analysis revealed increased early apoptosis (AnnexinV+Draq7-) in CD71+Ter119-Kit+ cells from Samd14-Enh-/- vs. WT spleen (2.5 fold, p = 0.033), with a commensurate reduction in live (AnnexinV-Draq7-) cells (Figure 4J, Figure S4C). No differences in apoptotic cells were detected between WT and Samd14-Enh-/- in other c-Kit+ splenic populations (Figure S4D). Thus, Samd14-Enh conferred survival to CD71+Ter119-Kit+ splenic erythroid stress progenitors in PHZ-induced acute anemia.
As c-Kit signaling can positively autoregulate Kit expression (Hewitt et al., 2015; Zhu et al., 2011), and Samd14 downregulation attenuates SCF/c-Kit signaling in fetal liver erythroid precursors (Hewitt et al., 2015), we tested whether Samd14-Enh deletion impacts Kit signaling during stress erythropoiesis in the spleen. Phospho-flow cytometry was used to quantitate SCF/c-Kit-induced AKT and ERK phosphorylation (pAKT and pERK). Using splenic cells from control and PHZ-treated mice (3 days), magnetic beads were used to deplete Ter119+ cells expressing low-level c-Kit. SCF did not increase pAKT in CD71+Ter119-c-Kit+ splenic cells from untreated WT mice (Figure 5A). Whereas SCF increased pAKT and pERK in WT CD71+Ter119-c-Kit+ cells from PHZ-treated mice, signaling was unaffected in CD71+Ter119-c-Kit- cells (Figure 5B). To compare SCF/c-Kit signaling in PHZ-treated WT and Samd14-Enh-/- mice, pAKT and pERK were quantitated in SCF-treated CD71+Ter119-c-Kit+ splenic cells. SCF-mediated induction of pAKT and pERK was attenuated in Samd14-Enh-/- cells (Figure 5C). Whereas SCF increased pAKT and pERK in WT cells from PHZ-treated mice 4.8 (p = 0.0003) and 12 fold (p = 0.0006), pAKT and pERK were 2.2 (p = 0.004) and 2.1 fold (p = 0.01) lower, respectively, in SCF-treated Samd14-Enh-/- cells (Figure 5D). In bone marrow, SCF-induced pAKT and pERK indistinguishably in WT and Samd14-Enh-/- cells (Figure S5), revealing a selective Samd14 function to promote anemia-dependent c-Kit signaling in the spleen.
Figure 5. Samd14-Enh Promotes Anemia-Dependent SCF/c-Kit Signaling.
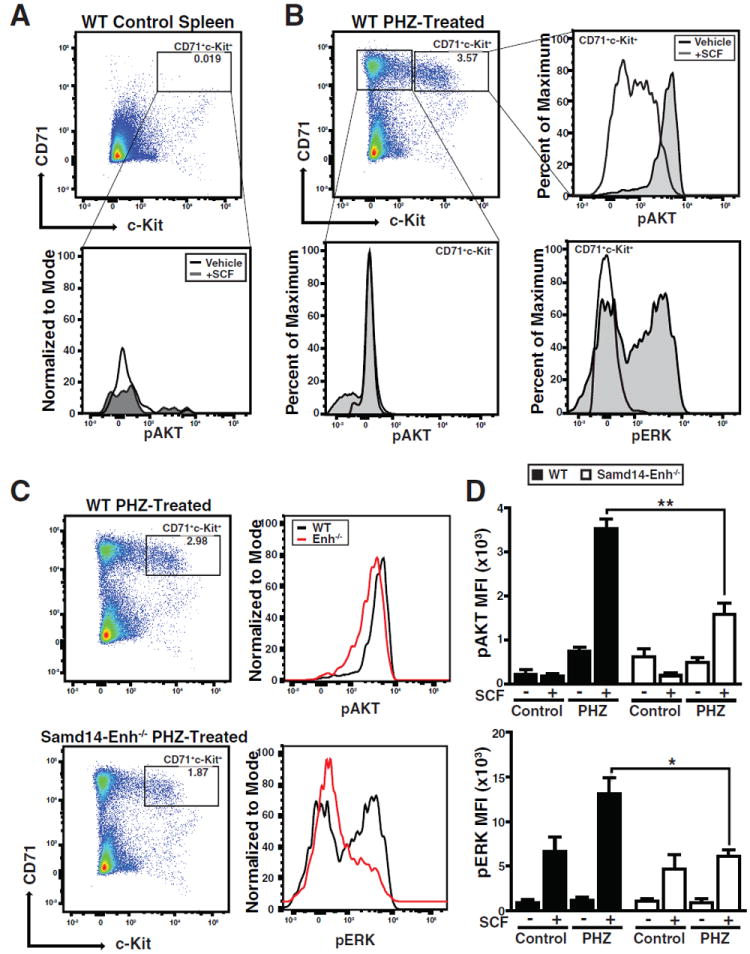
(A) Flow cytometric analysis of Ter119-depleted WT control spleen stained with CD71 and c-Kit (left) and quantitation of pAKT in the CD71+Kit+ cell population. All histograms were normalized to the percentage of the maximal cell number. (B) Flow cytometric analysis of WT PHZ-treated spleen stained with CD71 and c-Kit (top left) and histograms of pAKT in CD71+Kit- cells (bottom left), and pAKT and pERK in CD71+Kit+ cells (right). (C) Flow cytometric analysis of WT and Samd14-Enh-/- PHZ-treated spleen stained with CD71 and c-Kit (left) and comparative histogram plots of pAKT and pERK in WT and Samd14-Enh-/- CD71+Kit+ SCF-stimulated cells (right). (D) Quantitation of pAKT and pERK MFI in WT and Samd14-Enh-/- cells (n = 3). Statistical significance represented by mean +/- SEM.; *p<0.05; **p<0.01.
GATA-2- and Anemia-Activated (G2A) Enhancers Constitute a Vital Sector of the Hematopoietic Stem and Progenitor Cell Cistrome
Samd14-Enh promotes red blood cell regeneration, thereby conferring survival in severe anemia. The E-box-GATA composite element at Samd14-Enh nucleates a multi-protein complex comprised of GATA-1/2, Scl/TAL1, LDB1 and LMO2 to mediate transcription of hematopoietic loci (Hewitt et al., 2016). In CD71+Ter119- splenic erythroid precursors, anemia upregulates Gata1, Gata2, Tal1, Lmo2 and Ldb1, expression (Figure 4D, 4E, 4G, S6A). Enhancer hallmarks include open chromatin that facilitates transcription factor access and/or ensures complex stability. We tested whether anemia increases chromatin accessibility, or if accessibility preexists in the steady-state, which would imply that anemia induces transcription post-chromatin opening (Figure 6A). To establish whether anemia influences Samd14-Enh chromatin accessibility, we analyzed a 1 kb region surrounding the composite element. Formaldehyde-assisted isolation of regulatory elements (FAIRE) (Hewitt et al., 2015; Sanalkumar et al., 2014; Simon et al., 2012) was used to quantify chromatin accessibility in CD71+Ter119- splenic cells from control and PHZ-treated mice. This analysis revealed a striking increase (p = 0.0003) in chromatin accessibility directly upstream of the composite element (Figure 6B), which was abrogated by Samd14-Enh deletion (Figure 6C).
Figure 6. GATA-2-Regulated, Anemia-Responsive +9.5-Like Composite Elements.
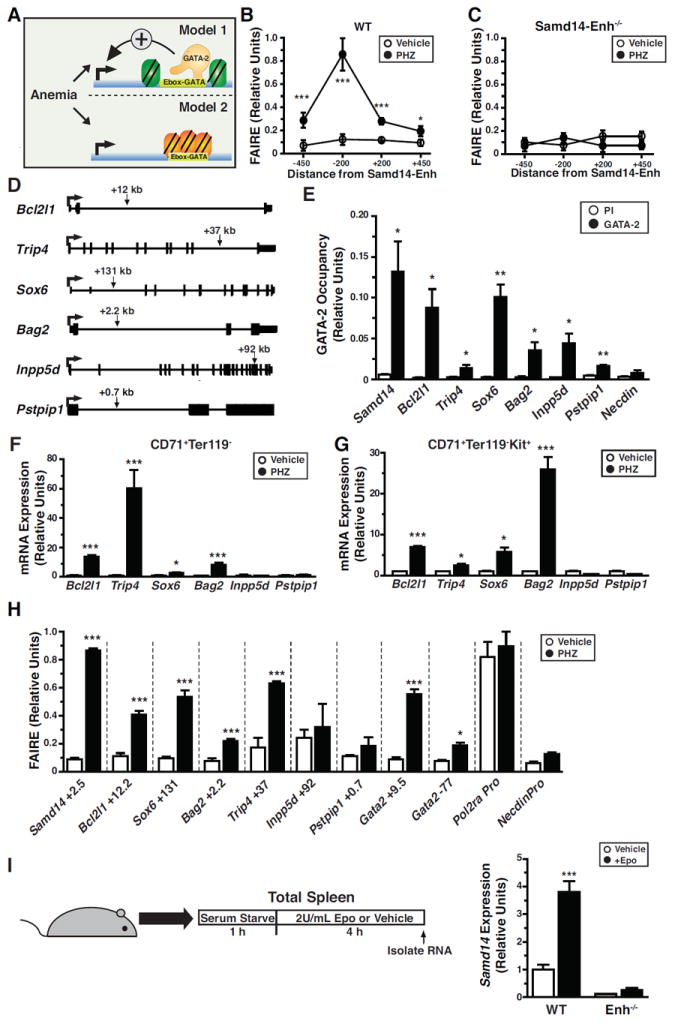
(A) Models of how anemia impacts transcription of loci containing +9.5-like composite elements. Model 1 assumes that anemia-responsive enhancers are accessible in the absence of anemia, and anemia increases transcription post-chromatin accessibility. Model 2 assumes that anemia increases chromatin accessibility as a regulatory step in anemia-dependent transcriptional activation. (B) Formaldehyde-assisted isolation of regulatory elements (FAIRE) analysis using 4 primer sets at Samd14 in WT cells from vehicle- or PHZ-treated spleen (n = 4). (C) FAIRE analysis using 4 primer sets at Samd14 in Samd14-Enh-/- cells from vehicle- or PHZ-treated spleen (n = 3). (D) Diagrams illustrating the intronic location of +9.5-like composite elements (arrow) in Bcl2l1, Sox6, Trip4, Bag2, Inpp5d and Pstpip1 (kb, kilobases). (E) GATA-2 ChIP in G1E cells at composite elements. The Necdin promoter served as a negative control (n = 3). (F) Relative mRNA expression at loci containing +9.5-like elements in control and PHZ-treated CD71+Ter119- cells from spleen. (G) Relative mRNA expression at loci containing +9.5-like elements in CD71+Ter119-c-Kit+ cells from spleens of control and PHZ-treated mice. (H) FAIRE analysis of +9.5-like loci in CD71+Ter119- cells isolated from vehicle- or PHZ-treated spleen. Pol2ra promoter represents a control with constitutively open chromatin, while the Necdin promoter represents a control with constitutively closed chromatin (n = 3). (I) Relative Samd14 expression in splenic cells treated with Vehicle or Epo (2U/mL) for 4 hours (n = 6) from PHZ-treated WT and Samd14-Enh-/- mice. Statistical significance represented by mean +/- SEM.; *p<0.05 **p<0.01***p<0.001.
We tested whether other +9.5-like components of the HSPC cistrome are also regulated by anemia (Figure 6D). Loci (Bcl2l1, Sox6, Trip4, Bag2, Inpp5d, and Pstpip1) were selected based on a prioritized ranking of chromatin features and biological criteria (Hewitt et al., 2015). Although anemia was reported to increase Bcl2l1 and Sox6 expression (Dumitriu et al., 2010; Koulnis et al., 2012), the underlying mechanisms are unclear. Other loci harboring +9.5-like sites have not been studied in erythropoiesis. Thyroid hormone receptor interactor 4 (Trip4) encodes a subunit of the tetrameric nuclear activating signal cointegrator 1 (ASC-1) complex (Jung et al., 2002). BCL2-associated athanogene-2 (Bag2) is a co-chaperone that opposes Hsp70 function (Takayama et al., 1999). Inositol polyphosphate 5-phosphatase (Inpp5d or Ship1) modulates signaling through the B-cell receptor (Ono et al., 1996) and promotes myeloid cell proliferation (Chen et al., 2015). Proline-serine-threonine phosphatase-interacting protein 1 gene (Pstpip1) encodes a cytoskeletal adapter (Spencer et al., 1997), and mutations cause Pyogenic Arthritis, Pyoderma gangrenosum, and Acne (PAPA) Syndrome (Wise et al., 2002).
Bcl2l1, Trip4, Sox6, Bag2, Inpp5d and Pstpip1 +9.5-like sites (Figure 6D) exhibit mouse-human conservation, DNase hypersensitivity and GATA-2 regulation, based on analyses of WT vs. +9.5-/- fetal liver (Hewitt et al., 2015). In G1E cells, GATA-2 occupied these sites, but not the negative control Necdin promoter (Figure 6E). Scl/TAL1 and Lmo2 occupied E-box-GATA composite elements at Gata2, Samd14, Bcl2l1 and Trip4 in Lin-Kit+ hematopoietic progenitors; Scl/TAL1 occupied Sox6 and Bag2 composite elements (Figure S6B). By contrast, Inpp5d and Pstpip1 composite elements were not GATA-2-, Scl/TAL1- or Lmo2-occupied (Figure S6B). Consistent with the established paradigm in which only certain GATA-2-regulated genes are GATA-1-regulated (Dore et al., 2012; Fujiwara et al., 2009; Katsumura et al., 2017), a subset of +9.5-like loci (Samd14, Bcl2l1, and Bag2) are GATA-1-regulated in G1E cells; others (Sox6, Trip4, Inpp5d and Pstpip1) are GATA-1-insensitive (Figure S7A). To test whether the +9.5-like loci are anemia-responsive, we quantified gene expression in spleen from control and PHZ-treated mice. PHZ increased Bcl2l1, Sox6, Trip4 and Bag2 expression, but not Inpp5d or Pstpip1, in sorted CD71+Ter119- (Figure 6F) and CD71+Ter119-Kit+ cells (Figure 6G), indicating that expression was altered in erythroid progenitors and did not reflect anemia-dependent cellularity changes. In addition to Samd14-Enh, PHZ increased accessibility at Gata2 +9.5, Bcl2l1, Bag2, Trip4 and Sox6 +9.5-like sites, but not Pstpip1 or Inpp5d (Figure 6H). Thus, GATA-2 and anemia increase chromatin accessibility and activity of a subset of E-box-GATA element-containing enhancers.
Anemia dramatically increases (up to 1000 fold) blood erythropoietin (Epo) levels (Bunn, 2013). Epo is commonly used to treat anemia and is a vital erythropoiesis-stimulating agent physiologically and upon hypoxia. To test whether anemia-dependent Epo generation constitutes a component of the Samd14-Enh anemia-responsive mechanism, we stimulated serum-starved PHZ-treated splenocytes with Epo for 4 hours. Epo increased Samd14 expression in WT, but not Samd14-Enh-/- cells (Figure 6I). Thus, Samd14-Enh is required for Epo-dependent Samd14 expression. Expression of Bcl2l1, Bag2, Trip4 and Sox6, but not Pstpip1 or Inpp5d, was also stimulated by Epo (Figure S7B). These findings implicate Epo signaling as one component of the multi-component anemia-responsive enhancer mechanism.
DISCUSSION
Mono-methylated histone H3 at lysine 4, acetylated histone H3 at K27, p300 and accessible chromatin are canonical enhancer attributes used to infer the existence of enhancers in genomes (Bulger and Groudine, 2011; Smith and Shilatifard, 2014). In the context of GATA factors, many prospective enhancers bearing these attributes are either not GATA factor-occupied or can be deleted from a genome with little to no functional consequence (Bresnick et al., 2010; DeVilbiss et al., 2014; Fujiwara et al., 2009; Sanalkumar et al., 2014; Snow et al., 2010; Snow et al., 2011; Tanimura et al., 2015). Previously, we used a multi-parametric prioritization strategy to identify GATA-2-regulated enhancers containing E-box-GATA elements in G1E cells (Hewitt et al., 2015). Herein, we analyzed one such enhancer in vivo and demonstrated that anemia activates Samd14-Enh, which promotes red blood cell regeneration in stress erythropoiesis, thereby conferring survival in severe anemia. Samd14-Enh represents the founding member of a G2A enhancer family.
Although Samd14-Enh has attributes resembling the Gata2 +9.5 enhancer required for HSPC genesis, steady-state hematopoiesis is normal in Samd14-Enh-/- mice. The +9.5 enhancer controls Gata2 expression in HSPCs (Gao et al., 2013; Johnson et al., 2012; Johnson et al., 2015), and the resulting GATA-2 protein functions in a cell type-specific manner to activate an ensemble of +9.5-like elements, including Samd14-Enh. Samd14-Enh cell type-specificity is illustrated by its importance for Samd14 transcription in bone marrow and splenic myelo-erythroid progenitors, but not in multipotent LSK cells nor in brain. The essential Samd14-Enh activity controls stress erythropoiesis, which extends E-box-GATA element functionality beyond that of developmental and steady-state hematopoiesis. Three independent modes of acute anemia activate Gata2 and Samd14 transcription in the spleen, and GATA-2 regulates Samd14 expression. This anemia-protective circuit conforms to a Type I coherent feed-forward loop, which predicts that establishment of the circuit requires a persistent stimulus, while stimulus loss rapidly inactivates the circuit (Figure 7A) (Shoval and Alon, 2010).
Figure 7. GATA-2- and Anemia-Activated (G2A) Enhancers that Oppose Lethal Anemia Define a Vital Sector of the Hematopoietic Stem and Progenitor Cell Cistrome.

(A) Anemia upregulates Samd14 and Gata2 expression in splenic erythroid progenitors, which conforms to a Type I coherent feed-forward loop. Targeted deletion of the intronic GATA-2-regulated Samd14-Enh enhancer disrupts this mechanism, impairs red blood cell regeneration in the context of anemic stress and is lethal. (B) Anemia induces expression of additional genes containing +9.5-like enhancers. Bcl2l1 and Sox6 have established functions to promote stress erythropoiesis (Dumitriu et al., 2010; Koulnis et al., 2012).
Mass spectrometric and Western blotting analyses provided evidence that Samd14-Enh controls expression of the SAM domain protein Samd14. Samd14-Enh was required for anemia-dependent induction of SCF/c-Kit signaling in spleen. Mammalian proteomes contain several hundred SAM domain proteins, and SAM domains are implicated in protein, RNA and lipid binding (Kim and Bowie, 2003). Kit signaling-defective knock-in mice can have phenotypes resembling Samd14-Enh-/- mice (Agosti et al., 2009; Perry et al., 2007), suggesting that Samd14 activity to enhance SCF/c-Kit signaling in vivo confers survival in severe anemia.
Our results indicate that anemia activates a subset of GATA-2-regulated E-box-GATA element enhancers to elevate transcription of the linked genes. Since the GATA-2 target gene Samd14 promotes red blood cell regeneration in severe anemia, and GATA-2 target genes Bcl2l1 (Koulnis et al., 2012) and Sox6 (Dumitriu et al., 2010) also likely favor regeneration, GATA-2 is a vital determinant of stress erythropoiesis. Mechanistically, anemia increased GATA-2 and Scl/TAL-1 expression, which function through E-box-GATA elements at these enhancers, and the classic anemia responsive cytokine, Epo, stimulated transcription of the respective genes. Furthermore, mitogen-activated protein kinases (p38 and ERKs) induce GATA-2 phosphorylation, which increases transcriptional activation of select target genes (Katsumura et al., 2016; Katsumura et al., 2014). Scl/TAL1 can also be phosphorylated by mitogen-activated protein kinases (Tang et al., 1999).
Our results point to a multi-component anemia-responsive mechanism involving: (i) anemia-dependent induction of transcription factors critical for composite element-containing enhancer function; (ii) anemia-dependent alterations in signaling circuitry, which increases enhancer component activities; and (iii) acute Epo-responsiveness. Samd14-Enh represents a foundational G2A enhancer and related G2A enhancers constitute an important sector of the HSPC cistrome (Figure 7B). Based on the results described herein and functional insights of G2A enhancer-containing genes, we propose that G2A enhancers establish a genetic network that uniquely orchestrates red blood cell regeneration in anemia, thereby conferring survival. Samd14-Enh is not required for steady-state hematopoiesis, illustrating how certain E-box-GATA composite elements can have essential functions in the context of stress. It will be instructive to obtain a global perspective of how anemia regulates this sector to generate transcriptional circuits/networks that enable and/or drive massive red blood cell regeneration and survival in anemia. One would predict that these networks are defective in pathologies of ineffective erythropoiesis and will provide opportunities to overcome limitations to red blood cell production in these contexts.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Emery Bresnick (ehbresni@wisc.edu)
Experimental Model and Subject Details
Mutant Mice Generation
TALENs were generated as described (Sung et al., 2013), functionally validated in G1E cells (Hewitt et al., 2015) and transcribed in vitro using the mMESSAGE mMACHINE® T7 Transcription Kit (Ambion) to generate mRNA. Pronuclear injection of TALEN mRNA (20 ng/μl) was conducted at the 1-cell stage at the UW-Madison Biotech. Center. Samd14-Enh mutations were detected in 6 of 11 live pups by T7 endonuclease I (New England Biolabs) treatment of PCR products from 3-week tail clip DNA, and positive hits were sequenced. Genotyping for a 34-bp deletion in Samd14 intron 1 (Samd14-Enh) was performed using tail clip DNA and forward (5’-CAGTCTGAAGGGAGGGCAC) and reverse (5’-GTCTAAAGCTACTCCAATTCTGAG) primers generating a 110 base pair wild-type (WT) product and 76 base pair mutant product. Animal experiments were performed with the ethical approval of the AAALAC International (Association for the Assessment and Accreditation of Laboratory Animal Care) at UW–Madison.
Method Details
Flow Cytometry
BM, FL or spleen cells from WT or Samd14-Enh-/- mice were dissociated, resuspended in PBS with 2% fetal bovine serum (FBS) and passed through a 35 μm nylon filter to obtain single-cell suspensions before antibody staining. All antibodies from eBioscience unless otherwise stated. Lineage markers were stained with fluorescein isothiocyanate (FITC)–conjugated B220 (11-0452), CD3 (11-0031), CD4 (11-0041), CD5 (11-0051), CD8 (11-0081), CD41 (11-0411), CD48 (11-0481), Gr-1 (11-5931), and TER-119 (11-5921) antibodies. Other surface proteins were detected with phycoerythrin (PE)–conjugated CD71 (R17217) and Fc receptor (12-0161); peridinin chlorophyll protein (PerCP)–Cy5.5–conjugated Sca1 (45-5981); APC-conjugated Ter119 (17-5921); Alexa Fluor 647–conjugated CD34 (BDB560230, BD Biosciences), and PE-Cy7-conjugated c-Kit (2B8) (25-1171) antibodies. Analysis of BM LSKs (Lin- Kit+Sca1+), CMPs (Lin- CD34+FcgRlowKit+Sca1-), GMPs (Lin-CD34+FcgRhighKit+Sca1-), and MEPs (Lin-CD34-FcgRlowKit+Sca1-) was conducted as described (Johnson et al., 2015). Analysis of erythroid maturation using CD71 and Ter119 was conducted as described (McIver et al., 2014; McIver et al., 2016). Data was collected on a LSRII flow cytometer. For applications requiring cell isolation, FACS was conducted on a FACSAriaII (BD Biosciences) and analyzed using FlowJo v9.0.2 software (TreeStar).
Acute Anemia
Hemolytic anemia was induced by PHZ (Sigma; 114715) administered subcutaneously with either a single dose of 100 mg/kg, or two doses (one per day) of 60 mg/kg at 0 and 24 h. Blood samples were collected by small-volume retro-orbital bleeding (~10 μl per collection), and hematologic parameters measured on a HemaVet complete blood count (CBC) instrument. Phlebotomy was induced by 3 daily bleedings (400 μl) collected retro-orbitally, in conjunction with intraperitoneal injections of 400 μl PBS. Unless otherwise stated, spleens were harvested 3 days post-PHZ injection or post-phlebotomy. Bone marrow transplantation assays were conducted using 5×105 bone marrow mononuclear cells transplanted into the retro-orbital sinus of lethally-irradiated (9.5-Gy) B6.SJL-Ptprca Pep3b/BoyJ recipient mice (Harandi et al., 2010). CFU assays in PHZ-treated spleens were conducted using 0.3 ml spleen suspension at 1 × 106 cells/ml mixed with 3 ml of Epo-only MethoCult™ M3334, or Epo, SCF, IL-3 and IL-6 Methocult M3434 (STEMCELL Technologies), and 1.1 ml was plated in replicate 35 mm dishes. BFU-E colonies were counted 5 days after culturing. CFU-E colonies were counted 2 days after plating.
Quantitation of mRNA and Allele-Specific Primary Transcripts
Total RNA was purified from 2-30 × 106 cells in 1 ml TRIzol (Life Technologies). For primary transcripts, RNA was subjected to on-column DNase treatment at 25°C for 10 min followed by column purification (Qiagen RNeasy). To synthesize cDNA, 2 μg RNA was incubated with 250 ng of random hexamer and oligo(dT) primers preheated at 68°C for 10 min. RNA was incubated with reverse transcriptase (Life Technologies) with 10 mM dithiothreitol (DTT), RNAsin (Promega), and 0.5 mM deoxynucleoside triphosphates at 42°C for 1 h, and then heat inacti vated at 98°C for 5 min. Real-time RT-PCR reactions contain 2 μl of cDNA, 10 μl SYBR Green (Applied Biosystems), and 100-500 nM of the appropriate primers, and each biological replicate sample was analyzed in duplicate on a ViiA 7 real-time PCR instrument (Applied Biosystems). Relative expression levels were determined using a standard curve of serial dilutions from appropriate control cDNA (for mRNA analysis) or sonicated genomic DNA (for allele-specific primary transcript analysis) for each primer pair. Negative control cDNA reactions were performed on samples lacking reverse transcriptase. cDNA samples lacking the mutant or WT allele sequence were used as negative controls for each allele-specific primer pair.
Apoptosis and proliferation analysis
To assess apoptosis, 1×107 cells isolated from control or PHZ-treated spleen were washed with PBS, stained using PE-conjugated CD71 and APC-conjugated Ter119, followed by a wash with 1x Annexin binding buffer (Life Technologies). Cells were resuspended in 100 μl Annexin binding buffer and labeled with 5 μl Annexin V-Alexa Fluor 350 conjugate (Life Technologies) and DRAQ7TM (eBiosciences). To assess proliferation, total spleen was bead sorted into Ter119+ and Ter119- fractions using biotin-conjugated anti-Ter119 antibody and magnetic beads (STEMCELL Technologies). Cells were fixed in 2% paraformaldehyde for 10 min at 37°C and permeabilized in 95% methanol overnight at -20°C. Cells were washed in PBS, blocked in 4%FBS, and stained using PE-conjugated CD71, APC-conjugated Ki67, and DAPI. Data was collected on an LSRII cytometer (BD Biosciences).
Phospho-Flow Cytometry
Splenocytes were Ter119-depleted using biotin-conjugated anti-Ter119 antibody and magnetic beads (STEMCELL Technologies), serum-starved for 1 h in 1% BSA/IMDM at 37°C and treated with 10ng/uL SCF or vehicle 10 min . Cells were immediately fixed in 2% paraformaldehyde for 10 min at 37°C and permeabi lized in 95% methanol overnight at -20°C. Cells were stained with rabbit antibodies against phospho (S473)-AKT (p-AKT) and phospho (Thr202/Tyr204) p44/42 ERK1/2 (p-ERK) (9271, 9101; Cell Signaling) for 30 min, then incubated in APC-conjugated goat anti-rabbit (1:200), PE-Cy7-conjugated c-Kit (1:100) and PE-conjugated CD71 (1:100) for 30 min at room temperature. Zombie UV (Biolegend) dye discriminated dead cells. Samples were analyzed using a BD LSR II (BD Biosciences).
Mass Spectrometry
1 × 106 cells from WT and Samd14-Enh-/- spleen were resuspended in 300 μl 8 M urea, 100 mM Tris pH 8.0, 10 mM TCEP, and 40 mM chloroacetamide and incubated overnight in 50:1 trypsin prior to fractionation. 48 fractions were collected, each comprising 40 seconds of eluted peptides. Each fraction from the first bioreplicate was analyzed by LC-MS/MS to empirically determine the appropriate targeting window. For each analysis, 30% of the sample was loaded onto a 75 μm i.d. 30 cm long capillary with an imbedded electrospray emitter and packed with 1.7 μm C18 BEH stationary phase. A 3 m/z isolation window was centered on the most abundant isotope, fragmented with 30 NCE HCD collision energy and analyzed in the Orbitrap at 30,000 resolving power. Injection volumes of each sample was normalized based on peptide content. Each raw file was analyzed in Excalibur. The peak height from 3 transitions was recorded, and the mean normalized log2 peak intensity was averaged for each of three Samd14-specific peptides.
Antibody Generation
Anti-SAMD14 antibodies were generated from purified recombinant Samd14. Full-length mouse Samd14 cDNA was cloned into pET28a for expression of recombinant 6His-SAMD14 in BL21 E. coli induced with 1 mM IPTG for 5 h at 37°C. Recombinan t 6His-SAMD14 protein was purified using Ni-NTA agarose (Qiagen) and injected into rabbits (Cocalico Biologicals). Antibodies were purified from serum using a 6His-SAMD14 affinity column and dialyzed overnight into PBS for immediate use.
ChIP and FAIRE
GATA-2 ChIP was conducted as described (Im et al., 2004). G1E cells (Weiss et al., 1997) containing 1 × 107 cells were crosslinked with 1% formaldehyde for 10 min. Anti-GATA-2 rabbit polyclonal antibodies were used with protein A-Sepharose (Sigma) to adsorb immune-specific complexes. Samples were analyzed by real-time PCR. FAIRE was conducted as described (Hewitt et al., 2015; Sanalkumar et al., 2014; Simon et al., 2012). Cells were fixed with 1% formaldehyde and sonicated to shear the DNA. Following phenol:chloroform extraction of protein-bound DNA, samples were analyzed by real-time PCR relative to input controls.
Epo Stimulation
WT and Samd14-Enh-/- mice were treated with PHZ or vehicle for 3 days. Splenic cells were dissociated, washed in PBS, and serum-starved for 2 h in 1% BSA/IMDM at 37°C. 1 × 107 cells were treated with 2U/ml rhEPO or vehicle for 2 , 4 or 12 h. RNA was purified in TRIzol (Invitrogen).
Quantification and Statistical Analysis
For quantitation of mRNA, primary transcript, cell number, cell colonies, CBC and median fluorescence intensities, the results are presented as mean +/- standard error of the mean. Multiple independent cohorts were used in each experiment. Statistical comparisons between two groups were performed using the Students t-test, with a significance cutoff of p < 0.05 (GraphPad Prism). A log-rank test was performed on the Kaplan-Meier survival curve (n = 8 in each group) (GraphPad Prism).
Supplementary Material
HIGHLIGHTS.
A GATA-2-dependent enhancer controls expression of a novel SAM domain protein.
The Samd14 enhancer (Samd14-Enh) confers survival in severe anemia.
Samd14-Enh governs SCF/c-Kit signaling in erythroid stress progenitors.
Samd14-Enh and an ensemble of related enhancers are anemia-regulated in the spleen.
Acknowledgments
The work was supported by NIH grants DK50107 (EHB) and DK68634 (EHB), a pilot grant from the UW Skin Disease Research Center (EHB), P41 GM108538 (JJC) and Cancer Center Support Grant P30CA014520. JSK is supported by a grant from Institute for Basic Science (IBS-R021-D1). KJH is supported by an American Heart Association Postdoctoral Fellowship and the Training Program in Translational Cardiovascular Science (T32 HL-007936-16). We thank Lily Zemelko and Elsa Davids for technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS:
K.J.H., S.K., R.F.P., and E.H.B. designed the study. K.J.H., K.R.K, D.R.M., P.D., and A.S.H conducted the experiments. J-S.K. contributed genome editing reagents. A.S.H. and J.J.C. conducted mass spectrometry analysis. K.J.H. and E.H.B. wrote the paper with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosti V, Karur V, Sathyanarayana P, Besmer P, Wojchowski DM. A KIT juxtamembrane PY567 -directed pathway provides nonredundant signals for erythroid progenitor cell development and stress erythropoiesis. Exp Hematol. 2009;37:159–171. doi: 10.1016/j.exphem.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429 e1419. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzini CE, Barrio Rendo ME, Devoto FC, Epper CE. Studies on medullary and extramedullary erythropoiesis in the adult mouse. Am J Physiol. 1970;219:724–728. doi: 10.1152/ajplegacy.1970.219.3.724. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broudy VC, Lin NL, Priestley GV, Nocka K, Wolf NS. Interaction of stem cell factor and its receptor c-kit mediates lodgment and acute expansion of hematopoietic cells in the murine spleen. Blood. 1996;88:75–81. [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn HF. Erythropoietin. Cold Spring Harbor perspectives in medicine. 2013;3:a011619. doi: 10.1101/cshperspect.a011619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Krisman K, Theobald EH, Xu J, Akutagawa J, Lauchle JO, Kogan S, Braun BS, Shannon K. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest. 2013;123:335–339. doi: 10.1172/JCI63193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Lodish HF. Global analysis of induced transcription factors and cofactors identifies Tfdp2 as an essential coregulator during terminal erythropoiesis. Exp Hematol. 2014 doi: 10.1016/j.exphem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Shojaee S, Buchner M, Geng H, Lee JW, Klemm L, Titz B, Graeber TG, Park E, Tan YX, et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015;521:357–361. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVilbiss AW, Sanalkumar R, Johnson KD, Keles S, Bresnick EH. Hematopoietic transcriptional mechanisms: from locus-specific to genome-wide vantage points. Exp Hematol. 2014;42:618–629. doi: 10.1016/j.exphem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M, Lakey JH, Rahman T, Wang XN, McGovern N, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu B, Bhattaram P, Dy P, Huang Y, Quayum N, Jensen J, Lefebvre V. Sox6 is necessary for efficient erythropoiesis in adult mice under physiological and anemia-induced stress conditions. PloS one. 2010;5:e12088. doi: 10.1371/journal.pone.0012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS genetics. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Johnson KD, Chang YI, Boyer ME, Dewey CN, Zhang J, Bresnick EH. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210:2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim S-I, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, Babic M, Lin M, Carmagnac A, Lee YK, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harandi OF, Hedge S, Wu DC, McKeone D, Paulson RF. Murine erythroid short-term radioprotection requires a BMP4-dependent, self-renewing population of stress erythroid progenitors. J Clin Invest. 2010;120:4507–4519. doi: 10.1172/JCI41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Russell ES. The response of W-W v and Sl-Sl d anaemic mice to haemopoietic stimuli. British journal of haematology. 1972;22:155–168. doi: 10.1111/j.1365-2141.1972.tb08797.x. [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Johnson KD, Gao X, Keles S, Bresnick EH. The Hematopoietic Stem and Progenitor Cell Cistrome: GATA Factor-Dependent cis-Regulatory Mechanisms. Curr Top Dev Biol. 2016;118:45–76. doi: 10.1016/bs.ctdb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt KJ, Kim DH, Devadas P, Prathibha R, Zuo C, Sanalkumar R, Johnson KD, Kang YA, Kim JS, Dewey CN, et al. Hematopoietic Signaling Mechanism Revealed from a Stem/Progenitor Cell Cistrome. Mol Cell. 2015;59:62–74. doi: 10.1016/j.molcel.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Lambert JA, Martin R. SCL/TAL1 in Hematopoiesis and Cellular Reprogramming. Curr Top Dev Biol. 2016;118:163–204. doi: 10.1016/bs.ctdb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, Frucht DM, Vinh DC, Auth RD, Freeman AF, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- Inra CN, Zhou BO, Acar M, Murphy MM, Richardson J, Zhao Z, Morrison SJ. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature. 2015;527:466–471. doi: 10.1038/nature15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Hsu AP, Ryu MJ, Wang J, Gao X, Boyer ME, Liu Y, Lee Y, Calvo KR, Keles S, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122:3692–3704. doi: 10.1172/JCI61623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Kong G, Gao X, Chang Y-I, Hewitt KJ, Sanalkumar R, Prathibha R, Ranheim EA, Dewey CN, Zhang J, et al. Cis-regulatory mechanisms governing stem and progenitor cell transitions. Science Advances. 2015 doi: 10.1126/sciadv.1500503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DJ, Sung HS, Goo YW, Lee HM, Park OK, Jung SY, Lim J, Kim HJ, Lee SK, Kim TS, et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol. 2002;22:5203–5211. doi: 10.1128/MCB.22.14.5203-5211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Bresnick EH, Group GFM. The GATA factor revolution in hematology. Blood. 2017 doi: 10.1182/blood-2016-09-687871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Ong IM, DeVilbiss AW, Sanalkumar R, Bresnick EH. GATA Factor-Dependent Positive-Feedback Circuit in Acute Myeloid Leukemia Cells. Cell reports. 2016;16:2428–2441. doi: 10.1016/j.celrep.2016.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura KR, Yang C, Boyer ME, Li L, Bresnick EH. Molecular basis of crosstalk between oncogenic Ras and the master regulator of hematopoiesis GATA-2. EMBO Rep. 2014;15:938–947. doi: 10.15252/embr.201438808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Koulnis M, Porpiglia E, Porpiglia PA, Liu Y, Hallstrom K, Hidalgo D, Socolovsky M. Contrasting dynamic responses in vivo of the Bcl-xL and Bim erythropoietic survival pathways. Blood. 2012;119:1228–1239. doi: 10.1182/blood-2011-07-365346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenox LE, Perry JM, Paulson RF. BMP4 and Madh5 regulate the erythroid response to acute anemia. Blood. 2005;105:2741–2748. doi: 10.1182/blood-2004-02-0703. [DOI] [PubMed] [Google Scholar]
- Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, Bresnick EH, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver SC, Kang YA, DeVilbiss AW, O’Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang CJ, Yang D, Bouhassira EE, et al. The exosome complex establishes a barricade to erythroid maturation. Blood. 2014;124:2285–2297. doi: 10.1182/blood-2014-04-571083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver SC, Katsumura KR, Davids E, Liu P, Kang YA, Yang D, Bresnick EH. Exosome complex orchestrates developmental signaling to balance proliferation and differentiation during erythropoiesis. eLife. 2016;5 doi: 10.7554/eLife.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Kapur R. Role of c-Kit and erythropoietin receptor in erythropoiesis. Crit Rev Oncol Hematol. 2005;54:63–75. doi: 10.1016/j.critrevonc.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Nocka K, Majumder S, Chabot B, Ray P, Cervone M, Bernstein A, Besmer P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice--evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 1989;3:816–826. doi: 10.1101/gad.3.6.816. [DOI] [PubMed] [Google Scholar]
- Ono M, Bolland S, Tempst P, Ravetch JV. Role of the inositol phosphatase SHIP in negative regulation of the immune system by the receptor Fc(gamma)RIIB. Nature. 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- Paulson RF, Shi L, Wu DC. Stress erythropoiesis: new signals and new stress progenitor cells. Curr Opin Hematol. 2011;18:139–145. doi: 10.1097/MOH.0b013e32834521c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JM, Harandi OF, Paulson RF. BMP4, SCF, and hypoxia cooperatively regulate the expansion of murine stress erythroid progenitors. Blood. 2007;109:4494–4502. doi: 10.1182/blood-2006-04-016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanalkumar R, Johnson KD, Gao X, Boyer ME, Chang YI, Hewitt KJ, Zhang J, Bresnick EH. Mechanism governing a stem cell-generating cis-regulatory element. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1400065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitt HJ, Schafers S, Deiwick A, Eckardt KU, Pietsch T, Ebell W, Nashan B, Ringe B, Wonigeit K, Pichlmayr R. Extramedullary erythropoiesis in human liver grafts. Hepatology. 1995;21:689–696. doi: 10.1002/hep.1840210314. [DOI] [PubMed] [Google Scholar]
- Shen Y, Takahashi M, Byun HM, Link A, Sharma N, Balaguer F, Leung HC, Boland CR, Goel A. Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer Biol Ther. 2012a;13:542–552. doi: 10.4161/cbt.19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012b;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval O, Alon U. SnapShot: network motifs. Cell. 2010;143:326–e321. doi: 10.1016/j.cell.2010.09.050. [DOI] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nature protocols. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. A single cis element maintains repression of the key developmental regulator Gata2. PLoS genetics. 2010;6:e1001103. doi: 10.1371/journal.pgen.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JW, Trowbridge JJ, Johnson KD, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. Context-dependent function of “GATA switch” sites in vivo. Blood. 2011;117:4769–4772. doi: 10.1182/blood-2010-10-313031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA. PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Hegde S, Porayette P, Yon M, Hankey P, Paulson RF. Friend virus utilizes the BMP4-dependent stress erythropoiesis pathway to induce erythroleukemia. J Virol. 2008;82:382–393. doi: 10.1128/JVI.02487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Iijima T, Kano J, Kobayashi H, Li D, Morishita Y, Okubo C, Anami Y, Noguchi M. Frequent aberrant methylation of the promoter region of sterile alpha motif domain 14 in pulmonary adenocarcinoma. Cancer science. 2008;99:2177–2184. doi: 10.1111/j.1349-7006.2008.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nature biotechnology. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- Tang T, Prasad KS, Koury MJ, Brandt SJ. Mitogen-activated protein kinase mediates erythropoietin-induced phosphorylation of the TAL1/SCL transcription factor in murine proerythroblasts. Biochem J. 1999;343(Pt 3):615–620. [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Miller E, Igarashi K, Yang D, Burstyn JN, Dewey CN, Bresnick EH. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2015 doi: 10.15252/embr.201541465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S, Lovett M. Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum Mol Genet. 2002;11:961–969. doi: 10.1093/hmg/11.8.961. [DOI] [PubMed] [Google Scholar]
- Wlodarski MW, Hirabayashi S, Pastor V, Stary J, Hasle H, Masetti R, Dworzak M, Schmugge M, van den Heuvel-Eibrink M, Ussowicz M, et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood. 2016;127:1387–1397. doi: 10.1182/blood-2015-09-669937. quiz 1518. [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- Zhu HH, Ji K, Alderson N, He Z, Li S, Liu W, Zhang DE, Li L, Feng GS. Kit-Shp2-Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood. 2011;117:5350–5361. doi: 10.1182/blood-2011-01-333476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


