Abstract
Developmental flexibility under stress conditions largely relies on the interactions between hormones that mediate stress responses and developmental processes. In this study, we showed that the stress hormone jasmonic acid (JA) induces formation of extra xylem in the roots of wild-type Arabidopsis thaliana (Col-0). JA signaling mutants such as coronatine insensitive1-1 and jasmonate resistant1-1 did not form extra xylem in response to JA, but the JA biosynthesis mutant oxophytodienoate-reductase3 did form extra xylem. These observations suggested that the JA response promotes xylem development. To understand the mechanism, we examined the regulatory interaction between JA and cytokinin, a negative regulator of xylem development. JA treatment reduced cytokinin responses in the vasculature, and exogenous cytokinin nullified the effect of JA on formation of extra xylem. A time-course experiment showed that suppression of cytokinin responses by JA does not occur rapidly, but the JA-mediated xylem phenotype is tightly linked to the suppression of the cytokinin response. Further analysis of arabidopsis histidine phosphotransfer protein6-1 and myc2-3 mutants revealed that the JA-responsive transcription factor MYC2 regulates the expression of AHP6 in response to JA and expression of AHP6 is involved in the JA-mediated xylem phenotype.
Introduction
Molecular and genetic studies have identified many phytohormones and have shown that the activities of these hormones largely overlap, although each hormone has specific signaling pathways that act non-redundantly. These findings suggest that the interplay between phytohormones dynamically regulates plant development and physiology1. For example, cytokinin interacts with auxin in the regulation of all aspects of plant development2, 3.
JA regulates plant responses to abiotic and biotic stresses and modulates plant development, including stamen filament growth, root growth, and senescence4–8. JA is biosynthesized from linolenic acid via the octadecanoid pathway, and then further metabolized to a JA-isoleucine conjugate (JA-Ile)9, 10. The interaction between JA-Ile and the CORONATINE INSENSITIVE1 (COI1) receptor provokes proteolysis of transcriptional repressor JASMONATE ZIM-DOMAIN (JAZ) proteins, and the degradation of JAZs leads to release of the MYC2 transcription factor11–13. MYC2 regulates JA responses by controlling the expression of JA-responsive genes and plays an essential role in modulating plant defense and development in response to JA. For example, myc2 mutant plants exhibit enhanced resistance to pathogens such as Pseudomonas syringae pv. tomato DC3000, Botrytis cinerea, and Fusarium oxysporum compared to wild-type plants, and JA-mediated inhibition of root growth is suppressed in myc2 mutant plants14, 15. These observations suggest that MYC2 regulates the expression of key genes responsible for the modulation of defense and development in response to JA.
Previous studies showed that crosstalk between JA and other hormones modulates plant defense and development. JA interacts with ethylene for defense against necrotrophic fungi and herbivorous insects or for development of apical hook16–18. JA interacts with gibberellic acid for the regulation of trichome and stamen development19–22. In these crosstalks, the direct interaction between MYC2 and ethylene-stabilized transcription factor ETHYLENE INSENSITIVE3 (EIN3) or between JAZs and repressor of gibberellin signaling DELLA proteins are deeply involved. JA also interacts with auxin in root growth and flower development23–25. MYC2 also plays a role in the interaction between JA and auxin. For example, Chen et al. showed that apical root growth inhibition by JA is caused by the suppression of proliferative activity in root meristematic cells, and MYC2 regulates this process by repressing expression of the auxin-responsive gene PLETHORA, which is responsible for stem cell maintenance and cell division25.
Cytokinin governs plant growth and development26 and recent studies demonstrated that cytokinin plays a key role in the development of root vascular tissue27–29. In cytokinin signal transduction, hybrid histidine protein kinases perceive the cytokinin signal at the plasma membrane, and histidine phosphotransfer proteins (AHPs) transmit this signal to response regulators (ARRs) through a phosphorelay30. ARRs can be categorized into Type-B and Type-A ARRs encoding transcriptional activators and repressors responsible for modulating the expression of cytokinin-responsive genes. ARR1, 2, 10, 11, and 12 belong to Type-B and ARR3, 4, 5, 6, 8, and 9 belong to Type-A. In root vascular tissues, the cytokinin response occurs specifically in procambial cells. A strong cytokinin response in the procambium promotes polar auxin transport toward protoxylem precursors by controlling expression and localization of PIN-FORMED (PIN) proteins, leading to the establishment of auxin maxima in these cells29. The auxin response promotes xylem differentiation and suppresses the cytokinin response by inducing the expression of ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6), which encodes a pseudo-histidine phosphotransfer protein and functions as an inhibitor of cytokinin signaling. Expression of AHP6 is essential for regulation of the cytokinin response. For example, overexpression or knock-out of AHP6 affects the cytokinin response and plant development31. Several genetic studies support the essential role of the cytokinin response in vascular tissue develoment. For example, the wooden leg (wol) mutants have severe defects in cytokinin responses and display all-xylem phenotypes in their vasculature. Also, mutant plants that lack expression of Type-B ARRs such as ARR1, ARR10, and ARR12 produce extra xylem28, 29, 32. Furthermore, treatment with exogenous cytokinin strongly suppresses the formation of xylem. These observations suggested that cytokinin is a negative regulator of xylem development28, 29.
Crosstalk between JA and cytokinin remains largely unknown. However, previous studies showed that environmental stresses that provoke JA responses can affect the expression of cytokinin-responsive genes33–35. These findings suggested that JA might interact with cytokinin to coordinate plant stress responses and growth. In this study, we showed that JA promotes xylem differentiation and a reduction of the cytokinin response underlies this process. Further molecular and genetic analysis suggested that the JA-responsive transcription factor MYC2 and the cytokinin signaling inhibitor AHP6 participate in JA-induced xylem development.
Results
JA interacts antagonistically with cytokinin
Previous studies have proposed that JA and cytokinin act antagonistically in the regulation of plant development and immunity36–38. JA inhibits apical root growth by suppressing the activity of meristematic cells in roots25. To understand the regulatory interaction between JA and cytokinin, we examined root growth in Col-0 plants treated with JA, cytokinin, or both (Fig. 1a; see Supplementary Fig. S1). Treatment with JA or cytokinin alone inhibited apical growth of roots and the combined treatment inhibited root growth more severely than either treatment alone. These observations suggested that cytokinin does not nullify the negative effect of JA on apical growth of roots.
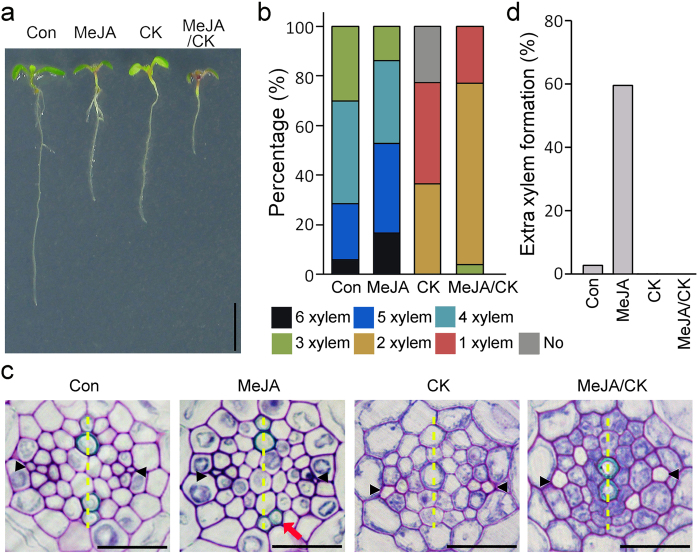
Figure 1.
Antagonistic interaction between JA and cytokinin in xylem development. (a) Root growth of Col-0 grown in the indicated conditions for 7 days (MeJA, 10 μM MeJA; cytokinin, 50 nM BAP; MeJA/cytokinin, 10 μM MeJA and 50 nM BAP). (b) Quantification of the number of xylem cells in the xylem axis of these plants (n > 20). (c) Vasculature morphology of Col-0 grown in the indicated conditions for 7 days. Black arrows and arrowheads indicate extra xylem cells and phloem cells, respectively. The yellow dotted line indicates a xylem axis. (d) Quantification of extra xylem formation in these plants. Percentages were calculated by dividing the number of plants with extra xylem by the number of plants observed. Scale bar = 0.5 cm in (a) and 20 μm in (c).
To further understand the interaction between JA and cytokinin, we analyzed the morphology of the root vasculature because cytokinin suppresses xylem formation whereas JA promotes xylem formation39. When we quantified the number of xylem cells in the roots of these plants by transverse sectioning and toluidine blue staining, we found that JA and cytokinin act antagonistically in xylem development (Fig. 1b). In normal growth conditions, 30%, 42%, 23%, and 5% of Col-0 roots showed 3, 4, 5, and 6 xylem cells in a xylem axis, respectively; by contrast, 15%, 33%, 36%, and 16% of JA-treated Col-0 roots formed 3, 4, 5, and 6 xylem cells, respectively, indicating that JA promotes xylem development. Unlike JA-treated or -untreated Col-0 roots, which generally had more than three xylem cells in a xylem axis, 23% of cytokinin-treated roots formed no xylem cells in the axis, 40% formed only one xylem cell, and 37% of roots formed two xylem cells, indicating that cytokinin strongly suppresses the formation of xylem. In JA/cytokinin-treated roots, all roots formed one or two xylem cells in the xylem axis, and the no-xylem phenotype was not observed, unlike Col-0 plants treated with cytokinin alone. Furthermore, we found that JA promotes formation of extra xylem adjacent to the xylem axis, which was rarely observed in JA-untreated wild-type plants (Fig. 1 c and d). The JA-induced xylem phenotype was rarely observed in the plants treated with both JA and cytokinin. These observations indicated that cytokinin diminishes the effect of JA on xylem development, suggesting an antagonistic interaction between JA and cytokinin.
To get an overview of how these hormones affect gene expression in the root, we performed RNA-sequencing (RNA-seq) using total RNA extracted from Col-0 roots treated with JA, cytokinin, or both. Results can be found in the GEO database under accession number GSE80188. When the expression patterns of 4,401 genes satisfying the criterion |fold change| ≥ 2 in at least one data set were displayed as a heat map, we found that gene expression patterns induced by cytokinin differed substantially from those induced by JA alone or by JA and cytokinin together (see Supplementary Fig. S2a). Quantitative RT-PCR analysis of cytokinin-induced gene expression partially supported this (see Supplementary Fig. S2b). JA reduced the expression of cytokinin-induced genes such as ARRs and PINs, but the roots treated with both JA and cytokinin did not show a reduction in ARR and PIN expression.
JA promotes differentiation of xylem
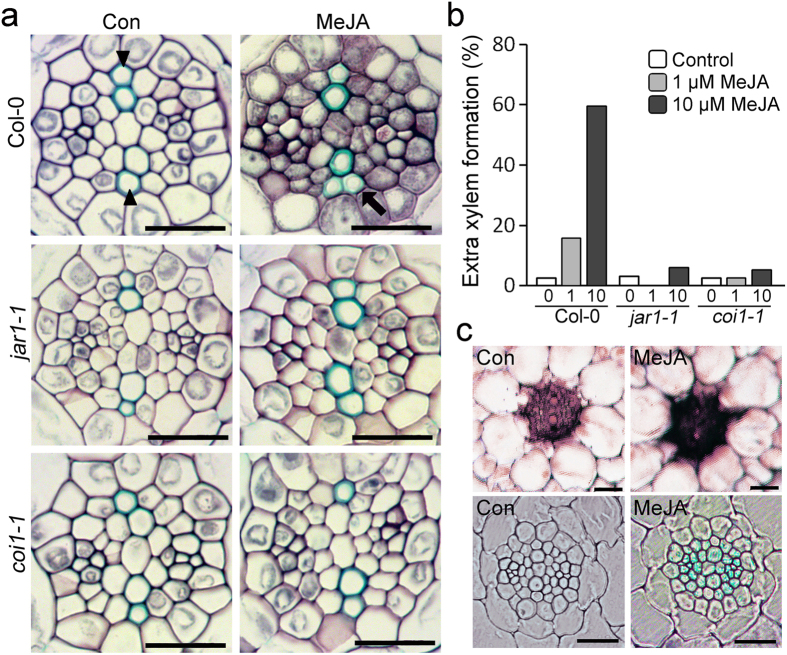
To further understand the effect of JA on the development of extra xylem, we examined the development of root vascular tissue in wild type and the JA signaling-defective mutants jar1-1 and coi1-1 (Fig. 2a and b). Transverse sectioning and toluidine blue staining showed that the morphology of root vascular tissue was almost identical between wild type and JA-signaling defective mutants grown in the absence of exogenous JA. However, around 15% of wild-type plants treated with 1 μM JA (Col-0, n = 32) developed extra xylem neighboring the protoxylem of a xylem axis. Around 60% of Col-0 plants (n = 37) developed extra xylem when grown in 10 μM JA. However, unlike Col-0 plants, in the jar1-1 and coi1-1 mutants, JA did not affect the morphology of root vascular tissues, and the extra-xylem phenotype was rarely detected in these mutant plants. However, the JA-biosynthesis mutant opr3 did form extra xylem in response to JA (see Supplementary Fig. S3). These observations suggested that JA signaling affects vascular tissue morphology in response to JA.
Figure 2.
JA affects development of root vascular tissues. (a) Morphology of root vascular tissues of Col-0, jar1-1, and coi1-1 grown in 10 μM MeJA or not treated for 7 days. Maturation regions of the indicated roots were transversely sectioned and stained with toluidine blue for detection of xylem cells. (b) Quantification of extra xylem formation in these plants (n > 30). Percentages were calculated by dividing the number of plants with extra xylem by the number of plants observed. (c) RNA in situ hybridization and GUS staining showing JA responses in vascular tissues. Spatial expression pattern of the JA-responsive gene VSP1 in roots (top) grown in 10 μM MeJA (right) or not treated (left) for 7 days. GUS staining analysis (bottom) was performed using 4XJARE::GUS plants in which GUS expression was controlled by the activity of 4 copies of the Jasmonic Acid Response Element (JARE). These plants were treated with 100 μM MeJA for 6 hrs. GUS staining solution with 1 mM ferrocyanide/ferricyanide was used. Arrow and arrowheads indicate extra xylem and protoxylem, respectively. Scale bar = 20 μm.
Examination of the JA response domain in the roots supported this (Fig. 2c). RNA in situ hybridization of the JA-induced gene VEGETATIVE STORAGE PROTEIN1 (VSP1) in Col-0 roots showed that vascular tissues had a stronger signal for the VSP1 transcripts, compared with other tissues. JA treatment increased accumulation of the VSP1 transcript in the vascular tissues. Additionally, transgenic plants expressing a GUS reporter gene under the control of the Jasmonic Acid Response Element (4XJARE::GUS) displayed GUS staining mainly in the vascular tissues in response to JA.
To characterize the effect of JA on xylem development, we counted the number of vascular tissue cells, and found that there was no significant difference in the number of vascular tissue cells between Col-0 roots grown with and without JA (see Supplementary Fig. S4a). However, the number of xylem cells in Col-0 roots grown in 10 μM JA was significantly higher than that of Col-0 grown without JA (see Supplementary Fig. S4b). These observations suggested that the JA response promotes xylem differentiation.
JA reduces the cytokinin response in root vascular tissue
Cytokinin negatively regulates xylem development28, 29. Because JA promotes xylem development, we hypothesized that JA might affect cytokinin responses in the root vasculature. To address this, we tested whether JA causes changes in the cytokinin response in transgenic plants containing the TCS::GFP reporter for the cytokinin response2 (Fig. 3a). In JA-untreated conditions, we detected strong fluorescent signals in TCS::GFP root caps, but JA treatment decreased the intensity of the fluorescent signal in a dosage-dependent manner. Unlike wild-type plants, coi1-1 mutants did not respond to JA, and the suppression of GFP signals by JA was not observed in the mutant plants. These observations suggested that the JA response suppresses the cytokinin response. However, changes in the cytokinin response in root vasculature were not obvious in this system because the fluorescent signals in the tissues were too weak to visualize.
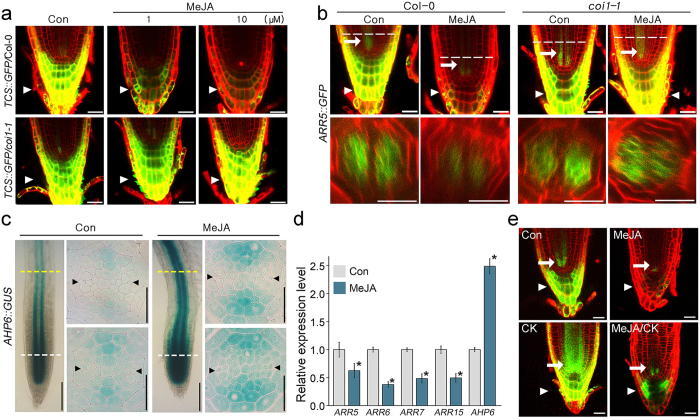
Figure 3.
JA reduces the cytokinin response. (a) Effect of JA on the cytokinin response was analyzed by visualizing the fluorescent signals in TCS::GFP/Col-0 (top) and TCS::GFP/coi1-1 (bottom) plants grown in 1 and 10 μM MeJA for 7 days. (b) Confocal longitudinal (top) and cross (bottom) section images showing that JA suppressed cytokinin-induced ARR5 expression in root vascular tissues of Col-0, but not in coi1-1 mutants. The indicated plants were grown in 10 μM MeJA or not treated for 7 days. (c) GUS staining of AHP6::GUS plants grown in 10 μM MeJA or not treated for 7 days using GUS staining solution without ferrocyanide/ferricyanide. (d) Expression levels of cytokinin-responsive genes measured by qRT-PCR. Total RNA was extracted from Col-0 roots grown in the indicated conditions for 7 days. GAPDH was used as a reference gene for normalization. Error bars represent S.D. and asterisks indicate statistically significant differences between the corresponding samples and their control (p < 0.01, t-test). (e) Visualization of the fluorescent signals in ARR5::GFP/Col-0 grown in the indicated conditions (MeJA, 10 μM MeJA; cytokinin, 50 nM BAP; MeJA/cytokinin, 10 μM MeJA and 50 nM BAP). White arrows and arrowheads indicate cytokinin response in the vascular tissue and root cap, respectively. Dotted lines indicate the longitudinal position where confocal optical cross-sectioning was performed. Scale bar = 100 μm in whole-mounted images of (c) and 20 μm in others.
We also tested the ARABIDOPSIS RESPONSE REGULATORS5 (ARR5) and WOODEN LEG (WOL) promoters, two other cytokinin-responsive markers28, 40. In ARR5::GFP transgenic plants not treated with JA, fluorescent signals were observed in root caps and vascular tissues. In ARR5::GFP plants grown with JA, the GFP signals were much weaker than in ARR5::GFP plants grown without JA (Fig. 3b). Furthermore, optical sectioning showed that JA treatment decreased the intensity of fluorescent signals in vascular tissues of ARR5::GFP plants. However, in the coi1-1 background, JA did not affect the fluorescent signals. Similar to ARR5::GFP transgenic plants, WOL::GFP/Col-0 plants displayed suppressed fluorescent signals in response to JA but WOL::GFP/coi1-1 plants did not (see Supplementary Fig. S5). These results suggested that the JA response suppresses the cytokinin response in root vascular tissues. AHP6 inhibits the cytokinin response and AHP6 expression is negatively correlated with cytokinin responses28, 29. In contrast to ARR5 and WOL, expression of AHP6 increased in response to JA. In JA-untreated AHP6::GUS seedlings, GUS staining was predominantly detected in the protoxylem and neighboring pericycle cells. However, JA treatment expanded the AHP6 expression domain to the procambial cells (Fig. 3c; see supplementary Figs S6 and S7). These results suggested that JA suppresses the cytokinin response in the root vasculature. Quantitative RT-PCR analysis supported this idea (Fig. 3d). Expression levels of Type-A ARRs such as ARR5, 6, 7, and 15 were lower in Col-0 roots grown with JA than those grown without JA, whereas the expression level of AHP6 was higher in the JA-treated roots. Unlike Type-A ARRs, expression of Type-B ARRs in JA-treated Col-0 plants tended to be slightly upregulated compared to those in JA-untreated Col-0 (see Supplementary Fig. S8). Previous studies showed that expression levels of Type-A ARRs increased in response to cytokinin, but expression of Type-B ARRs tended to decrease41. These results suggested that the decreased expression of Type-A ARRs and the increased expression of Type-B ARRs might be caused by suppression of the cytokinin response by JA. When the fluorescent signals were visualized in ARR5::GFP plants treated with JA, cytokinin, or both, the intensity of fluorescent signal in JA/cytokinin-treated ARR5::GFP plants was higher than that in the JA-treated ARR5::GFP plants, but lower than that in cytokinin-treated ARR5::GFP plants. These observations supported the negative effect of JA on cytokinin responses and the antagonistic relationship between JA and cytokinin (Fig. 3e).
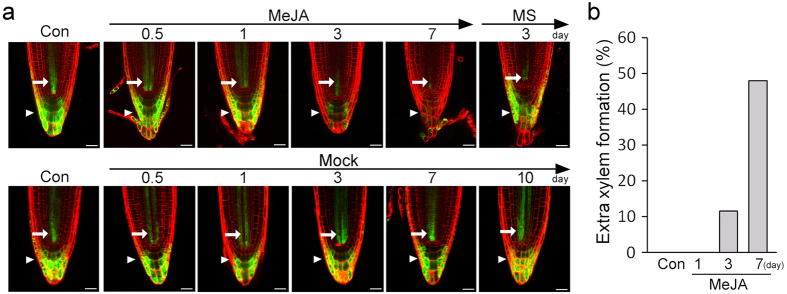
To further understand the reduction of the cytokinin response by JA, we analyzed changes in the cytokinin response in a time course in the ARR5::GFP roots treated with JA (Fig. 4a). The fluorescent signals in the roots exposed to JA for 0.5 or 1 day were almost identical to those in JA-untreated roots. However, as the exposure time increased, the intensity of the fluorescent signals gradually decreased. In the roots exposed to JA for 3 days, the fluorescent signals were obviously weaker than those in control roots and the signals almost disappeared at 7 days. When these roots were transferred to media without JA and grown for 3 days, the fluorescent signals reappeared. To understand the relationship between cytokinin responses and extra xylem formation, we quantified formation of extra xylem in these plants (Fig. 4b). The extra-xylem phenotype was rarely detected in the roots exposed to JA for 1 day, but the roots exposed to JA for 3 days or 7 days showed the extra-xylem phenotype. These observations suggested that suppression of the cytokinin response by JA does not occur rapidly, but the JA-mediated xylem phenotype is caused by suppression of the cytokinin response.
Figure 4.
Gradual suppression of the cytokinin response by JA. (a) Suppression of the cytokinin response by JA was analyzed in a time course by monitoring fluorescent signals in ARR5::GFP plants. Five-day-old ARR5::GFP plants grown without JA were transferred to MS media containing 10 μM MeJA, and grown for the indicated times. Mock indicates MS media containing 0.05% EtOH. White arrows and arrowheads indicate cytokinin response in the vascular tissue and root cap, respectively. (b) Quantification of extra xylem formation in these plants (n > 20). Percentages were calculated by dividing the number of plants with extra xylem by the number of plants observed. Scale bar = 20 μm.
Overexpression of AHP6 promotes formation of extra xylem
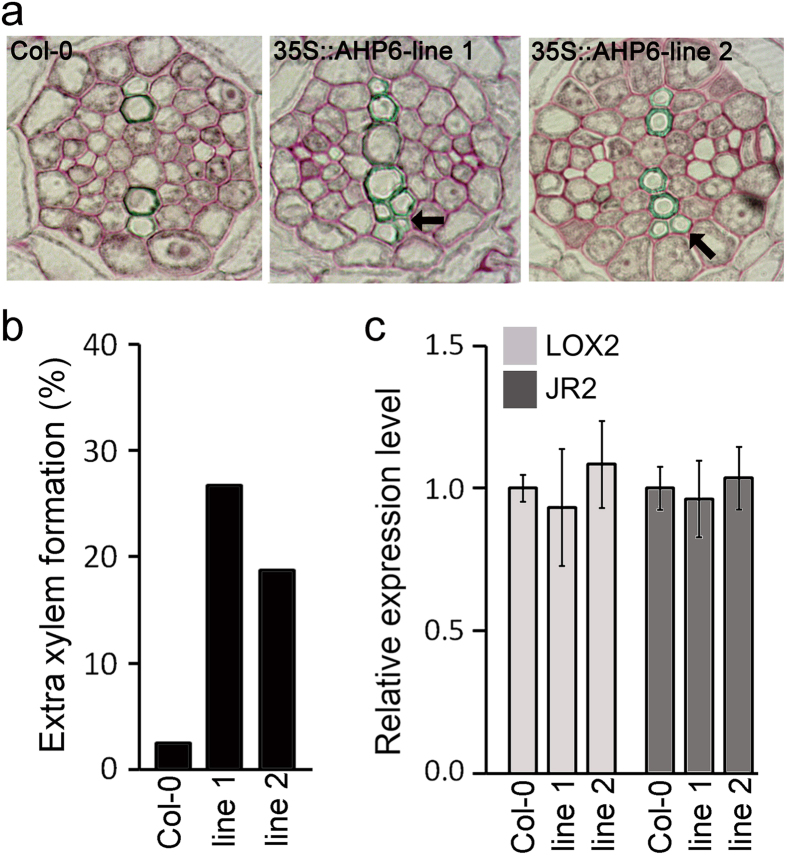
To understand whether the low cytokinin response can promote the development of extra xylem, we analyzed xylem development in AHP6-overexpressing transgenic plants, which have reduced cytokinin responses (see Supplementary Fig. S9). Further characterization showed that around 20–27% of 35S::AHP6 plants form extra xylem, even in the absence of exogenous JA, suggesting that a reduction of cytokinin can induce the formation of extra xylem (Fig. 5a and b). The 35S::AHP6 and wild-type plants also showed similar numbers of vascular cells; however, the 35S::AHP6 plants tended to have more xylem cells, compared to wild-type plants (see Supplementary Fig. S10). We then checked the JA response in AHP6-overexpressing transgenic plants by analyzing expression levels of the JA-induced genes LIPOXYGENASE2 (LOX2) and JASMONATE-RESPONSIVE2 (JR2). We found no difference in the expression levels of LOX2 and JR2 between wild-type and AHP6-overexpressing plants (Fig. 5c). These findings indicated that a low cytokinin response promotes xylem development without affecting JA responses.
Figure 5.
Reduction of the cytokinin response promotes the formation of extra xylem. (a) Cross section images of 35S::AHP6 roots grown in MS media for 7 days. (b) Quantification of extra xylem formation in these plants (n > 30). Percentages were calculated by dividing the number of plants with extra xylem by the number of plants observed. (c) Expression levels of the JA-induced genes LOX2 and JR2 in these plants. Line 1 and 2 indicate individual lines of 35S::AHP6 transgenic plants. Error bars represent S.D. GAPDH was used as a reference gene. Black arrows indicate extra xylem cells. Scale bar = 20 μm.
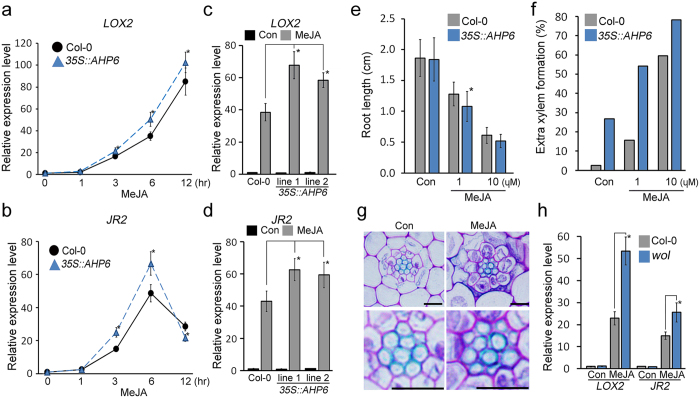
To understand the regulatory interaction between JA and cytokinin, we checked JA-induced changes in the expression of JA-induced genes in 35S::AHP6 plants in a time course (Fig. 6a–d). In JA-untreated conditions, LOX2 and JR2 expression was almost identical between wild-type and AHP6-overexpressing plants. However, LOX2 and JR2 expression in JA-treated 35S::AHP6 plants was higher than that in JA-treated Col-0 plants, and the difference tended to increase with increasing treatment time. These results suggested that 35S::AHP6 plants show higher JA responses when treated with JA. To explore this, we analyzed root growth inhibition and extra xylem formation in JA-treated 35S::AHP6. In JA-untreated conditions, the root length of 35S::AHP6 plants was similar to that of wild type. However in JA-treated conditions, the root length of 35S::AHP6 plants was shorter than that of wild-type plants (Fig. 6e). When extra xylem formation was quantified, approximately 54% and 78% of 35S::AHP6 plants showed extra xylem in response to 1 µM and 10 µM JA, respectively, while around 15% and 60% of wild-type plants did (Fig. 6f). These results suggested that the 35S::AHP6 plants might be more sensitive to JA.
Figure 6.
JA responses in plants with reduced cytokinin responses. LOX2 (a) and JR2 (b) expression was analyzed in 35S::AHP6 plants by qRT-PCR. Seven-day-old 35S::AHP6 and Col-0 seedlings were transferred to media containing 100 μM MeJA and incubated for the indicated time. Total RNA was extracted from these roots. Expression levels of LOX2 (c) and JR2 (d) in two independent lines of 7-day-old 35S::AHP6 plants treated with 100 μM JA for 6 hrs. (e) Root growth of 35S::AHP6 plants grown in JA-treated and -untreated conditions for 7 days (n > 30). (f) Quantification of extra xylem formation in these plants (n > 29). Percentages were calculated by dividing the number of plants with extra xylem by the number of plants observed. (g) Root vasculature (top) and high-resolution images (bottom) of wol mutants grown in 10 μM MeJA-untreated (left) or -treated (right) conditions for 12 days. (h) Expression levels of LOX2 and JR2 in wol mutant roots. Ten-day-old wol and Col-0 seedlings were transferred to MS media containing 100 μM MeJA and incubated for 3 hrs. Total RNA was extracted from the roots of these plants. GAPDH was used as a reference gene. Error bars represent S.D. and asterisks indicate statistically significant differences between the corresponding samples and their control (p < 0.01, t-test). Scale bar = 10 μm.
The wol mutants have severely compromised cytokinin responses and defects in root growth27, 29. The wol mutants have fewer vascular cells compared with wild-type plants, and all root vascular cells differentiate into xylem cells27, 29. As expected, the root vascular morphology of JA-treated wol mutants was almost identical to JA-untreated wol mutants, and the number of vascular tissue cells all of which were xylem cells was similar between them (Fig. 6g; see Supplementary Fig. S11). The expression of LOX2 and JR2 was almost identical between JA-untreated wild type and wol mutants. However, in JA-treated conditions, expression levels of LOX2 and JR2 were higher in in wol mutants than in wild-type, suggesting that wol mutants show higher JA responses when treated with JA (Fig. 6h).
We also checked cytokinin responses in the JA-signaling mutant jar1-1 by analyzing the expression level of ARR5 (see Supplementary Fig. S12). In cytokinin-untreated or -treated conditions, the expression levels of ARR5 were similar between wild-type and jar1-1 mutant plants grown in the same conditions. To verify this, we quantified the expression level of ARR5 in another JA-signaling mutant, myc2-3. Similar to jar1-1, the myc2-3 mutants exhibited similar cytokinin responses to wild-type plants in cytokinin-untreated or -treated conditions. These observations suggested that JA signaling mutants and wild-type plants have similar sensitivities to cytokinin.
Expression of AHP6 and MYC2 is involved in the JA-mediated xylem phenotype
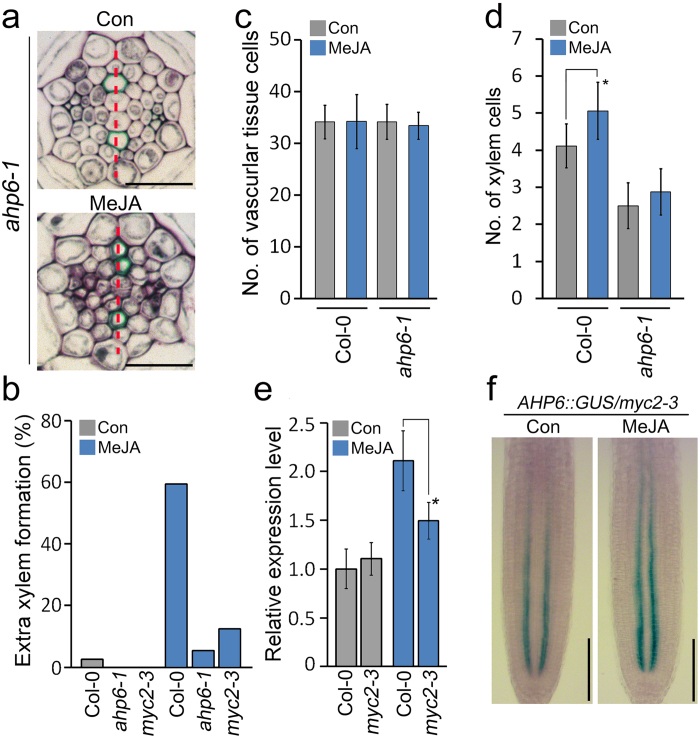
Since JA expanded the AHP6 expression domain to the procambium, where extra xylem formed, we explored the possible involvement of AHP6 in JA-mediated xylem development. To do this, we analyzed xylem development in ahp6-1 mutants grown with and without JA. Consistent with a previous study by Mähönen et al., development of protoxylem was suppressed in ahp6-1 mutants grown without JA28 (Fig. 7a and b). Furthermore, unlike wild-type plants that formed extra xylem cells in response to JA, ahp6-1 mutants did not produce, or rarely produced extra xylem cells adjacent to a xylem axis in response to JA. Counting the number of vascular tissue cells and xylem cells in ahp6-1 mutants showed no significant difference between JA-untreated and JA-treated ahp6-1 mutants. These findings suggested that AHP6 is deeply involved in the JA-induced extra-xylem phenotype (Fig. 7c and d).
Figure 7.
AHP6 and MYC2 are involved in JA-mediated xylem development. (a) Root vasculature of ahp6-1 plants grown in 10 μM MeJA-untreated (top) or -treated (bottom) conditions for 7 days. The red dotted line indicates a xylem axis. (b) Quantification of extra xylem formation in the roots of Col-0, ahp6-1, and myc2-3 mutants grown in 10 μM MeJA or not treated for 7 days (n > 22). (c and d) The number of vascular tissues cells (c) and xylem cells (d) in JA-treated and -untreated ahp6-1 mutant plants. (e) Analysis of AHP6 expression in myc2-3 mutants by qRT-PCR. Total RNA was extracted from Col-0 and myc2-3 roots grown in 10 μM MeJA or not treated for 7 days. GAPDH was used as a reference gene. Error bars represent S.D. and asterisks indicate statistically significant differences between the corresponding samples and their control (p < 0.01, t-test). (f) GUS staining of AHP6::GUS/myc2-3 plants grown in 10 μM MeJA or not treated for 7 days using 1 mM ferrocyanide/ferricyanide-containing GUS staining solution. Scale bar = 20 μm in (a) and 100 μm (d).
We also found that MYC2 is involved in JA-mediated xylem development. In JA-untreated conditions, MYC2-overexpressing transgenic plants did not show an extra-xylem phenotype, similar to Col-0. However in 1 µM JA-treated conditions, approximately 50% of 35S::MYC2 transgenic plants exhibited an extra-xylem phenotype and the rate was around 3-fold higher than in wild-type plants grown in the same conditions. This suggested that overexpression of MYC2 promotes the formation of extra xylem in response to JA, and MYC2 is involved in JA-mediated modulation of xylem development (see Supplementary Fig. S13). To explore this, we examined xylem development in myc2-3 mutants and found that formation of extra xylem is inhibited in myc2-3 mutants (Fig. 7b), indicating that MYC2 mediates xylem development in response to JA.
When we compared AHP6 expression between Col-0 and myc2-3 mutants grown without JA, we observed no significant difference in AHP6 expression. However, in JA-treated conditions, the expression of AHP6 in myc2-3 mutants was lower than that in wild type (Fig. 7e). These results suggested that MYC2 is involved in the regulation of AHP6 expression in response to JA. GUS staining of AHP6::GUS/myc2-3 plants supported this, as myc2-3 plants grown with and without JA showed similar GUS staining (Fig. 7f). Promoter analysis predicted that the AHP6 promoter contains a putative MYC2-binding sequence, (CACATG, at -1623 bp) (see Supplementary Table S1). A yeast one-hybrid assay suggested that MYC2 possibly binds to the AHP6 promoter region containing the MYC2-binding sequence (see Supplementary Fig. S14). However, ChIP analysis using 35S::MYC2-GFP transgenic plants showed that MYC2 does not directly interact with the AHP6 promoter.
Discussion
Developmental flexibility under stress conditions largely occurs via the interaction between hormones that mediate stress responses and developmental processes. JA coordinates the dynamics of plant stress responses and growth through interactions with other phytohormones such as gibberellic acid and auxin21, 25, 42. Growing numbers of studies have proposed that JA also interacts with cytokinin in the regulation of plant development and physiology. For example, JA prevents cytokinin-induced soybean callus growth43 and also inhibits the effect of cytokinin on chlorophyll degradation and the plant immune system36, 37. Expression of genes involved in these processes is regulated differently by JA and cytokinin. Furthermore, each treatment nullifies the effect of the other. A recent study of Nitschke et al. also supports the interaction of JA and cytokinin in circadian stress responses38. In this study, we showed that JA signaling promotes xylem differentiation in response to JA, and the antagonistic interaction with cytokinin is involved in this process. Cytokinin is a key negative regulator of xylem development27–29. For example, cytokinin signaling-defective wol plants develop small numbers of procambial cell files due to suppression of procambial cell division27. Moreover, this mutant produces all protoxylem in its root vascular tissues29. In this study, we showed that JA induces formation of extra xylem in wild-type plants, but not in JA-signaling mutants such as coi1-1 and jar1-1. These suggest that JA response promotes xylem development in response to JA.
Analysis of the cytokinin response showed that the reduction of the cytokinin response is responsible for the JA-mediated xylem phenotype. Expression of cytokinin-induced genes was downregulated by JA, and cytokinin treatment diminished the effect on JA on formation of extra xylem and expression of cytokinin-induced genes. Additionally, 35S::AHP6 plants with reduced cytokinin responses formed extra xylem even in the absence of JA. When expression levels of JA-responsive genes were analyzed in the 35S::AHP6 and wild-type grown in JA-untreated conditions, we found no difference between them. However 35S::AHP6 plants exhibited increased expression of these genes compared to wild type in JA-treated conditions, suggesting that 35S::AHP6 plants with reduced cytokinin responses show higher JA responses than wild-type plants when treated with JA. Furthermore, root growth inhibition and extra xylem development is promoted in these transgenic plants compared to wild-type plants. Previous studies reported that plants with higher JA responses show enhanced tolerance to environmental stresses such as drought and salt44–48. Additionally, a study by Nishiyama et al. showed that cytokinin-deficient transgenic plants are also resistant to these environmental stresses49. Collectively, these suggest that JA antagonistically interacts with cytokinin and the plants with reduced cytokinin responses are more sensitive to JA.
In this study we also showed that the expression level of AHP6 in two independent 35S::AHP6 lines, was approximated 36- and 28-fold higher than in wild-type plants, respectively. Extra xylem was observed in around 22% and 17% of the transgenic plants, respectively, suggesting a positive relationship between AHP6 expression and extra xylem formation. Together with the results that ahp6-1 rarely formed extra xylem in response to JA, these observations suggest that AHP6 is involved in the JA-mediated xylem phenotype. When the expression level of AHP6 was compared between JA-untreated 35S::AHP6 and JA-treated Col-0 plants, the 35S::AHP6 plants showed about 15-fold higher expression of AHP6 than JA-treated Col-0. However, the extra-xylem phenotype was observed in around 20% of 35S::AHP6 and 60% of wild-type plants, indicating that AHP6 expression is not tightly linked to the JA-mediated formation of extra xylem although expression of AHP6 is essential for the formation of extra xylem in response to JA. These findings suggest that other JA-regulated factors are involved in this process together with AHP6.
MYC2 plays a key role in the regulation of JA responses11, 50. Chen et al. reported that MYC2 mediates the inhibition of apical root growth in response to JA by suppressing expression of the auxin-responsive gene PLETHORA 25, suggesting that MYC2 plays an essential role in modulation of root development in response to JA. In this study, we suggest that MYC2 is also responsible for JA-mediated xylem development. Indeed, formation of extra xylem was strongly suppressed in myc2-3 mutants. These findings suggest that the MYC2 transcription factor mediates xylem differentiation as well as apical growth of roots in response to JA. When expression of endogenous AHP6 was analyzed in myc2-3 and wild-type plants exposed to JA, the myc2-3 mutants exhibited reduced expression of AHP6 compared to wild-type plants. These findings suggest that the MYC2 transcription factor promotes AHP6 expression in response to JA. However, based on our ChIP results, it is not likely that MYC2 directly regulates expression of AHP6 and downstream regulators of MYC2 might be involved in the direct regulation of AHP6 expression.
Plants dynamically coordinate their growth and defenses in response to changes in environmental conditions, and it has been thought that growth inhibition is one of the adaptations that help plants survive environmental stresses51. In this study we showed that JA promotes differentiation of meristematic procambial cells to xylem cells. When we analyzed root development in wild-type plants grown under drought stress, drought inhibited root growth and promoted the formation of extra xylem, as JA does (see Supplementary Fig. S15). This suggests that promotion of xylem differentiation from meristematic cells might be one of the developmental mechanisms that plants use to inhibit their growth and thus to survive environmental stresses. Previous studies showed that JA or JA-dependent environment stresses can affect the expression of genes involved in cytokinin responses33–35 partially support this hypothesis. Collectively, our study proposes that the interaction of JA and cytokinin is involved in coordinating the dynamics of plant growth and defense under environmental stresses. Further molecular and genetic approaches will expand our understanding of the mechanisms of the regulatory interaction between JA and cytokinin.
Materials and Methods
Plant materials, growth and treatment
Plants of the Arabidopsis thaliana ecotype Columbia (Col-0) were used as controls in this study. The jar1-1, coi1-1, myc2-3, wol, ahp6-1, 4XJARE::GUS, ARR5::GFP, TCS::GFP, AHP6::GFP and 35S::MYC2-GFP plants have been described previously2, 14, 27–29, 40, 52–54. These seeds were obtained from the Nottingham Arabidopsis Stock Centre (NASC) or kind donation from Dr. Helariutta, Dr. Mähönen, and Dr. Chua. Seeds were surface sterilized, and plated on ½-strength Murashige and Skoog (1/2x MS) solid media. After 2 days of vernalization at 4 °C in darkness, plants were grown vertically with a light regime of 16/8 hours (light/dark) at 22 °C. For drought treatment, PEG-containing media was prepared as previously described55.
Plasmid construction
For overexpression of AHP6, the 35S::AHP6 construct was generated using the GATEWAY system (Invitrogen). The AHP6 cDNA fragment was amplified by PCR using total RNA extracted from 7-day-old Arabidopsis roots. The pENTRY-AHP6 plasmid was generated by inserting the amplified cDNA fragments into the pDONR221 vector (Invitrogen) using the BP reaction. The pENTRY-AHP6 construct was then recombined into the modified pMDC plant binary vector carrying the 35S promoter through the LR reaction. For the AHP6::GUS construct, the AHP6 promoter (1877 bp) was isolated by PCR from Arabidopsis genomic DNA. This DNA fragment was inserted into the PstI/EcoRI-digested pCAMBIA vector containing β-glucuronidase (GUS) by the Gibson Assembly Cloning system (New England BioLabs). Both constructs were introduced into Arabidopsis Col-0 plants by the floral dip method.
Embedding, sectioning, and staining
Technovit embedding and sectioning were performed as described56 with slight modifications. Arabidopsis roots were fixed in 4% paraformaldehyde for 2 h and then washed in ddH2O three times for 1 h each. The fixed samples were dehydrated in an ethanol series (20, 40, 60, 80, and 100% (v/v) in ddH2O). The dehydrated samples were sequentially incubated in a series of Technovit 7100 cold-polymerizing resin (33, 66, and 100% (v/v) in EtOH) for 3 h each. Samples were further incubated in 100% Technovit resin for 1 day and solidified with a 15:1 (v/v) mixture of Technovit and hardener solution II at room temperature in a mold for 1 day. Sections (3–4 μm) were taken from the maturation zone of roots (around 2.5–3 mm above the root tip for JA-untreated plants, and around 2–2.5mm above the root tip for JA-treated plants with shorten roots). Dehydrated sections were stained with 0.05% toluidine blue solution to detect xylem cells (pH 4.5).
GUS staining
GUS staining was performed as described57 with slight modifications. AHP6::GUS/Col-0 and AHP6::GUS/myc2-3 grown in JA-treated or -untreated conditions for 7 days were incubated in GUS staining solution with or without 1 mM ferrocyanide/ferricyanide (100 mM NaPO4 pH 7.0, 0.5 mM 5-bromo-4-chloro-3-indolyl-glucuronide, and 0.2% Triton X-100) at 37 °C for 3 or 8 h. The samples were then washed with 100 mM NaPO4 (pH 7.0) and incubated in 70% ethanol at 4 °C overnight.
Quantitative RT-PCR
Quantitative RT-PCR analyses were performed using total RNA extracted from roots. Total RNA extraction was carried out using the RNeasy plant mini-prep kit (Qiagen) according to the manufacturer’s instructions. For the first-strand cDNA synthesis, 20 µL of reverse transcription reaction was performed using 2 µL of total RNA and Superscript III reverse transcriptase (Invitrogen). For quantitative PCR, a master mix was prepared using a LightCycler 480 SYBR GREEN I Master (Roche). PCR reactions and fluorescence detection were performed using a LightCycler NANO Real-Time PCR machine (Roche). PCR conditions were programmed according to the manufacturer’s instructions (initial denaturation at 95 °C for 5 min, denaturation at 95 °C for 10 sec, annealing at 58 °C for 10 sec, and extension at 72 °C for 10 sec with 45 cycles). Expression levels were analyzed using at least two biological and three technical replicates. AtGAPDH (At1G13440) was used as an internal control. Primer sequence information is available in Supplementary Table S2.
RNA in situ hybridization
RNA in situ hybridization was carried out as described by Takechi et al. with slight modification58. The sections (4–6 mm thick) were collected using a rotary microtome and these sections were stretched on the surface of a glass slide using DEPC-treated water. VSP1-specific sense and antisense probes were labeled using the DIG RNA Labeling Kit (Roche) according to the manufacturer’s protocol. Ten ng of DIG-labeled probe was used for the hybridization.
Yeast one-hybrid assay
To investigate the direct interaction between MYC2 and the AHP6 promoter, a yeast one-hybrid assay (Clontech) was performed according to the manufacturer’s instructions. The AHP6 promoter and full-length MYC2 cDNA were amplified by PCR and inserted into the pAbAi bait vector and pGADT7 prey plasmid, respectively. The bait plasmids were digested with BbsI (NEB) and transformed into Saccharomyces cerevisiae Y1HGold. Transformed colonies were selected on synthetic dropout glucose medium (SD) without uracil (SSD). The pGADT7-MYC2 prey plasmid was introduced into the bait-integrated Y1HGold, and transformed colonies were selected on SSD media without leucine. To test the interaction between MYC2 and the AHP6 promoter, transformed yeast lines (OD280 = 0.1) were dropped on SSD-Leu media containing 200 ng/ml aureobasidin A (Abs A). The dropped cells were grown at 30 °C for 4 days.
Chromatin immunoprecipitation assay
To investigate the direct interaction between MYC2 and the AHP6 promoter, ChIP assays were performed using 35S::MYC2-GFP transgenic plants54. Around 0.5 g of roots of 7-day-old 35S::MYC2-GFP seedlings were harvested and then immediately immersed in fixing solution (0.4 M sucrose, 10 mM Tris-HCl (pH 8.0), 5 mM β-mercaptoethanol, and 1% formaldehyde) for 15 min. After washing, the roots were ground in liquid nitrogen and nuclei were isolated. For fragmentation, the nuclei were sonicated (Ultrasonic Processor, GE-50). Immunoprecipitation was performed as described by Gendrel et al. except that a GFP antibody (ab290, ABcam) was used59. For qPCR analysis, 18S rRNA was used as an internal control. The JAZ1 promoter was used for a positive control of the MYC2 interaction. Primer sequences are listed in Supplementary Table S2.
Microscopy
For the visualization of root vascular tissues, whole roots of 7-day-old Arabidopsis seedlings were dipped in propidium iodide (PI) solution (10 µg/ml) for 1 min. After staining, the roots were mounted on glass slides in ddH2O. For the detection of PI and green fluorescent protein (GFP), fluorescence was visualized with wavelengths of 591–635 nm for PI and 505–530 nm for GFP, using a Leica SP8 STED laser scanning confocal microscope. Photographs of plants and tissue sections were taken with a Nikon SMZ-U stereomicroscope and an Olympus BX41 light microscope.
RNA sequencing analysis
Col-0 plants were grown in 1/2× MS solid media containing 10 μM MeJA, 50 nM BAP, or 10 μM MeJA plus 50 nM BAP for 7 days. Total RNA was extracted from the roots of these plants together with those of Col-0 plants grown in 1/2x MS solid media. To generate cDNA libraries with the TruSeq RNA library kit, 1 μg of total RNA was used. Library construction consisted of polyA-selection of RNA, RNA fragmentation, random hexamer primed reverse transcription and 100 nt paired-end sequencing with the Illumina HiSeq. 2000. The libraries were quantified using qPCR according to the qPCR Quantification Protocol Guide and qualified using an Agilent Technologies 2100 Bioanalyzer. Raw data were calculated as Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) of each transcript in each sample by Cufflinks software. The transcripts with zero FPKM values were removed from each data set. A combined data set of 22,291 genes was obtained by merging the data sets for each condition. For generation of the heat map of hierarchical clustering to visualize expression patterns of differentially expressed transcripts, 4,401 genes satisfying |fold change| ≥ 2 in at least one data set were collected. The GEO accession number of these datasets is GSE80188 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80188).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: WOL (At2G01830), ARR6 (At5G62920), ARR7 (At1G19050), ARR15 (At1G74890), AHP6 (At1G80100), LOX2 (At3G45140), JR2 (At4G23600), MYC2 (At1G32640), and GAPDH (At1G13440).
Electronic supplementary material
Acknowledgements
We heartily thank Dr. Helariutta, Dr. Mähönen, and Dr. Chua for donating mutant and transgenic plant seeds. This work was carried out with the support of Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0112152017 to Y.D.C.) Rural Development Administration, Republic of Korea, through the National Center for GM Crops, and the National Research Foundation of Korea Grant funded by the Korean Government (MOE) [NRF-2016R1D1A1B03931167].
Author Contributions
Y.C. and G.J. conceived the original screening and research plans; Y.C. supervised the experiments. G.J. and S.C. performed most of the experiments; T.U. and S.L. provided technical assistance to G.J.; T.U. analyzed the data; G.J., J.K. and Y.C. wrote the article with contributions of all the authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10634-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vanstraelen M, Benková E. Hormonal interactions in the regulation of plant development. Annual review of cell and developmental biology. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- 2.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller GE, Bishopp A, Kieber JJ. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annual review of plant biology. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 5.Creelman, R. A. & Mulpuri, R. The oxylipin pathway in Arabidopsis. The Arabidopsis Book, e0012 (2002). [DOI] [PMC free article] [PubMed]
- 6.Qi T, Huang H, Song S, Xie D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. The Plant Cell. 2015;27:1620–1633. doi: 10.1105/tpc.15.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schommer C, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber H, Vick BA, Farmer EE. Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proceedings of the National Academy of Sciences. 1997;94:10473–10478. doi: 10.1073/pnas.94.19.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kombrink E. Chemical and genetic exploration of jasmonate biosynthesis and signaling paths. Planta. 2012;236:1351–1366. doi: 10.1007/s00425-012-1705-z. [DOI] [PubMed] [Google Scholar]
- 11.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448 doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 12.Thines B, et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JP, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song S, et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. The Plant Cell. 2014;26:263–279. doi: 10.1105/tpc.113.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. The Plant Cell. 2014;26:1105–1117. doi: 10.1105/tpc.113.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng H, et al. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS genetics. 2009;5 doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi T, et al. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. The Plant Cell. 2014;26:1118–1133. doi: 10.1105/tpc.113.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Yang D-L, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagpal P, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 24.Sun J, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. The Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. The Plant Cell. 2011;23:3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaller GE, Street IH, Kieber JJ. Cytokinin and the cell cycle. Current opinion in plant biology. 2014;21:7–15. doi: 10.1016/j.pbi.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Mähönen AP, et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes & development. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 29.Bishopp A, et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Current Biology. 2011;21:917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Sheen J. Phosphorelay and transcription control in cytokinin signal transduction. Science. 2002;296:1650–1652. doi: 10.1126/science.1071883. [DOI] [PubMed] [Google Scholar]
- 31.Besnard F, et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014;505 doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama A, et al. Type-B ARR Transcription Factors, ARR10 and ARR12, are Implicated in Cytokinin-Mediated Regulation of Protoxylem Differentiation in Roots of Arabidopsis thaliana. Plant and Cell Physiology. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- 33.Hare P, Cress W, Van Staden J. The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regulation. 1997;23:79–103. doi: 10.1023/A:1005954525087. [DOI] [Google Scholar]
- 34.Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Current opinion in plant biology. 2002;5:230–236. doi: 10.1016/S1369-5266(02)00250-9. [DOI] [PubMed] [Google Scholar]
- 35.Argueso CT, Ferreira FJ, Kieber JJ. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant, cell & environment. 2009;32:1147–1160. doi: 10.1111/j.1365-3040.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 36.Naseem M, Kaltdorf M, Hussain A, Dandekar T. The impact of cytokinin on jasmonate-salicylate antagonism in Arabidopsis immunity against infection with Pst DC3000. Plant signaling & behavior. 2013;8 doi: 10.4161/psb.26791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L, et al. Exogenous jasmonic acid and cytokinin antagonistically regulate rice flag leaf senescence by mediating chlorophyll degradation, membrane deterioration, and senescence-associated genes expression. Journal of Plant Growth Regulation. 2016;35:366–376. doi: 10.1007/s00344-015-9539-0. [DOI] [Google Scholar]
- 38.Nitschke S, et al. Circadian Stress Regimes Affect the Circadian Clock and Cause Jasmonic Acid-Dependent Cell Death in Cytokinin-Deficient Arabidopsis Plants. The Plant Cell. 2016;28:1616–1639. doi: 10.1105/tpc.16.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghuge SA, et al. The MeJA-inducible copper amine oxidase AtAO1 is expressed in xylem tissue and guard cells. Plant signaling & behavior. 2015;10 doi: 10.1080/15592324.2015.1073872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Current Biology. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 41.Kim K, et al. Cytokinin‐facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR 2 attenuates signaling output in two‐component circuitry. The Plant Journal. 2012;69:934–945. doi: 10.1111/j.1365-313X.2011.04843.x. [DOI] [PubMed] [Google Scholar]
- 42.Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends in plant science. 2012;17:22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Ueda J, Kato J. Inhibition of cytokinin‐induced plant growth by jasmonic acid and its methyl ester. Physiologia Plantarum. 1982;54:249–252. doi: 10.1111/j.1399-3054.1982.tb00255.x. [DOI] [Google Scholar]
- 44.Wang S. Methyl jasmonate reduces water stress in strawberry. Journal of plant growth regulation. 1999;18:127–134. doi: 10.1007/PL00007060. [DOI] [PubMed] [Google Scholar]
- 45.Bandurska H, Stroiński A, Kubiś J. The effect of jasmonic acid on the accumulation of ABA, proline and spermidine and its influence on membrane injury under water deficit in two barley genotypes. Acta physiologiae plantarum. 2003;25:279–285. doi: 10.1007/s11738-003-0009-0. [DOI] [Google Scholar]
- 46.Qiu Z, Guo J, Zhu A, Zhang L, Zhang M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicology and environmental safety. 2014;104:202–208. doi: 10.1016/j.ecoenv.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Pedranzani H, et al. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regulation. 2003;41:149–158. doi: 10.1023/A:1027311319940. [DOI] [Google Scholar]
- 48.Kang DJ, et al. Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt‐tolerant and salt‐sensitive rice cultivars. Journal of Agronomy and Crop Science. 2005;191:273–282. doi: 10.1111/j.1439-037X.2005.00153.x. [DOI] [Google Scholar]
- 49.Nishiyama R, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. The Plant Cell. 2011;23:2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dombrecht B, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huot B, Yao J, Montgomery BL, He SY. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 53.Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants ofArabidopsisare susceptible to the soil fungusPythium irregulare. The Plant Journal. 1998;15:747–754. doi: 10.1046/j.1365-313X.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 54.Jung C, et al. Plant U-box protein10 regulates MYC2 stability in Arabidopsis. The Plant Cell. 2015;27:2016–2031. doi: 10.1105/tpc.15.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verslues PE, Bray EA. LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiology. 2004;136:2831–2842. doi: 10.1104/pp.104.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang G, Dolan L. Auxin promotes the transition from chloronema to caulonema in moss protonema by positively regulating PpRSL1and PpRSL2 in Physcomitrella patens. New Phytologist. 2011;192:319–327. doi: 10.1111/j.1469-8137.2011.03805.x. [DOI] [PubMed] [Google Scholar]
- 57.Jang G, et al. Volatile methyl jasmonate is a transmissible form of jasmonate and its biosynthesis is involved in systemic jasmonate response in wounding. Plant biotechnology reports. 2014;8:409–419. doi: 10.1007/s11816-014-0331-6. [DOI] [Google Scholar]
- 58.Takechi K, Sakamoto W, Katsuhara M, Murata M, Motoyoshi F. In situ RNA hybridization using Technovit resin in Arabidopsis thaliana. Plant Molecular Biology Reporter. 1999;17:43–51. doi: 10.1023/A:1007503912661. [DOI] [Google Scholar]
- 59.Gendrel A-V, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.