Abstract
Introduction
The emergence and mass utilization of high-throughput (HT) technologies, including sequencing technologies (genomics) and mass spectrometry (proteomics, metabolomics, lipids), has allowed geneticists, biologists, and biostatisticians to bridge the gap between genotype and phenotype on a massive scale. These new technologies have brought rapid advances in our understanding of cell biology, evolutionary history, microbial environments, and are increasingly providing new insights and applications towards clinical care and personalized medicine.
Areas covered
The very success of this industry also translates into daunting big data challenges for researchers and institutions that extend beyond the traditional academic focus of algorithms and tools. The main obstacles revolve around analysis provenance, data management of massive datasets, ease of use of software, interpretability and reproducibility of results.
Expert Commentary
The authors review the challenges associated with implementing bioinformatics best practices in a large-scale setting, and highlight the opportunity for establishing bioinformatics pipelines that incorporate data tracking and auditing, enabling greater consistency and reproducibility for basic research, translational or clinical settings.
Keywords: High throughput sequencing, bioinformatics pipelines, bioinformatics best practices, RNAseq, ExomeSeq, variant calling, reproducible computational research, genomic data management, analysis provenance
1. Introduction
The emergence and global utilization of high throughput (HT) technologies, including HT sequencing technologies (genomics) and mass spectrometry (proteomics, metabolomics, lipids), has allowed geneticists, biologists, and biostatisticians to bridge the gap between genotype and phenotype on a massive scale (Figure 1). We have witnessed ultra-high-throughput or deep sequencing of DNA transition from development to widespread use and acceptance [1-3]. The adoption of these novel technologies has been met by a paradigm shift in how biological assays are designed and executed [4]. Deep sequencing has reduced the cost per base of DNA sequencing by several orders of magnitude, and has effectively introduced genome sequencing center capability into every laboratory [1].
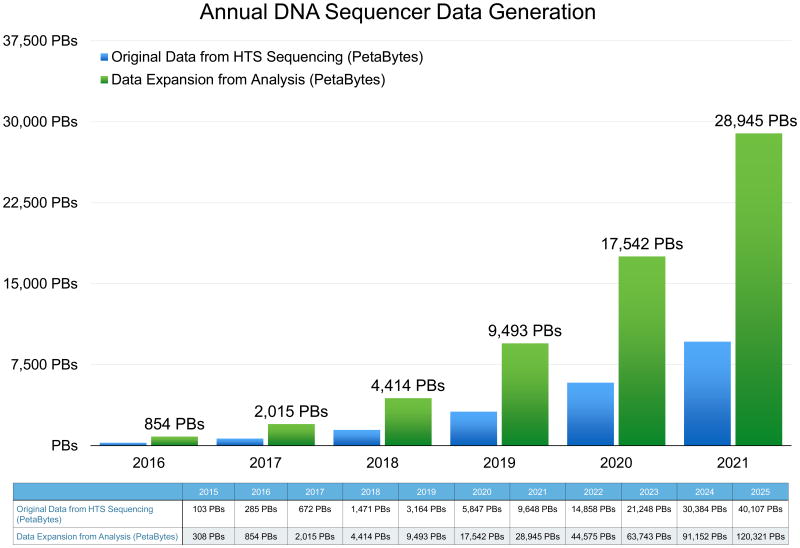
Figure 1. Annual DNA Sequencer Data Generation.
A: Graphical presentation of annual HTS data generation is presented for 2016, and extrapolated through 2021. B: Tabular presentation of HTS for 2015 and 2016 and extrapolation through 2025. Both original data from HTS sequencing (Petabytes) and data expansion from analysis (Petabytes) are presented.
These new genomics technologies have brought swift advances to our understanding of cell biology, evolutionary history, microbial environments, and are increasingly unlocking new insights and applications towards clinical care and personalized medicine. Yet the very success of this industry also translates into daunting challenges for researchers and institutions (Figure 2). While the scientific community has responded with novel tools and algorithms to process and interpret this data, many challenges remain in analysis provenance, data management of massive datasets, ease of use of software, interpretability and reproducibility of results, as well as privacy and security of protected health information. Our focus in this article is genomics data and the establishment of consistent, reproducible analysis pipelines based on traditional scientific tools enabled with comprehensive data tracking software.
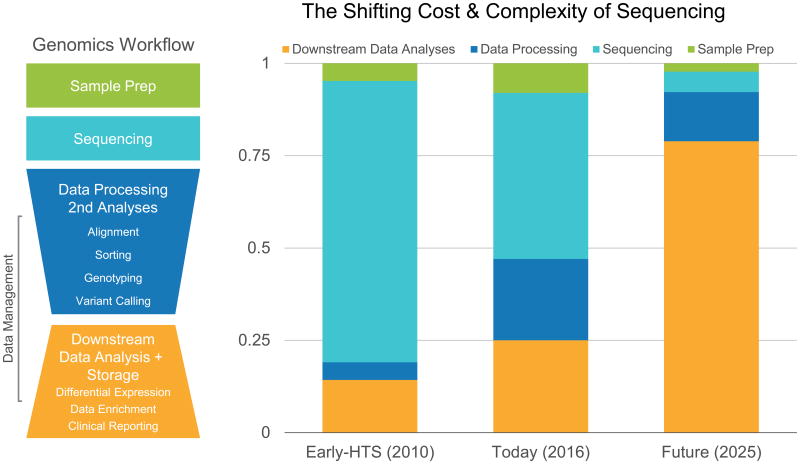
Figure 2. The Shifting Cost & Complexity of Sequencing.
The shifting cost & complexity of sequencing (sample preparation, data processing, downstream analysis and data management) from the early days of HTS through the present and extrapolation through 2025.
2. The Genomic Promise
The wide application of HT genomics technologies, including microarray and deep sequencing technologies has allowed scientists to bridge the gap between gene sequence and function on a scale that was not possible until recently [4]. This has enabled the era of precision medicine (PM), a medical model that aims to customize healthcare to individuals, with medical decisions, treatments and therapies tailored to individual patients. In his 2015 State of the Union address, President Obama announced the launch of the Precision Medicine Initiative –to transform health care and disease treatment. PM is an innovative approach that encompasses the patient's genetic background, environments, and lifestyle choices. PM exploits diagnostic testing to select optimal therapies guided by the genetic blueprint of an individual. Such diagnostics include advances in molecular diagnostics (microarray, targeted sequencing), imaging techniques and advanced analytics/software [5]. To support the import and standardization of genomic and clinical data from cancer research programs, NCI has established the Genomics Data Commons (GDC) [6] as a next generation cancer knowledge network addressing the receipt, quality control, integration, storage, and redistribution of standardized cancer genomic data sets. Enabling standardization, consistency and reproducibility is fundamental for future medical professionals to utilize PM in targeting the treatment of disease.
Cancer genomics is a rapidly expanding area of PM. Patients with cancer face an ever-widening chasm between the exponential technology advances and the linear rate at which these breakthroughs are implemented in the clinic [7]. The racial disparities in cancer detection, incidence, and mortality that exist in the US provide additional challenges for PM approaches. The American Cancer Society recently provided estimates of the number of new cancer cases and deaths for African Americans and the most recent data on incidence, mortality, survival, screening, and risk factors for cancer [8]. Approximately 189,910 new cases of cancer and 69,410 cancer deaths will occur among African Americans in 2016. Although African Americans continue to have higher cancer death rates than whites, the disparity has narrowed for all cancers combined in men and women, and for lung and prostate cancers in men. In contrast, the racial gap in death rates has widened for breast cancer in women and remained level for colorectal cancer in men [8].
At the core of PM approaches is the ability to leverage novel approaches in data science to assemble, mine and exploit the large Omics data generated during deep genome and transcriptome sequencing experiments, often in a rapid timeframe. In the case of cancer genomics initiatives, big data approaches have the potential to provide cancer researchers with novel biomarkers particularly the identification of single nucleotide variants (SNV) and point mutations that may serve as therapeutic targets. In addition to expediting personalized genome sequencing, novel applications and innovative assay concepts have emerged in the research setting (including diverse applications such as RNA-Seq, micro-RNAseq, ChIP-Seq Exome-Seq and MethylC-seq experiments) that are vastly increasing our ability to understand genome function [9].
3. Challenges in Genomics Data Analyses
These novel and ever-expanding applications bring with them a host of challenges that create a daunting barrier to their adoption beyond the research setting. HT technologies are in a state of flux due to constant improvements and upgrades, with new platforms, innovations and algorithms being added frequently, thereby creating complexity in the choice of the most appropriate data analysis pipelines. As observed with DNA microarray analysis over a decade ago, better analytical tools are emerging over time.
There are many challenges associated with selecting and implementing the right set of tools in basic research, translational or clinical settings. Bioinformatics analyses are complex, multi-step processes comprised of multiple software applications. Most of the applications specific to HT sequencing data have been developed in the last decade at multiple academic institutions, by graduate students and postdoctoral researchers. These applications are often incredibly sophisticated and intricate in their statistical and algorithmic approaches. Many academic efforts are spent improving the statistical accuracy of particular approaches leading to constant changes in the most appropriate pipeline that can be assembled at any given point in time. The result from these converging trends is a vast landscape of mostly open-source tools: at time of publication, there are over 11,600 genomic, transcriptomic, proteomic, and metabolomic tools listed at OMICtools (www.omictools.com). Furthermore, each assay requires a unique set & sequence of logic to be properly analyzed, and thus require their own set of software and algorithms to be assembled into a sequence of execution, or analysis pipeline. This complexity is often left to the researcher to reconstruct (Figure 2). The resulting combination of open source applications, algorithms and custom scripts resemble more a spaghetti code, than a repeatable, accurate clinical analysis.
Although the scientific community has long recognized the requirements for improving the reproducibility of computational research [10], these fundamentals of computer science are often eschewed for the more enticing challenge of developing novel algorithms. Consequently, bioinformatics pipelines developed with mainstream scientific tools often fall short in addressing these basic rules required for analysis provenance, including the tracking of metadata for the production of every result and the associated application versioning. Another challenge in traditional pipelines is the handling of all intermediate data, including temporary and log files. Such files contribute to the massive expansion of data during processing, and are often stored in obscure folders within each bioinformatics application. Such metadata tracking is rarely found in typical bioinformatics pipelines, yet is now required for all data sharing with the GDC.
One of the greatest challenges to overcome for broader adoption of HT technologies is the user experience complexity. Currently, the majority of these tools require intricate command-line instructions to operate and set analysis parameters. This level of required technical experience precludes broader audiences from properly utilizing these tools. Hence, there is a need to achieve a new level of simplicity and ease of use in the user experience of HTS analysis tools, such that non-technical audiences, who lack the computer science background, such as the clinicians and wet lab researchers, are afforded the opportunity to interact with these software tools and explore HTS datasets. Enabling such improvements in Ease of Use will expand the breadth of analyses and scope of data interpretation, potentially unlocking new insights into important biomedical research questions.
One of the most notorious challenges in genomics lies in the sheer number of databases and knowledge bases provided by the community and commercial vendors: In fact there is such a preponderance of these resources that Nucleic Acid Research publishes a yearly summary of them [11] – the latest count was 1685, as of January 2016. Keeping up with the latest database, developing integration methods, and tracking changes to formats remains a daunting challenge. Recent, promising efforts to create standards and enable sharing of knowledge have focused on the use of APIs as a universal communication protocol (e.g. the Global Alliance for Genomics and Health (GA4GH)) [12]. In this constantly evolving landscape, reproducibility of research can only be achieved by a comprehensive policy to manage the volume of knowledge and data passing through the virtual laboratory.
Data management is a key challenge that must be addressed at the onset of large-scale projects, rather than as an afterthought. HTS experiments generate massive raw data files known as FASTQ files (Figure 1), which are text based files containing nucleotide sequence and quality score information. These files are usually considered the ‘raw’ data and are therefore treated as precious as the biological samples from which they are derived. To generate useful knowledge from this data, trimmed and cleaned raw data files are then subject to secondary analysis, usually including alignment to a reference genome, de-novo assembly, or k-mer counting. Such analyses generate equally massive secondary and intermediate files describing the alignment, assembly or quantification of the raw data. In turn, these derived files may often be sorted, filtered, annotated, or analyzed in any number of ways that generate even more data. All of these data, whether stored as file, objects or elements in a database, amount to a massive expansion, in some cases 3× - 5× expansion, in the number and total footprint of the initial data, resulting in significant storage management challenges.
All too often, research institutions lack a comprehensive policy and data tracking mechanism to ascertain at any time how much computer storage space is being utilized by these files. It is not uncommon to hear that research institutions have Terabytes of HTS data scattered across hundreds of directories and folders, while the only expansion ‘backup’ strategy consists of removing old hard drives and placing them on a bookshelf, or on the other extreme that teams of researchers are required to meet weekly to assess which files can be deleted from overloaded data volumes. It is, therefore, paramount to incorporate data management systems and retention policies with traditional bioinformatics pipelines to track analyses and other pertinent information so that users and administrators of the system can find their data, preserve analysis provenance and enable consistent reproducibility of results.
4. Bioinformatics Best Practices
The BioInformatics Shared Resource at the Medical University of South Carolina Center (MUSC) for Genomic Medicine sought to overcome the aforementioned challenges in traditional tools and pipelines through the implementation of a new bioinformatics system incorporating best practices with mainstream tools enhanced by a scalable commercial platform for genomics data management. MUSC's new system was designed and implemented over the past two years to incorporate mechanisms for addressing the typical pitfalls in scientific pipelines developed with freely available open source software tools, in the areas of data tracking, storage management, analysis provenance, workflow automation, as well as system infrastructure.
Prior to these enhancements, an older high-performance computing cluster similar to those used at research institutions across the country was utilized. Bioinformatics pipelines were run by users creating their own scripts and manually submitting jobs to the system scheduler. Data management, if implemented, was user driven, and in the majority of cases, performed manually. This system featured multiple compute nodes, a specialized storage array and network which had reached its capacity of 30 terabytes (TB), and a job scheduler to manage and distribute bioinformatics workloads from traditional user-defined pipelines across the computational cluster.
In establishing the BioInformatics Shared Resource a strategic collaboration with OnRamp BioInformatics was implemented (San Diego, CA) and the Genomics Research Platform™ (GRP) was chosen as the basis for genomics research and data management. The OnRamp BioInformatics platform provides advanced genomics data management software and scalable big data system architecture. This unified bioinformatics system with comprehensive data tracking, analysis provenance and a rich portfolio of bioinformatics pipelines extends the capabilities of mainstream open source analysis tools.
As shown in Figure 3, the GRP converges genomic analysis software with highly scalable hardware based on big data architecture. This approach distributes data storage across 10 hyperscale compute nodes, rather than employing a large, expensive storage array or network. Hyperscale compute nodes were originally designed to provide highly efficient infrastructure for cloud-computing, and are well suited for bioinformatics due to their high-performance computation, high-throughput networking and high-capacity internal storage. The system then creates multiple protected data volumes that span all computational nodes utilizing a commercial-grade, high-performance, Distributed File System (DFS) from MapR Technologies. Proprietary OnRamp software integrates MUSC-specified bioinformatics pipelines with cluster management software, including a job scheduler and metadata database for all tracking and analytics.
Figure 3. OnRamp BioInformatics Genomics Research Platform Architecture.
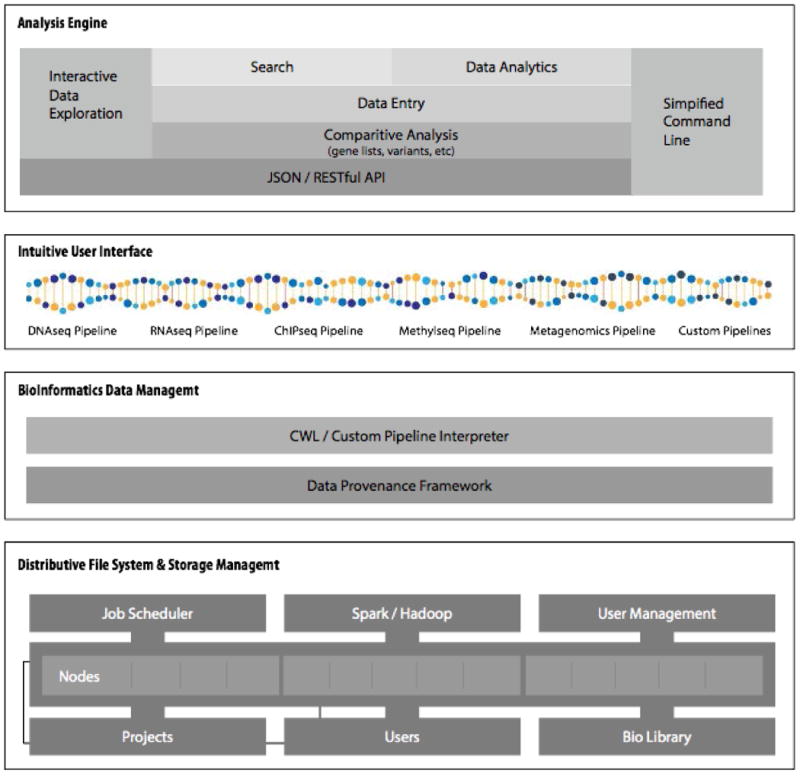
The architecture of the Genomics Research Platform provides a unified system for genomic analysis and data exploration. The GRP consists of four major modules, Intuitive User Interface, Analysis Engine, BioInformatics Data Management, and Distributed File System & Storage Management.
4.1 Intuitive User Interface
As a major advancement in Ease of Use, the GRP provides an intuitive user interface, based on the latest web standards, that enables non-technical audiences, such as clinicians and wet lab researchers, to interact with bioinformatic tools and, most importantly, explore HTS datasets. Though this interface, sample and experiment data is tracked, sequence data is easily uploaded, automated pipelines are launched and resulting data is interactively visualized and downloaded. Search capabilities enable all metadata to be searched for any file, pipeline or command within the GRP. Comparative meta-analysis enables researchers to dynamically contrast the output from multiple pipelines within interactive Venn diagrams to identify and explore overlapping areas of interest within gene expression or variant calling.
4.2 Laboratory information management system (LIMS)
The management of laboratory samples and the associated analysis (data analysis pipelines and files) and reporting are time-consuming manual processes often riddled with human transcription errors [13]. We sought to streamline the collection of data (sample names, quality control metrics on samples) and how this was ultimately recorded. This included capturing information on Sample Name, Species (of origin), Age, Gender, Genotype (wild type, gain or loss of function), Tissue (of origin), Sample Volume (ul), Concentration (ng/ul), Disease (if from a disease tissue), information on whether the data derived from a time course experiment (Time) or compound treatment (Drug), and Cell line (or other biological source).
To integrate the collection of data in this way, we employed the GRP's built-in LIMS, which is specifically designed to import both single-end and paired-end FASTQ files generated by Illumina sequencing instruments. The LIMS system identifies the FASTQ files and pairs each unique sample with each matching FASTQ file so they are tracked throughout the analytics pipeline and downstream data manipulations. Once the LIMS system has been populated with the essential data described above the FASTQ files are imported into a project folder within the GRP, and quality control subroutines are automatically launched.
5. Bioinformatics Pipelines
MUSC bioinformatics researchers, like many academic institutions, perform many ‘standard’ analyses using HTS technology, including RNA expression (mRNA-seq/miRNA-seq), DNA resequencing (targeted panel, whole genome sequencing (WGS), whole exome sequencing (WES), and epigenetic assays (ChIP-seq, Methyl-seq). These pipelines can be quite complex, yet often follow the same outline, so it is naturally desirable to automate this process. We leveraged the OnRamp GRP to deploy these pipelines and analyses in a highly reproducible and traceable way.
5.1 RNA-Seq pipeline
RNAseq Analysis is a powerful technique that allows comparison of the transcriptomes of different cell states and/or populations [14]. The output of the sequencing instrument is the FASTQ file which contains the reads and a quality score for each base [15]. A read is a sequence of nucleotides 30-400 base pairs in length, depending of the sequencing technology utilized. The FASTQ files represents the raw data, or the input, of RNAseq Analysis. A summary flowchart of the analysis is shown in Figure 4 and is discussed below. Although the specific steps of the analysis are complex, the logic is straightforward. The researcher needs to evaluate the quality of their data, to select and analyze only data with quality scores above threshold. Any adapter sequences that remain from the library preparation step need to be located and removed while at the same time filtering out the low quality reads. Next the reads are aligned to the genome determine the genes that the reads derived from. Finally the expression of each gene is quantified, enabling the comparison of the transcriptome in different cellular states [16].
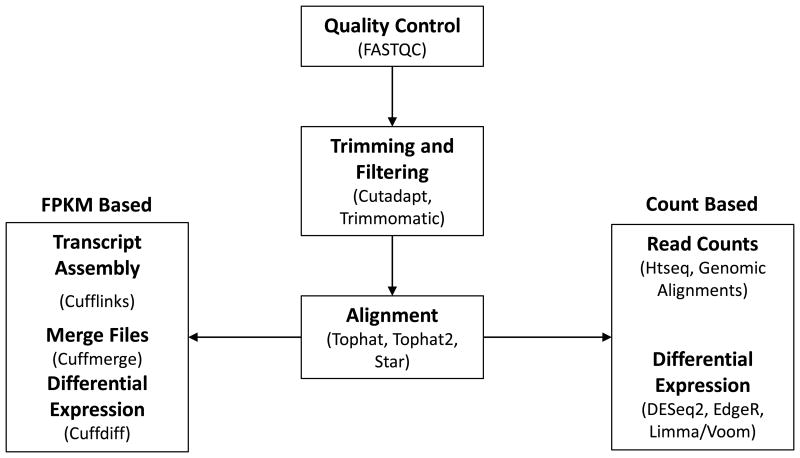
Figure 4. Flow Chart summarizing the analyses of RNAseq data.
In each box, the upper part in bold describes the analytical step, and the bottom part in plain text notes the programs or services that can be used. FPKM = Fragments Per Kilobase of transcript per Million mapped reads, THP Atlas = The Human Protein Atlas, GSEA = Gene Set Enrichment Analysis).
To focus on mRNAs, polyadenylated (poly A+) RNA is enriched from total RNA and reverse-transcribed and converted to a strand-specific sequencing library [17-20]. Single end (SE) or paired end (PE) sequencing is carried out to depths of 50 to 100 million reads and 50 to 125 cycles respectively. In projects for which isoform-specific expression or novel transcript identification is desired, longer 150 to 250 bp-paired end reads or greater are sequenced. Each sample is sequenced to a minimum depth of 100 million reads.
FASTQC is one of the most widely used programs to check the quality of raw sequence data [21]. FASTQC output provides graphical representations of the input FASTQ file with metrics including “Per Base Sequence Quality” and “Overrepresented sequences”. FASTQC outputs graphics which highlight FASTQ input files that can be considered acceptable or unacceptable (Figure 5). Running FASTQC before and after adaptor trimming allows the user to make a decision as to whether the FASTQs are of high enough quality for downstream analyses [21]. Trimming and Filtering are part of the pre-processing step in which low quality bases and overrepresented sequences are removed [22]. Figure 5 illustrates this issue. The sample in Figure 5A, is of high quality whereas the sample in Figure 5B has several quality issues owing to the library preparation step but overall represents high quality sequencing. The researcher would most likely trim and filter overrepresented sequences, and rerun FASTQC to ensure that the sample was acceptable for downstream analyses. These processes increase the quality and reliability of the analyses and diminish the computational cost and execution time [23]. Cutadapt and Trimmomatic are two frequently cited software suites which perform this step [24]. Although the two have differences in their approach their results are similar [23]. Trimmomatic runs in Java, can work simultaneously with paired end data but only supports Illumina sequencing data [23, 25]. Cutadapt runs in Python and C, and paired ended samples have to be analyzed separately. However sequencing data is not restricted to Illumina data. Cutadapt works with SOLID sequencing data [23, 24].
Figure 5. FASTQC Report.
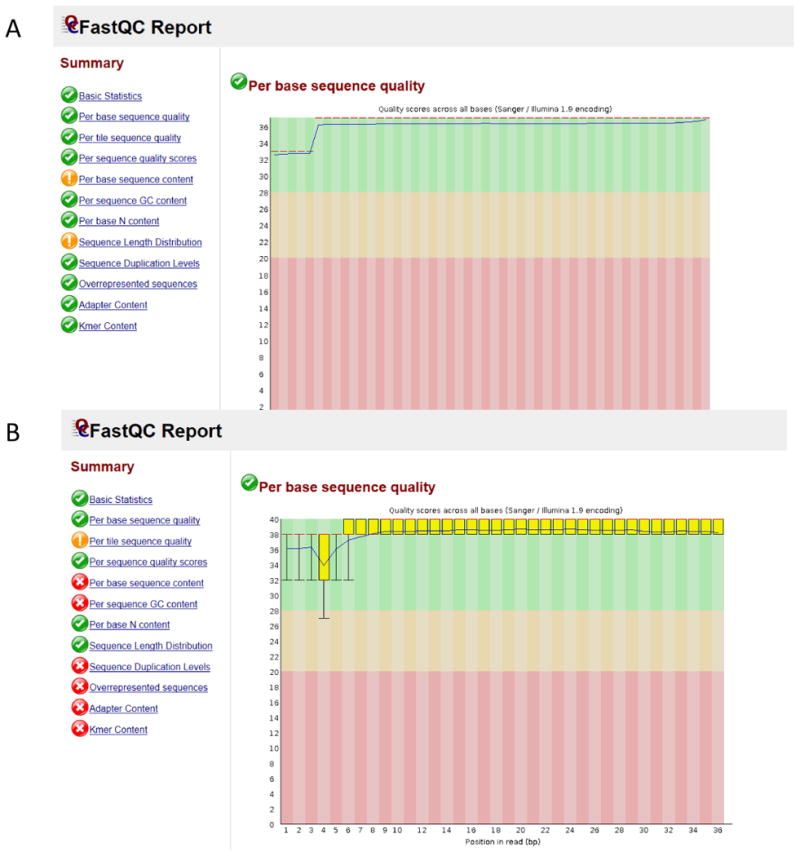
A: High Quality Data. B: Data with potential RNAseq library preparation issues, overrepresented sequences and duplication that should be removed prior to any downstream analysis.
The reads that pass QC, are aligned to the genome to identify genomics features and quantify gene expression. The average mature mRNA transcript size in the human genome, based on the GRCh37 annotation, is 2,227bp, with each exon an average of 235bp in length, and each transcript containing an average of 9.5 exons [26]. As the read length can vary from 30 to 400bp and differential splicing, mismatches, insertions, deletions and mutations can occur [14], it is necessary to take this into consideration during the alignment process [26-28]. Sequence reads are mapped to the mouse (GRCm38/mm10) or human genome (GRCh37/38 or hg19/38) depending on the species. The alignment is considered successful if 70 to 90% of the reads map to the genome [29].
STAR, TopHat and TopHat2 are frequently used alignment tools for RNAseq data [26-28]. STAR (Spliced Transcripts Alignment to a Reference) aligns all the sequences, including reads containing junctions, directly to the reference genome in two steps, first the “searching seed” step aligns all possible parts of the reads and then the “clustering/stitching/scoring” step aligns the remaining parts [28]. STAR is one of fastest aligners in use, and can align both short and long reads and is the ideal choice for large datasets [28]. Tophat first employs bowtie to align all the non-junction reads and then aligns the unmapped reads to possible splice regions via a seed-and-extend algorithm. It is designed to align reads up to 50bp in length [27]. Tophat2, an improved version of Tophat that exploits the BowTie2 aligner, employs the same strategy in that it aligns short reads. TopHat2 however is better suited to aligning reads greater than 50bp. Furthermore It is more sensitive to insertions, deletions and other structural variations than previous version and more sensitive to splicing regions than STAR [26]. Both TopHat versions are less computationally intensive than STAR but are slower [28].
There are basically two methods to define the differential expression of genes in RNAseq analysis: 1) methods based on raw count data or 2) methods based on transformed counts i.e. FPKM (Fragments Per Kilobase of transcript per Million mapped reads) [30]. Raw count data is a measure of how many reads for that gene exist in the file [31]. FPKM on the other hand is a transformation that normalizes the counts by taking into consideration the different library sizes and the length of the transcripts. Since a long transcript is expected to obtain more reads than a short transcript with the same expression level [30], this correction is not necessary when the intention is to compare the expression of the same gene across multiple samples, but is necessary if the objective is ranking genes by their expression level or abundance [29].
The Cufflinks package is one of the most used FPKM based method for gene expression analysis [30, 32]. With Tophat and Bowtie it forms the basis of the “Tuxedo Pipeline” [32]. The Cufflinks package contains the Cufflinks program itself, and Cuffmerge and CuffDiff. Cufflinks assemble the reads into transcripts, Cuffmerge merge these assemblies together to provide a uniformed basis for gene expression calculation and CuffDiff use these merged assemblies to calculate differential gene expression and tests the statistical significance of the observation [32]. Compared with DESeq2 and Limma/Voom the Tuxedo pipeline has lower precision and sensitivity [30, 33].
Several programs exist that count reads from RNAseq, the most widely used is HTseq [13]. HTseq is designed to map reads unambiguously to a single gene. Reads that align to more than one gene are discarded [31]. GenomicAlignments, an R package, has the function ‘summarize overlaps’ that can be used for count reads with similar results [31].
For differential expression calculations, DESeq2, EdgeR and Limma/voom are widely utilized [33-36]. DESeq2 and EdgeR both use the negative binomial distribution model and an empirical Bayes method for calculation. Both require medium computational times [30, 33, 37]. DESeq2 should be used carefully when only two replicates per group are available, however as sample size increases DESeq2 has robust False Discovery Rate (FDR) control and can accommodate outlier samples [30, 33, 37]. EdgeR can be also be used with small sample size, for example 2 biological replicate samples per condition, but the FDR control is poor and not optimal when outliers are present [30, 37].
Limma bases its calculation on linear models and empirical Bayes analyses, is computationally fast, and has been the method of choice for gene expression analysis for microarray data for more than a decade [36]. The function ‘voom’ transforms HTS count values to logarithmic values, adding a precision weight for each observation and allowing exploitation of the Limma pipeline with this data [34]. Limma/voom can be used with small samples sizes, such as 2 biological replicates per condition, allows complex multi-factorial designs and permits both RNAseq and microarray data to be analyzed in similar pipelines facilitating comparisons [30, 34, 36].
5.2 miRNA-Seq
Analysis of miRNA sequencing data is challenging as it requires significant computational resources. Many user friendly web based analytical tools are available but they often lack flexibility and are usually restricted to processing one or a pair of samples at time. Furthermore they are not suitable for large scale studies. The Comprehensive Analysis Pipeline for microRNA Sequencing data (CAP-miRSeq) from the Mayo Clinic is a robust tool which integrates read pre-processing, alignment, mature/precursor/novel miRNA detection and quantification, data visualization, variant detection in miRNA coding regions, and flexible differential expression analysis between given experimental conditions. A significant advantage of this tool over many of the web based tools is its scalability from a few to hundreds of samples [38].
5.3 ChIP-seq
ChIP-seq defines the genomic locations of transcription factors and histone modifications associated with distinct transcriptional states [39-42]. Following base-calling, reference genome mapping is performed using Bowtie2 [19, 43]. Other data processing steps, such as quality control, read counting, peak finding, motif analysis and visualization are performed using the HOMER software suite [44], Model-based Analysis for ChIP-seq (MACS) [45, 46] and MOSAiCS [47-49]. Differential binding analysis of ChIP-seq peak data is performed with the Bioconductor package DiffBind [50, 51].
5.4 Methyl-C sequencing and Reduced Representation Bisulfite Sequencing (RRBS)
Methyl-C and RRBS libraries are constructed from the bisulfite-treated DNA and sequenced using an established single-end sequencing SE50 protocol. Bismark is utilized for both read mapping and methylation calling [52]. Following alignment, the percentage of methylation at any site can be determined by comparing the ratio of T to C detected at any site. Any sample that shows a bisulfite conversion efficiency of less than 95% is repeated. The BiSeq R package is used to detect differentially methylated regions (DMRs) [53]. RnBeads, an R package for the comprehensive analysis of genome-wide DNA methylation data with single basepair resolution presents the analysis results in a highly interpretable fashion [54].
5.5 Metagenomics analysis
Insights into commensal complex microbiota are provided via HTS. Interactions taking place between microbes in these diverse environments and the implications their interactions may have for pathogenesis is uncovered via metagenomic approaches [55-58]. Over the past decade, cultivation independent molecular techniques, particularly those based on the small subunit ribosomal RNA (16S rRNA) gene, have provided a broader unbiased biased view of diversity and abundance of the microbiota [59-61]. The prokaryotic 16S rRNA is approximately 1500 bp in size and contains nine interspersed conserved and variable regions that facilitate sequencing and phylogenetic analyses. The analytical strategy that we employ for community analyses is based on Illumina MiSeq sequencing and targets the V3 and V4 regions (an amplicon of approximately 459 bp). The Illumina sequencing approach generates paired end 250 bp reads overlapping the entire V3 and V4 regions. Up to 96 samples can be multiplexed into a single library, for high throughput sequencing. Initial data QC and preliminary analysis is performed using Illumina MiSeq Reporter proprietary software. The ‘Quantitative Insights Into Microbial Ecology’ (QIIME) software suite is used to perform microbial community analysis. It facilitates analysis and interpretation of nucleic acid sequence data from fungal, viral, bacterial, and archaeal communities [62-64]. For shotgun metagenomics sequencing data, we utilize SUPER-FOCUS, SUbsystems Profile by databasE Reduction using FOCUS, a homology-based approach using a reduced reference database to report the subsystems present in metagenomic datasets and profile their abundances [65, 66]. SUPER-FOCUS accurately predicts the subsystems present in the profiled microbial communities, and is up to 1,000 times faster than other tools.
5.6 Whole Genome and Exome Sequencing
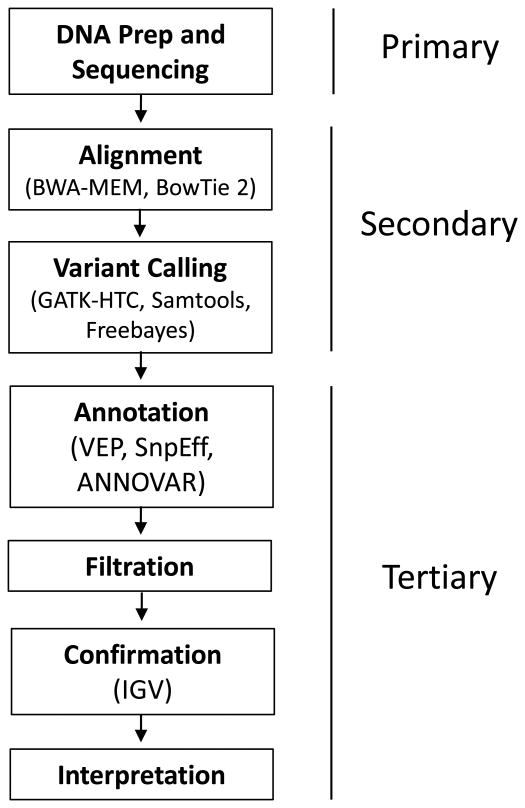
There are many techniques for identifying an individual's genetic variation for diagnostic purposes. Here we focus the bioinformatics methods used in the analysis of whole genome and exome sequencing. HTS is a complex process which is frequently divided into three large steps: primary, secondary, and tertiary (Figure 6). Primary analysis is essentially generation of the FASTQ files as described above by the sequencing instrument. Secondary analysis takes the raw read sequences, associates these smaller reads with the overall target genome, and identifies specific sites of genetic variation. Tertiary analysis is the process of gathering information about the individual genetic variation identified, and using this information to generate a subset of relevant variants. Below, we will explore the processes in the secondary and tertiary analysis.
Figure 6. Flow Chart summarizing the analyses of DNAseq data (whole genome and exome).
In each box, the upper part in bold describes the analytical step, and the bottom part in plain text notes specific programs or services that can are employed. Labels on the right indicate a general categorization of the analysis into primary, secondary, and tertiary.
In order to derive variant information from the individual reads, they must first be aligned to a genome. In general, this process involves comparing each read to a reference genome (though it is also possible to align the reads to themselves via de novo assembly). The development of human reference genomes is an ongoing process. The most current version is the Genome Reference Consortium human version 38/University of Santa Cruz human genome 38 (GRCh38/hg38) though the slightly older version (GRCh37/hg19) is still much more commonly used. A variety of algorithms have been developed for the purpose of sequence alignment and the selection of the aligner will be based upon many factors, including the type of sequencer, the DNA library preparation, and the expected mutations. For short read sequences, two of the most popular options freely available are the Burrows-Wheeler Aligner (BWA) [67] and Bowtie 2 [68]. The choice of aligner will impact speed, performance, accuracy, indel detection, and alignment algorithm is an area of active development. The output of these files is a sequence alignment map (SAM) or its more common binary counterpart, the BAM. This file contains most of the basic information from the FASTQ file, as well as information including the position at which each read was aligned, how closely the read matched the reference, and a quality score for the alignment.
The next major step of secondary analysis is variant calling. This is the process by which the aligned reads are looked at in aggregate to identify locations at which an individual has genetic variation, and determine the nature of that variation. Modern variant callers are very complex pieces of software which incorporate the various quality scores generated thus far (one for each base for each read, and one for the alignment of each read), depth over coverage at a given site, and probabilities of various kinds of mutations. Furthermore, experimental design can play a critical role in the assumptions utilized to improve variant calling. For instance, when performing sequencing on a single individual, the caller makes assumptions of biallelic distribution. However, when sequencing a tumor sample, tissue heterogeneity and subclonal populations dramatically alter the kinds of assumptions possible about allelic frequency. Similarly, if sequencing related individuals, the genetic relationships between individuals can be taken into consideration during variant calling. Similarly, when performing a tumor-normal analysis, the calls made on the normal sample can influence calls on the tumor sample. Three of the most popular variant calling algorithms freely available are the Genome Analysis Toolkit Haplotype Caller [69-71], Samtools mpileup [72], and Freebayes [73]. The general output of a variant caller is a variant call file (VCF). In its simplest form, a variant call file specifies the genomic location of a variant, the alternate observed, various statistical measures related to the call, and an assigned quality score. As we discuss in the tertiary analysis, it is possible to annotate these VCFs with many sources of information on each variant.
Even more so than aligners, variant calling is a rapidly developing area with many new variant callers on the horizon, which incorporate both advances in bioinformatics and computer science (such as Apache Hadoop/Spark), including GATK 4.0 (with MUTECT 2) and ADAM. Specialized variant callers are also in development for special cases, such as detection of large insertions and deletions and the detection of copy number variants ie. Amplicon Indel Hunter [74].
The need for comparison of secondary pipelines is a growing concern. While many excellent reviews have tackled the issue [75, 76], a number of online resources have begun to tackle the issue in a more exhaustive way. For instance the online resource ‘Genome Comparison and Analytic Testing’ [14] provides validated test data for users to run through their various pipelines and share results [77]. The Federal Drug Administration has a similar initiative in development.
As sequencing and variant identification has become commonplace, the real challenge lies in finding meaning and significance in the vast data sets. For HTS data, this largely involves annotating a VCF against many annotation sources. There are three popular open source tools for directly annotating a VCF file, incorporating the annotations directly into the “INFO” field of the VCF: SnpEff [78], Variant Effect Predictor [79], and ANNOVAR [80, 81]. At their most basic, these tools determine the relationship between a variant, determine the gene and “canonical” transcript this variant impacts and determine the expected impact of the variant at the codon level (if it is in a coding region). From this the variant can be assigned a standardized human genome variation society (HGVS) nomenclature. However, these tools also come with frequently updated “databases” of information including resources describing the variant (dbSNP) its clinical significance (ClinVar [82], OMIM [83], COSMIC [84]), its estimated impact on protein function (PROVEAN [85], PolyPhen-2 [86]), and its frequency of expression in “normal healthy” individuals (EXAC [87]). Then, having annotated a VCF, one can begin to deal with the list. Usually, the first step in this process is to filter out variants. This filtration process will in part be based on annotations and in part on quality controls. There are also many commercial products for acquiring and visualizing annotations on a mutation, and other open source tools attempting to move away from a strict dependence on the VCF file format (GEMINI [88]).
Once a candidate list of variants is identified, it is important to confirm the list by examining the individual variants as a sequence pileup in a product such as the Integrated Genome Viewer [89, 90] to make sure no erroneous calls have been made. Ultimately, the final variants are evaluated for their biological significance. For instance, in a clinical cancer laboratory, an individual's variants will be examined first for known actionable variants which enable precision medicine approaches, i.e. impact current treatment options or predict patient outcome. Again, many resources exist for determining the actionability of a variant, including COSMIC [84], My Cancer Genome [91], JAX Clinical Knowledgebase [92], CIVIC [93], and Cornell-Weill PMKB [94]. It may also be desirable to attempt to identify cancer drivers within the individual by performing pathway analysis or utilizing computational software for identifying drivers. In any case, the annotations and downstream analysis of a given patient are greatly determined by the application and are rapidly evolving.
6. Conclusions
Remarkably, while massively parallel sequencing instrumentation was not even commercially available a decade ago we have transitioned through several generations of commercial DNA Sequencers. This instrumentation has been and will continue to be transformative for genomics research. It has revolutionized the analysis of gene expression, enabling the implementation of a variety of assays such as ChIP-Seq and RNA-Seq that provide global and multi-dimensional views of the transcriptional landscapes in diverse cell types and tissues. Such information is contributing to rapid advances in our understanding of gene regulation in development that are directly relevant to unanswered questions in biomedical research. At the core of genomic medicine approaches is the ability to leverage novel approaches in data science to assemble, mine and exploit the large Omics data generated during deep genome and transcriptome sequencing experiments, often in a rapid timeframe, particularly in clinical settings.
7. Expert Commentary
Implementing technical infrastructure and workflows for high throughput sequencers requires a significant amount of storage, network bandwidth, and computational support. HTS datasets can easily reach terabyte sizes per instrument run, excluding secondary analyses. In addition to the computational burden resulting from the need to store and process gigabytes of data streaming from sequencing machines, collecting metadata and providing data to end users will continue to grow in complexity. Additionally, there are many challenges associated with selecting and implementing the right set of analytical tools for both the analysis and interpretation of HT datasets. To overcome these, we formed a strategic collaboration with OnRamp BioInformatics and implemented several best practices, in the areas of analysis provenance, data management, workflow automation, user interface as well as scalable system infrastructure and storage. The impact of this is still being realized as we continue to expand the scope of our research while accumulating an ever greater genomic data store that will be accessible to our wider research community.
7. Five Year View
We expect scientific focus in the field of genomic analysis to shift downstream from the current study of bioinformatics pipelines for secondary and tertiary analysis. As more and more large countries and institutions begin to collect vast datasets (e.g. 100K Genomes UK), both the amount, and cost, of genomic data stored in the cloud are projected to increase. Driven by national privacy laws, the cost of cloud computing/storage and huge data upload requirements, institutions are already moving toward private cloud environments. Private clouds will offer advantages over commercial cloud subscriptions, but will come at a cost: data may become difficult to aggregate and standards may diverge between platforms. Successful solutions will offer some flavor of data beacons that allow users to advertise the presence of potentially useful data without revealing much about the data itself. Furthermore, the Big Data and machine learning fields will continue to advance in sophistication and increasingly will have the ability to analyze large data stores that may be geographically distributed. The role of machine learning will shift from exploratory to fundamentally required, in order to draw relationships and insights from ever growing genomic data stores. As best practices, such as those outlined in this paper, enable streamlined analysis of high-throughput sequence data sets, commercial and clinical institutions will join the race to accumulate ever greater, differentiated genomic data sets with the objective of improving precision medicine programs. With the growth of these data sets, our scientific focus will center on training the machine learning to help us explore, exchange and interpret datasets measured in exabytes. Accessing and analyzing these treasure troves of data will provide a competitive battleground for the future of precision medicine.
Key issues.
HTS datasets can easily reach terabyte sizes per sequencing instrument run, excluding secondary analyses. In addition to the computational burden resulting from the need to store and process gigabytes of data streaming from sequencing machines, collecting metadata and providing data to end users is complex.
The main challenges revolve around ease of use of software, data management of massive datasets, interpretability and reproducibility of results, and privacy and security of protected health information.
As observed with DNA microarray analysis over a decade ago, better analytical tools are emerging over time for HT genomic analyses.
As sequencing and variant identification has become commonplace, the real challenge lies in finding meaning and significance in the vast data sets. For HTS data, this largely involves annotating a VCF against many annotation sources.
A sound data management and retention policy to track analyses and other pertinent information so that users and administrators of the system can find their data is critical.
One of the greatest challenges to overcome for broader adoption of HT technologies is the user experience complexity. Currently, the majority of these tools require intricate command-line instructions to operate and set analysis parameters.
There is a need to achieve greater simplicity and ease of use in the user experience of HTS analysis tools, such that non-technical audiences, who lack the computer science background, such as the clinicians and wet lab researchers, are afforded the opportunity to interact with these software tools and HTS data. This will expand the breadth of analyses, potentially unlocking new insights into important biomedical research questions.
Acknowledgments
Funding This work was supported by the US National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant P60 AR062755 (Bethany J. Wolf), from the National Institutes of Health (NIH), and the startup grants from the Department of Public Health Sciences (Dongjun Chung) and the Department of Medicine (Gary Hardiman) at the Medical University of South Carolina. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health.
Footnotes
Declaration of Interest: J.D. Turak and T. Wesselman are employees of OnRamp Bioinformatics, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed
References
Papers of special note have been highlighted as:
• Of interest
• • of considerable interest
- 1.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nature reviews Genetics. 2004;5(5):335–344. doi: 10.1038/nrg1325. Review of HTS technologies, and the potential impact of a ‘personal genome project’ on both the research community and on society. [DOI] [PubMed] [Google Scholar]
- 4.Bhasker CR, Hardiman G. Advances in pharmacogenomics technologies. Pharmacogenomics. 2010;11(4):481–485. doi: 10.2217/pgs.10.10. [DOI] [PubMed] [Google Scholar]
- 5.Lu YF, Goldstein DB, Angrist M, Cavalleri G. Personalized medicine and human genetic diversity. Cold Spring Harb Perspect Med. 2014;4(9):a008581. doi: 10.1101/cshperspect.a008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossman RL, Heath AP, Ferretti V, et al. Toward a Shared Vision for Cancer Genomic Data. The New England journal of medicine. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blau CA, Liakopoulou E. Can we deconstruct cancer, one patient at a time? Trends in Genetics. 29(1):6–10. doi: 10.1016/j.tig.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desantis CE, Siegel RL, Sauer AG, et al. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA: a cancer journal for clinicians. 2016 doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 9.Wold B, Myers RM. Sequence census methods for functional genomics. Nature methods. 2008;5(1):19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 10••.Sandve GK, Nekrutenko A, Taylor J, Hovig E. Ten simple rules for reproducible computational research. PLoS computational biology. 2013;9(10):e1003285. doi: 10.1371/journal.pcbi.1003285. Ten rules that provide QC for computational research are presented. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigden DJ, Fernandez-Suarez XM, Galperin MY. The 2016 database issue of Nucleic Acids Research and an updated molecular biology database collection. Nucleic acids research. 2016;44(D1):D1–6. doi: 10.1093/nar/gkv1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawler M, Siu LL, Rehm HL, et al. All the World's a Stage: Facilitating Discovery Science and Improved Cancer Care through the Global Alliance for Genomics and Health. Cancer discovery. 2015;5(11):1133–1136. doi: 10.1158/2159-8290.CD-15-0821. [DOI] [PubMed] [Google Scholar]
- 13.Gibbon GA. A brief history of LIMS. Laboratory Automation & Information Management. 1996;32(1):1–5. [Google Scholar]
- 14••.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484. RNA-Seq provides a far more precise measurement of levels of transcripts and their isoforms than other methods This review article examines challenges associated with RNAseq analysis and progress with several eukaryote transcriptomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic acids research. 2010;38(6):1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. To infer differential signal in DE data estimation of data variability throughout the dynamic range and a suitable error model are required. DESeq is based on the negative binomial distribution, with variance and mean linked by local regression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S, Mccarthy DJ, Chen Y, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature protocols. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ten Bosch JR, Grody WW. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. The Journal of molecular diagnostics: JMD. 2008;10(6):484–492. doi: 10.2353/jmoldx.2008.080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S. FastQC: A quality control tool for high throughput sequence data. Reference Source. 2010 [Google Scholar]
- 22.Williams CR, Baccarella A, Parrish JZ, Kim CC. Trimming of sequence reads alters RNA-Seq gene expression estimates. BMC bioinformatics. 2016;17:103. doi: 10.1186/s12859-016-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PloS one. 2013;8(12):e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–12. [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. TopHat2 is a newer alignment tool that can align reads of various lengths while allowing for variable-length indels with respect to the reference genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29 doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome biology. 2016;17(1):1. doi: 10.1186/s13059-016-1047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC bioinformatics. 2013;14:91. doi: 10.1186/1471-2105-14-91. Extensive comparison of eleven methods for differential expression analysis of RNA-seq data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2014:btu638. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA- sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Z, Evans J, Bhagwate A, et al. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson G, Hirst M, Bainbridge M, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature methods. 2007;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Tanasa B, Tyurina OV, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irish JC, Mills JN, Turner-Ivey B, et al. Amplification of WHSC1L1 regulates expression and estrogen- independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol Oncol. 2016 doi: 10.1016/j.molonc.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Escoubet-Lozach L, Benner C, Kaikkonen MU, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 2011;7(12):e1002401. doi: 10.1371/journal.pgen.1002401. Application of ChIP seq which demonstrates that nearly all immediate/early and late Toll Like Receptor target genes exhibit characteristics of active genes under basal conditions regardless of CpG content and direct detectable levels of expression of mature mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B. Aligning short sequencing reads with Bowtie. Current protocols in bioinformatics. 2010:11.17.11–11.17.14. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J, Liu T, Zhang Y. Using MACS to identify peaks from ChIP-Seq data. Current Protocols in Bioinformatics. 2011:2.14.11–12.14.14. doi: 10.1002/0471250953.bi0214s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuan PF, Chung D, Pan G, Thomson JA, Stewart R, Keleş S. A statistical framework for the analysis of ChIP-Seq data. Journal of the American Statistical Association. 2011;106(495):891–903. doi: 10.1198/jasa.2011.ap09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun G, Chung D, Liang K, Keleş S. Statistical analysis of ChIP-seq data with MOSAiCS. Deep Sequencing Data Analysis. 2013:193–212. doi: 10.1007/978-1-62703-514-9_12. [DOI] [PubMed] [Google Scholar]
- 49.Chung D, Zhang Q, Keleş S. Statistical Analysis of Next Generation Sequencing Data. Springer; 2014. MOSAiCS-HMM: A model-based approach for detecting regions of histone modifications from ChIP-seq data; pp. 277–295. [Google Scholar]
- 50.Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark R, Brown G. DiffBind: differential binding analysis of ChIP-Seq peak data. R package version. 2011;100 [Google Scholar]
- 52.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics (Oxford, England) 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hebestreit K, Klein HU. BiSeq: A package for analyzing targeted bisulfite sequencing data. 2013 [Google Scholar]
- 54.Assenov Y, Muller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Meth. 2014;11(11):1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38(7):779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 56.Chapple IL, Hamburger J. The significance of oral health in HIV disease. Sex Transm Infect. 2000;76(4):236–243. doi: 10.1136/sti.76.4.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13(12):589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. The New England journal of medicine. 2002;346(5):334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 59.Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta'omics for microbial community studies. Mol Syst Biol. 2013;9:666. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 61.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current protocols in microbiology. 2012:1E.5.1–1E.5.20. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi1007s36. Chapter 10 Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Methods in enzymology. 2013;371 doi: 10.1016/B978-0-12-407863-5.00019-8. 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva GGZ, Green KT, Dutilh BE, Edwards RA. SUPER-FOCUS: A tool for agile functional analysis of shotgun metagenomic data. Bioinformatics (Oxford, England) 2015 doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva GG, Green KT, Dutilh BE, Edwards RA. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics (Oxford, England) 2016;32(3):354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25 doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Depristo MA, Banks E, Poplin RE, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. The Genome Analysis Toolkit (GATK), a unified analytic framework to discover and genotype variation among multiple samples simultaneously. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mckenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. Genome Analysis Toolkit (GATK), a structured programming framework designed to ease the development of efficient and robust analysis tools for HTS using the functional programming philosophy of MapReduce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Current protocols in bioinformatics. 2013:11.10.11–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England) 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. 2012 [Google Scholar]
- 74.Kadri S, Zhen CJ, Wurst MN, et al. Amplicon Indel Hunter Is a Novel Bioinformatics Tool to Detect Large Somatic Insertion/Deletion Mutations in Amplicon-Based Next-Generation Sequencing Data. The Journal of molecular diagnostics: JMD. 2015;17(6):635–643. doi: 10.1016/j.jmoldx.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 75••.Cornish A, Guda C. A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. BioMed Research International. 2015;2015;11 doi: 10.1155/2015/456479. NIST Genome in a Bottle data is used as a resource for exome analysis pipeline validation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Scientific reports. 2015;5:17875. doi: 10.1038/srep17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Highnam G, Wang JJ, Kusler D, et al. An analytical framework for optimizing variant discovery from personal genomes. Nature communications. 2015;6:6275. doi: 10.1038/ncomms7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cingolani P, Platts A, Wang Le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mclaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome biology. 2016;17(1):1. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80••.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164–e164. doi: 10.1093/nar/gkq603. ANNOVAR tool to annotate single nucleotide variants (SNVs) and insertions/deletions, such as examining their functional consequence on genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nature protocols. 2015;10(10):1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82••.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic acids research. 2014;42(D1):D980–D985. doi: 10.1093/nar/gkt1113. Freely available archive of reports of relationships among medically important variants and phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic acids research. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84••.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic acids research. 2015;43(D1):D805–D811. doi: 10.1093/nar/gku1075. The Catalogue Of Somatic Mutations In Cancer is the world's largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England) 2015;31(16):2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS computational biology. 2013;9(7):e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nature biotechnology. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Micheel CM, Lovly CM, Levy MA. My Cancer Genome. Cancer Genetics. 2014;207(6):289. [Google Scholar]
- 92.Patterson SE, Liu R, Statz CM, Durkin D, Lakshminarayana A, Mockus SM. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Human genomics. 2016;10:4. doi: 10.1186/s40246-016-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Griffith M, Spies NC, Krysiak K, et al. CIViC: A knowledgebase for expert-crowdsourcing the clinical interpretation of variants in cancer. bioRxiv. 2016;072892 doi: 10.1038/ng.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang L, Fernandes H, Zia H, et al. The Precision Medicine Knowledge Base: an online application for collaborative editing, maintenance and sharing of structured clinical-grade cancer mutations interpretations. bioRxiv. 2016 doi: 10.1101/059824. [DOI] [Google Scholar]