Abstract
Archaeal swimming motility is driven by archaella: rotary motors attached to long extracellular filaments. The structure of these motors, and particularly how they are anchored in the absence of a peptidoglycan cell wall, is unknown. Here, we use electron cryotomography to visualize the archaellar basal body in vivo in Thermococcus kodakaraensis KOD1. Compared to the homologous bacterial type IV pilus (T4P), we observe structural similarities as well as several unique features. While the position of the cytoplasmic ATPase appears conserved, it is not braced by linkages that extend upward through the cell envelope as in the T4P, but rather by cytoplasmic components that attach it to a large conical frustum up to 500 nm in diameter at its base. In addition to anchoring the lophotrichous bundle of archaella, the conical frustum associates with chemosensory arrays and ribosome‐excluding material and may function as a polar organizing center for the coccoid cells.
Keywords: archaeal motility, archaella, electron cryotomography, flagella, Thermococcus kodakarensis
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Introduction
Many single‐celled organisms are motile by means of extracellular appendages. In archaea, swimming motility is driven by archaella: rotary motors attached to long extracellular fibers. Archaella are functionally analogous to bacterial flagella, but evolutionarily homologous to the type IV pilus (T4P) and type II secretion system (T2SS) machineries of bacteria 1. Recently, an atomic structure of the archaellum fiber purified from the euryarchaeon Methanospirillum hungatei revealed differences compared to the bacterial T4P fiber, including lack of a central pore and more extensive inter‐subunit interactions 2. The structure of the archaellar basal body, and its similarity to the T4P basal body, remains unknown.
Unlike T4P fibers that only assemble and disassemble, archaella assemble and then can rotate in both directions to either push or pull the cell 3, 4. Light microscopy of Halobacterium salinarum revealed discrete steps during rotation, likely corresponding to ATP hydrolysis events by the basal body ATPase, FlaI 5. While the bacterial T4P contains two distinct ATPases for assembly and disassembly of the pilus fiber, the single ATPase FlaI drives both assembly and rotation of the archaellum 6. The N‐terminal domain of the archaellum/T2SS/T4P superfamily ATPases is the most variable, and the first 29 residues of FlaI, located on the outer edge of the hexamer, were found to be essential for motility but not assembly, although the basis of this functional separation remains unclear 6.
FlaI is predicted to interact with the integral membrane protein FlaJ 7. Structural studies of the bacterial T4P suggest that ATPase‐driven rotation of the FlaJ homolog, PilC, incorporates pilin subunits from the membrane into the growing fiber 8. This is possible because the ATPase itself is clamped in an integrated structure that spans the bacterial inner and outer membranes and periplasm and anchors on the cell‐encompassing peptidoglycan cell wall 8. A similar cell‐wall attached structure anchors the bacterial flagellar basal body 9. Without knowing the structure of the archaellar basal body, it is unclear how similar anchoring could occur in the envelope of archaea, which consists of a single membrane and a proteinaceous surface (S‐) layer. It was recently proposed that FlaF might anchor the archaellum through interactions with the S‐layer 10. Others have suggested that a cytoplasmic structure mechanically stabilizes the basal body 3. Supporting this idea, cytoplasmic structures underlying the archaella have been observed by traditional electron microscopy (EM) of Halobacteria 11, 12.
Electron cryotomography (ECT) can image intact cells in a frozen, fully hydrated state, providing macromolecular resolution (~4–6 nm) details about native cellular structures 13. Here, we used ECT to visualize the structure of the archaellar basal body in vivo in Thermococcus kodakaraensis cells. T. kodakaraensis (originally designated Pyrococcus sp. strain KOD1 and later identified as belonging to the Thermococcus genus 14; also known as T. kodakarensis) is one of the best‐studied archaeal species. It was isolated from a Japanese solfatara in 1994 15, and has been a useful source of thermostable enzymes (e.g., high‐fidelity DNA polymerase for PCR 16). In addition to revealing the overall structure of the archaellar basal body in vivo, we discovered a novel cytoplasmic conical structure in T. kodakaraensis associated with archaellar motility and potentially other polar organizing activities.
Results
Electron cryotomography of the T. kodakaraensis archaellum and associated structures
We imaged T. kodakaraensis KOD1 cells by ECT in a native, frozen‐hydrated state. Many cells appeared to be lysed prior to plunge‐freezing for ECT, but out of 18 apparently intact cells, we observed a lophotrichous bundle of archaella in 13. Each bundle contained between four and 14 archaella. Due to the size of T. kodakaraensis cells (cells are irregular cocci ~1.5 μm in diameter), only a portion of the cell was visible in the limited field of view of our high‐magnification cryotomograms. Given this limited field of view, we think it likely that all cells contained an archaellar bundle, but it was not visible in the remaining five cells. In addition, we observed well‐preserved archaellar bundles in eight apparently lysed cells.
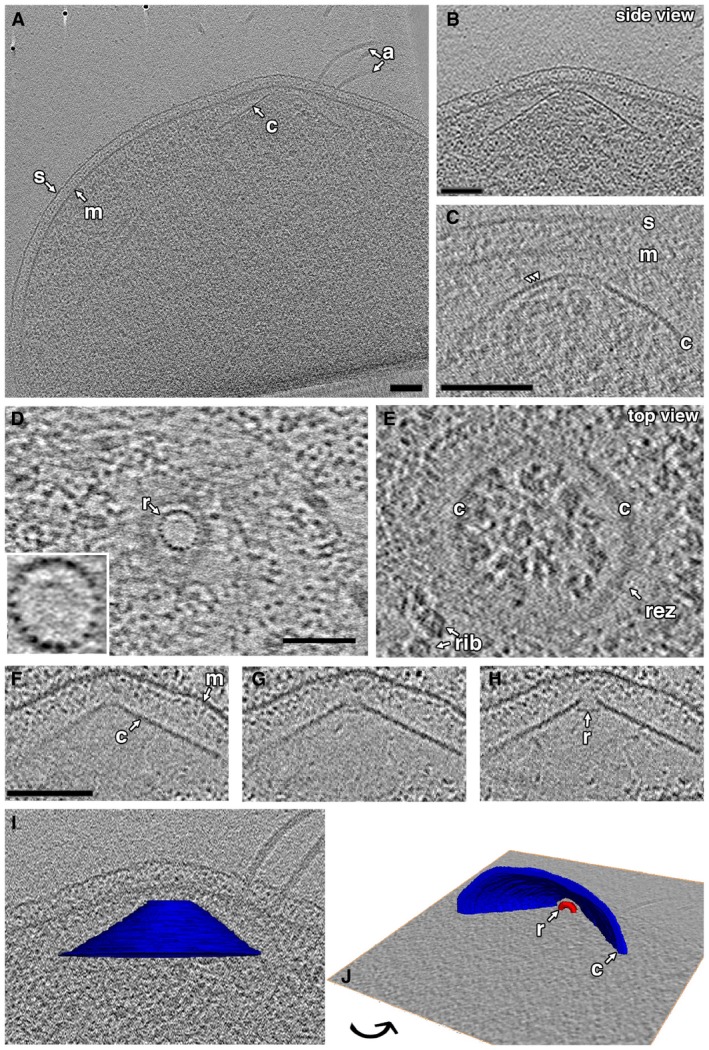
We consistently observed a prominent conical structure associated with the archaellar bundle in the cytoplasm (Fig 1). In intact cells, all conical structures were associated with archaella. In some lysed cells, however, we observed cones without archaella, suggesting that the conical structure remains intact upon loss of archaella. The conical structure showed a consistent morphology and localization inside the cell: closely associated with, but not touching, the cytoplasmic membrane at its narrow end and expanding a variable length to a wide base, which varied from 220 to 525 nm in diameter. The central axis (an imaginary line through the center of the conical frustum) was perpendicular to the membrane, as seen in cross‐sectional side views (Fig 1A–C, additional examples in Fig EV1). The edges, seen in cross‐section, frequently exhibited periodic densities suggestive of individual protein subunits (Fig 1C), with a thickness of 3–4 nm. We observed that while cones in different cells had different heights, the opposite side walls of each cone were symmetric (had similar lengths). Tomographic slices capturing the central axis of the conical structure in side view showed an angle of 109 ± 6° (mean ± s.d., n = 5) between opposite edges. The structures were not complete cones but rather conical frusta: They did not taper fully to a point, but exhibited a blunt tip. In top views, we observed a ring situated in the throat of the frustum, just below the tip (Fig 1D and E). These rings comprised 19 subunits (Fig 1D inset), each again 3–4 nm thick, with an overall ring diameter of 31 ± 2 nm (mean ± s.d., n = 10). The position of the ring in the conical frustum was clearest in tomograms of lysed cells, which were thinner and contained less cytoplasmic material (Fig 1F–H). Even in such tomograms, however, we could not visualize a well‐defined connection between the two portions of the structure, so it is unclear whether and how the components are connected.
Figure 1. Cytoplasmic conical structures in Thermococcus kodakaraensis .
-
A, BA tomographic slice shows a side view of a conical structure (c) in the cytoplasm, rotated and enlarged in (B).
-
CA tomographic slice shows a side view of the cone in another cell, highlighting the subunit texture along the edge of the cone (arrowheads).
-
D, ETop views of a cone at different heights show the inner ring (r; enlarged in inset to highlight 19‐subunit structure) and outer cone.
-
F–HSequential slices through a side view of a cone in a lysed cell show the relative location of the ring in the cone.
-
I, JDifferent views of a 3D segmentation of the cone shown in (A), embedded in a tomographic slice.
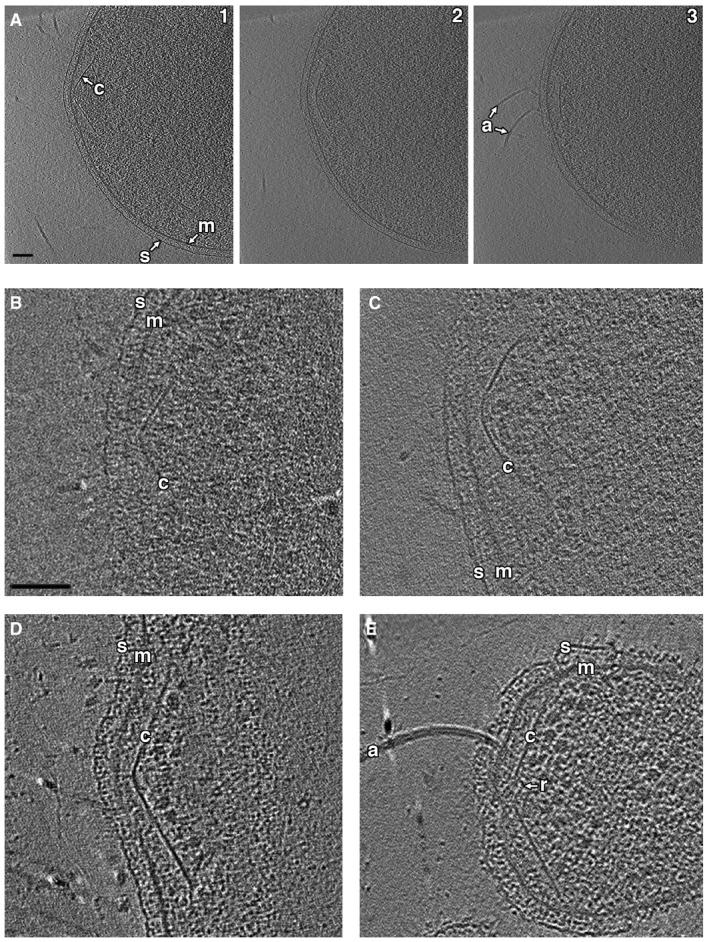
Figure EV1. Additional examples of conical structures in Thermococcus kodakaraensis cells.
-
ASequential tomographic slices (1–3) at different heights through a side view of the cone.
-
B–EAdditional examples of cones in intact (B and C) and lysed (D and E) cells are shown.
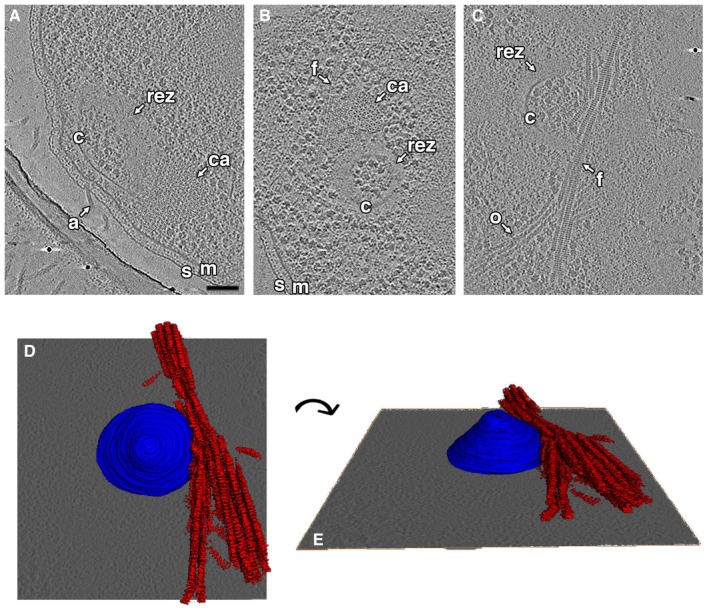
Conical structures were surrounded by an ~30‐ to 45‐nm‐wide ribosome‐excluding zone (REZ; Figs 1E and 2). In nearly all cells, both intact and lysed, we observed filament bundles near or associated with this REZ (Fig 2B–E). The bundles were more extensive in lysed cells. Each filament was ~12 nm wide and made up of a series of disk‐like densities spaced ~7 nm apart. Chemosensory arrays were also consistently observed near the conical structures (Fig 2A and B). In one cell, we observed two attached conical structures, each associated with archaella and each ~250 nm in diameter at its base (Fig EV2).
Figure 2. Cones are associated with chemosensory arrays, ribosome‐excluding zones, and filament bundles.
-
A–CTomographic slices show side (A) and top (B, C) views of cones (c) in three cells, highlighting associated chemosensory arrays (ca), ribosome‐excluding zones (rez), and filament bundles (f). For a slice‐by‐slice view through the tomogram shown in (A), see Movie EV1. s, S‐layer; m, membrane; a, archaellum; o, other filaments. Scale bar, 100 nm (applies to A–C); segmentation not to scale.
-
D, EDifferent views of a 3D segmentation of the structures shown in (C), with the conical structure in blue and the filament bundle in red.
Figure EV2. Double‐cone structure observed in Thermococcus kodakaraensis .

A tomographic slice through a side view shows two associated conical structures (c1 and c2), both associated with archaella (a). s, S‐layer; m, membrane. Scale bar, 100 nm.
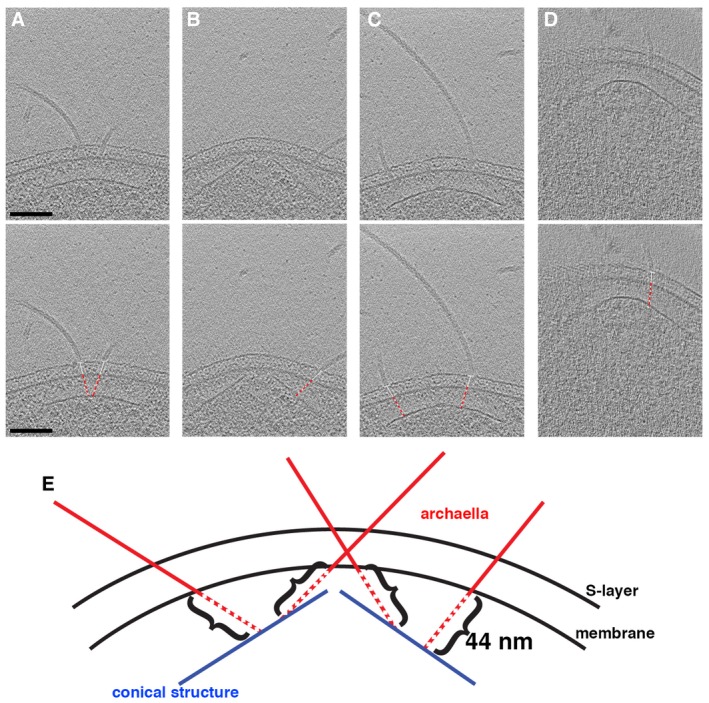
To characterize the interaction between the archaellar bundle and the conical structure, we measured the distance from the base of each archaellum in the membrane to the cone. The orientation of the measurement line was determined by the orientation of the archaellar filament between the S‐layer and the membrane (Fig 3A–D). Due to the geometric shape of the cone, the distance between it and the membrane varies—shortest at the tip of the cone and longest at the base. Since archaella were located at various radial positions along the cone, we expected their distance to vary similarly. Interestingly, however, we measured a much more consistent distance of 44 ± 5 nm (mean ± s.d., n = 29) from the cone to the base of each archaellum in the membrane (Fig 3). Consistent with this, we observed a variety of orientations of archaella in the cell envelope, frequently not perpendicular to the S‐layer, allowing the conserved distance to the cone (Figs 3 and EV3). In a few cases, we observed continuous densities connecting the archaella and the cone (Fig EV3E).
Figure 3. Archaellum orientation with respect to the cell envelope.
-
A–DTomographic slices through side views of cones. In the bottom row, white lines show the angle of the archaellum with respect to the surface layer, and red dashed lines show the conserved distance from the archaellum at the membrane to the cone. Scale bar, 100 nm (applies to A–D).
-
ESchematic depicting the 44 nm distance from the cone to the basal body in the membrane for archaella at different radial positions along the cone. Since different radial positions on the cone are located at different distances from the membrane (shorter at the tip and longer at the base), this results in a range of archaellar orientations in the cell envelope.
Figure EV3. Association of cones with the archaellar bundle.
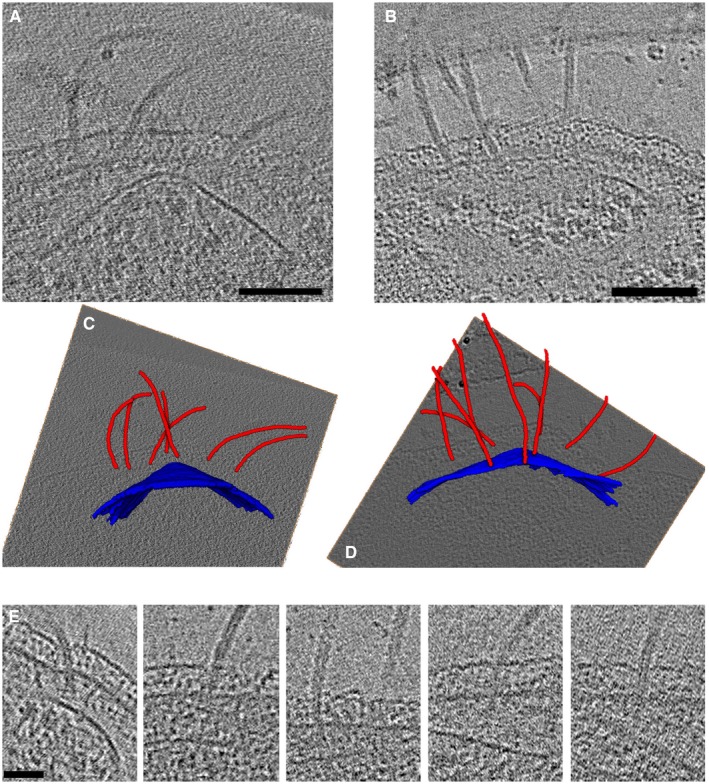
-
A, BTomographic slices through two cells, highlighting the association between the cone and the archaella.
-
C, D3D segmentations of the cells in (A) and (B), respectively, with cones in blue and archaella in red, embedded in tomographic slices.
-
ETomographic slices of individual archaella show the varying orientations of archaella with respect to the cell envelope, as well as apparent connections to the cone.
To determine the structure of the archaellar basal body, we calculated a subtomogram average (Fig 4). Thirty particles were used, and an axial twofold symmetry was applied. The resulting average revealed several layers of density extending into the cytoplasm. Immediately adjacent to the membrane‐embedded density was a ring‐like structure (L1 in Fig 4A). Below the ring was a disk of similar diameter (L2), followed by a larger diameter component (L3) and finally, at a greater distance, a less well‐defined density. This density was 44 nm away from the membrane, corresponding to the cone. Consistent with our observation that archaella exhibited various orientations with respect to the S‐layer, we did not observe a strong density corresponding to the S‐layer in the average. As seen in individual particles, the component in L3 does not appear to be a ring, but rather comprises distinct legs, seen on one or both sides, that appear symmetric in the average (Fig EV4). Similarly, the density of the cone is more prominent in individual particles; due to the different angles of the structure in different particles, the density becomes indiscernible in the average (Fig EV4).
Figure 4. Structure of the Thermococcus kodakaraensis archaellum.
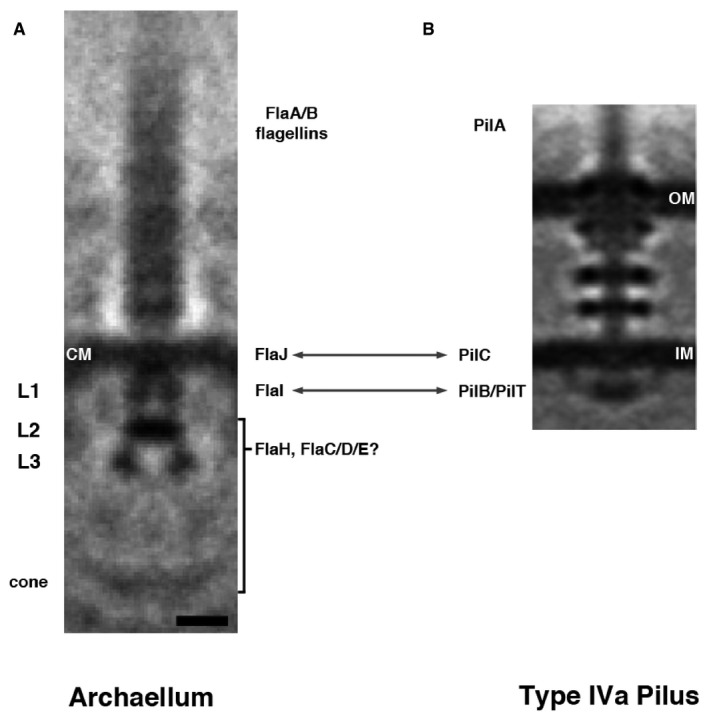
- A subtomogram average of the archaellum reveals structural features, including four layers of density in the cytoplasm (L1–L3, cone). CM, cytoplasmic membrane. The speculated identity of densities in the archaellum is proposed: archaellum fiber = FlaA/B flagellins; integral membrane density = FlaJ; L1 = FlaI; L2/L3/cone = FlaH/FlaC/D/E.
- For comparison, a subtomogram average of the type IVa pilus machine from Myxococcus xanthus is shown (adapted with permission from 8). Arrows indicate components with recognized homology. OM, outer membrane; IM, inner membrane.
Figure EV4. Individual particles from the subtomogram average show heterogeneity in the L3 density and angle of cone density.
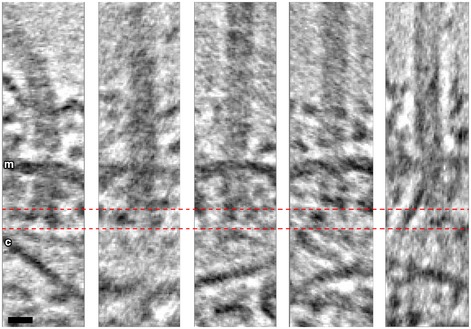
The L3 density appears as either two dots of similar (first two panels) or different intensity (third panel), a single dot (fourth panel), or a dot and an extended line (fifth panel). m, membrane; c, conical structure. Scale bar, 10 nm (applies to all panels).
Protein composition of the T. kodakaraensis archaellum
Inspection of the T. kodakaraensis genome 17 revealed the presence of a typical archaellar 13‐gene cluster (TK0038–TK0050) containing five paralogous flaB archaellin genes plus other genes needed for archaellar biogenesis and basal body synthesis, flaCDFGHIJ. To establish which of the FlaB archaellin proteins compose the archaellar filament, we performed a quantitative proteomic survey of T. kodakaraensis cells (Materials and Methods). All five of the FlaB paralogs were detected, with the most abundant being FlaB1, FlaB2, and FlaB3 at 0.4–0.25% of total cell protein (Table 1), and the FlaB4 and FlaB5 paralogs fivefold lower in abundance. Therefore, the T. kodakaraensis flagellar filament is heterogeneous in composition. The FlaHI basal body proteins that compose the motor, as well as the FlaCD proteins of unknown function, were also detected but at lower abundance than the FlaB archaellin proteins.
Table 1.
Abundance of the Thermococcus kodakaraensis KOD1 archaellar and chemotaxis proteins
| Accessiona | Gene | Protein annotation | Sizea | % Ab | % B | Average, % |
|---|---|---|---|---|---|---|
| TK0038 | flaB1 | Archaeal flagellin B1 precursor | 294 | 0.40 | 0.43 | 0.41 |
| TK0039 | flaB2 | Archaeal flagellin B2 precursor | 580 | 0.26 | 0.27 | 0.27 |
| TK0040 | flaB3 | Archaeal flagellin B3 precursor | 258 | 0.24 | 0.24 | 0.24 |
| TK0041 | flaB4 | Archaeal flagellin B4 precursor | 218 | nd | 0.05 | 0.03 |
| TK0042 | flaB5 | Archaeal flagellin B5 precursor | 274 | 0.05 | 0.05 | 0.05 |
| TK0043 | flaC | Archaeal flagella‐related protein C | 158 | 0.03 | 0.03 | 0.03 |
| TK0044 | flaD | Archaeal flagella‐related protein D | 474 | 0.11 | 0.11 | 0.11 |
| TK0045 | flaF | Archaeal flagella‐related protein F | 174 | nd | nd | |
| TK0046 | flaG | Archaeal flagella‐related protein G | 161 | nd | nd | |
| TK0047 | flaH | Archaeal flagella‐related protein H | 232 | nd | 0.06 | 0.03 |
| TK0048 | flaI | Archaeal flagella‐related protein I | 540 | 0.02 | 0.02 | 0.02 |
| TK0049 | flaJ | Archaeal flagella‐related protein J | 579 | nd | nd | nd |
| TK0050 | hyp | Hypothetical protein | 664 | 0.01 | nd | nd |
| TK0629 | cheW | Chemotaxis signal transduction | 145 | 0.12 | 0.11 | 0.12 |
| TK0630 | mcp1 | Methyl‐accepting chemotaxis | 438 | 0.11 | 0.12 | 0.11 |
| TK0631 | cheR | Chemotaxis methyltransferase | 279 | nd | nd | nd |
| TK0632 | cheY | Chemotaxis response regulator | 120 | 0.06 | 0.05 | 0.06 |
| TK0633 | cheB | Chemotaxis methylesterase | 355 | 0.02 | 0.02 | 0.02 |
| TK0634n | cheA | Chemotaxis histidine kinase | 548 | nd | nd | |
| TK0636 | cheC | Chemotaxis inhibitor protein | 207 | nd | nd | |
| TK0637 | cheC | Chemotaxis protein cheC | 207 | nd | nd | |
| TK0638 | mcp2 | Methyl‐accepting chemotaxis | 747 | 0.02 | 0.02 | 0.02 |
| TK0639 | cheD | Chemotaxis protein cheD | 161 | nd | nd | |
| TK0640 | hyp | Hypothetical protein | 251 | nd | nd | |
| TK0641 | cheF | Hypothetical protein | 341 | nd | nd | |
| TK0156 | mcp3 | Methyl‐accepting chemotaxis | 742 | 0.05 | 0.05 | 0.05 |
| TK1606 | mcp4 | Methyl‐accepting chemotaxis | 251 | 0.02 | 0.01 | 0.02 |
| TK2147 | mcp5 | Methyl‐accepting chemotaxis | 769 | 0.01 | 0.02 | 0.02 |
nd, not detected.
Accession numbers, gene names, annotations, and protein size in number of amino acids are from the IMG‐JGI database (Materials and Methods).
Protein values are expressed as percent of total cell protein detected for replicates A and B in addition to the average value.
Source data are available online for this table.
Our proteomic survey also allowed us to investigate the presence and abundance of chemotaxis‐related proteins in T. kodakaraensis (Table 1). These proteins are encoded by one 12‐gene che‐mcp cluster plus three unlinked mcp genes. Of the five methyl‐accepting chemotaxis proteins (MCPs), Mcp1 was the highest in abundance, but all were expressed. These chemoreceptors are candidates for the T. kodakaraensis chemosensory arrays previously observed by ECT 18. No candidates for the protein(s) composing the conical structure could be identified based on the presence of nearby genes with related or hypothetical annotations in either the fla or che‐mcp gene clusters.
Discussion
Structure of the basal body of the T. kodakaraensis archaellum
Here, we describe the morphology of the archaellar basal body in T. kodakaraensis (Fig 4). We think it is almost certain that density L1 in the T. kodakaraensis basal body corresponds to the ATPase, FlaI, since its size and shape match those of the homologous ATPases in the T4P. Also, like the T4P ATPases, FlaI is predicted to interact directly with an integral membrane protein 6, 7. More specifically, FlaI shares domain homology with the assembly/disassembly ATPases, PilB and PilT, of the bacterial T4P. The size of L1 is comparable to that of the PilB/PilT ring in the bacterial T4P (Fig 4B), consistent with their conserved hexameric oligomerization 7 and similar sizes of the protein monomers (540 amino acids for FlaI and 566 for PilB). FlaI is predicted to interact directly with the polytopic integral membrane protein FlaJ 6, 7. FlaJ shares sequence homology with the ATPase‐interacting inner membrane protein PilC of the T4P 19. The relative locations of these components are therefore predicted to be the same in the basal bodies of the archaellum and the bacterial T4P (Fig 4) 8, 20, and the size and shape and position of density L1 seen here support that expectation, and the corollary that these two systems likely share a similar assembly mechanism.
The identities of the proteins making up L2, L3, and the cone remain unclear. In the T4P, no structures were observed in the cytoplasm below the ATPase 8, 20, 21. In Crenarchaeota, only one accessory component is not membrane‐bound (FlaH). In Euryarchaeota like T. kodakaraensis, however, additional soluble proteins, FlaC/D/E, are thought to be components of the archaellum that receive switching signals from the chemotaxis machinery 22. All of these proteins, and potentially others, are candidates for the densities we observed. It will therefore be of great interest to obtain a structure of the crenarchaeal basal body, which lacks FlaC/D/E, for comparison, and/or to dissect the T. kodakaraensis basal body structure through analysis of deletion mutants.
Conical structures anchor T. kodakaraensis archaella
We observed that T. kodakaraensis archaella associate with a large conical structure in the cytoplasm. In a few cases, we observed direct connections between archaella and cone. The fact that we did not see such a connection for every archaellum may simply reflect variations in image clarity and orientation of the structures between cells in different cryotomograms. The conserved distance from the cone to the archaellar basal body in the membrane suggests a rigid interaction. It is an interesting question how archaella are attached to the cone. We did not observe strong densities connecting L3 and the cone in the averaged basal body structure, but in individual particles we observed heterogeneity. Also, the resolution of the average may be too low to detect such connections. If, for example, the links are thin (such as coiled‐coils), they would not be resolved; similar coiled‐coil linkages in the bacterial flagellar basal body between FlaH and the C‐ring were not resolved even in higher‐resolution subtomogram averages 23.
While it is possible that the conical structure and the archaellar bundle are independent (though colocalizing) structures, we propose that the T. kodakaraensis cone anchors the archaellar basal body in part to provide leverage for rotation. In the bacterial T4P, the ATPase is clamped by extensive interactions up through the cell envelope that anchor it to the peptidoglycan cell wall 8 (Fig 5B). Signals governing disassembly are thought to be processed by sensory elements in the periplasm 8. In the absence of a peptidoglycan cell wall and outer membrane, the T. kodakaraensis archaellum appears to turn the system upside down, with components stacking nearly 50 nm into the cytoplasm to anchor onto a large cone (Fig 5A). Signals governing rotation and direction are likely integrated by sensory components in the cytoplasm. Even thin linkages between the basal body and cone could provide leverage, as long as there were multiple linkages (as is likely due to the multimeric nature of the basal body). While each archaellar filament is likely more massive than the cone and might therefore induce it to rotate, the presence of multiple archaella (and additional associated structures, as described below) would allow the cone to leverage them against one another, providing a massive anchor.
Figure 5. Schematic comparing organization of the related archaellum and type IVa pilus basal bodies.
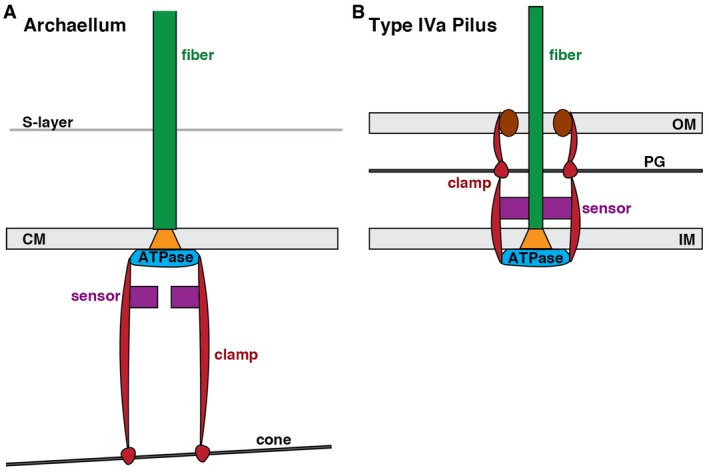
-
A, BIn the bacterial T4P (B), an integrated system of components spanning the outer and inner membranes (OM, IM) uses the peptidoglycan cell wall (PG) to clamp the ATPase, allowing rotation of PilC (orange) in the membrane to assemble the pilus fiber. In the Thermococcus kodakaraensis archaellum (A), our results suggest that an integrated system of components extends from the single membrane (CM) inward to a large conical structure in the cytoplasm to similarly brace the ATPase. Sensory components (purple) are proposed to be located in the periplasm for the T4P (integrating signals for disassembly) and the cytoplasm for the archaellum (integrating signals for rotation).
Similar leveraging structures may exist in other archaea. More than 20 years ago, it was observed that when archaella are dissociated from lophotrichous H. salinarum cells by detergent, the bundles remain intact, connected to a large (~500 nm diameter) structure 11. A similar structure was also observed below the cell membrane in cell ghosts 12. More recently, a spherical structure was observed anchoring Iho670 fibers, T4P‐like filaments in Ignicoccus hospitalis. This structure is thought to be located in the cytoplasm of the cell and contains a central ring of similar dimensions to the one observed here 24. It is possible that either or both of these structures are related to the T. kodakaraensis cone. Large cytoplasmic structures have not been described in other motile archaeal species to date, however, so it will be interesting to determine how archaella may be anchored in those systems.
It will also be of great interest to identify the proteins that form the T. kodakaraensis cone and associated ring. These subunits must be capable of interacting both circumferentially around the cone and radially with subunits making up the next (larger or smaller diameter) ring. While it is possible that the conical structure is an assembly of stacked rings, we think it more likely that the subunits assemble into a filament spiral, similar to what has been proposed for ESCRT‐III polymers 25, 26. Interestingly, an architecturally similar spiral has been observed in the basal body of the bacterial flagellar motor: In Wolinella succinogenes, an Archimedean spiral forms a bushing for the motor in the periplasm, allowing the flagellum to rotate in the cell wall. This spiral is formed by protein subunits interacting both circumferentially and laterally through nonspecific interactions 27. While that spiral takes the form of a disk, similar protein interactions may give rise to a cone in T. kodakaraensis.
Thermococcus kodakaraensis cones are potential polar organizing structures
In addition to a potential role in rigidly anchoring the basal body of the archaellum, the T. kodakaraensis cone may function to gather the archaellar bundle to maximize efficiency, either by concentrating molecules for assembly or signaling, or by concentrating force at one point on the coccoid cell for directional swimming. The cone's structure may also help distribute the force from archaellar rotation to the larger bulk of the cell's contents. This might be more efficient in a pushing than a pulling mode; swimming speed in another euryarchaeon, H. salinarum, was found to be approximately twice as fast when the archaella push as when they pull the cell body 5. The structurally similar spiral basal disk in the bacterial flagellar basal body of W. succinogenes was suggested to play a role in dispersing lateral forces created by flagellar rotation 27.
Our results suggest a further role for the cone in breaking the symmetry of the coccoid cell. In many rod‐shaped bacterial cells, proteins and other macromolecules are specifically localized to the cell pole for various purposes ranging from cell motility and adhesion to differentiation and division 28. One well‐studied example of this polar organization occurs in Caulobacter crescentus, where the oligomeric protein PopZ defines an asymmetric pole, localizing many cytoplasmic proteins and tethering the chromosomal centromere to facilitate division 29, 30, 31. In Vibrio cholerae, the HubP protein organizes the polar localization of the chromosomal origin, chemotaxis machinery, and flagella 32. Perhaps the T. kodakaraensis cone similarly defines a pole in the spherical cells, anchoring the chemotaxis and motility machinery. An intriguing feature observed in our cryotomograms is the cone‐associated REZ. In bacterial cells, such REZs are commonly interpreted to be the nucleoid 33, 34. Supporting this assignment, we observed bundles of filaments (most extensive in lysed cells) associated with the REZ (Fig 2). Such filaments are reminiscent of nucleoprotein filaments formed by various bacterial DNA‐binding proteins in stress conditions 35, 36, 37.
A spatial organizer analogous to PopZ may be especially important for a polyploid species like T. kodakaraensis (chromosome copy number varies depending on growth phase, from 7 to 19 copies 38). Fluorescence imaging suggests that the nucleoid is relatively compact in log phase growth, and nucleoids appear to separate before the cells are deeply constricted 39. Perhaps the cones segregate attached structures, including the archaella and possibly chromosomes. This function is consistent with the duplicated cone structure we observed in one cell (Fig EV2), which could represent an intermediate after replication and prior to segregation, or may simply represent an aberrant structure. Further studies imaging cells throughout the cell cycle could shed light on whether, and how, cones function to coordinate archaellar and chromosomal segregation.
Understanding the prevalence of this structure among Euryarchaeota and across different archaeal kingdoms may illuminate its function. If it is restricted to lophotrichous species, it may simply be an anchoring mechanism for the archaella in the absence of a peptidoglycan cell wall. In that case, monotrichous or peritrichous species may exhibit a less extensive plate underneath the basal bodies of individual archaella. If it is more widely found in coccoid, and/or highly polyploid cells, it may serve an added role in polar specification.
In a very recent publication, Daum et al 40 report the structure and organization of the archaellum machinery of another Euryarchaeon, Pyrococcus furiosus. The authors observe a very similar basal body core structure and assign similar densities to FlaJ and FlaI. They further assign FlaH to a cytoplasmic ring interacting with FlaI. They observe an additional density forming a wide cytoplasmic ring surrounding the inner core, which we did not resolve. The positions of FlaC/D/E remain unclear, and finding each component in diverse motor structures will be a focus of further studies.
Similarly to our findings in T. kodakaraensis, Daum et al 40 also show that the archaella in P. furiosus co‐localize with a “polar cap”, a cytoplasmic structure similar to the cone described here. This finding, along with previous observations (see above), suggests that this archaellar anchoring mechanism may be a widespread feature of Euryarchaeota.
Materials and Methods
Growth
Thermococcus kodakaraensis strain KOD1 [JCM 12380] was grown anaerobically in MA‐YT medium supplemented with elemental sulfur as previously described 14, 18.
Protein extraction and digestion
Cells were lysed and digested by the eFASP method 41. Tryptic peptides were pre‐fractionated using strong cation exchange with StageTips 42, with a modification in the peptide elution steps. Peptides were eluted into nine instead of four fractions, using ammonium acetate solution diluted in water containing 0.5% acetic acid, 30% acetonitrile at the following final concentrations: 17.5, 20, 35, 50, 65, 80, 100, 300, and 500 mM. Fractions were analyzed by liquid chromatography–tandem mass spectrometry (LC‐MS/MS) on an EASY nLC1000 (Thermo Scientific) coupled to a hybrid mass spectrometer (Q‐Exactive, Thermo Scientific). Peptides were desalted on an Acclaim PepMap100 C18 nano‐trap column (Dionex, 75 μm × 2 cm, Product # 164535) and separated on an Acclaim PepMap RSLC analytical column (Dionex, 75 μm × 25 cm, Product # ES802).
Database search
Mass spectral data were searched with Mascot version 2.4.1 (Matrix Science, Boston, MA, USA) through Thermo Proteome Discoverer version 1.4.0.288 (Thermo Scientific). All of the sample fractions were combined in a single search against the Thermococcus kodakaraensis protein database downloaded from https://img.jgi.doe.gov/ (version 05/15/2015) concatenated with a reverse decoy database and protein sequences of common contaminants. Carbamidomethylation of cysteine was specified as a fixed modification. Oxidation of methionine was specified as a variable modification. Trypsin/P was specified as the cleavage enzyme, and the search allowed for up to two missed cleavages. The precursor mass error was set to ±10 ppm and the product mass error to ±0.02 Da. Target false discovery rate (FDR) was specified to be 0.01 (strict) and 0.05 (relaxed).
Electron cryotomography and image analysis
Samples of cell cultures in growth media were mixed with bovine serum albumin‐treated colloidal gold fiducial markers (Sigma) and applied to Quantifoil R2/2 200 copper EM girds (Quantifoil Micro Tools). After blotting excess liquid, grids were plunge‐frozen in a mixture of liquid ethane and propane 43 and subsequently kept at liquid nitrogen temperature. Images were acquired using either an FEI Polara G2 or Titan Krios 300 keV transmission electron microscope (FEI Company) equipped with a field emission gun, image corrector for lens aberration, energy filter (Gatan), and K2 Summit direct electron detector (Gatan). Cumulative electron dose was 160 e−/Å2 or less for each tilt series. Tilt series were acquired using UCSF Tomography software 44. Images were contrast‐transfer‐function‐corrected, aligned, and reconstructed by weighted back projection with the IMOD software package 45. SIRT (Simultaneous Iterative Reconstruction Technique) reconstructions were calculated with TOMO3D 46, subtomogram averages generated using PEET 47, and segmentations generated with Amira software (FEI Company).
Accession codes
The subtomogram average of the T. kodakaraensis archaellar basal body was deposited into the Electron Microscopy Data Bank (entry number EMD‐8603).
Author contributions
AB and GJJ conceived the experiments; AB, ANH, KWK, DG, HHN, DK and RROL conducted the experiments; AB, CMO, YWC, AK, ANH, HHN, DK, RROL and RPG performed analysis; CMO and GJJ wrote the manuscript; all authors revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Movie EV1
Review Process File
Source Data for Table 1
Acknowledgements
This work was supported by the NIH (grant R01AI127401 to G.J.J.) and the UCLA‐DOE Institute (grant DE‐FC03‐02ER26342 to R.P.G.).
EMBO Reports (2017) 18: 1660–1670
References
- 1. Pohlschroder M, Ghosh A, Tripepi M, Albers SV (2011) Archaeal type IV pilus‐like structures–evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol 14: 357–363 [DOI] [PubMed] [Google Scholar]
- 2. Poweleit N, Ge P, Nguyen HH, Loo RR, Gunsalus RP, Zhou ZH (2016) CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat Microbiol 2: 16222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alam M, Oesterhelt D (1984) Morphology, function and isolation of halobacterial flagella. J Mol Biol 176: 459–475 [DOI] [PubMed] [Google Scholar]
- 4. Shahapure R, Driessen RP, Haurat MF, Albers SV, Dame RT (2014) The archaellum: a rotating type IV pilus. Mol Microbiol 91: 716–723 [DOI] [PubMed] [Google Scholar]
- 5. Kinosita Y, Uchida N, Nakane D, Nishizaka T (2016) Direct observation of rotation and steps of the archaellum in the swimming halophilic archaeon Halobacterium salinarum . Nat Microbiol 1: 16148 [DOI] [PubMed] [Google Scholar]
- 6. Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche AL, Albers SV, Tainer JA (2013) Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol Cell 49: 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh A, Albers SV (2011) Assembly and function of the archaeal flagellum. Biochem Soc Trans 39: 64–69 [DOI] [PubMed] [Google Scholar]
- 8. Chang YW, Rettberg LA, Treuner‐Lange A, Iwasa J, Sogaard‐Andersen L, Jensen GJ (2016) Architecture of the type IVa pilus machine. Science 351: aad2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murphy GE, Leadbetter JR, Jensen GJ (2006) In situ structure of the complete Treponema primitia flagellar motor. Nature 442: 1062–1064 [DOI] [PubMed] [Google Scholar]
- 10. Banerjee A, Tsai CL, Chaudhury P, Tripp P, Arvai AS, Ishida JP, Tainer JA, Albers SV (2015) FlaF Is a beta‐sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S‐layer protein. Structure 23: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kupper J, Marwan W, Typke D, Grunberg H, Uwer U, Gluch M, Oesterhelt D (1994) The flagellar bundle of Halobacterium salinarium is inserted into a distinct polar cap structure. J Bacteriol 176: 5184–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metlina AL (2004) Bacterial and archaeal flagella as prokaryotic motility organelles. Biochemistry (Mosc) 69: 1203–1212 [DOI] [PubMed] [Google Scholar]
- 13. Oikonomou CM, Chang YW, Jensen GJ (2016) A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol 14: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T (2004) Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1: 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T (1994) Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol 60: 4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y (2001) Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J Mol Biol 306: 469–477 [DOI] [PubMed] [Google Scholar]
- 17. Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T (2005) Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15: 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Briegel A, Ortega DR, Huang A, Oikonomou C, Gunsalus RP, Jensen GJ (2015) Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ Microbiol Rep 7: 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peabody CR, Chung YJ, Yen MR, Vidal‐Ingigliardi D, Pugsley AP, Saier MH Jr (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149: 3051–3072 [DOI] [PubMed] [Google Scholar]
- 20. Chang YW, Kjaer A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ (2017) Architecture of the Vibrio cholerae toxin‐coregulated pilus machine revealed by electron cryotomography. Nat Microbiol 2: 16269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gold VA, Salzer R, Averhoff B, Kuhlbrandt W (2015) Structure of a type IV pilus machinery in the open and closed state. Elife 4: e07380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schlesner M, Miller A, Streif S, Staudinger WF, Muller J, Scheffer B, Siedler F, Oesterhelt D (2009) Identification of Archaea‐specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol 9: 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abrusci P, Vergara‐Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM et al (2013) Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meyer C, Heimerl T, Wirth R, Klingl A, Rachel R (2014) The Iho670 fibers of Ignicoccus hospitalis are anchored in the cell by a spherical structure located beneath the inner membrane. J Bacteriol 196: 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL, Bell SD, Jensen GJ (2013) Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell 24: 2319–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wollert T, Wunder C, Lippincott‐Schwartz J, Hurley JH (2009) Membrane scission by the ESCRT‐III complex. Nature 458: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Engelhardt H, Schuster SC, Baeuerlein E (1993) An archimedian spiral: the basal disk of the Wolinella flagellar motor. Science 262: 1046–1048 [DOI] [PubMed] [Google Scholar]
- 28. Laloux G, Jacobs‐Wagner C (2014) How do bacteria localize proteins to the cell pole? J Cell Sci 127: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ebersbach G, Briegel A, Jensen GJ, Jacobs‐Wagner C (2008) A multimeric pole‐ organizing protein critical for chromosome attachment, division and protein localization in Caulobacter . Cell 134: 956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L (2008) A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowman GR, Perez AM, Ptacin JL, Ighodaro E, Folta‐Stogniew E, Comolli LR, Shapiro L (2013) Oligomerization and higher‐order assembly contribute to sub‐cellular localization of a bacterial scaffold. Mol Microbiol 90: 776–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK (2012) A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26: 2348–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, Subramaniam S, Milne JL (2011) Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus . J Bacteriol 193: 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J (2012) Three‐dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo‐electron tomography. J Bacteriol 194: 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A (1999) DNA protection by stress‐induced biocrystallization. Nature 400: 83–85 [DOI] [PubMed] [Google Scholar]
- 36. Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A (2010) A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res 38: 3605–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim CJ, Lee SY, Teramoto J, Ishihama A, Yan J (2013) The nucleoid‐associated protein Dan organizes chromosomal DNA through rigid nucleoprotein filament formation in E. coli during anoxia. Nucleic Acids Res 41: 746–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spaans SK, van der Oost J, Kengen SW (2015) The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles 19: 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeon SJ, Fujiwara S, Takagi M, Fukui K, Imanaka T (2001) Unique nucleoid structure during cell division of Thermococcus kodakaraensis KOD1. J Biosci Bioeng 91: 40–43 [DOI] [PubMed] [Google Scholar]
- 40. Daum B, Vonck J, Bellack A, Chaudhury P, Reichelt R, Albers SV, Rachel R, Kuhlbrandt W (2017) Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. Elife 6: e27470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erde J, Loo RRO, Loo JA (2014) Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res 13: 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- 43. Tivol W, Briegel A, Jensen GJ (2008) An improved cryogen for plunge freezing. Microsc Microanal 14: 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng QS, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA (2007) UCSF tomography: an integrated software suite for real‐time electron microscopic tomographic data collection, alignment and reconstruction. J Struct Biol 157: 138–147 [DOI] [PubMed] [Google Scholar]
- 45. Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional data using Imod. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- 46. Agulleiro JI, Fernandez JJ (2011) Fast tomographic reconstruction on multicore computers. Bioinformatics 27: 582–583 [DOI] [PubMed] [Google Scholar]
- 47. Nicastro D, Schwartz CL, Pierson J, Gaudette R, Porter ME, McIntosh JR (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Movie EV1
Review Process File
Source Data for Table 1


